Abstract
Introgressive hybridization may cause substantial discordances among phylogenies based on different genetic markers. Such discordances have been found in diverse mammal species including primates. A recent study of mitochondrial DNA (mtDNA) revealed several poly- and paraphyletic relationships in African green monkeys (Chlorocebus), suggesting contemporary and/or ancient introgressive hybridization among almost all parapatric species of the genus. However, mtDNA analyses alone do not allow us to draw conclusions concerning introgression events. In this study we analyzed two Y chromosomal (Y-chr) markers for 30 African green monkey samples and compared the resulting genetic relationships to those based on published mtDNA data. In line with the results for mtDNA, we found no Y-chr evidence of hypothesized hybridization among Chlorocebus sabaeus and C. tantalus in the northern part of the contact zone in West Africa, and we found two distinct and distantly related Y-chr haplotypes within the range of C. tantalus, suggesting possible cryptic genetic diversity rather than ancient introgressive hybridization in this species. In contrast, Y-chr data revealed monophyletic relationships within Chlorocebus pygerythrus from East Africa, suggesting that mtDNA paraphylies found in this species are most likely to be the result of ancient introgressive hybridization and subsequent cytonuclear extinction of an earlier taxon. Our results accentuate the importance of analyzing sex chromosomal data in addition to mtDNA to obtain more information on the potential outcomes of hybridization with respect to genetic and species diversity. Analysis of more diverse nuclear marker sets is needed to obtain a more complete picture of the African green monkey evolution.
Keywords: Ancient hybridization, Nuclear swamping, Old World monkeys, Outcomes of hybridization
Introduction
Molecular genetic studies, often based on mitochondrial DNA (mtDNA), have provided evidence of hybridization in diverse animal taxa in the last two decades (Abbott et al. 2013; Mallet 2005; Zinner et al. 2011). Introgressive hybridization, or the transmission of genetic material from one species into the genome of another, closely related species, accounts for a large proportion of reticulated phylogenies and apparent para- or polyphyletic relationships (Mallet 2005). As the heterogametic sex is often inviable or sterile in hybrids (Haldane 1922), one common result of introgressive hybridization in mammals is the transmission of the maternally inherited mitochondrial genome (Mallet 2005; Orr 1997; Schilthuizen et al. 2011). However, factors such as sex-biased dispersal or unequal population sizes may lead to backcrossing with only one of the parental species, which may cause nuclear swamping, mitochondrial or Y-chromosomal (Y-chr) capture (Zinner et al. 2011).
Evidence of frequent introgressive hybridization has been found in diverse African savannah mammals, and this has been attributed to recurrent changes in the extent and distribution of the savannah biome, and hence in range expansions and retractions of populations with the formation of secondary contact zones due to climatic changes in the last million years, e.g., hartebeests (Flagstad et al. 2001) and warthogs (Muwanika et al. 2003). Among the primates, baboons (Papio) are widely distributed in sub-Saharan African savannahs, and hybridization is ongoing in most places where the ranges of two species meet (Charpentier et al. 2012; Jolly et al. 2011; Keller et al. 2010; Tung et al. 2008). Moreover, discordant patterns between mtDNA and morphotypes also occur in regions where no current hybrid zone exists (Zinner et al. 2009). As in most mammalian taxa, males are predominantly the dispersing sex in baboons, suggesting that the discordant patterns are due to nuclear swamping through male-mediated introgressive hybridization (Zinner et al. 2009). In some of these cases introgression presumably led to the cytonuclear extinction of earlier baboon taxa or populations, whose mitochondrial genomes persist only as a trace in extant species (Keller et al. 2010; Zinner et al. 2009).
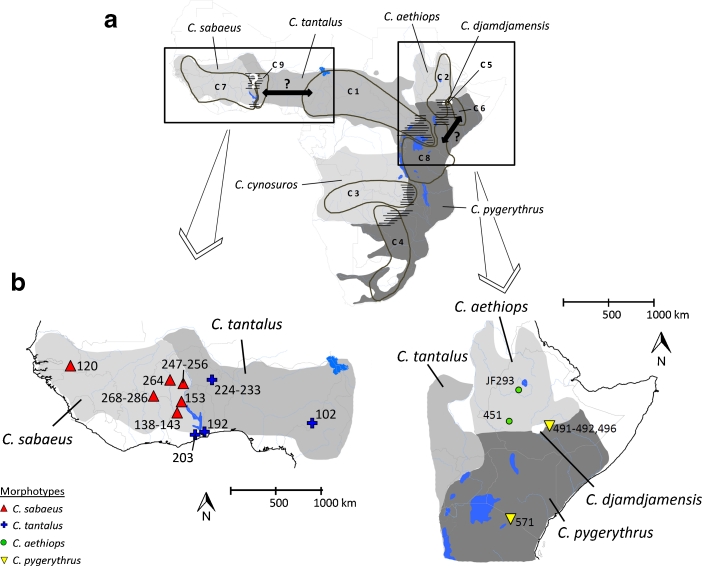
Like baboons, African green monkeys of the genus Chlorocebus constitute parapatric species that inhabit sub-Saharan savannahs (Hill 1966; Kingdon 1997; Lernould 1988). Hybridization has been assumed between most of the species in East Africa based on morphological data such as the occurrence of intermediate morphotypes (Kingdon 1997; Mekonnen et al. 2012; Napier 1981). Further, a recent study of mtDNA including samples from almost all contact zones throughout the genus’ range found several discordant phylogenetic patterns, suggesting that introgressive hybridization might be common among all African green monkeys (Haus et al. 2013). Most of these discordances support previous observations of ongoing hybridization in contemporary contact zones between Chlorocebus aethiops and C. djamdjamensis and between C. aethiops and C. pygerythrus in Ethiopia as well as between C. tantalus and C. pygerythrus in Uganda and Kenya (Fig. 1a; Haus et al. 2013; Kingdon 1997; Napier 1981). In addition, Haus et al. (2013) found some inconsistencies between morphology and mtDNA beyond current contact zones. Their analysis revealed two distinct clades within the range of Chlorocebus tantalus: a western clade including samples from Ghana, Burkina Faso, and Togo (C 9, Fig. 1a) and an eastern clade with samples originating from Nigeria, Cameroon, Uganda, and northwest Kenya (C 1, Fig. 1a). Similarly, samples of Chlorocebus pygerythrus cause polyphyletic relationships in the mtDNA tree with distinct clades found in southeast Ethiopia and Somalia (C 6), in Kenya and Tanzania (C 8), and in Southern Africa (C 4, Fig. 1a; Haus et al. 2013). Because these mtDNA clades are geographically localized, they are unlikely to be explained by random sorting failures such as incomplete lineage sorting (Avise 2004; Funk and Omland 2003). A possible alternative reason for the discordant patterns might be nuclear swamping and subsequent cytonuclear extinction through ancient introgression, as suggested for some baboon taxa (Zinner et al. 2009).
