Background: Fbxw7 is the F-box protein component of an SCF-type ubiquitin ligase that plays key roles in tumor suppression and stem cell maintenance.
Results: The transcription factors OASIS and BBF2H7 were identified as new substrates of Fbxw7.
Conclusion: Fbxw7 controls osteogenesis and chondrogenesis by targeting OASIS and BBF2H7 for degradation.
Significance: Fbxw7 is an important regulator of osteogenesis and chondrogenesis.
Keywords: Mesenchymal Stem Cells, Protein Degradation, Proteomics, Transcription Factors, Ubiquitin Ligase, F-box Protein
Abstract
Although identification of substrates for an enzyme is a key step in elucidation of its biological functions, detection of the interaction between enzymes and substrates remains challenging. We recently developed a new approach, termed differential proteomics-based identification of ubiquitylation substrates (DiPIUS), for the discovery of substrates of ubiquitin ligases. We have now applied this approach to Fbxw7, the F-box protein component of an Skp1-Cul1-F-box protein-type ubiquitin ligase and, thereby, identified two similar transcription factors, old astrocyte specifically induced substance (OASIS) and BBF2 human homolog on chromosome 7 (BBF2H7), as candidate substrates. Coimmunoprecipitation analysis confirmed that the α and γ isoforms of Fbxw7 interact with OASIS and BBF2H7 in vivo. Sustained overexpression of Fbxw7 resulted in marked down-regulation of OASIS and BBF2H7, whereas RNAi-mediated Fbxw7 depletion stabilized both proteins. Mutation of a putative Cdc4 phosphodegron in OASIS and BBF2H7 attenuated their association with Fbxw7 and resulted in their stabilization. Depletion of Fbxw7 promoted the differentiation of mouse C2C12 mesenchymal cells into osteoblasts in association with the accumulation of OASIS. Conversely, overexpression of Fbxw7 in C2C12 cells resulted in down-regulation of Col1A1 mRNA, a target of OASIS. Conditional ablation of Fbxw7 in primary mouse mesenchymal cells promoted chondrogenesis in association with up-regulation of BBF2H7, whereas overexpression of Fbxw7 inhibited chondrogenesis in ATDC5 cells. Collectively, our results suggest that OASIS and BBF2H7 are bona fide substrates of Fbxw7 and that Fbxw7 controls osteogenesis and chondrogenesis by targeting OASIS and BBF2H7, respectively, for degradation.
Introduction
The Skp1-Cul1-F-box protein (SCF)2 complex is one of the most well characterized types of ubiquitin ligase. Each SCF complex consists of four subunits: Skp1, Cul1, Rbx1 (also known as Roc1 or Hrt1), and an F-box protein (1–3). F-box proteins are responsible for substrate recognition by each complex and constitute a large family of > 70 eukaryotic proteins (4). The F-box protein Fbxw7 was initially identified as a negative regulator of LIN-12 (Notch)-mediated signaling in Caenorhabditis elegans by genetic analysis (5, 6). It was shown subsequently to play a pivotal role in the control of cell proliferation through ubiquitylation of cyclin E (7–9), c-Myc (10, 11), Notch (5, 12, 13), c-Jun (14, 15), and Krüppel-like factor (KLF) 5 (16, 17). Given its function in the degradation of proliferation-related proteins, Fbxw7 is thought to act as a tumor suppressor protein. Indeed, ∼6% of primary human tumors have been found to harbor mutations in FBXW7 (18), with the mutations being detected most frequently in cholangiocarcinoma (35%) and T cell acute lymphocytic leukemia (31%).
In addition to tumor suppression, Fbxw7 has been shown recently to contribute to stem cell maintenance and cell differentiation. Thus, it regulates the quiescence and self-renewal of hematopoietic stem cells and neural stem cells (19–23) as well as of leukemia stem cells (24–26). The loss of Fbxw7 in neural stem cells also impairs their differentiation into neurons as a result of the up-regulation of Notch (22, 23). Fbxw7 deficiency in intestinal crypts also leads to the accumulation of progenitor cells and impairment of their differentiation into goblet cells (27, 28). Conditional ablation of Fbxw7 in the liver results both in a shift in the differentiation of liver stem cells from the hepatocyte lineage to cholangiocytes as well as in increased cell proliferation (29). Liver-specific deficiency of Fbxw7 also results in lipid accumulation in hepatocytes as a consequence of the accumulation of its targets KLF5 and sterol response element-binding proteins (SREBPs) (29, 30). In cultured cells, Fbxw7 deficiency promotes the formation of lipid droplets as a result of the accumulation of SREBPs and CCAAT/enhancer-binding protein α (c/EBPα) (31, 32). Collectively, these observations indicate that Fbxw7 regulates various developmental and differentiation processes by targeting multiple substrate molecules for degradation.
OASIS (CREB3L1) and BBF2H7 (CREB3L2) are basic leucine zipper-type transcription factors that belong to the cAMP response element-binding protein (CREB)/activating transcription factor family. Both proteins possess a transmembrane domain that allows them to associate with the endoplasmic reticulum (ER) and is cleaved by site 1 and site 2 proteases (S1P and S2P) in response to ER stress (33, 34). Expression of OASIS is restricted to certain tissues and cells, including astrocytes and osteoblasts (35, 36). OASIS-deficient mice manifest severe osteopenia that is associated with a decrease in the amount of type I collagen in the bone matrix and a reduced activity of osteoblasts (37). Expression of OASIS in osteoblasts is induced by bone morphogenetic protein 2 (BMP2), signaling by which is required for bone formation, and OASIS up-regulates transcription of the gene for the type I collagen Col1A1 directly. BBF2H7 is highly expressed in the proliferating zone of cartilage in developing long bones (38). Mice deficient in BBF2H7 manifest pronounced chondrodysplasia and die from choking after birth as a result of immaturity of the chest cavity. Type II collagen (Col2) and cartilage oligomeric matrix protein accumulate in the ER lumen of BBF2H7-deficient chondrocytes. BBF2H7 directly activates transcription of the gene for Sec23a, a component of coat protein complex II responsible for protein transport from the ER to the Golgi apparatus, suggesting that BBF2H7 controls the secretion of extracellular matrix molecules in cartilage by regulating vesicle transport.
We now show that OASIS and BBF2H7 are targets of SCFFbxw7. Fbxw7 controls osteoblast and chondrocyte differentiation by targeting OASIS and BBF2H7 for proteasome-mediated degradation. Thus, our results suggest that Fbxw7 is an important regulator of osteogenesis and chondrogenesis.
EXPERIMENTAL PROCEDURES
Cell Culture
Neuro2A cells, mHepa cells, C2C12 cells, and HeLa cells stably expressing murine cationic amino acid transporter type 1 (mCAT-HeLa cells) were maintained in DMEM supplemented with 10% FBS (Invitrogen), 1 mm sodium pyruvate, penicillin (100 units/ml, Invitrogen), streptomycin (100 mg/ml, Invitrogen), 2 mm l-glutamine, and nonessential amino acids (10 ml/l, Invitrogen). Differentiation of C2C12 cells toward the osteoblast lineage was induced by their exposure for 6 days to recombinant human BMP2 (300 ng/ml, Shenandoah Biotechnology) in DMEM supplemented with 2.5% FBS. The differentiated cells were washed with PBS, fixed in Bouin's solution for 1 h, washed with distilled H2O, stained with 0.1% Sirius red (Direct Red 80, Sigma) for 1 h at 37 °C, and then washed with 10 mm HCl. The murine chondrogenic cell line ATDC5 was cultured in a 1:1 (v/v) mixture of DMEM and Ham's F12 medium (Invitrogen) that was supplemented with 5% FBS, human transferrin (10 μg/ml, Sigma), 30 nm sodium selenite (Sigma), 1 mm sodium pyruvate, penicillin (100 units/ml), streptomycin (100 mg/ml), 2 mm l-glutamine, and nonessential amino acids (10 ml/l). For induction of chondrogenesis, ATDC5 cells were plated at an initial density of 6250 cells/cm2 and cultured for 16 days in medium supplemented with bovine insulin (10 μg/ml, Sigma). For inhibition of Notch signaling, the cells were also cultured with the γ-secretase inhibitor N-[N-(3,5-difluorophenacetyl)-l-alanyl]-S-phenylglycine t-butyl ester (DAPT, Calbiochem). For Alcian blue staining, the cells were washed twice with PBS, fixed in 3.7% formaldehyde for 10 min, and stained overnight with 0.1% Alcian blue in 0.1 m HCl. The cells were then solubilized with 6 m guanidine hydrochloride for measurement of A595 with a spectrophotometer. Sf21 cells were cultured at 27 °C in Sf-900 II SFM medium (Invitrogen) supplemented with 5% FBS (Invitrogen), penicillin (100 units/ml, Invitrogen), and streptomycin (100 mg/ml, Invitrogen).
