Abstract
Skin stem cells (SCs) are specified and rapidly expanded to fuel body growth during early development. However, molecular mechanisms that govern the amplification of skin SCs remain unclear. Here we report an essential role for miR-205, one of the most highly expressed miRNAs in skin SCs, in promoting neonatal expansion of these cells. Unlike most mammalian miRNAs, genetic deletion of miR-205 causes neonatal lethality with severely compromised epidermal and hair follicle growth. In the miR-205 KO skin SCs, phospho-Akt is significantly downregulated, and the SCs prematurely exit the cell cycle. In the hair follicle, this accelerates the transition of the neonatal SCs towards quiescence. We identify multiple miR-205 targeted negative regulators of PI3K signaling that mediate the repression of phospho-Akt and restrict the proliferation of SCs. Our findings reveal an essential role for miR-205 in maintaining the expansion of skin SCs by antagonizing negative regulators of PI3K signaling.
Tissue stem cells (SCs) are specified during early development. In mouse skin, hair follicle stem cells (HFSCs) emerge between embryonic day 14 (E14) and postnatal day 0.5 (P0.5) at the initiation of hair follicle (HF) morphogenesis1. These newly specified SCs must undergo proliferative expansion to accommodate body growth and enlargement of individual tissues. In adults, however, HFSCs form a quiescent cell population that infrequently cycle and maintain the SC pool throughout life2, 3. Therefore, there is a transition leading proliferative SCs to exit the cell cycle and become quiescent. During this transition, gene regulatory networks balancing output from multiple pathways regulating proliferation, quiescence, survival and cell death must be precisely coordinated4, 5. In skin, the molecular mechanisms governing the transition of SCs from the expansion phase to quiescence remain poorly understood.
MicroRNAs (miRNAs), a class of tiny regulatory RNAs, have been proposed to play important roles in diverse biological processes6. However, few studies have specifically addressed the role played by individual miRNAs in the expansion and maintenance of tissue SCs during mammalian development in vivo7. Although global ablation of the miRNA pathway through genetic deletion of either Dicer or Dgcr8 has provided insights into the requirement of miRNAs in embryonic SCs and tissue development8–13, the pleiotropic defects caused by the loss of hundreds of miRNAs make it impossible to elucidate the underlying mechanisms. Surprisingly, it has not yet been reported that the loss of a single miRNA leads to severe defects either in animal development or in tissue SCs, in contrast to lethal phenotypes often observed in Dicer or Dgcr8 conditional knockout (cKO) animals7, 14, 15. Thus, it remains a major challenge to characterize the functional significance of individual miRNAs in tissue SCs and elucidate their molecular mechanisms.
Here we report that miR-205 is one of the most highly expressed miRNAs in skin SCs. We demonstrate by genetic deletion of miR-205 that miR-205 is not only essential for mouse development but also specifically required for the expansion of the progenitor and SC populations in both epidermis and HFs during neonatal skin development. Furthermore, we provide mechanistic insights into miR-205 function by demonstrating potent regulation of the PI3K pathway. Together, this study establishes a paradigm for the role of individual miRNAs in balancing the proliferation and quiescence of tissue SCs during early development.
Results
miR-205 is highly enriched in skin progenitors and stem cells
We have previously shown that both Dicer and Dgcr8 skin cKO animals exhibit much smaller body size and shortened HFs (Supplementary Fig. S1a)10, 13. When we monitored the dynamics of the HFSCs, we observed that HFSC specification was intact in the absence of Dicer in the newborn skin, as determined by immunofluorescence (IF) staining of Nfatc11, 16 (Supplementary Fig. S1b). However, in P4.5 Dicer cKO skin, the Nfatc1+ HFSC population was absent (Supplementary Fig. S1c) and the Sox9+ HFSCs and progenitor cells1, 17 were also significantly reduced in the bulge (Supplementary Fig. S1d). These observations indicate that global miRNA expression is required for the expansion and self-renewal of HFSCs during neonatal development.
We then performed in situ hybridization to determine the expression patterns for 10 most highly expressed miRNAs in neonatal skin13. We found that miR-205 is highly enriched in skin progenitors and SCs during skin development (Fig. 1a–e). At E10, when the skin still comprises a single layer of multipotent embryonic skin progenitors, miR-205 was already detectable in the cytoplasm of these p63+ progenitors18 (Fig. 1a). At E14, when the basal cells begin to stratify, miR-205 was restricted to the basal progenitors, and was absent from the differentiating suprabasal cells (Fig. 1b). When the HF lineage begins to emerge from the basal layer, miR-205 was further upregulated in the HF placode (Fig. 1c). When visualized in both neonatal and adult skin, miR-205 was expressed in interfollicular progenitor cells but most highly enriched in the bulge, where HFSCs reside (Fig. 1d,e and Supplementary Fig. S2a).
Figure 1.
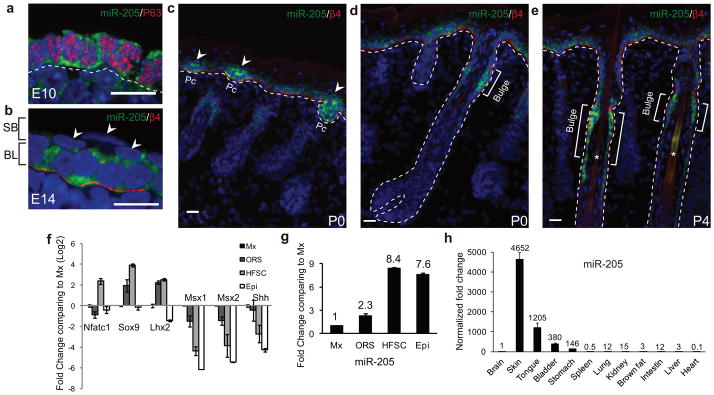
miR-205 is highly expressed in skin progenitors and SCs. (a) At E10, miR-205 is expressed in embryonic skin SCs that also express high levels of p63. Note the cytoplasmic localization of miR-205 signals. (b) At E14, miR-205 is restricted in the basal SCs. Arrowheads point to the differentiated, suprabasal cells that are negative for miR-205. The basement membrane is marked by β4 integrin. SB, suprabasal layer; BL, basal layer. (c) miR-205 is upregulated in the HF placode (Pc, arrowheads) but also expressed in interfollicular progenitors in the newborn skin (P0). Note more intense signal of miR-205 in the HF placode than in the interfollicular epidermis. (d and e) miR-205 is most highly expressed in the HFSCs located in the bulge of mature HFs. Brackets denote the bulge region. (f) FACS-purified populations are validated by qRT-PCR. Data shown are mean ± s.d. collected from 3 independent experiments. (g) Comparing to Matrix (Mx), miR-205 is highly enriched in HFSCs, followed by interfollicular progenitors (Epi) and ORS, as determined by qRT-PCR. Data shown are mean ± s.d. collected from 3 independent experiments. (h) miR-205 is highly enriched in organs containing stratified epithelial tissue, in contrast to a very low or non-detectable level in most of other vital organs. Data shown are mean ± s.d. collected from 3 independent experiments. All of the data are normalized to Sno25. Numbers indicate normalized fold change. Statistics source data for Fig. 1f–h can be found in Table S2. White dotted lines mark the epidermal/dermal boundary. a, b, confocal images; c, d, e, epifluorescence images. Scale bars, 20 μm.