Fig. 1.
Distribution map of African green monkeys (Chlorocebus spp.). (a) Distribution of mitochondrial DNA (mtDNA) clades and presumed hybridization in contemporary contact zones (hatched areas) according to Haus et al. (2013). Black arrows indicate possible ancient introgression events we test in this study. Possible hybridization in the West African contact zone is indicated with a white question mark. (b) Collection sites for samples of morphotypes from West and East Africa used in this study.
There is conflicting information on the occurrence of hybridization between Chlorocebus sabaeus and C. tantalus. These taxa seem to be separated by the White Volta and the Oti River in Ghana, with Chlorocebus sabaeus west and C. tantalus east of the rivers (Fig. 1; Booth 1956, 1958; Haus et al. 2013). However, hybridization has been assumed to occur in the northern part of the contact zone in Burkina Faso, where confluents of the Volta River are unlikely to represent geographical barriers for the monkeys (Fig. 1a; Lernould 1988). The analysis of samples collected east and west of the Volta River and its tributaries in northern Ghana and in Burkina Faso yielded concordant results between mt-haplotype variation and morphotype distribution, suggesting that no gene flow exists in the northern part of the contact zone (Haus et al. 2013). An exceptional population of Chlorocebus tantalus population occurs west of the Volta River in south Ghana (Booth 1956, 1958; Groves 2001). Despite its morphology, which clearly resembles that of Chlorocebus tantalus, mtDNA of this population falls within the C. sabaeus clade (C 7), suggesting introgressive hybridization in the south (Haus et al. 2013; Fig. 1). However, mtDNA analysis alone cannot detect hybridization processes such as Y-chr capture, and the possibility remains that hybridization also occurred in the northern range.
We here analyze two Y-chr markers for 30 African green monkey samples and compare genetic relationships based on Y-chr sequences to those based on previously published mtDNA sequences and morphology. We focus 1) on potential ancient introgression events within Chlorocebus pygerythrus from East Africa and within C. tantalus, which may have caused the observed polyphyletic relationships in the mtDNA phylogeny (Haus et al. 2013); and 2) on the hypothesized hybridization in the West African contact zone between Chlorocebus sabaeus and C. tantalus (Fig. 1b). If some mtDNA clades in Chlorocebus pygerythrus and C. tantalus represent relict mitochondrial genomes resulting from ancient introgression, then we predict a pattern of Y-chr DNA relationships that is concordant with the species morphology, but not with the native mtDNA. If male-mediated introgressive hybridization has occurred or is ongoing among African green monkey species in the West African contact zone, we predict that our analysis will reveal discordant phylogenetic relationships based on Y-chr and mtDNA data.
Methods
Data Sampling
We collected fecal samples of Chlorocebus aethiops and of the population of C. pygerythrus from southeast Ethiopia. We also collected samples at eight different sites on both banks of the Volta River and its confluents in Ghana and Burkina Faso covering the distribution of the western clade of Chlorocebus tantalus as well as the potential contact zone of C. sabaeus and C. tantalus (Fig. 1b, Table I). We preserved feces in >90% ethanol for ≥24 h, dried them, and transferred them into tubes with silica beads for further storage (Nsubuga et al. 2004). Whenever we observed a green monkey we recorded information on external morphological characters based on Hill (1966), Napier (1981), and Groves (2001) to determine the species (Table II). We recorded all geographic localities using GPS (Table I). We also analyzed fecal samples from Chlorocebus tantalus (East Nigeria) and C. sabaeus (Senegal) (Haus et al. 2013), and blood samples from captive C. sabaeus and C. pygerythrus from the Paul Ehrlich Institute (PEI) in Langen, Germany. These individuals were offspring of green monkeys originating from Barbados and from East Africa (Kenya or Tanzania), respectively (Fig. 1b, Table I).