Antibodies
Antibodies to KLF5 (BTEB2, H-300) and to c-Myc (N-262) were obtained from Santa Cruz Biotechnology. Antibodies to Skp1 and to Hsp90 were from BD Biosciences. Antibodies to 14-3-3ϵ were from BD Transduction Laboratories. Antibodies to Cul1 were from Zymed Laboratories Inc.. Antibodies to the HA epitope (HA11) were from Babco. Antibodies to the FLAG epitope (M2) were from Sigma. Antibodies to GAPDH (1D4) were from Assay Designs. Antibodies to protein disulfide isomerase were from Thermo Scientific. Antibodies to cleaved Notch1 (Val1744) were from Cell Signaling Technology. Antibodies to OASIS and BBF2H7 were described previously (37, 38).
Plasmids
Complementary DNAs encoding mouse Fbxw7α or its ΔF mutant were subcloned into p3×FLAG-CMV 7.1 (Sigma), and those for HA epitope- or FLAG epitope-tagged human OASIS or BBF2H7 were subcloned into pcDNA3 (Invitrogen). The resulting vectors were introduced into mCAT-HeLa cells with the use of the FuGENE 6 or FuGENE HD transfection reagents (Roche or Promega). For retroviral expression, cDNAs for FLAG-tagged mouse Fbxw7α, Fbxw7β, Fbxw7γ, or corresponding mutants or those for HA epitope-tagged human OASIS, BBF2H7, or corresponding mutants were subcloned into pMX-puro (provided by T. Kitamura). The resulting vectors were introduced into Plat E cells with the use of FuGENE HD. The recombinant retroviruses generated thereby were used to infect mCAT-HeLa or HEK293T cells, which were then subjected to selection in medium containing puromycin (10 μg/ml). For baculoviral expression, cDNAs for FLAG-tagged mouse Fbxw7α, Myc epitope-tagged mouse Cul1 and Skp1, and HA epitope-tagged NH2-terminal fragments of human OASIS and BBF2H7 were subcloned into pFASTBacHT (encoding a His6 tag), whereas that for human Rbx1 containing an NH2-terminal Myc epitope tag was subcloned into pFASTBac1.
DiPIUS and DiPIUS-NL
DiPIUS and DiPIUS-NL were performed as described (39). For DiPIUS on the basis of stable isotope labeling with amino acids in cell culture (SILAC), WT and ΔF mutant forms of Fbxw7α with an NH2-terminal FLAG tag were expressed in light isotope- and heavy isotope-labeled mCAT-HeLa cells, respectively. The cells (2 × 107) were incubated for 6 h in the presence of the proteasome inhibitor MG132 (10 μm, Peptide Institute) and were then lysed in 8 ml of a solution containing 20 mm HEPES-NaOH (pH 7.5), 150 mm NaCl, 1% digitonin, 10 mm NaF, 10 mm Na4P2O7, 0.4 mm Na3VO4, 0.4 mm EDTA, 20 μg/ml leupeptin, 10 μg/ml aprotinin, and 1 mm PMSF. The lysates were centrifuged at 2200 × g for 20 min at 4 °C to remove debris, and the resulting supernatants were adjusted with lysis buffer to a protein concentration of 2.5 mg/ml. The supernatants (20 mg of protein in 8 ml of solution) from light-labeled cells and heavy-labeled cells were then combined and incubated for 1 h at 4 °C with 120 μl of beads conjugated with M2 antibodies to FLAG (Sigma). The beads were washed three times with 4 ml of a solution containing 10 mm HEPES-NaOH (pH 7.5), 150 mm NaCl, and 0.1% Triton X-100, and bead-bound proteins were then eluted with the FLAG peptide (500 μg/ml, Sigma), precipitated with ice-cold 20% trichloroacetic acid, and washed with acetone.
The concentrated proteins were digested with Lys-C and trypsin, and the resulting peptides were analyzed with an LTQ Orbitrap Velos LC-MS/MS system (Thermo Finnigan). The peak lists were generated with Mascot Distiller (version 2.3.0) and were compared with the “Target-decoy” Human IPI database (version 3.1.6, see below) with the use of the MASCOT algorithm (version 2.2.1). Trypsin was selected as the protease, the allowed number of missed cleavages was set to one, and carbamidomethylation of cysteine was selected as the fixed modification. Oxidized methionine, pyroglutamine, 13C6-lysine, and 13C6-arginine were searched as variable modifications. Precursor mass tolerance was 20 ppm, and tolerance of MS/MS ions was 0.8 Da. Assigned high-scoring peptide sequences (MASCOT score > 20) were processed with in-house software. If the MASCOT score was < 45, assigned sequences were confirmed manually as described below for spectral counting, and the false discovery rate at the peptide level was estimated. SILAC quantitation was performed with the use of the Mascot Distiller Quantitation Tool (version 2.3.0). Extracted ion chromatogram peak areas for the heavy and light peptides were measured, and the results were verified by manual inspection of MS spectra. SILAC ratios were calculated by comparison of the extracted ion chromatogram peak areas of light peptides with those of the heavy peptides. The method “Auto” was selected for the removal of outlier peptide pairs (peptides with a number between 4 and 25 by Dixon's method or of > 25 by Rosner's method). SILAC ratios for different peptides of a protein were averaged to give protein abundance ratios, which were then normalized with the heavy/light ratio for Fbxw7α.
For DiPIUS-NL on the basis of spectral counting, the concentrated immunoaffinity-purified proteins derived separately from lysates of nonlabeled cells expressing FLAG-tagged WT or ΔF mutant forms of Fbxw7α were dissolved in SDS sample buffer, fractionated by SDS-PAGE, and stained with silver. Individual lanes of the stained gel were sliced into 8 to 16 pieces, and proteins within these pieces were subjected to in-gel digestion with trypsin as described previously (40). The resulting peptides were dried and then dissolved in a solution containing 0.1% trifluoroacetic acid and 2% acetonitrile before analysis with an ion-trap mass spectrometer (LTQ-XL, Thermo Finnigan). Peak lists were generated with lcq_dta.exe (Thermo Finnigan) and compared with the use of the MASCOT algorithm (version 2.2.1) either with the “Target-decoy” Human IPI database (version 3.1.6, released in November 2006, with 62,322 target sequences, searched against a total of 124,644 sequences (target and reverse/decoy)) or with the “Target-decoy” Mouse IPI database (version 3.4.4, released in June 2008, with 55,078 target sequences, searched against a total of 110,156 sequences (target and reverse/decoy)), both maintained by the European Bioinformatics Institute. Trypsin was selected as the protease, the allowed number of missed cleavages was set to one, and carbamidomethylation of cysteine was selected as the fixed modification. Oxidized methionine and pyroglutamine were searched as variable modifications. Precursor mass tolerance was 1.5 Da, and tolerance of MS/MS ions was 0.8 Da. Assigned high-scoring peptide sequences (MASCOT score > 35) were processed with in-house software. If the MASCOT score was < 45 (peptides for which the MS2 score was above the 95th percentile of significance), assigned sequences were manually confirmed by comparison with the corresponding collision-induced dissociation spectra on the basis of the following criteria: 1) a Δ score of > 15 or 2) at least six successive matches for y- or b-ions or at least three blocks of three successive matches for y- or b-ions. Identified peptides from independent experiments were integrated and regrouped by IPI accession number. When multiple accession numbers were obtained with the same set of peptides, a representative IPI accession was chosen according to the following order of priorities: 1) IPI accession containing NCBI GeneID, 2) IPI accession with the use of Swiss-Prot as the master sequence, and 3) IPI accession with the use of Translated European Molecular Biology Laboratory (TrEMBL) as the master sequence. Proteins identified in only one experiment or with a single-peptide assignment were removed from spectral counting data. Estimated false discovery rates were zero at the protein level.