To quantify the differential expression of miR-205 in the epidermal and HF lineages, we isolated interfollicular progenitors, HFSCs, outer root sheath (ORS) progenitors and transient amplifying Matrix (Mx) cells with a K14-RFP/Sox9-GFP mouse model19 in P4 skin (Supplementary Fig. S2c). We validated each population by qRT-PCR for well-established maker genes (Fig. 1f). We then determined that miR-205 was expressed at the highest level in the HFSCs, followed by the interfollicular progenitors and the ORS, when compared to the Mx cells (Fig. 1g). When we quantified miR-205 in telogen (P19) and anagen (P28) bulge as α6hiCD34hi populations20, we observed higher expression in the telogen HFSCs (Supplementary Fig. S2b). Together, our in situ and qRT-PCR analyses identify miR-205 as one of the most highly expressed miRNAs in skin progenitors and SCs.
We next determined miR-205’s expression among different murine tissues. In a comprehensive survey of miRNA expression patterns in mouse and human, miR-205 was only detected in epithelial tissues that include skin, prostate, and mammary gland21. Similarly, when we quantified miR-205 expression in 12 murine tissues obtained from P7.5 animals with qRT-PCR, we found that miR-205 was strongly enriched in stratified epithelial tissues including skin, tongue, bladder and stomach and largely absent from other tissues (Fig. 1h). Furthermore, in situ hybridization for miR-205 together with a well-established basal epithelial cell marker, Krt5 (K5) revealed that miR-205 was expressed only in K5+ cells in all tissues examined (Supplementary Fig. S3a–b). Although it remains a possibility that miR-205 may be expressed in rare cell populations in non-epithelial tissues, these data suggest that miR-205 is highly specific for K5+ basal cells within stratified epithelial tissues.
Generation of miR-205 knockout mouse
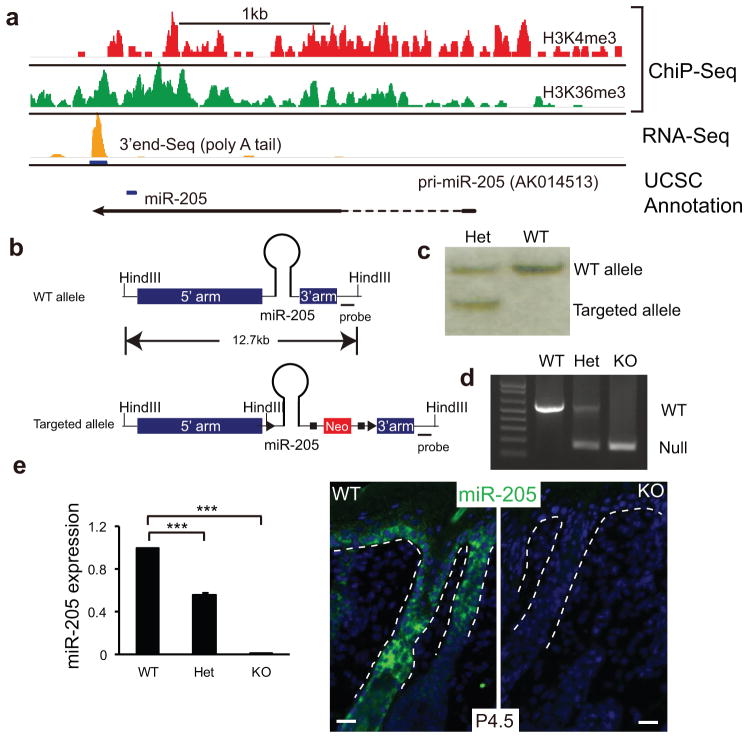
The miR-205 gene is located in an intergenic region on mouse chromosome 1. The primary transcript of miR-205 has been previously detected in mouse neonatal skin cDNA as a non-coding RNA (AK014513, UCSC genome browser) (Fig. 2a and Supplementary Fig. S4a). By using ChIP-Seq for H3K4me3 and H3K36me3 marks that define actively transcribed genomic regions by RNA polymerase II22, we determined that the primary transcript of miR-205 was transcribed in E14 basal cells (Fig. 2a). With an RNA-Seq specifically for the polyA tail23, we determined that the primary transcript was polyadenylated using a canonical signal (AUUAAA) (Fig. 2a). Although mature miR-205 sequences are highly conserved in vertebrates, the primary transcript shows poor conservation outside of the miR-205 hairpin (Supplementary Fig. S4). We then generated a miR-205 KO mouse model (Fig. 2b–d) and validated the complete ablation of miR-205 by qRT-PCR and in situ (Fig. 2e).
Figure 2.
Generation of miR-205 knockout mice. (a) Genomic analysis of the miR-205 locus in embryonic skin progenitor cells: H3K4me3 and H3K36me3 ChIP-Seq data define the actively transcribed region. 3′end sequencing of polyA RNAs detects the polyadenylation site of the primary transcript of miR-205. The blue bar denotes the position of the pre-miR-205 hairpin in the pri-miR-205 transcript. (b) Structure of the WT and targeted miR-205 locus. (c) Southern blot analysis confirms the generation of the targeted allele. (d) PCR analysis confirms genotypes. (e) Depletion of miR-205 is determined by qRT-PCR and in situ hybridization. Data shown are mean ± s.d. collected from five independent experiments. ***, P < 0.001 by Student’s t-test. White dotted lines mark the epidermal/dermal boundary.