Table I.
Sample identities, collection sites, and GenBank accession numbers of the two Y-chr markers SRY and ZFY and the mitochondrial cytochrome b gene (cyt b) for Chlorocebus samples used in this study
| ID | Species | Country | Location | Longitude latitude | GenBank accession no. | ||
|---|---|---|---|---|---|---|---|
| SRY | ZFY | Cyt b | |||||
| JF293 | aet | — | — | — | JF293182b | JF293267b | — |
| 102 | tan | Nigeria | Gashaka Gumti NP | 11.19677 | KC843980 | KC844009 | JX983843a |
| 31.73892 | |||||||
| 120 | sab | Senegal | Niokolo Koba NP | 13.02577 | KC843991 | KC844020 | JX983828a |
| 13.23736 | |||||||
| 138 | sab | Ghana | Bui NP | 8.29083 | KC844002 | KC844031 | JX983804a |
| –2.28465 | |||||||
| 139 | sab | Ghana | Bui NP | 8.29114 | KC843997 | KC844026 | JX983805a |
| –2.28431 | |||||||
| 142 | sab | Ghana | Bui NP | 8.29114 | KC843992 | KC844021 | JX983806a |
| –2.28431 | |||||||
| 143 | sab | Ghana | Bui NP | 8.29090 | KC843990 | KC844019 | JX983807a |
| –2.28388 | |||||||
| 153 | sab | Ghana | Mole NP | 9.26008 | KC843993 | KC844022 | JX983811a |
| –1.86058 | |||||||
| 192 | tan | Ghana | Kalapka RR | 6.45293 | KC843985 | KC844014 | JX983840a |
| 0.38055 | |||||||
| 203 | tan | Ghana | Shai Hills RR | 5.89777 | — | — | JX983823a |
| 0.06897 | |||||||
| 224 | tan | Burkina Faso | Park D'Arly | 11.60094 | KC843984 | KC844013 | JX983831a |
| 1.39187 | |||||||
| 225 | tan | Burkina Faso | Park D'Arly | 11.60094 | KC843981 | KC844010 | JX983832a |
| 1.39187 | |||||||
| 226 | tan | Burkina Faso | Park D'Arly | 11.58281 | KC843982 | KC844011 | JX983833a |
| 1.46103 | |||||||
| 233 | tan | Burkina Faso | Park D'Arly | 11.58087 | KC843983 | KC844012 | JX983829a |
| 1.45781 | |||||||
| 247 | sab | Burkina Faso | Ranch Nazinga | 11.14870 | KC843999 | KC844028 | JX983796a |
| –1.62334 | |||||||
| 255 | sab | Burkina Faso | Ranch Nazinga | 11.1554 | KC843987 | KC844016 | JX983801a |
| –1.61030 | |||||||
| 256 | sab | Burkina Faso | Ranch Nazinga | 11.15547 | KC843988 | KC844017 | JX983802a |
| –1.61030 | |||||||
| 264 | sab | Burkina Faso | Deux Bale Foret | 11.55079 | KC843998 | KC844027 | JX983792a |
| –2.95757 | |||||||
| 268 | sab | Burkina Faso | Comoe—Leraba NP | 9.90260 | KC844004 | KC844033 | JX983778a |
| –4.65454 | |||||||
| 269 | sab | Burkina Faso | Comoe—Leraba NP | 9.90260 | KC843989 | KC844018 | JX983779a |
| –4.65454 | |||||||
| 271 | sab | Burkina Faso | Comoe—Leraba NP | 9.90260 | KC844003 | KC844032 | JX983780a |
| –4.65454 | |||||||
| 275 | sab | Burkina Faso | Comoe—Leraba NP | 9.84896 | KC843995 | KC844024 | JX983783a |
| –4.62302 | |||||||
| 277 | sab | Burkina Faso | Comoe—Leraba NP | 9.85291 | KC844000 | KC844029 | JX983784a |
| –4.61415 | |||||||
| 284 | sab | Burkina Faso | Comoe—Leraba NP | 9.86207 | KC843996 | KC844025 | JX983788a |
| –4.66974 | |||||||
| 285 | sab | Burkina Faso | Comoe—Leraba NP | 9.86207 | KC844001 | KC844030 | JX983789a |
| –4.66974 | |||||||
| 286 | sab | Burkina Faso | Comoe—Leraba NP | 9.86207 | KC843994 | KC844023 | JX983790a |
| –4.66974 | |||||||
| 451 | aet | Ethiopia | Bonga | 7.26645 | KC843976 | KC844005 | JX983743a |
| 36.23131 | |||||||
| 491 | pyg | Ethiopia | Sof Omar Caves | 6.91205 | KC843977 | KC844006 | — |
| 40.84583 | |||||||
| 492 | pyg | Ethiopia | Sof Omar Caves | 6.91205 | KC843978 | KC844007 | — |
| 40.84583 | |||||||
| 496 | pyg | Ethiopia | Sof Omar Caves | 6.91205 | — | — | JX983764a |
| 40.84583 | |||||||
| 569 | sab | PEI, Barbados | — | — | KC843986 | KC844015 | — |
| 571 | pyg | PEI, TZ/KY | — | — | KC843979 | KC844008 | — |
Table II.