RNAi
Construction of shRNA vectors and RNAi were performed as described previously (41). The sequences targeted for human or mouse Fbxw7 and mouse OASIS were 5′-GCAAATGTCTGAGAACATTAG-3′ (human Fbxw7 RNAi-1), 5′-GGTTGTTAGTGGAGCATATGA-3′ (human Fbxw7 RNAi-2), 5′-GGTTCTGAAGTTCGTTCCTTT-3′ (mouse Fbxw7), and 5′-GCATTCTCTTCCGGCTCAATG-3′ (mouse OASIS). An RNAi vector for enhanced GFP was used as a control.
RT and Real-time PCR Analysis
Total RNA (1 μg) isolated from cells with the use of Isogen (Nippon Gene) was subjected to RT with a QuantiTect reverse transcription kit (Qiagen), and the resulting cDNA was subjected to real-time PCR analysis with SYBR Green PCR Master Mix and specific primers in a StepOnePlus real-time PCR system (Applied Biosystems). The sequences of the various primers (sense and antisense, respectively) were 5′-TCTGAGGTCCGCTCTTTTTCTT-3′ and 5′-TGAGGTCCCCAAAAGTTGTTG-3′ for human Fbxw7; 5′-TGAAGTTCGTTCCTTTTCTTTGG-3′ and 5′-CAAAAGTTGTTGGTGTTGCTGAA-3′ for mouse Fbxw7; 5′-GCCTAAAGACGGTGGAAACG-3′ and 5′-GGCATGGACCTAGGAGGTTTG-3′ for human OASIS; 5′-CTGAGCACAGCTACTCCCTG-3′ and 5′-CCGTGTTCCACATCTTGAGT-3′ for mouse OASIS; 5′-AGAACTTGGAGCTTCGGAAGAA-3′ and 5′-GCTAACTTGCAGGTTCGAGAAAC-3′ for human BBF2H7; 5′-CTCATGCCAGACGCTTATTCC-3′ and 5′-ACTGGGTGGTGTTGGTGGTAA-3′ for mouse BBF2H7; 5′-TGGTCCTGCTACCCAGGGGC-3′ and 5′-TCTGGTCCAACGCACAGGCG-3′ for mouse Sec23a; 5′-CCCCAACCCTGGAAACAGAC-3′ and 5′-GGTCACGTTCAGTTGGTCAAAGG-3′ for mouse Col1A1; 5′-CAGGGCTGCTTTTAACTCTGGTA-3′ and 5′-GGGTGGAATCATATTGGAACATG-3′ for human GAPDH; and 5′-AGGTTGTCTCCTGCGACTTCA-3′ and 5′-CCAGGAAATGAGCTTGACAAAGTT-3′ for mouse GAPDH. The amounts of target mRNAs were normalized by that of GAPDH mRNA.
Immunoprecipitation, Cycloheximide Chase, and Immunoblot Analyses
Cells were incubated for 6 h in the presence of MG132 (10 μm) before lysis for immunoprecipitation. The cells were lysed by incubation for 20 min at 4 °C with a solution containing 50 mm Tris-HCl (pH 7.5), 150 mm NaCl, 0.5% Triton X-100, 10 mm NaF, 10 mm Na4P2O7, 0.4 mm Na3VO4, 0.4 mm EDTA, leupeptin (20 μg/ml), and aprotinin (10 μg/ml). The lysates were centrifuged at 20,400 × g for 20 min at 4 °C, and equal amounts of protein from the resulting supernatants were subjected to immunoprecipitation for 1 h at 4 °C with anti-FLAG (M2)-agarose affinity gel (Sigma). For cycloheximide chase experiments, cycloheximide (50 μg/ml) was added to culture medium, and cells were harvested at the indicated times thereafter. Immunoprecipitates and cell lysates were subjected to immunoblot analysis as described (42), with Hsp90, GAPDH, and 14-3-3ϵ used as loading controls. For Phos-tag SDS-PAGE, a standard discontinuous 8% polyacrylamide gel was prepared with the modification that 10 μm Phos-tag reagent (Wako) and 40 μm MnCl2 were added to the separation gel mix before polymerization. After electrophoresis, the gel was incubated for 40 min in Western transfer buffer containing 10 mm EDTA and for 10 min in transfer buffer.
Immunofluorescence Staining
mCAT-HeLa cells grown on glass coverslips were transfected with expression vectors with the use of the FuGENE HD reagent and subsequently prepared for immunostaining. The cells were fixed for 10 min at 37 °C with 4% paraformaldehyde in PBS and then incubated first overnight at 4 °C with primary antibodies in PBS containing 0.5% Triton X-100 and then for 1 h at room temperature with Alexa Fluor 488- or Alexa Fluor 546-labeled secondary antibodies (dilution of 1:2000, Molecular Probes). The cells were finally stained with Hoechst 33258 (Wako), covered with a drop of Fluoromount (Diagnostic BioSystems), and examined with a laser-scanning confocal microscope (LSM510, Carl Zeiss).
In Vitro Ubiquitylation Assays
Recombinant ubiquitin and purified rabbit Uba1 (E1) were obtained from Sigma and Lifesensors, respectively. Human UbcH5C (E2) was expressed in and purified from Escherichia coli as described (42). Recombinant baculoviruses were generated with the Bac-to-Bac (Invitrogen) baculovirus systems. Sf21 cells that had been infected with baculoviruses encoding Fbxw7α, Cul1, Skp1, and Rbx1 were lysed on ice with the lysis buffer used for immunoprecipitation, and the expressed proteins were purified with the use of beads conjugated with M2 antibodies to FLAG. The beads were washed three times with a solution containing 10 mm HEPES-NaOH (pH 7.5), 150 mm NaCl, and 0.1% Triton X-100, and the bead-bound proteins were then eluted with the FLAG peptide (500 μg/ml). Sf21 cells that had been infected with baculoviruses encoding His6-tagged HA-OASIS(N) or HA-BBF2H7(N) were lysed on ice with a solution containing 50 mm Tris-HCl (pH 8.0), 500 mm NaCl, 10 mm imidazole, 1% Nonidet P-40, 20 μg/ml leupeptin, and 10 μg/ml aprotinin, and the recombinant proteins were purified by Ni2+-agarose chromatography as described (43). To determine the ability of the purified recombinant SCFFbxw7α complex to mediate the ubiquitylation of OASIS(N) and BBF2H7(N), we incubated the complex (400 ng) for 2 h at 30 °C with 100 ng of Uba1, 500 ng of UbcH5C, 1 μg of ubiquitin, and 400 ng of either OASIS(N) or BBF2H7(N) in a final volume of 20 μl containing 25 mm Tris-HCl (pH 7.5), 120 mm NaCl, 2 mm ATP, 1 mm MgCl2, 1 mm DTT, 0.5 units of phosphocreatine kinase, and 1 mm creatine phosphate. The reaction mixtures were then subjected to immunoblot analysis with antibodies to HA.
Identification of Phosphorylation Sites by LC-MS/MS Analysis
FLAG-tagged OASIS and BBF2H7 were expressed, purified, and concentrated as described above for FLAG-Fbxw7α in DiPIUS-NL. The protein preparations were fractionated by SDS-PAGE and stained with silver, and bands corresponding to the full-length forms of OASIS and BBF2H7 were excised and subjected to in-gel digestion with trypsin. The resulting peptides were dried, dissolved in a mixture of 0.1% trifluoroacetic acid and 2% acetonitrile, and then applied to a nanoflow LC system (Michrom BioResources) equipped with an L-column (C18, 0.1 by 160 mm, particle size of 3 μm, CERI, Tokyo, Japan). The peptides were fractionated with a linear gradient of solvent A (0.1% formic acid in water) and solvent B (100% acetonitrile), with 5–45% solvent B over 40 min, 45–95% over 1 min, 95% over 2 min, and 95–5% over 2 min at a flow rate of 0.2 μl/min. They were then sprayed directly into a Q-Exactive mass spectrometer (Thermo Fisher). MS and MS/MS spectra (top 10 peaks) were obtained automatically in a data-dependent scan mode with a dynamic exclusion option. For protein identification, tandem mass spectra were extracted, and the charge state deconvoluted with Proteome Discoverer 1.3.0.339 (Thermo Fisher). All MS/MS spectra were compared with protein sequences in the IPI database (human version 3.26, released February 2007, 67,665 sequences, 28,353,548 residues) with the use of the MASCOT algorithm (version 2.3.02). An error of 10 ppm or 0.01 Da was set for full searches of MS and MS/MS spectra, respectively. Trypsin was selected as the enzyme used, the allowed number of missed cleavages was set at two, and carbamidomethylation of cysteine was selected as a static modification. Oxidized methionine, NH2-terminal pyroglutamine, NH2-terminal acetylation, and phosphorylation of Ser, Thr, or Tyr were specified as dynamic modifications. The ions score cutoff was 10. The phosphoRS mode of Proteome Discoverer was used for phosphorylation site localization.