miR-205 knockout mouse is neonatal lethal
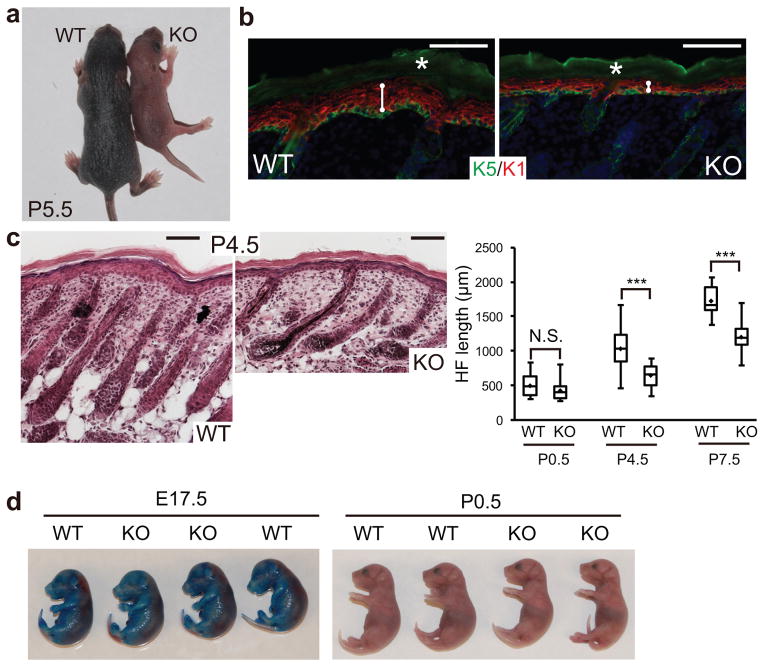
miR-205 KO animals were born with the expected Mendelian ratio [WT:het:KO=137(22%):328(53%):156(25%)]. Whereas the KO animals were indistinguishable in appearance from their WT or heterozygous littermates at birth, by P5 they became weaker and had less well-developed hair coats (Fig. 3a). miR-205 KO animals usually died ~10 days after birth, whereas the heterozygous animals showed no sign of any defects and were indistinguishable from their WT littermates. Therefore, the loss of miR-205 has dire consequences leading to neonatal lethality, unlike many individual miRNA KOs that are apparently normal and lack obvious developmental defects14. We investigated miR-205 in the skin because of the clear similarities in the miR-205 KO and the Dicer skin cKO animals, and because of the high miR-205 levels in skin. At P4.5, the miR-205 KO skin developed thinner epidermis (Fig. 3b). The HFs were short and also mis-angled, similar to the HFs that grow downward in the Dicer cKO (Fig. 3c). The terminal differentiation of both epidermal and HF lineages in the miR-205 KO was not dramatically altered, as judged by IF staining of skin lineage markers (Supplementary Fig. S5). Furthermore, these animals did not exhibit any discernible barrier defects (Fig. 3d). We did not observe any evaginating HFs and apoptotic cells, hallmarks of the Dicer cKO skin9, 10, suggesting that the absence of other miRNAs may be responsible for these defects.
Figure 3.
Genetic deletion of miR-205 causes neonatal lethality with severe skin defects. (a) At P5.5, the miR-205 KO pup is smaller and shows less developed hair coat, indicated by less pigmentation, compared with the WT littermate. (b) miR-205 KO skin shows thinner epidermis. K5 labels the basal layer and K1 labels the spinous layer. White lines measure the thickness of the epidermis; white asterisks denote autofluorescence. (c) Shorter HFs are developed in the KO skin. Box and whisker plots quantify the HF length: minimum and maximum lengths are marked by the whiskers; upper and lower boundaries indicate 25 and 75 quartile divisions; central lines indicate median; diamonds indicate average. Data shown are mean ± s.d., n = 20 HFs from 5 pairs of WT and KO littermates. ***, P < 0.001 by Student’s t-test. (d) miR-205 deletion does not cause barrier defects as indicated by the toluidine blue dye penetration assay. Scale bars in b and c, 100 μm.
Ablation of miR-205 impairs the proliferation of interfollicular progenitors
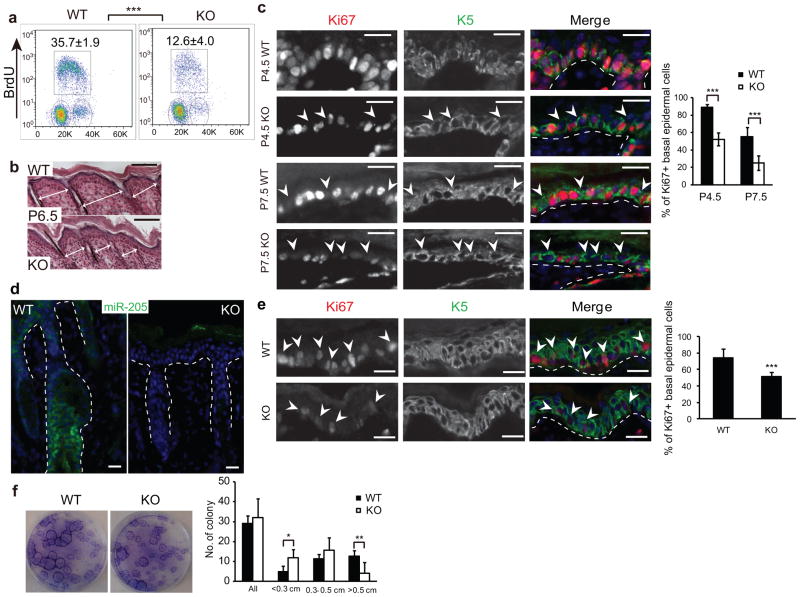
To examine the interfollicular progenitors24, we bred miR-205+/− to K14-RFP mice so that we could identify the progenitors by α6hi/RFPhi expression in the epidermal fraction after separating the epidermis from HFs (Supplementary Fig. S2c). The miR-205 KO progenitors exhibited significantly reduced proliferation compared to their WT counterparts using BrdU cell cycle analysis. Whereas an average of 35.7% of the progenitors were BrdU+ in the WT, only 12.6% were BrdU+ in the miR-205 KO (Fig. 4a). Therefore, the compromised proliferation likely contributes to the thinner epidermis, as well as the smaller skin surface of miR-205 KO animals (Fig. 4b). To further document the proliferation defects in vivo, we used Ki67 staining to distinguish cells that were in active cell cycle from those that exited the cell cycle. At P4.5, 88.9% of interfollicular progenitors were positive for Ki67 in the WT skin. In contrast, only 52.3% of progenitors were positive for Ki67 in the miR-205 KO skin. By P7.5, 55.1% of interfollicular progenitors were positive for Ki67 in the WT, whereas the Ki67 positive population dropped to 24.9% in the miR-205 KO skin (Fig. 4c). To distinguish whether these defects were intrinsic to the loss of miR-205 in the skin or due to systemic effects, we grafted WT and KO skin onto nude mice (Supplementary Fig. S6). In agreement with the cell-autonomous role of miR-205, the transplanted KO skin showed much less Ki67+ cells than the WT skin (Fig. 4d,e).
Figure 4.