External morphological characters used to identify different Chlorocebus species in West and East Africa in this study
| Species | Diagnostic morphological featuresa | IDs of assigned groups/samples | ||
|---|---|---|---|---|
| Face | Tail | Extremities | ||
| aethiops | Narrow white brow band, elongated white cheek whiskers, fine white moustache | Paracaudal white tuft, whitish tale tip | Gray limbs, hands and feet pale | 451 |
| pygerythrus | White brow band, very short whiskers form a complete face-ring | Red subcaudal patch, no tuft, black tail tip | Color of limbs like dorsum, black hands and feet | 491, 492, 496, 571 |
| sabaeus | Brow band poorly expressed or absent, yellow whiskers directed upward in front of ears, whorl in front of ears | No tuft, yellow golden tail tip | Grizzled golden limbs, hands and feet pale | 120, 138, 139, 142, 143, 153, 255, 256, 264, 247, 268, 269, 271, 275, 277, 284, 285, 286, 569 |
| tantalus | Long but tiff whiskers, yellowish, brushed up, tapered white brow band, black bar from whiskers | Paracaudal white tuft, pale or creamy tail tip | Gray limbs, pale feet | 102, 192, 203, 224, 225, 226, 233 |
We conducted data collection in compliance with the animal care regulations and the principles of the American Society of Primatologists for the ethical treatment of nonhuman primates. We collected all fecal samples from wild, nonhabituated groups without threatening or harming the subjects. We obtained blood samples as part of regular diagnostic checks by veterinarians from the PEI. The research complied with protocols approved by the German Primate Center and the PEI in Germany, the Ethiopian Wildlife and Conservation Authority (EWCA) in Ethiopia, the Centre National de la Recherche Scientifique et Technologique (CNRST) in Burkina Faso, and the Forestry Commission (FC) of Ghana, and adhered to the legal requirements of the countries in which the research was conducted.
DNA Extraction, Amplification, and Sequencing
We extracted total genomic DNA from fecal and blood samples using the QIAamp DNA Stool Mini Kit and the DNeasy Blood & Tissue Kit from Qiagen (Qiagen, Germany), respectively. We applied standard protocols as provided by the company with only minor changes for the extraction of fecal samples (Haus et al. 2013). We stored blood sample extracts at 4°C and fecal sample extracts at –20°C in 50-μl aliquots for up to 24 mo before further processing.
Only samples from males can be used in the analyses of Y-chr markers. Because the collection of fecal samples from unhabituated wild animals does not allow the assignment of samples to individuals, we obtained information on the individuals’ sex using a gonosomal polymerase chain reaction (PCR) –based sexing method (Roos unpubl. data).
We amplified and sequenced fragments of two Y-chr genes: 695 bp of the last intron of the zinc finger (ZFY) and a 783 bp long fragment of the sex determining region (SRY). We amplified both markers via two overlapping fragments, each measuring ca. 450–500 bp length, using the primer pairs ZFY-F1: CCTGATTCCAGGGAGTACC and ZFY-R1: AGTAAAGCTTAACTGCACCTAT, ZFY-F2: AGGACAGATTACTATCCTGTG and ZFY-R2: CAGTATGAGTGCTTAATCAAAC, SRY-F1: CTTGAGAATGAATACATTGTCAGGG and SRY-R1: GTATCCCAGCTGCTTGCTG, and SRY-F2: GATCAGAGGCGCAAGATGG and SRY-R2: AGGTCTTTGTAGCCAATGTTACCCG. We used 1 U BiothermTaq 5000 (Genecraft, Germany) in a 30-μl PCR mix (1× reaction buffer, 0.16 mM for each dNTP, 0.33 μM for each primer, and 0.6 mg/ml bovine serum albumin), with the following thermo cycler conditions: 94°C for 2 min, followed by 60 cycles of 94°C for 1 min, 60°C (SRY) and 58°C (ZFY) for 1 min, 72°C for 1 min, and 72°C for 5 min. We conducted all PCR reactions with at least one PCR blank (HPLC-purified water). We ran and checked PCR products on 1–2% agarose gels, excised DNA fragments of relevant lengths, and purified PCR products with the Qiagen Gel Extraction Kit (Qiagen, Germany). We used the BigDye Terminator Cycle Sequencing Kit (Applied Biosystems, Germany) and the respective forward and reverse primers to run sequences on an ABI 3130 xL sequencer. We deposited all sequences in GenBank (Table I).
Analysis of Genetic Structures
To compare the results obtained from the Y-chr data with those for mtDNA in African green monkeys, we used published cytochrome b (cyt b) gene sequences from Haus et al. (2013). We selected cyt b sequences from the same samples for which we sequenced Y-chr markers. In cases where mitochondrial sequences were not available we used sequence information from samples that were collected at the same site, assuming that indivduals from the same social group carry similar mtDNA (Fig. 1, Table I).
We assembled and aligned all Y-chr sequences with the program Geneious Pro 5.0.4 (Drummond et al. 2011) and added an additional sequence from GenBank representing Chlorocebus aethiops (Roos et al. 2011; Table I). We also used Geneious Pro 5.0.4 to prepare two separate alignments, one concatenated alignment of the two Y-chr markers ZFY and SRY, and a second alignment including all cyt b sequences. To remove deletions in the Y-chr data set we used Gblocks (Talavera and Castresana 2007), leading to a final length of the Y-chr alignment of 1476 bp, for which we measured variable and parsimony-informative sites with Mega 5.0 (Tamura et al. 2011). To compare genetic patterns of Y-chr and mtDNA we calculated median-joining networks for both data sets separately, using Network 4.610 (Bandelt et al. 1999). Finally, we displayed and edited resulting networks with Network Publisher.