Mice and Primary Culture of Chondrocytes
For generation of CAG-CreERT2;Fbxw7F/F mice, CAG-CreERT2 mice (44) were crossed with Fbxw7F/F mice (45). For primary chondrocyte culture, mesenchymal cells were prepared from the limbs of mouse embryos at embryonic day 11.5 by digestion with 0.1% trypsin and 0.1% collagenase D. Cells were plated at a density of 2 × 107 cells/ml and maintained in α-minimum essential medium supplemented with 10% FBS, ascorbic acid (50 μg/ml), and β-glycerophosphate (5 mm). Tamoxifen (2 μm, Sigma) was added to the culture medium to induce expression of Cre recombinase, and the cells were analyzed at the indicated times thereafter.
Statistical Analysis
Data are presented as mean ± S.D. and were analyzed with the Tukey-Kramer multiple comparison test. p < 0.05 was considered statistically significant.
RESULTS
Isolation of Substrates for SCFFbxw7 by DiPIUS
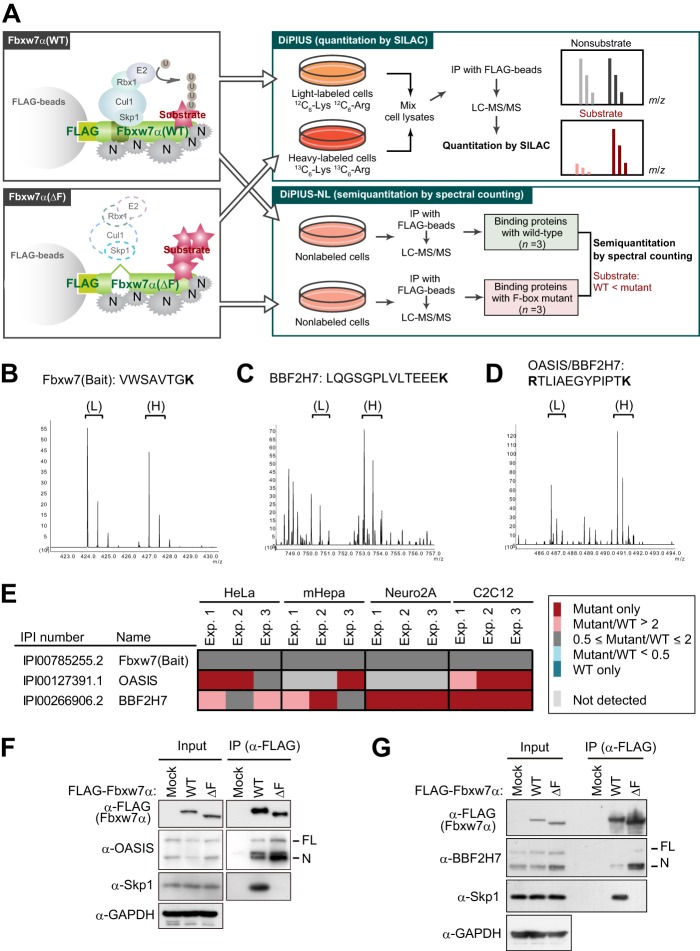
To comprehensively isolate the substrates for a given F-box protein, we developed a differential proteomics approach termed differential proteomics-based identification of ubiquitylation substrates (DiPIUS) (39). The DiPIUS system is on the basis of a difference in binding affinity for substrates between WT and mutant F-box proteins. Wild-type F-box proteins bind weakly to their substrates, which are then ubiquitylated and undergo subsequent degradation, whereas F-box proteins with a deletion in the F-box domain might be expected to associate stably with substrates and thereby allow them to accumulate in the cell (Fig. 1A).
FIGURE 1.
Identification of OASIS and BBF2H7 as candidate substrates of SCFFbxw7 by DiPIUS analyses. A, strategy for comprehensive identification of substrates for Fbxw7α. Schematics of DiPIUS and DiPIUS-NL are shown. Proteins that bind to WT or ΔF mutant forms of Fbxw7α are analyzed by quantitative SILAC (DiPIUS) or semiquantitative spectral counting (DiPIUS-NL). U, ubiquitin; N, nonspecific binding protein; IP, immunoprecipitation. B–D, representative mass spectra for DiPIUS analysis of Fbxw7α in mCAT-HeLa cells subjected to SILAC. Peak areas of light (L) and heavy (H) tryptic peptides derived from Fbxw7 (bait) (B), BBF2H7 (C), or OASIS and BBF2H7 (D) represent the abundance of proteins associated with the WT or ΔF mutant forms of Fbxw7α. E, OASIS and BBF2H7 were detected by DiPIUS-NL in mCAT-HeLa, mHepa, Neuro2A, or C2C12 cells. The ratios of the number of spectral counts for WT and mutant F-box proteins are indicated by the color scale. Proteins with a mutant/WT ratio of > 2 in at least two of three independent experiments (Exp.) were considered binding proteins with a higher affinity for the mutant F-box protein than for the WT protein. F and G, coimmunoprecipitation of endogenous OASIS (F) and BBF2H7 (G) as well as of Skp1 with FLAG-tagged WT or ΔF mutant forms of Fbxw7α from mCAT-HeLa cell lysates. Immunoprecipitates (IP) prepared with antibodies to FLAG from cells expressing the recombinant Fbxw7α proteins (or from those transfected with the corresponding empty vector, Mock) and treated with 10 μm MG132 for 6 h were subjected, together with the original cell lysates (Input), to immunoblot analysis with the indicated antibodies (α-).
Both WT Fbxw7α and a mutant form of the protein lacking the entire F-box domain (ΔF mutant) were tagged at their NH2 termini with the FLAG epitope and expressed separately in HeLa cells. Cell lysates were subjected to immunoprecipitation with antibodies to FLAG, and the immunoprecipitated proteins were analyzed by LC-MS/MS. The abundance of proteins that bound to the WT or mutant F-box proteins was compared by quantitative SILAC analysis (original DiPIUS) or semiquantitative spectral counting (DiPIUS-NL) (Fig. 1A). We identified OASIS (CREB3L1) and BBF2H7 (CREB3L2) as candidate substrates of Fbxw7α with both the DiPIUS (Fig. 1, B–D) and DiPIUS-NL (Fig. 1E) systems in HeLa cells (39). OASIS was also detected as a candidate substrate in C2C12 cells, whereas BBF2H7 was so identified in mHepa, Neuro2A, and C2C12 cells. We also examined the interaction between Fbxw7α and endogenous OASIS or BBF2H7 in HeLa cells by coimmunoprecipitation analysis. We treated the cells with the proteasome inhibitor MG132 for 6 h before immunoprecipitation to prevent the degradation of OASIS and BBF2H7 and, thereby, maximize their interaction with Fbxw7α. Such an analysis verified that endogenous OASIS and BBF2H7 interacted with Fbxw7α(ΔF) more efficiently than with Fbxw7α(WT) in HeLa cells (Fig. 1, F and G).