Ablation of miR-205 compromises the proliferation of interfollicular progenitors. (a) Ablation of miR-205 results in a marked decline in S-phase cells that incorporate BrdU in the basal epidermis. Values are mean ± s.d. from five independent experiments; ***, P < 0.001 by Student’s t-test. (b) The KO skin shows shortened distance between HFs, an indication of less proliferative epidermis. White arrows measure the distance between adjacent HFs. (c) miR-205 KO interfollicular progenitors prematurely exit the cell cycle, as indicated by significantly decreased Ki67+ cells in the basal epidermis. White dotted lines mark the epidermal/dermal boundary; arrowheads indicate K5+/Ki67− interfollicular progenitors. Data shown are mean ± s.d. from five independent experiments; ***, P < 0.001 by Student’s t-test. (d) In situ hybridization confirms the complete loss of miR-205 in the grafted KO skin. (e) Fifteen days after grafting onto nude mice, the miR-205 KO epidermis is much less proliferative as indicated by less Ki67+ interfollicular progenitors and thinner epidermis. Data shown are mean ± s.d. from five independent experiments; ***, P < 0.001 by Student’s t-test. (f) miR-205 KO primary epidermal keratinocytes show a significant reduction in the formation of large holoclones in vitro. Data shown are mean ± s.d. from five independent experiments. *, P < 0.05; **, P < 0.01 by Student’s t-test. Scale bar is 100 μm in b and 20 μm in c, d and e.
We next isolated the interfollicular progenitors from both WT and KO skin and compared their colony formation capacity in vitro. We observed a strong reduction in the formation of large, holoclonal colonies by miR-205 KO progenitors even though the total number of colonies was similar (Fig. 4f). Collectively, these data suggest that the loss of miR-205 directly compromises the proliferation and leads to premature cell cycle exit in the interfollicular progenitors.
Ablation of miR-205 compromises the proliferation of progenitors and stem cells in hair follicles
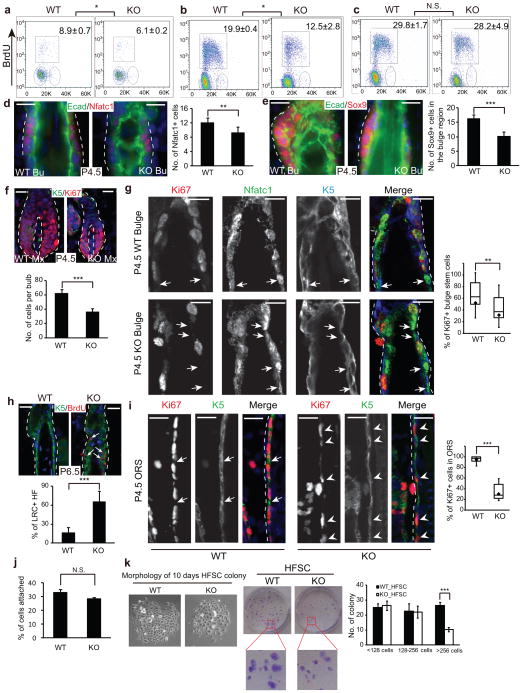
In the HF lineage, BrdU analysis revealed that the HFSCs and ORS progenitor cells were also less proliferative in the KO skin (Fig. 5a,b). In contrast, the transient amplifying Mx cells were equally proliferative between the WT and KO samples (Fig. 5c). Because miR-205 is highly enriched in the HFSCs and ORS but not in the Mx cells, these data indicate that the loss of miR-205 specifically compromises the proliferation of the progenitors and HFSCs. Consistent with the impaired cell cycle progression, when we quantified the HFSC population by IF staining for Nfatc1 or Sox9, miR-205 KO showed a mild but statistically significant reduction in the number of HFSCs (Fig. 5d,e). This indicates that the compromised proliferation leads to a smaller HFSC population in the absence of miR-205. The diminished SC and ORS progenitor population likely reduced the supply for transient amplifying Mx cells, leading to smaller hair bulbs, even though all Mx cells were still actively cycling in the KO (Fig. 5f).
Figure 5.
miR-205 is required for the expansion of HFSCs during neonatal development. (a–c) Ablation of miR-205 results in a marked decline in S-phase cells that incorporate BrdU in the HFSCs (a) and ORS progenitors (b), but does not affect the matrix cells (c). Values are mean ± s.d. from 5 independent experiments. *, P < 0.05. N.S., not significant. (d and e) Reduced HFSC population in the miR-205 KO bulge is shown by Nfatc1 and Sox9 staining. Bu, bulge. Data shown are mean ± s.d., n = 10 HFs from 5 pairs of WT/KO littermates. **, P < 0.01; ***, P < 0.001. (f) Matrix cells are equally proliferative in the KO and WT, despite the overall reduction of the matrix size. Data shown are mean ± s.d., n = 10 HFs from 5 pairs of WT/KO littermates. ***, P < 0.001. (g) Nfatc1+ HFSCs prematurely exit the cell cycle and become quiescent upon miR-205 deletion, determined by Ki67 staining. Arrows indicate Nfatc1+/Ki67− quiescent HFSCs. n = 10 HFs from 5 pairs of WT/KO littermates. **, P < 0.01. (h) BrdU pulse/chase experiments show increased label-retaining cells (LRC) in the miR-205 KO bulge. Data shown are mean ± s.d., n = 30 HFs from 3 pairs of WT/KO littermates. ***, P < 0.001. (i) Many ORS progenitors in the miR-205 KO exit the cell cycle prematurely. Arrows indicate K5+/Ki67− ORS cells in WT; arrowheads indicate K5+/Ki67+ ORS cells that are actively cycling in KO. n = 10 HFs from 5 pairs of WT/KO littermates. ***, P < 0.001. (j) Similar plating efficiency between WT and miR-205 KO HFSCs. (k) miR-205 KO HFSCs show a significant reduction in the formation of large holoclones in vitro. Data shown are mean ± s.d. from 5 independent experiments. ***, P < 0.001. For g and i, box and whisker plots: minimum and maximum are marked by the whiskers; upper and lower boundaries indicate 25 and 75 quartile divisions; central lines indicate median; diamonds indicate average. Statistics for all assays is calculated by Student’s t-test. Scale bars, 20 μm.
Unlike interfollicular progenitors, HFSCs in adult skin are quiescent and only become activated to fuel hair cycle periodically or in response to epidermal wound repair3, 24, 25. However, HFSCs are specified during hair morphogenesis1 and they must undergo neonatal expansion before entering quiescence. Because the deletion of miR-205 causes impaired HFSC proliferation, we asked whether miR-205 plays a key role in the transition from proliferation to quiescence. We first monitored the dynamics of newly specified HFSCs as marked by Nfatc1 during embryonic skin development1, 16, 17. At E17.5, all of these nascent HFSCs were actively cycling as indicated by strong Ki67 signals in the newly formed hair germ (Supplementary Fig. S7a). At P0.5, all of these HFSCs were still Ki67+, reflecting a continuous expansion of the HFSC population to fuel HF growth (Supplementary Fig. S7b). By P1.5, however, a few Nfatc1+ HFSCs especially those in mature HFs began to lose Ki67 signals, marking the onset of the transition towards quiescence (Supplementary Fig. S7c). By P4.5, 47.5% of the WT HFSCs lost Ki67 signal and transitioned towards quiescence. In contrast, 68.4% of the miR-205 KO HFSCs lost Ki67 signal and became quiescent (Fig. 5g). Furthermore, reduced HFSC proliferation led to the accumulation of label-retaining cells in the KO bulge in a BrdU pulse/chase experiment (Fig. 5h). Together, these analyses suggest the possibility that miR-205 KO HFSCs prematurely exit the cell cycle and transition towards quiescence.