Results
The samples in our analysis represent four morphological species: Chlorocebus aethiops, C. pygerythrus, C. sabaeus, and C. tantalus (Fig. 1). PCR-based sexing indicated that our fecal sample data set includes 28 samples from male individuals originating from all sampling sites except the population of Chlorocebus tantalus west of the Volta River in south Ghana, for which we were not able to sample a male. For this site we have only mtDNA information (ID 203 in Fig. 1b).
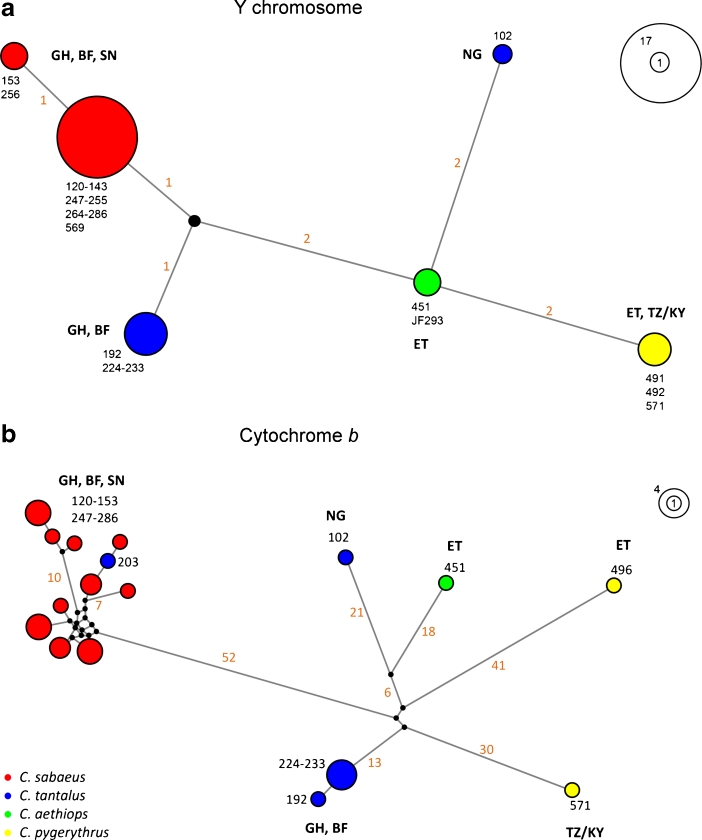
The analysis of Y-chr markers showed a low genetic variability among the 30 samples analysed, with nine variable and seven parsimony-informative sites within the concatenated 1476 bp data set of ZFY and SRY. Accordingly, we refrained from reconstructing a rooted phylogenetic tree, and instead used median-joining networks to depict haplotype diversity and genetic distance among haplotypes and clades, respectively. We found 6 Y-chr haplotypes, whereas the corresponding cyt b sequences represent 17 haplotypes forming 6 mtDNA clades. Y-chr sequences of Chlorocebus pygerythrus from southeast Ethiopia are identical to the C. pygerythrus sequence from Kenya/Tanzania and thus constitute a single haplotype, whereas mtDNA reveals two distinct clades, one for southeast Ethiopia and one for Kenya/Tanzania (Fig. 2). In contrast, the analysis of samples of Chlorocebus tantalus from east of the Volta River and its confluents in Ghana and Burkina Faso indicates a distinct Y-chr haplotype, which differs from the haplotype obtained from East Nigerian samples of C. tantalus. This is in agreement with the mtDNA results, where two distinct clades are also found. Y-chr sequences of Chlorocebus sabaeus and C. tantalus from Ghana and Burkina Faso represent two different haplotypes, which are clearly separated east and west of the Volta River, again reflecting the same pattern as the mtDNA data.
Fig. 2.
Median-joining networks of (a) Y-chr and (b) cyt b haplotypes of samples of Chlorocebus from West and East Africa. Circle sizes specify haplotype frequencies as shown by the scales. Colors represent morphotypes, orange numbers indicate mutated positions of major branches, and black numbers correspond to sample IDs in Fig. 1b and Table I. GH = Ghana; BF = Burkina Faso; SN = Senegal; NG = Nigeria; ET = Ethiopia; TZ/KY = Tanzania/Kenya.
Discussion
East Africa
Based on mtDNA two distantly related pygerythrus clades occur in East Africa, one in southeast Ethiopia (C 6), which is closely related to Chlorocebus djamdjamensis, and a second in Kenya and Tanzania (C 8, Fig. 1; Haus 2013). Our results show that both clades possess the same Y-chr haplotype despite their large mtDNA divergence. Because the social system of African green monkeys is characterized by male-biased dispersal (Jaffe and Isbell 2011; Kingdon 1997), male-mediated introgressive hybridization is the most probable explanation for this discordant pattern between maternal (mtDNA) and paternal (Y-chr) inherited markers. Given the large mtDNA divergence (Haus 2013), it is unlikely that this discordance has been caused by geographic isolation and the higher rate of mtDNA evolution in comparison to Y-chr DNA, which can induce a similar discordance. Therefore, ancient introgressive hybridization and nuclear swamping might have led to a morphotype of Chlorocebus pygerythrus in southeast Ethiopia that carries Y-chr DNA of C. pygerythrus (and likely autosomal DNA), but the mtDNA of a possible now cytonuclear extinct taxon.