OASIS and BBF2H7 Interact with Fbxw7α and Fbxw7γ but not with Fbxw7β
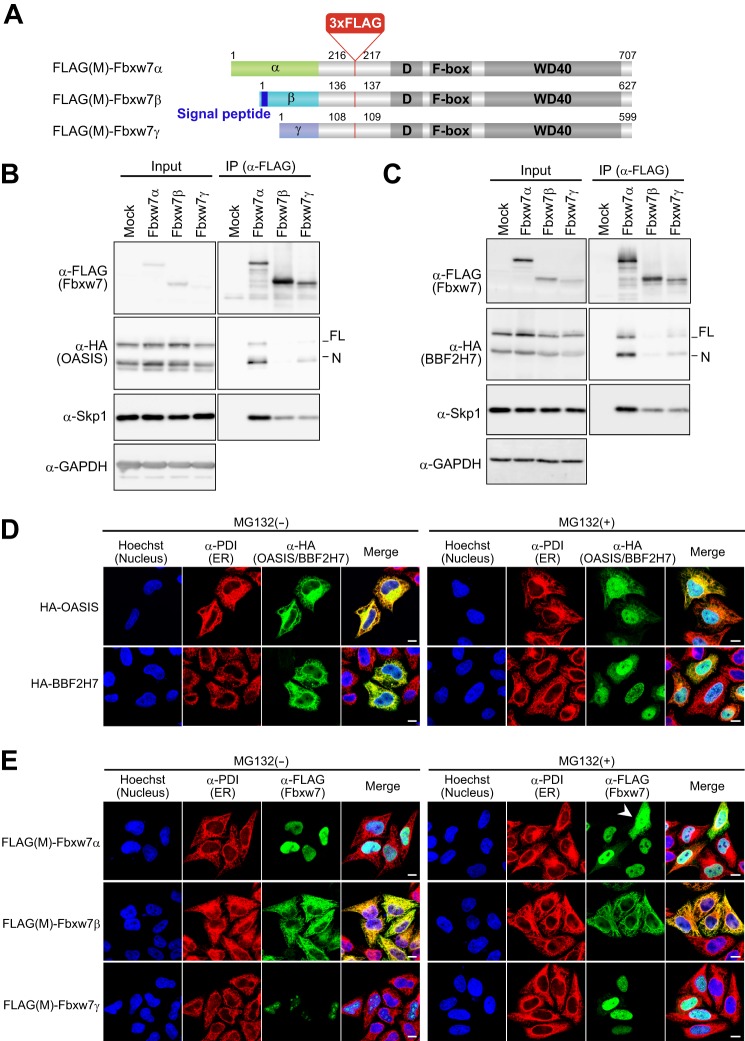
The full-length (FL) forms of OASIS and BBF2H7 are cleaved in response to ER stress, and the released NH2-terminal cytoplasmic domain (N) of each protein activates the transcription of target genes. Fbxw7 exists in three isoforms (Fbxw7α, Fbxw7β, and Fbxw7γ) that differ in their NH2-terminal regions and subcellular distributions. Fbxw7α is localized to the nucleus, Fbxw7β shows a cytoplasmic distribution suggestive of localization to the ER and Golgi apparatus, and Fbxw7γ is predominantly nucleolar (3), suggesting that each isoform might interact with a different set of substrates. We constructed vectors for versions of the three Fbxw7 isoforms with a 3× FLAG tag inserted on the NH2-terminal side of the dimerization domain, designated FLAG(M)-Fbxw7α, FLAG(M)-Fbxw7β, and FLAG(M)-Fbxw7γ (Fig. 2A). To investigate the contribution of each isoform of Fbxw7 to the interaction with OASIS and BBF2H7, we performed coimmunoprecipitation experiments with the FLAG(M)-tagged Fbxw7 proteins expressed in HEK293T cells. Fbxw7α interacted efficiently with the N forms of both OASIS and BBF2H7 and, to a lesser extent, with the FL form of each protein (Fig. 2, B and C). Fbxw7γ interacted at a low level with both FL and N forms of OASIS and BBF2H7, but we did not detect any interaction between Fbxw7β and OASIS or BBF2H7. We speculate that the anchoring of Fbxw7β and the FL forms of OASIS and BBF2H7 to the ER membrane may structurally impede the interaction of these proteins.
FIGURE 2.
OASIS and BBF2H7 interact with Fbxw7α and Fbxw7γ but not with Fbxw7β. A, schematic of FLAG(M)-Fbxw7α, -Fbxw7β, and -Fbxw7γ. The isoform-specific, dimerization (D), F-box, and WD40 domains are indicated, as is the putative signal peptide of Fbxw7β. B and C, lysates of HEK293T cells expressing FLAG(M)-tagged Fbxw7α, Fbxw7β, or Fbxw7γ together with HA-tagged forms of OASIS (B) or BBF2H7 (C) were subjected to immunoprecipitation (IP) with antibodies to FLAG, and the resulting precipitates were subjected to immunoblot analysis with the indicated antibodies. D, mCAT-HeLa cells expressing HA-tagged OASIS or BBF2H7 were incubated in the absence or presence of 10 μm MG132 for 6 h, fixed, and processed for immunofluorescence staining with antibodies to HA (green) and protein disulfide isomerase (PDI, red). Nuclei were stained with Hoechst 33258 (blue), and merged images are also shown. Scale bars = 10 μm. E, mCAT-HeLa cells expressing FLAG(M)-tagged Fbxw7α, Fbxw7β, or Fbxw7γ were incubated in the absence or presence of 10 μm MG132 for 6 h, fixed, and processed for immunofluorescence staining with antibodies to FLAG (green) and to protein disulfide isomerase (red). Nuclei were stained with Hoechst 33258 (blue), and merged images are also shown. The arrowhead indicates a cell in which FLAG(M)-Fbxw7α had leaked into the cytosol. Scale bars = 10 μm.
We examined the subcellular localization of HA epitope-tagged OASIS and BBF2H7 by immunofluorescence analysis in cells incubated with or without MG132. In the absence of MG132 treatment, HA-OASIS and HA-BBF2H7 were colocalized with the ER marker protein disulfide isomerase, whereas MG132 promoted the cleavage of these proteins at the ER and stabilized the cleaved forms, resulting in the accumulation of HA-OASIS(N) and HA-BBF2H7(N) in the nucleus (Fig. 2D). We also examined the localization of FLAG(M)-tagged Fbxw7α, Fbxw7β, and Fbxw7γ (Fig. 2E). In the absence of MG132, FLAG(M)-Fbxw7α was localized to the nucleus, FLAG(M)-Fbxw7β was detected at the ER, and FLAG(M)-Fbxw7γ was predominantly nucleolar, consistent with previous findings (3). In the presence of MG132, however, FLAG(M)-Fbxw7α was detected in the cytosol, in addition to the nucleus in some cells, with a high expression level of the protein (Fig. 2E, arrowhead). Similarly, FLAG(M)-Fbxw7γ was detected in the nucleoplasm in addition to the nucleolus. These results suggested that the interaction between Fbxw7α and the FL forms of OASIS and BBF2H7 detected by coimmunoprecipitation analysis was an artifact of Fbxw7α overexpression and MG132 treatment.
Fbxw7 Controls the Stability of OASIS and BBF2H7
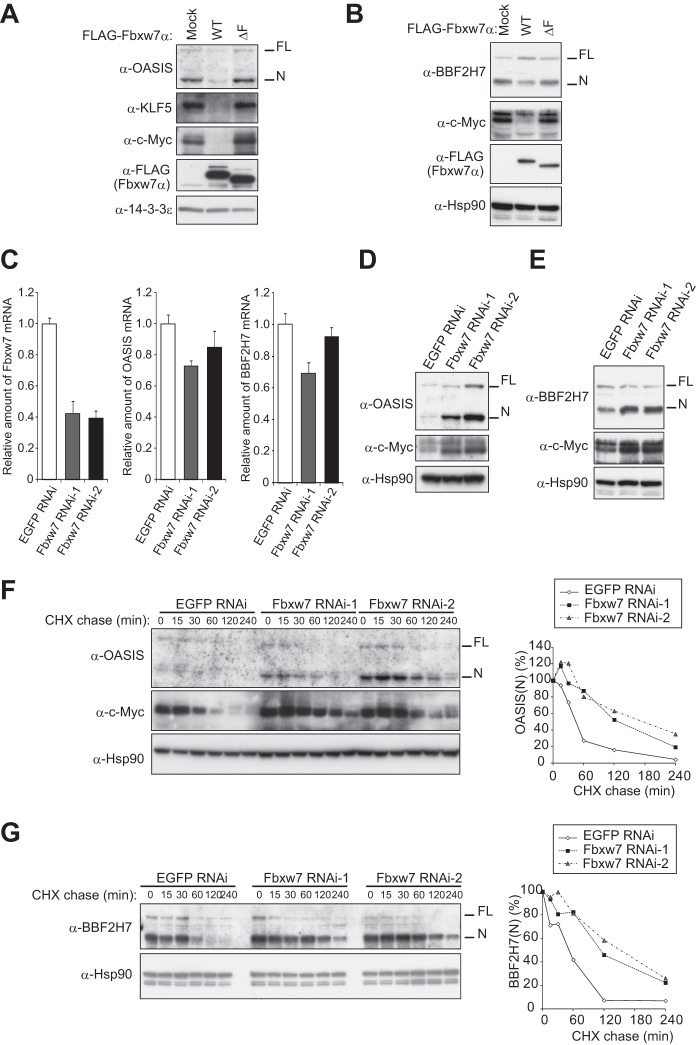
We next examined whether Fbxw7 affects the abundance of OASIS and BBF2H7 in cells. Sustained overexpression of Fbxw7α in HeLa cells resulted in a substantial decrease in the amounts of both OASIS(N) and BBF2H7(N) as well as of the positive controls KLF5 and c-Myc (Fig. 3, A and B). The abundance of FL forms of OASIS and BBF2H7 at the ER was not affected by overexpression of Fbxw7α. Although RNAi-mediated depletion of Fbxw7 did not affect the abundance of OASIS and BBF2H7 mRNAs (Fig. 3C), it resulted in a marked increase in the amounts of OASIS(N) and BBF2H7(N) proteins without substantially affecting the amounts of OASIS(FL) and BBF2H7(FL) (D and E). Furthermore, cycloheximide chase assays revealed that the half-lives of OASIS(N) and BBF2H7(N) were increased substantially in Fbxw7-depleted cells compared with those in mock-depleted cells (Fig. 3, F and G).