While miR-205 is most highly expressed in HFSCs, it is also expressed in the ORS. As expected, we observed a dramatic decrease in Ki67+ cells in the miR-205 KO ORS (Fig. 5i). The ORS cells constitute the major portion of the HF in length, such that their reduced proliferation likely contributed to the shortened HFs as seen in the KO skin. Taken together, these in vivo results provide insights for the requirement for miR-205 to maintain cell proliferation in neonatal HFSCs and progenitors.
We next isolated HFSCs from WT and miR-205 KO skin and compared their colony formation capacity in vitro. We first confirmed that both WT and KO cells had similar plating efficiency (Fig. 5j). Importantly, miR-205 KO HFSCs showed significantly compromised colony formation capacity such that the number of large holoclones was dramatically reduced (Fig. 5k). Altogether, these findings demonstrate that miR-205 is intrinsically required for the proliferation of HFSCs both in vivo and in vitro.
Ablation of miR-205 impairs the proliferation of basal cells in stratified epithelium
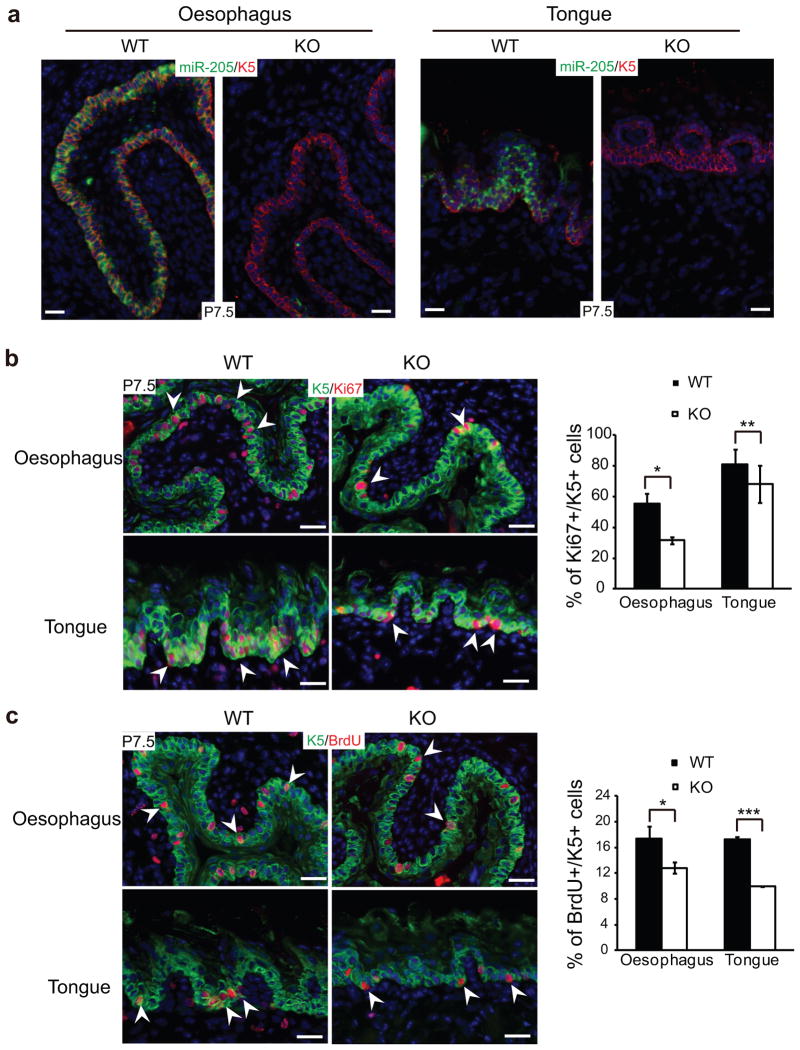
Because miR-205 was detected in K5+ basal cells in other stratified epithelium located in oesophagus and tongue (Supplementary Fig. S3a), we examined the cell proliferation in these tissues. As expected, the proliferation of K5+ basal cells in oesophagus and tongues was also compromised, as judged by both Ki67 and BrdU labeling (Fig. 6). These results indicate a general requirement for miR-205 in K5+ basal cells of the stratified epithelium. Furthermore, compromised proliferation in multiple stratified epithelial tissues could contribute to the lethality of miR-205 KO animals.
Figure 6.
Ablation of miR-205 impairs the proliferation of K5+ basal cells in stratified epithelium. (a) Confirmation of the complete loss of miR-205 in the KO oesophagus and tongue by in situ hybridization. (b and c) K5+ basal cells are less proliferative as indicated by Ki67 and BrdU staining in the KO oesophagus and tongue. Data shown are mean ± s.d. from five independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001 by Student’s t-test.
miR-205 directly targets multiple negative regulators of the PI3K pathway
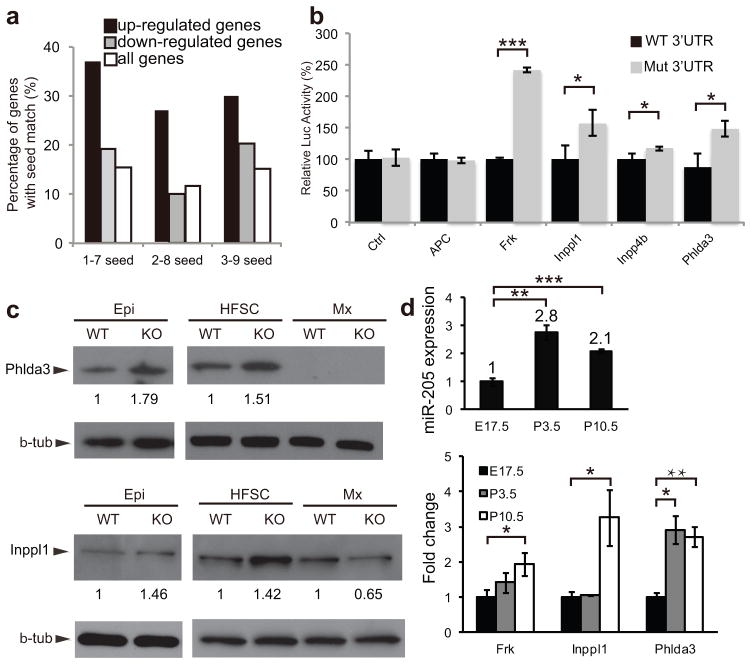
To identify direct targets of miR-205, we isolated the HFSCs from WT and miR-205 KO skin at P4 and compared mRNA expression by microarray analysis. We identified 201 genes that were consistently upregulated more than 20% in miR-205 KO samples in two sets of biological duplicates. We performed a motif search for all possible 7mers that are represented in the miR-205 sequences in the 5′ UTR, coding region and 3′UTR of these genes. We found that only the 1–7, 2–8 and 3–9 seed motifs, which are located at the 5′ end of miR-205, were significantly enriched only in the 3′UTRs of the upregulated genes (Fig. 7a and Supplementary Fig. S8a–c). As a control, we did not observe any enrichment for any 7mer motif represented by miR-1 (Supplementary Fig. S8d). These results indicate, as expected, that miR-205 selectively represses the expression of genes that bear its seed sequences in their 3′UTR. Out of 201 upregulated genes, 96 genes (47.8%) contain at least one miR-205 seed match in their 3′UTR (Supplementary Table 1). Collectively, these results support the cell-autonomous role of miR-205 in modulating gene expression in the HFSCs.