West Africa
We found a different picture in Chlorocebus tantalus. Both Y-chr and mtDNA data support the differentiation of Chlorocebus tantalus into a western and eastern clade despite their morphological similarity (Figs. 1 and 2). Here, the genetic concordance of maternal and paternal inherited markers is inconsistent with introgression patterns caused by male dispersal. However, a population genetic approach with more diverse nuclear markers and a larger data set from other parts of the range of Chlorocebus tantalus are needed to test for potential admixture in other regions of the genome. An alternative explanation for the discrepancy between genetic relationships and morphotype variation may be cryptic genetic, and thus, possibly taxonomic diversity. Although recent primate taxonomies recognize only the Chlorocebus tantalus tantalus subspecies for the region west of Cameroon, Schwarz reported a different morphotype from Togo in 1926 (Schwarz 1926), but this was not accepted by other authors (Booth 1956; Groves 2001, 2005; Hill 1966). Another tantalus-like morphotype (chrysurus) with unknown type locality was described by Blyth (1845), which is currently recognized as a synonym of tantalus (Hill 1966) or sabaeus (Groves 2001; Napier 1981). A detailed morphological analysis covering the whole range of Chlorocebus tantalus is required to determine whether the two detected genetic clades are similar or differ in morphology. Because there are no type localities for tantalus or chrysurus, further genetic and morphological analyses should include both type specimens to test whether they belong to one of the clades detected and whether further nuclear data also support two different genetic clades.
Hybridization between Chlorocebus sabaeus and C. tantalus was thought to occur only in the northern range of the Volta River and its tributaries in Ghana and Burkina Faso (Lernould 1988), but mtDNA results provided evidence for a contradicting picture with hybridization in the south but not in the north (Haus et al. 2013). Although our data set may not include all populations of Chlorocebus sabaeus and C. tantalus along the Volta River and its tributaries, the analysis of samples from the northern part of their potential contact zone showed no inconsistencies between the genetic patterns of Y-chr and mtDNA, nor between the Y-chr and morphological data. Accordingly, there is also no Y-chr evidence of interspecific gene flow across the Volta River in northern regions. In contrast, discordance between morphotype and mtDNA from a single population of Chlorocebus tantalus from the Accra plains west of the Volta River in Ghana suggests introgressive hybridization in the south (Fig. 1a; Haus et al. 2013). Unfortunately, our data set did not include male samples from this population, preventing us from analyzing Y-chr markers. However, there is no evidence of morphological individuals of Chlorocebus sabaeus in or close to this area and because this population of C. tantalus carries the mitochondrial genome of C. sabaeus (Booth 1956, 1958; Haus et al. 2013), introgressive hybridization remains the most plausible explanation here. The factors that led to this unexpected pattern remain unclear. One possible explanation is that limited hybridization across the Volta River occurred throughout the contact zone of Chlorocebus sabaeus and C. tantalus in the past, when African green monkey populations were distributed more commonly along the river banks than today. If this is the case, then the C. tantalus population west of the Volta River in South Ghana may represent the only remaining hybrid population. Alternatively, if the Volta River represents a geographic barrier to the distribution of both species, individuals of Chlorocebus tantalus from the eastern river bank may have been introduced into a population of C. sabaeus west of the Volta River by humans. Such anthropological impact is not unlikely, as African green monkeys are often kept as pets (Riley et al. 2011).
Conclusions
Our analysis of sex-linked markers indicates that two populations exist in East African Chlorocebus pygerythrus carrying distinct mtDNA haplotypes, but the same Y-chr haplotype, suggesting that male-mediated gene flow between these populations existed in the past or is still ongoing. In contrast, the two populations of Chlorocebus tantalus in West Africa, which were detected based on mtDNA, also carry distinct Y-chr haplotypes. Here it remains to be tested whether (cryptic) speciation is in progress or if genomic admixture due to ancient introgression has occurred in a part of the range of Chlorocebus tantalus. The analysis of the potential West African contact zone showed that Chlorocebus sabaeus and C. tantalus differ in mtDNA as well as Y-chr DNA. These results suggest that ongoing interspecific gene flow between these species across the Volta River is strongly reduced or even absent, with one exception in South Ghana, where introgressive hybridization may have led to a phenotypic population of C. tantalus west of the Volta River carrying mtDNA of C. sabaeus. Our study highlights the importance of the analysis of Y-chr data in addition to mtDNA to obtain information on hybridization patterns, and especially, on ancient events, which might have influenced the genetic diversity of some taxa.
Our results indicate that the evolution of African green monkeys was influenced by introgressive hybridization, most probably with the cytonuclear extinction of some populations or taxa. This aspect may also play an important role in the survival of the endemic species Chlorocebus djamdjamensis. Continuing asymmetric introgression between Chlorocebus aethiops and C. djamdjamensis in Ethiopia may also lead to the cytonuclear extinction of the restricted species C. djamdjamensis (Haus et al. 2013; Kingdon 1997; Mekonnen et al. 2012). A comprehensive genetic analysis of samples including all remaining populations of Chlorocebus djamdjamensis is needed to explore the progress and pattern of introgression.