FIGURE 3.
Fbxw7 controls the stability of OASIS and BBF2H7. A and B, immunoblot analysis of OASIS (A) and BBF2H7 (B) as well as of KLF5 and c-Myc in mCAT-HeLa cells stably infected with retroviruses encoding FLAG-tagged WT or ΔF mutant forms of Fbxw7α. C, RT and real-time PCR analysis of Fbxw7, OASIS, and BBF2H7 mRNAs in mCAT-HeLa cells stably infected with retroviruses encoding Fbxw7 or enhanced GFP (control) shRNAs. Normalized data are expressed relative to the corresponding value for control cells and are mean ± S.D. from three independent experiments. D and E, immunoblot analysis of OASIS (D) and BBF2H7 (E) as well as of c-Myc in mCAT-HeLa cells stably infected with retroviruses encoding Fbxw7 or enhanced GFP (control) shRNAs. F and G, cycloheximide (CHX) chase analysis of OASIS and c-Myc (F) and of BBF2H7 (G) in mCAT-HeLa cells depleted of Fbxw7. The percentage of endogenous OASIS(N) or BBF2H7(N) remaining after the various chase times was quantitated with ImageJ software.
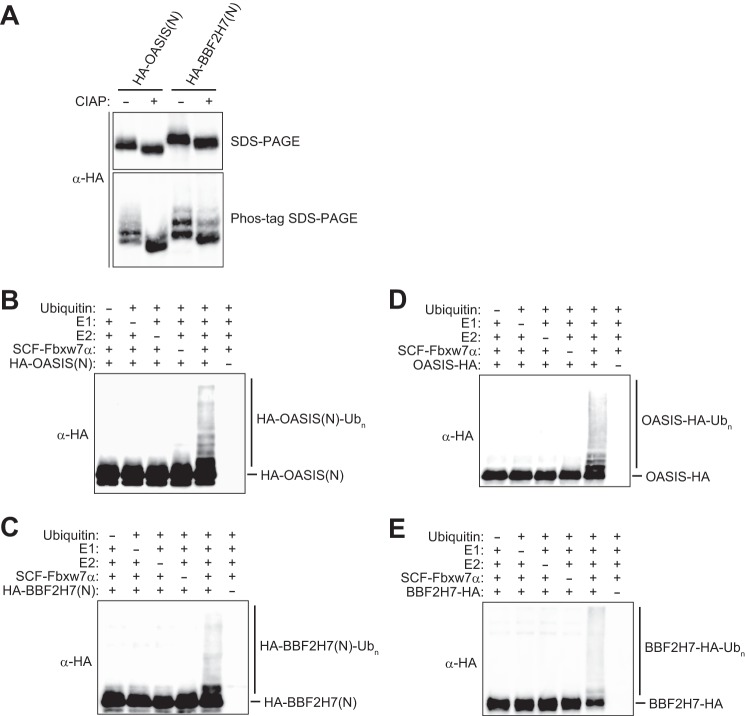
We also examined whether OASIS(N) and BBF2H7(N) are direct substrates of SCFFbxw7α in vitro. His6-HA-tagged forms of OASIS(N) and BBF2H7(N) were expressed in and purified from Sf21 insect cells. The purified proteins exhibited multiple bands on Phos-tag SDS-PAGE (Fig. 4A), and the lower-mobility bands were reduced in intensity after treatment of the proteins with calf intestinal alkaline phosphatase, supporting the notion that the recombinant proteins were phosphorylated. We also expressed His6-Myc-tagged Skp1, His6-Myc-tagged Cul1, Myc-tagged Rbx1, and His6-FLAG-tagged Fbxw7α in Sf21 cells and purified SCFFbxw7α from the cells with antibodies to FLAG. In vitro ubiquitylation assays with these recombinant proteins revealed that the ubiquitylation of OASIS(N) and BBF2H7(N) was apparent only in the presence of all reaction components, including Uba1 (E1), UbcH5C (E2), SCFFbxw7α (E3), and ubiquitin (Fig. 4, B and C). We also examined whether full-length recombinant OASIS and BBF2H7 underwent ubiquitylation in vitro. Both OASIS(FL)-HA and BBF2H7(FL)-HA were indeed ubiquitylated by SCFFbxw7α in vitro (Fig. 4, D and E). However, given that the subcellular localization of endogenous full-length OASIS and BBF2H7 differs from that of Fbxw7α under normal conditions (Fig. 2, D and E), these latter reactions are unlikely to occur in vivo. Collectively, these results suggested that SCFFbxw7 is an authentic ubiquitin ligase for OASIS(N) and BBF2H7(N).
FIGURE 4.
Fbxw7 mediates the ubiquitylation of OASIS and BBF2H7. A, HA-tagged forms of OASIS(N) or BBF2H7(N) purified from Sf21 insect cells were incubated with or without calf intestinal alkaline phosphatase (CIAP, 1 units/μl) for 3 h at 37 °C before conventional or Phos-tag SDS-PAGE followed by immunoblot analysis with antibodies to HA. B and C, HA-tagged forms of OASIS(N) (B) or BBF2H7(N) (C) were subjected to an in vitro ubiquitylation assay with immunopurified SCFFbxw7α as well as with E1, E2, ATP, and ubiquitin, as indicated. Reaction mixtures were subjected to immunoblot analysis with antibodies to HA. The positions of unmodified and polyubiquitylated (Ubn) forms of OASIS(N) and BBF2H7(N) are indicated. D and E, HA-tagged full-length forms of OASIS (D) or BBF2H7 (E) were subjected to an in vitro ubiquitylation assay as in B and C.
Phosphorylation of OASIS and BBF2H7 Is Necessary for Interaction with Fbxw7
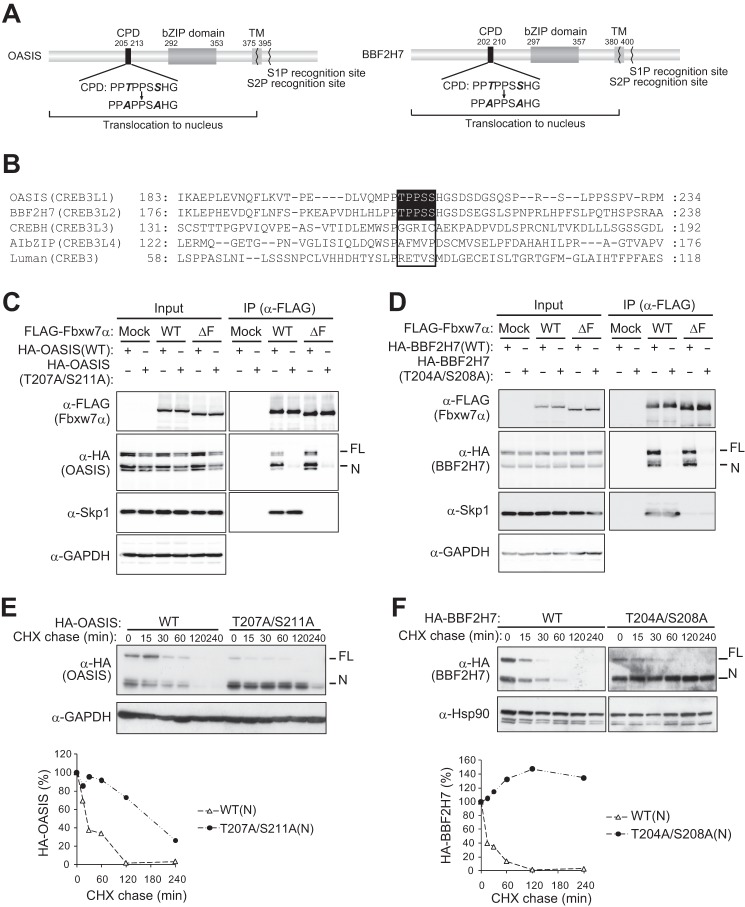
Many substrates of Fbxw7 contain a conserved amino acid sequence for phosphorylation, termed the Cdc4 phosphodegron (CPD) (3). OASIS(N) and BBF2H7(N) each contain an amino acid sequence that matches the CPD, whereas this sequence is not conserved in other CREB3 or CREB3L proteins, including CREBH (CREB3L3), AIbZIP (CREB3L4), and Luman (CREB3) (Fig. 5, A and B).