Figure 7.
miR-205 directly targets multiple negative regulators of the PI3K/Akt pathway. (a) The enrichment of miR-205 seed matches in the 3′UTR of the upregulated genes in the miR-205 KO HFSCs is shown by a higher percentage of genes containing the seed matches over that of the downregulated or all genes. (b) Target validation is shown by the 3′UTR luciferase assay. For control (Ctrl), black and grey bars represent the luciferase activity of pGL3-Ctrl with and without miR-205 transfection, respectively. For the target genes, black bars represent the normalized luciferase activity (set at 100%) of the pGL3-Luc reporter fused with the 3′UTR from each candidate; grey bars represent the derepressed luciferase activity of the pGL3-Luc reporter with mutated miR-205 seed matches in the 3′UTR. Data shown are mean ± s.d. from six independent experiments. *, P < 0.05; ***, P < 0.001 by Student’s t-test. (c) Derepression of miR-205 targets Phlda3 and Inppl1 in miR-205 KO interfollicular progenitors (Epi) and HFSCs. Phlda3 is not detected in either WT or KO matrix (Mx), whereas Inppl1 is not derepressed in miR-205 KO matrix. Numbers between panels represent the densitometry values of target protein normalized to β-tubulin. (d) Gradually increased expression of miR-205 and its targets from embryonic stage to postnatal stages is quantified by qRT-PCR. Data shown are mean ± s.d. from 5 independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001 by Student’s t-test. Uncropped images of blots are shown in Supplementary Fig. S9.
We performed a pathway analysis for these 96 genes26 and identified tumor suppressor as the most significantly enriched SwissProt/PIR keyword. Derepression of tumor suppressor genes (TSGs) is known to inhibit self-renewal and reduce stem cell populations27–29. In our list, APC, Frk, Inpp4b and Phlda3 are known TSGs30–32. We also identified Inppl1, a known negative regulator in the PI3K pathway33, which is a previously identified miR-205 target in human keratinocytes34, 35. To validate these targets, we first determined the 3′ UTR for each gene using our RNA-Seq database generated from the HFSCs and then tested their responsiveness to miR-205 by insertion 3′ to a luciferase indicator gene, with both WT and mutated miR-205 seed sequences (Supplementary Fig. S8e). Four of five genes were robustly regulated by miR-205 as demonstrated by derepressed luciferase activity when the miR-205 target site(s) was mutated (Fig. 7b). Among these targets, Phlda3 and Inppl1 were abundantly expressed and we further validated their upregulation at the protein level by Western blotting with FACS purified interfollicular progenitors and HFSCs (Fig. 7c). When we used the purified Mx cells as a control, we did not detect Phlda3 in either WT or KO cells and we did not observe upregulation of Inppl1 in the KO Mx cells. These data confirm the specific regulation of these targets by miR-205 in interfollicular progenitors and HFSCs.
To provide more insights into the expression pattern of miR-205 and its target mRNAs, we isolated HFSCs and measured their expression level by qRT-PCR during embryonic/neonatal development. Interestingly, the expression of both miR-205 and these targets gradually increased from E17.5 to P3.5 and P10.5 (Fig. 7d). This result suggests that miR-205 is upregulated to balance the enhanced expression of these TSGs.
miR-205 is required for the maintenance of pAkt levels
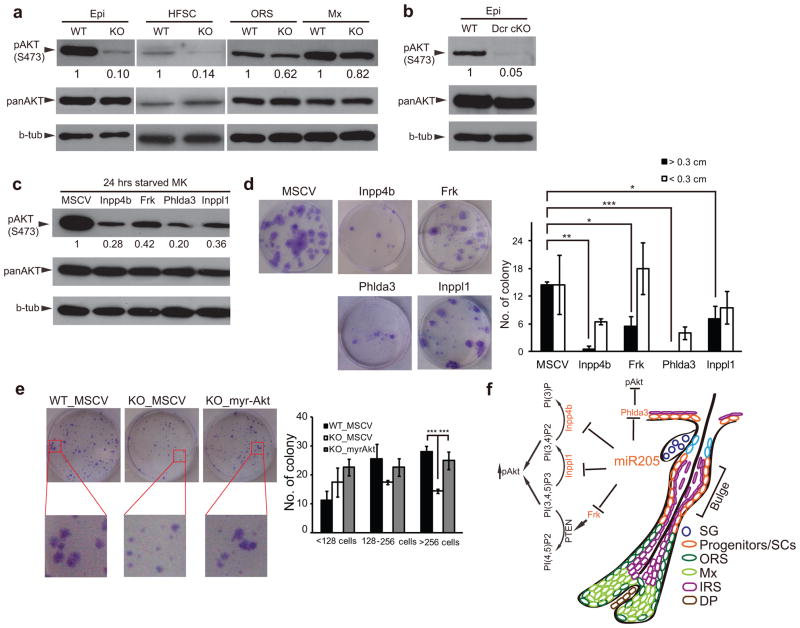
We noted that Frk, Inpp4b, Inppl1 and Phlda3 all negatively regulate the PI3K/Akt pathway30–33. This prompted us to examine pAkt levels in the skin. Strikingly, the pAkt levels were significantly downregulated in interfollicular progenitors (90%), HFSCs (86%) and ORS (38%) but only slightly in the Mx cells (18%) in miR-205 KO skin (Fig. 8a). To ensure that the downregulation of pAkt was specific to these skin lineages and not due to systemic defects in the miR-205 KO animals, we measured pAkt levels in other vital organs including brain, brown fat, heart and lung, and observed normal pAkt levels (Supplementary Fig. S8f). Intriguingly, pAkt was also significantly downregulated in the Dicer cKO epidermis (95%) (Fig. 8b). These results suggest that miR-205 is specifically required to maintain pAkt levels in the progenitors and SCs in skin.