Acknowledgments
We acknowledge the Ethiopian Wildlife and Conservation Authority (EWCA) in Ethiopia, the Centre National de la Recherche Scientifique et Technologique (CNRST) in Burkina Faso, and the Forestry Commission (FC) of Ghana for providing necessary research permits. Special thanks go to William Oduro and Emmanuel Akom of the Kwame Nkrumah University for Science and Technology (KNUST) in Kumasi for supporting our data collection in Ghana. We thank Colin Groves for providing important information on the tantalus–chrysurus problem. We are grateful to Kwaku Duah, Annika Simon, Ahandi Thiombiano, Charlotte Rahn, Teklu Tesfaye, Andreas Ploß, Peter Maciej, and Yama Ethiopia Tours as well as numerous local residents for their assistance with data collection in the field and to Roland Plesker (PEI), Elody Ey (DPZ), and Tabitha Price (DPZ) for providing further samples. For laboratory assistance we thank Anja Ebenau, Christiane Schwarz, and Teresa Kreusch. We thank Joanna Setchell and two anonymous reviewers for very helpful comments on a previous version of this manuscript. This project was financed by the German Volkswagen Foundation.
Footnotes
Christian Roos and Dietmar Zinner these authors contributed equally to this work.
References
- Abbott R, Albach D, Ansell S, Arntzen JW, Baird SJE, Bierne N, Boughman J, Brelsford A, Buerkle CA, Buggs R, Butlin RK, Dieckmann U, Eroukhmanoff F, Grill A, Cahan SH, Hermansen JS, Hewitt G, Hudson AG, Jiggins C, Jones J, Keller B, Marczewski T, Mallet J, Martinez-Rodriguez P, Möst M, Mullen S, Nichols R, Nolte AW, Parisod C, Pfennig K, Rice AM, Ritchie MG, Seifert B, Smadja CM, Stelkens R, Szymura JM, Väinölä R, Wolf JBW, Zinner D. Hybridization and speciation. Journal of Evolutionary Biology. 2013;26:229–246. doi: 10.1111/j.1420-9101.2012.02599.x. [DOI] [PubMed] [Google Scholar]
- Avise JC. Molecular markers: Natural history, and evolution. Sunderland, MA: Sinauer Associates; 2004. [Google Scholar]
- Bandelt H, Forster P, Rohl A. Median-joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- Blyth E. Notices of various Mammalia, with descriptions of many new species. The Annals and Magazine of Natural History. 1845;15:461. [Google Scholar]
- Booth A. The distribution of primates in the Gold Coast. Journal of the West African Science Association. 1956;2:122–133. [Google Scholar]
- Booth A. The zoogeography of West African primates: A review. Bull I’lFAN. 1958;20:587–622. [Google Scholar]
- Charpentier MJE, Fontaine MC, Cherel E, Renoult JP, Jenkins T, Benoit L, Barthès N, Alberts SC, Tung J. Genetic structure in a dynamic baboon hybrid zone corroborates behavioural observations in a hybrid population. Molecular Ecology. 2012;21:715–731. doi: 10.1111/j.1365-294X.2011.05302.x. [DOI] [PubMed] [Google Scholar]
- Drummond, A., Ashton, B., Buxton, S., Cheung, M., Cooper, A., Duran, C., Field, M., Heled, J., Kearse, M., Markowith, S., Moir, R., Stones-Havas, S., Sturrock, S., Thierer, T., & Wilson, A. (2011). Geneious v5.4. Available from http://www.geneious.com [DOI] [PMC free article] [PubMed]
- Flagstad O, Syvertsen PO, Stenseth NC, Jakobsen KS. Environmental change and rates of evolution: The phylogeographic pattern within the hartebeest complex as related to climatic variation. Proceedings of the Royal Society of London B: Biological Sciences. 2001;268:667–677. doi: 10.1098/rspb.2000.1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk DJ, Omland KE. Species-level paraphyly and polyphyly: Frequency, causes, and consequences, with insights from animal mitochondrial DNA. Annual Review of Ecology, Evolution, and Systematics. 2003;34:397–423. doi: 10.1146/annurev.ecolsys.34.011802.132421. [DOI] [Google Scholar]
- Groves CP. Primate taxonomy. Washington, DC: Smithonian Institution Press; 2001. [Google Scholar]
- Groves CP. Order primates. In: Wilson D, Reeder D, editors. Mammal species of the world: A taxonomic and geographic reference. 3. Baltimore: Johns Hopkins University Press; 2005. pp. 111–184. [Google Scholar]
- Haldane JBS. Sex ratio and unisexual sterility in hybrid animals. Journal of Genetics. 1922;12:101–109. doi: 10.1007/BF02983075. [DOI] [Google Scholar]
- Haus, T. (2013). Phylogeography and impact of hybridization on the evolution of African green monkeys (Chlorocebus Gray, 1870). Ph.D. thesis, University of Göttingen, Germany.