FIGURE 5.
Fbxw7 interacts with OASIS and BBF2H7 via a conserved phosphodegron. A, domain organization of human OASIS and BBF2H7. Potential CPD sequences, introduced mutations, basic leucine zipper (bZIP) and transmembrane (TM) domains, and recognition sites for S1P and S2P are shown. B, alignment of amino acid sequences of human CREB3 and CREB3L family members. Potential CPDs and their corresponding sequences are boxed. C and D, effects of CPD mutation on the binding of OASIS or BBF2H7 to Fbxw7α. Lysates of mCAT-HeLa cells expressing the HA-tagged forms of OASIS (C) or BBF2H7 (D), or of their CPD mutants, together with the FLAG-tagged WT or ΔF mutant forms of Fbxw7α were subjected to immunoprecipitation (IP) with antibodies to FLAG, and the resulting precipitates were subjected to immunoblot analysis with the indicated antibodies. E and F, cycloheximide (CHX) chase analysis of HA-tagged forms of OASIS (E) or BBF2H7 (F), or of their CPD mutants, in mCAT-HeLa cells. The percentages of HA-tagged OASIS(N) and BBF2H7(N) proteins remaining after the various chase times were quantitated with ImageJ software.
To investigate whether the CPD sequence is required for the degradation of OASIS and BBF2H7 dependent on SCFFbxw7, we introduced alanine mutations into this motif. Immunoprecipitation analysis revealed that a mutant (T207A/S211A) form of OASIS that contains two point mutations in the CPD (Fig. 5A) did not interact with Fbxw7α (Fig. 5C). Furthermore, cycloheximide chase analysis revealed that the N form of this mutant was more stable than that of WT OASIS in transfected HeLa cells (Fig. 5E). A similar mutant (T204A/S208A) of BBF2H7 also did not bind to Fbxw7α (Fig. 5D), and the N form of this mutant was more stable than that of the WT protein (Fig. 5F).
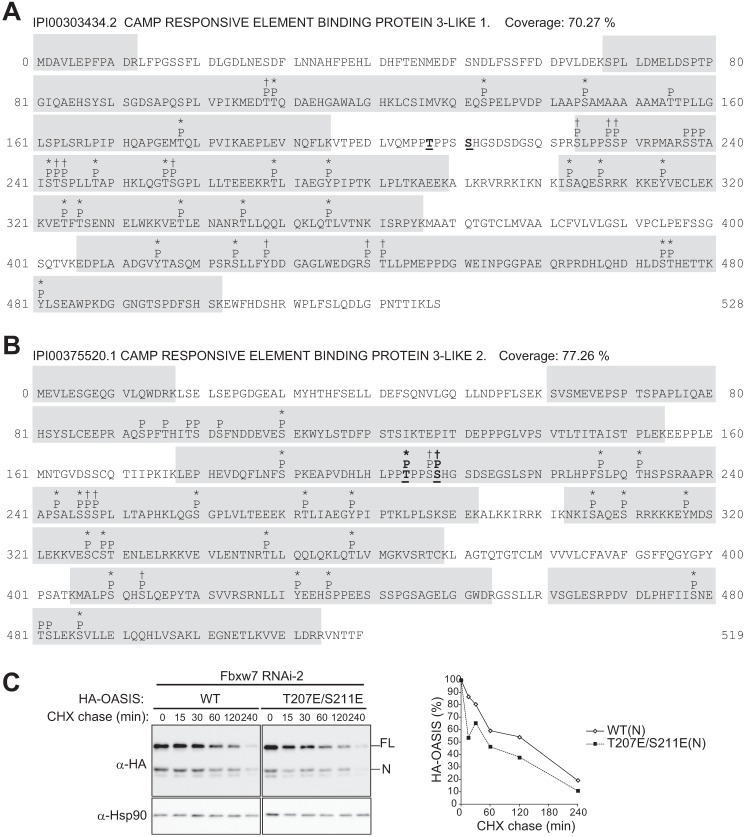
To verify that the CPD motifs of OASIS and BBF2H7 are phosphorylated, we investigated the phosphorylation status of FLAG-OASIS and FLAG-BBF2H7 in HeLa cells by LC-MS/MS analysis. Many peptides derived from FLAG-OASIS(FL) and FLAG-BBF2H7(FL) were highly phosphorylated (at least 22 phosphorylated sites in each protein) (Fig. 6, A and B, and supplemental Table S1). In particular, we detected phosphorylation of the CPD in BBF2H7, with Thr204 and either Ser207 or Ser208 being phosphorylated. We did not detect phosphorylation of the CPD of OASIS, however. Therefore, we generated a phosphomimetic mutant of OASIS by replacing Thr207 and Ser211 with Glu (T207E/S211E) and examined its stability in cells depleted of endogenous Fbxw7 by RNAi (Fig. 6C). We detected a small but reproducible increase in the rate of degradation for the N form of the HA-tagged mutant compared with that for the N form of the HA-tagged WT protein. Together, these observations suggested that phosphorylation of the CPD of OASIS and BBF2H7 is required for the interaction of these proteins with Fbxw7 and their consequent degradation.
FIGURE 6.
Phosphorylation of OASIS and BBF2H7 expressed in mCAT-HeLa cells. A and B, phosphorylation sites of OASIS (A) and BBF2H7 (B) identified by LC-MS/MS analysis of FLAG-tagged forms of the human proteins expressed in and purified from mCAT-HeLa cells. Potential phosphorylated residues (P) are indicated above the amino acid sequences. Asterisks and † indicate phosphorylation probabilities of > 99% and 45–99%, respectively. Phosphorylation sites within the CPD of OASIS or BBF2H7 are shown in boldface and underlined. C, cycloheximide (CHX) chase analysis of the HA-tagged WT and phosphomimetic mutant (T207E/S211E) forms of OASIS in mCAT-HeLa cells depleted of Fbxw7. The percentages of HA-tagged OASIS(N) proteins remaining after the various chase times were quantitated with ImageJ software.
Fbxw7 Inhibits Osteogenesis by Mediating the Degradation of OASIS
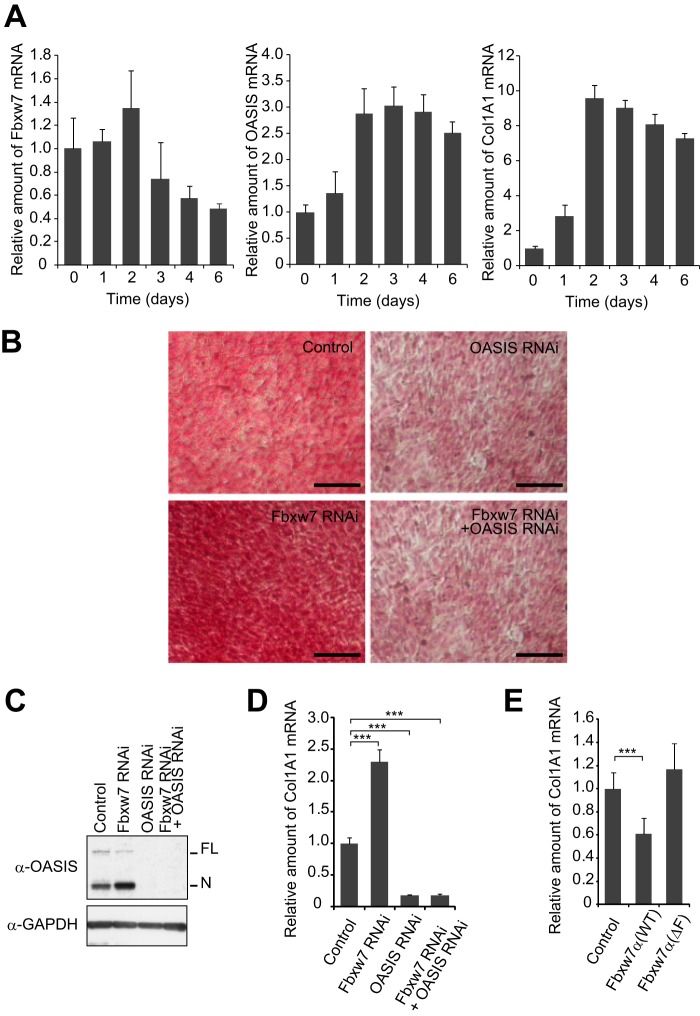
OASIS and BBF2H7 participate in mesenchymal differentiation, being essential for osteogenesis and chondrogenesis, respectively. OASIS controls osteoblast differentiation through activation of Col1A1 gene transcription (37). We examined changes in the abundance of Fbxw7, OASIS, and Col1A1 mRNAs during the osteogenic differentiation of mouse C2C12 mesenchymal cells induced by treatment with BMP2 (Fig. 7A). The amount of Fbxw7 mRNA increased slightly during the first 2 days of the differentiation process and then declined. In contrast, the abundance of OASIS and Col1A1 mRNAs was increased substantially after 2 days and remained at this high level until the end of the 6-day culture period.