Figure 8.
miR-205 is required to maintain pAkt levels by targeting multiple negative regulators in skin progenitors and SCs. (a) pAkt levels are significantly decreased in miR-205 KO epidermis (Epi), HFSCs and ORS, but only slightly decreased in matrix (Mx). (b) pAkt is significantly downregulated in Dicer cKO (Dcr cKO) epidermis (Epi). (c) Keratinocytes infected with miR-205 targets (Inpp4b, Frk, Phlda3 and Inppl1) show lower pAkt levels compared to control (MSCV) infected cells. (d) Forced expression of individual miR-205 targets compromises the capacity of primary keratinocytes to form productive colonies. Data shown are mean ± s.d. from five independent experiments. *, P < 0.05; **, P < 0.01; ***, P< 0.001 by Student’s t-test. (e) Constitutively active Akt (myr-Akt) rescues the growth defects of miR-205 KO HFSCs in vitro. Data shown are mean ± s.d. from five independent experiments. ***, P < 0.001 by Student’s t-test. (f) A model illustrates the role of miR-205 in the proliferation and expansion of skin progenitors and SCs by antagonizing multiple negative regulators of the PI3K/Akt pathway. Uncropped images of blots are shown in Supplementary Fig. S9.
The level of pAkt has been directly linked to the proliferation and self-renewal of skin SCs36–38. When we blocked pAkt using a small molecule inhibitor of PI3K in cultured keratinocytes, the growth of primary keratinocytes was abolished (Supplementary Fig. S8g). To test whether the derepression of miR-205 targets, including Inpp4b, Frk, Phlda3 and Inppl1, dampens the PI3K/Akt pathway, we infected primary keratinocytes with MSCV vectors expressing each of these genes. Upon enhanced expression of these targets, pAkt levels were downregulated (Fig. 8c) and cell proliferation was compromised (Fig. 8d). Interestingly, the ability to inhibit growth by these regulators correlated with their ability to reduce pAkt levels. This suggests that high pAkt levels are closely associated with proliferative capacity. Finally, to test whether the downregulation of pAkt contributes primarily to the growth defects caused by the loss of miR-205, we infected the miR-205 KO HFSCs with a constitutively active Akt mutant (myr-Akt) and evaluated the colony formation capacity. As expected, myr-Akt largely rescued the growth defects of freshly isolated miR-205 KO HFSCs (Fig. 8e). Taken together, these results argue that miR-205 co-represses multiple negative regulators of the PI3K/Akt pathway and, by doing so, miR-205 maintains a high pAkt level that promotes the proliferation and expansion of progenitors and SCs in developing skin (Fig. 8f).
Discussion
miRNAs have been implicated important roles in gene regulation in mammalian SCs7, 39–41. However, genetic KO models in either C. elegans or mouse have implicated very few individual miRNAs as indispensable for animal development14, 15, 42. Here we report that miR-205 plays an essential role in promoting their expansion during early development. This provides one of the first examples in which genetic deletion of a single miRNA causes severe developmental defects and compromises the function of tissue SCs.
Our study also provides additional insights into the transition of neonatal HFSCs from the active cycling state to quiescence. We identify a network of negative regulators of the PI3K pathway that are directly targeted by miR-205. During skin development, the expression of these negative regulators and miR-205 increases concurrently. Upon the loss of miR-205, negative regulators of the PI3K pathway become prematurely elevated and reduce pAkt levels. Reduced pAkt levels then compromises the proliferation of the progenitor and SC populations in the skin. However, the proliferation defect alone does not likely cause the lethality. We note that miR-205 has been implicated important roles in the regulation of cell adhesion and migration43, 44. Positive regulators of cell migration e.g. Arhgap5 and Cxcl12 are also identified as targets of miR-205 (Supplementary Table S1). It is an intriguing possibility that potential defects in cell migration could contribute to the severe developmental defects.
Methods
Methods and any associated references are available in the online version of the paper.
Supplementary Material
Acknowledgments
We are grateful to E. Fuchs for K14-RFP mice. We thank T. Blumenthal, T. Cech, B. Cullen, M. Han, M. Winey and X-J. Wang for comments on the manuscript. We thank C. Yang, J. Gao, D. Feng for the generation of miR-205 knockout; S. Ha and L. Greiner for assistance in the animal facility; Y. Han for FACS; and G. Voeltz for confocal microscopy. We also thank members of the Yi lab for their critical discussions. This publication was made possible by a start-up fund provided by the University of Colorado and Grant Number R00AR054704 and R01AR059697 (R.Y.) and R01GM083300 (E.C.L).
Footnotes
Note: Supplementary Information is available on the Nature Cell Biology Website
Author Contributions
R.Y. conceived the study. D.W. carried out most experiments and analyzed the data with assistance from Z.Z. (bioinformatic analysis), E.O. (in situ hybridization), L.W. (ChIP-Seq and RNA-Seq) and X.F. (target validation). E.C.L. provided critical resources. R.Y. and D.W. wrote the manuscript with the input from all authors.
Competing Financial Interests
The authors declare no competing financial interests.