- Haus T, Akom E, Agwanda B, Hofreiter M, Roos C, Zinner D. Mitochondrial diversity and distribution of African green monkeys (Chlorocebus Gray, 1870) American Journal of Primatology. 2013;75:350–360. doi: 10.1002/ajp.22113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill WCO. Primates: Comparative anatomy and taxonomy. VI. Catarrhini, Cercopithecoidea, Cercopithecinae. Edingburgh: Edingburgh University Press; 1966. [Google Scholar]
- Jaffe KE, Isbell LA. The guenons: Polyspecific associations in socioecological perspective. In: Campbell CJ, Fuentes A, MacKinnon KC, Bearder SK, Stumpf RM, editors. Primates in perspective. New York: Oxford University Press; 2011. pp. 277–300. [Google Scholar]
- Jolly CJ, Burrell AS, Phillips-Conroy JE, Bergey C, Rogers J. Kinda baboons (Papio kindae) and grayfoot chacma baboons (P. ursinus griseipes) hybridize in the Kafue river valley, Zambia. American Journal of Primatology. 2011;73:291–303. doi: 10.1002/ajp.20896. [DOI] [PubMed] [Google Scholar]
- Keller C, Roos C, Groeneveld LF, Fischer J, Zinner D. Introgressive hybridization in southern African baboons shapes patterns of mtDNA variation. American Journal of Physical Anthropology. 2010;142:125–136. doi: 10.1002/ajpa.21209. [DOI] [PubMed] [Google Scholar]
- Kingdon J. The Kingdon fieldguide to African mammals. London: Academic Press; 1997. [Google Scholar]
- Lernould J. Classification and geographical distribution of guenons: A review. In: Gautier-Hion A, Bourliere F, Gautier J, Kingdon J, editors. A primate radiation: Evolutionary biology of the African guenons. Cambridge, U. K.: Cambridge University Press; 1988. pp. 54–78. [Google Scholar]
- Mallet J. Hybridization as an invasion of the genome. Trends in Ecology & Evolution. 2005;20:229–237. doi: 10.1016/j.tree.2005.02.010. [DOI] [PubMed] [Google Scholar]
- Mekonnen A, Bekele A, Fashing PJ, Lernould J-M, Atickem A, Stenseth NC. Newly discovered Bale monkey populations in forest fragments in southern Ethiopia: Evidence of crop raiding, hybridization with grivets, and other conservation threats. American Journal of Primatology. 2012;74:423–432. doi: 10.1002/ajp.21999. [DOI] [PubMed] [Google Scholar]
- Muwanika VB, Nyakaana S, Siegismund HR, Arctander P. Phylogeography and population structure of the common warthog (Phacochoerus africanus) inferred from variation in mitochondrial DNA sequences and microsatellite loci. Heredity. 2003;91:361–372. doi: 10.1038/sj.hdy.6800341. [DOI] [PubMed] [Google Scholar]
- Napier PH. Catalogue of primates in the British Museum (Natural History) and elsewhere in the British Isles. Part II: Family Cercopithecidae, subfamily Cercopithecinae. London: British Museum (Natural History); 1981. [Google Scholar]
- Nsubuga AM, Robbins MM, Roeder AD, Morin PA, Boesch C, Vigilant L. Factors affecting the amount of genomic DNA extracted from ape faeces and the identification of an improved sample storage method. Molecular Ecology. 2004;13:2089–2094. doi: 10.1111/j.1365-294X.2004.02207.x. [DOI] [PubMed] [Google Scholar]
- Orr HA. Haldane’s rule. Annual Review of Ecology and Systematics. 1997;28:195–218. doi: 10.1146/annurev.ecolsys.28.1.195. [DOI] [PubMed] [Google Scholar]
- Riley EP, Wolfe LD, Fuentes A. Ethnoprimatology: Contextualizing human and nonhuman primate interactions. In: Campbell CJ, Fuentes A, MacKinnon KC, Bearder SK, Stumpf RM, editors. Primates in perspective. New York: Oxford University Press; 2011. pp. 676–686. [Google Scholar]
- Roos C, Zinner D, Kubatko LS, Schwarz C, Yang M, Meyer D, Nash SD, Xing J, Batzer MA, Brameier M, Leendertz FH, Ziegler T, Perwitasari-Farajallah D, Nadler T, Walter L, Osterholz M. Nuclear versus mitochondrial DNA: Evidence for hybridization in colobine monkeys. BMC Evolutionary Biology. 2011;11:e77. doi: 10.1186/1471-2148-11-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilthuizen M, Giesbers MCWG, Beukeboom LW. Haldane’s rule in the 21st century. Heredity. 2011;107:95–102. doi: 10.1038/hdy.2010.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz E. Die Meerkatzen der Cercopithecus aethiops-Gruppe. Zeitschrift für Säugetierkunde. 1926;1:28–47. [Google Scholar]
- Talavera G, Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Systematic Biology. 2007;56:564–577. doi: 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung J, Charpentier MJE, Garfield DA, Altmann J, Alberts SC. Genetic evidence reveals temporal change in hybridization patterns in a wild baboon population. Molecular Ecology. 2008;17:1998–2011. doi: 10.1111/j.1365-294X.2008.03723.x. [DOI] [PubMed] [Google Scholar]
- Zinner D, Arnold ML, Roos C. The strange blood: Natural hybridization in primates. Evolutionary Anthropology. 2011;20:96–103. doi: 10.1002/evan.20301. [DOI] [PubMed] [Google Scholar]
- Zinner D, Groeneveld LF, Keller C, Roos C. Mitochondrial phylogeography of baboons (Papio spp.): Indication for introgressive hybridization? BMC Evolutionary Biology. 2009;9:e83. doi: 10.1186/1471-2148-9-83. [DOI] [PMC free article] [PubMed] [Google Scholar]