References
- 1.Nowak JA, Polak L, Pasolli HA, Fuchs E. Hair follicle stem cells are specified and function in early skin morphogenesis. Cell Stem Cell. 2008;3:33–43. doi: 10.1016/j.stem.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cotsarelis G, Sun TT, Lavker RM. Label-retaining cells reside in the bulge area of pilosebaceous unit: implications for follicular stem cells, hair cycle, and skin carcinogenesis. Cell. 1990;61:1329–1337. doi: 10.1016/0092-8674(90)90696-c. [DOI] [PubMed] [Google Scholar]
- 3.Tumbar T, et al. Defining the epithelial stem cell niche in skin. Science. 2004;303:359–363. doi: 10.1126/science.1092436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.He S, Nakada D, Morrison SJ. Mechanisms of stem cell self-renewal. Annu Rev Cell Dev Biol. 2009;25:377–406. doi: 10.1146/annurev.cellbio.042308.113248. [DOI] [PubMed] [Google Scholar]
- 5.Hsu YC, Fuchs E. A family business: stem cell progeny join the niche to regulate homeostasis. Nat Rev Mol Cell Biol. 2012;13:103–114. doi: 10.1038/nrm3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yi R, Fuchs E. MicroRNAs and their roles in mammalian stem cells. J Cell Sci. 2011;124:1775–1783. doi: 10.1242/jcs.069104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murchison EP, Partridge JF, Tam OH, Cheloufi S, Hannon GJ. Characterization of Dicer-deficient murine embryonic stem cells. Proc Natl Acad Sci U S A. 2005;102:12135–12140. doi: 10.1073/pnas.0505479102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andl T, et al. The miRNA-processing enzyme dicer is essential for the morphogenesis and maintenance of hair follicles. Curr Biol. 2006;16:1041–1049. doi: 10.1016/j.cub.2006.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yi R, et al. Morphogenesis in skin is governed by discrete sets of differentially expressed microRNAs. Nat Genet. 2006;38:356–362. doi: 10.1038/ng1744. [DOI] [PubMed] [Google Scholar]
- 11.Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet. 2007;39:380–385. doi: 10.1038/ng1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Melton C, Judson RL, Blelloch R. Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature. 2010;463:621–626. doi: 10.1038/nature08725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yi R, et al. DGCR8-dependent microRNA biogenesis is essential for skin development. Proc Natl Acad Sci U S A. 2009;106:498–502. doi: 10.1073/pnas.0810766105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu N, Olson EN. MicroRNA regulatory networks in cardiovascular development. Dev Cell. 2010;18:510–525. doi: 10.1016/j.devcel.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Small EM, Olson EN. Pervasive roles of microRNAs in cardiovascular biology. Nature. 2011;469:336–342. doi: 10.1038/nature09783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horsley V, Aliprantis AO, Polak L, Glimcher LH, Fuchs E. NFATc1 balances quiescence and proliferation of skin stem cells. Cell. 2008;132:299–310. doi: 10.1016/j.cell.2007.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hsu YC, Pasolli HA, Fuchs E. Dynamics between stem cells, niche, and progeny in the hair follicle. Cell. 2011;144:92–105. doi: 10.1016/j.cell.2010.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Senoo M, Pinto F, Crum CP, McKeon F. p63 Is essential for the proliferative potential of stem cells in stratified epithelia. Cell. 2007;129:523–536. doi: 10.1016/j.cell.2007.02.045. [DOI] [PubMed] [Google Scholar]
- 19.Zhang L, Stokes N, Polak L, Fuchs E. Specific MicroRNAs Are Preferentially Expressed by Skin Stem Cells To Balance Self-Renewal and Early Lineage Commitment. Cell Stem Cell. 2011;8:294–308. doi: 10.1016/j.stem.2011.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blanpain C, Lowry WE, Geoghegan A, Polak L, Fuchs E. Self-renewal, multipotency, and the existence of two cell populations within an epithelial stem cell niche. Cell. 2004;118:635–648. doi: 10.1016/j.cell.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 21.Landgraf P, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guttman M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang L, Dowell RD, Yi R. Genome-wide maps of polyadenylation reveal dynamic mRNA 3′-end formation in mammalian cell lineages. RNA. 2013;19:413–425. doi: 10.1261/rna.035360.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blanpain C, Fuchs E. Epidermal stem cells of the skin. Annu Rev Cell Dev Biol. 2006;22:339–373. doi: 10.1146/annurev.cellbio.22.010305.104357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ito M, et al. Stem cells in the hair follicle bulge contribute to wound repair but not to homeostasis of the epidermis. Nat Med. 2005;11:1351–1354. doi: 10.1038/nm1328. [DOI] [PubMed] [Google Scholar]
- 26.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park IK, et al. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature. 2003;423:302–305. doi: 10.1038/nature01587. [DOI] [PubMed] [Google Scholar]
- 28.Ezhkova E, et al. EZH1 and EZH2 cogovern histone H3K27 trimethylation and are essential for hair follicle homeostasis and wound repair. Genes Dev. 2011;25:485–498. doi: 10.1101/gad.2019811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cheng T, et al. Hematopoietic stem cell quiescence maintained by p21cip1/waf1. Science. 2000;287:1804–1808. doi: 10.1126/science.287.5459.1804. [DOI] [PubMed] [Google Scholar]
- 30.Gewinner C, et al. Evidence that inositol polyphosphate 4-phosphatase type II is a tumor suppressor that inhibits PI3K signaling. Cancer Cell. 2009;16:115–125. doi: 10.1016/j.ccr.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yim EK, et al. Rak functions as a tumor suppressor by regulating PTEN protein stability and function. Cancer Cell. 2009;15:304–314. doi: 10.1016/j.ccr.2009.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kawase T, et al. PH domain-only protein PHLDA3 is a p53-regulated repressor of Akt. Cell. 2009;136:535–550. doi: 10.1016/j.cell.2008.12.002. [DOI] [PubMed] [Google Scholar]
- 33.Dyson JM, et al. The SH2 domain containing inositol polyphosphate 5-phosphatase-2: SHIP2. Int J Biochem Cell Biol. 2005;37:2260–2265. doi: 10.1016/j.biocel.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 34.Yu J, et al. MicroRNA-184 antagonizes microRNA-205 to maintain SHIP2 levels in epithelia. Proc Natl Acad Sci U S A. 2008;105:19300–19305. doi: 10.1073/pnas.0803992105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu J, et al. MicroRNA-205 promotes keratinocyte migration via the lipid phosphatase SHIP2. FASEB J. 2010;24:3950–3959. doi: 10.1096/fj.10-157404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alonso L, et al. Sgk3 links growth factor signaling to maintenance of progenitor cells in the hair follicle. J Cell Biol. 2005;170:559–570. doi: 10.1083/jcb.200504131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murayama K, et al. Akt activation induces epidermal hyperplasia and proliferation of epidermal progenitors. Oncogene. 2007;26:4882–4888. doi: 10.1038/sj.onc.1210274. [DOI] [PubMed] [Google Scholar]
- 38.Kobielak K, Stokes N, de la Cruz J, Polak L, Fuchs E. Loss of a quiescent niche but not follicle stem cells in the absence of bone morphogenetic protein signaling. Proc Natl Acad Sci U S A. 2007;104:10063–10068. doi: 10.1073/pnas.0703004104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gangaraju VK, Lin H. MicroRNAs: key regulators of stem cells. Nat Rev Mol Cell Biol. 2009;10:116–125. doi: 10.1038/nrm2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ivey KN, Srivastava D. MicroRNAs as regulators of differentiation and cell fate decisions. Cell Stem Cell. 2010;7:36–41. doi: 10.1016/j.stem.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 41.Martinez NJ, Gregory RI. MicroRNA gene regulatory pathways in the establishment and maintenance of ESC identity. Cell Stem Cell. 2010;7:31–35. doi: 10.1016/j.stem.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miska EA, et al. Most Caenorhabditis elegans microRNAs are individually not essential for development or viability. PLoS Genet. 2007;3:e215. doi: 10.1371/journal.pgen.0030215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gregory PA, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 44.Li C, Finkelstein D, Sherr CJ. Arf tumor suppressor and miR-205 regulate cell adhesion and formation of extraembryonic endoderm from pluripotent stem cells. Proc Natl Acad Sci U S A. 2013;110:E1112–1121. doi: 10.1073/pnas.1302184110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.