FIGURE 7.
Fbxw7 inhibits osteogenesis by promoting the degradation of OASIS. A, RT and real-time PCR analysis of Fbxw7, OASIS, and Col1A1 mRNAs in C2C12 cells at the indicated times after induction of differentiation into osteoblasts. B–D, C2C12 cells depleted of Fbxw7, OASIS, or both proteins by RNAi were induced to differentiate into osteoblasts and then either stained with Sirius red to visualize collagen accumulation (B), subjected to immunoblot analysis with antibodies to OASIS (C), or assayed for the relative amount of Col1A1 mRNA (D). Scale bars = 100 μm (B). E, relative abundance of Col1A1 mRNA in C2C12 cells expressing ectopic WT or ΔF mutant forms of Fbxw7α and induced to differentiate into osteoblasts. Data in A, D, and E are mean ± S.D. from three independent experiments. ***, p < 0.001 (Tukey-Kramer multiple-comparison test).
We next examined the effect of Fbxw7 depletion on the differentiation of C2C12 cells into osteoblasts. RNAi-mediated depletion of Fbxw7 resulted in the accumulation of OASIS in C2C12 cells and markedly increased both collagen deposition and Col1A1 gene expression (Fig. 7, B–D). Additional depletion of OASIS greatly attenuated these latter effects of Fbxw7 depletion, suggesting that they were mediated by up-regulation of OASIS expression. Conversely, overexpression of Fbxw7α in C2C12 cells resulted in a decrease in the abundance of Col1A1 mRNA, whereas forced expression of Fbxw7α(ΔF) had no such effect (Fig. 7E). Thus, these data suggest that Fbxw7 inhibits osteogenesis by promoting the degradation of OASIS and the consequent attenuation of collagen deposition.
Fbxw7 Controls Chondrogenesis by Targeting Notch and BBF2H7 for Degradation
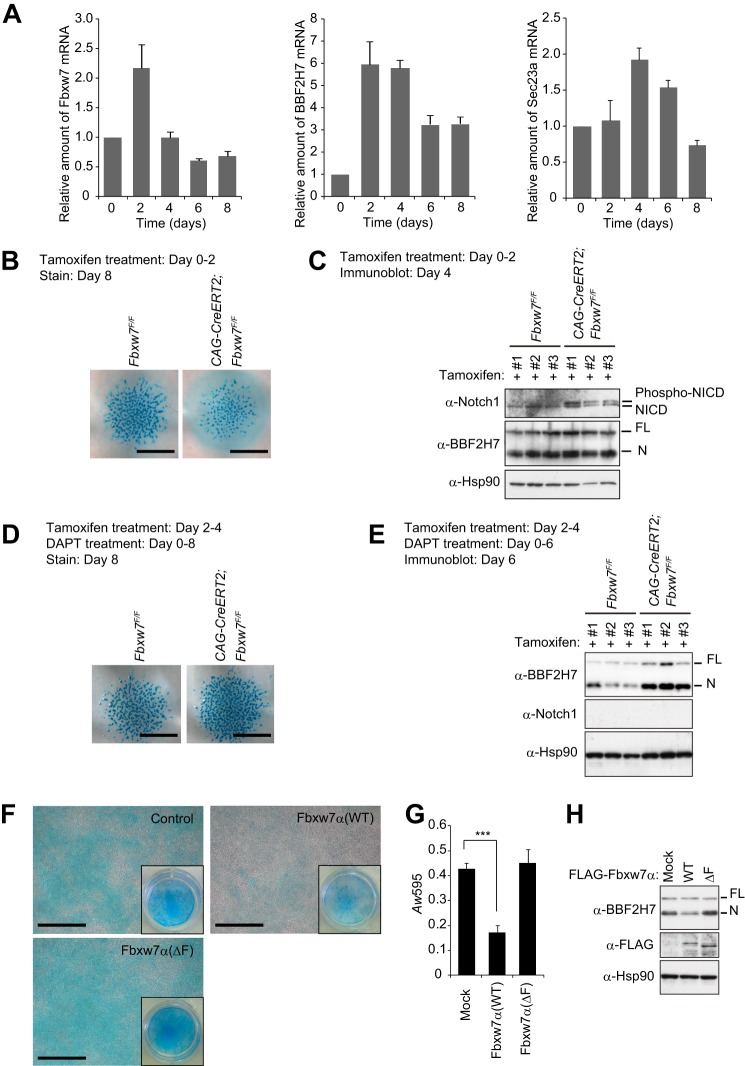
Similar to the role of OASIS in osteogenesis, BBF2H7 controls chondrogenesis through transcriptional activation of the Sec23a gene. We examined changes in the abundance of Fbxw7, BBF2H7, and Sec23a mRNAs during chondrogenesis in micromass cultures of mesenchymal cells isolated from mouse embryos (Fig. 8A). The amount of Fbxw7 mRNA was increased after 2 days of culture, but it had returned to the original level at day 4 and had declined slightly below this level at day 6. The abundance of BBF2H7 and Sec23a mRNAs was increased after days 2 and 4, respectively, but had declined substantially at days 6 and 8, respectively, consistent with previous observations (38).
FIGURE 8.
Fbxw7 controls chondrogenesis by targeting Notch and BBF2H7 for degradation. A, RT and real-time PCR analysis of Fbxw7, BBF2H7, and Sec23a mRNAs in mesenchymal cells isolated from mouse embryos and maintained as micromass cultures for the indicated times. Data are mean ± S.D. from three independent experiments. B and C, mesenchymal cells isolated from CAG-CreERT2;Fbxw7F/F or control (Fbxw7F/F) mouse embryos and maintained as micromass cultures were exposed to tamoxifen for the initial 2 days of culture and subjected to staining with Alcian blue after 8 days (B) or to immunoblot analysis of Notch1 and BBF2H7 after 4 days (C). Immunoblot data are shown for cultures derived from three different embryos. D and E, mesenchymal cells isolated from CAG-CreERT2;Fbxw7F/F or control (Fbxw7F/F) mice and maintained as micromass cultures in the presence of 1 μm DAPT were exposed to tamoxifen for 2 days, beginning 2 days after the onset of culture. The cells were stained with Alcian blue after 8 days (D) or subjected to immunoblot analysis of Notch1 and BBF2H7 after 6 days (E). F–H, ATDC5 cells expressing the FLAG-tagged WT or ΔF mutant forms of Fbxw7α were induced to differentiate into chondrocytes by exposure to 1 μm DAPT and were then either stained with Alcian blue for visualization (F) or absorbance-based measurement (G) of the accumulation of acid mucosubstances or subjected to immunoblot analysis with antibodies to BBF2H7 or to FLAG (H). The insets in F show low-magnification images of the culture plates. Quantitative data (G) are means ± S.D. from three independent experiments. ***, p < 0.001 (Tukey-Kramer multiple-comparison test). Scale bars = 1 mm (B and D) or 100 μm (F).
We next investigated the effect of Fbxw7 deficiency on the formation of cartilage by the micromass cultures. Conditional ablation of floxed (F) Fbxw7 alleles (45) by treatment of CAG-CreERT2;Fbxw7F/F cells with tamoxifen at the start of the culture resulted in attenuation of chondrogenesis, as revealed by staining with Alcian blue after culture for 8 days (Fig. 8B). Immunoblot analysis of Fbxw7-depleted chondrocytes after culture for 4 days revealed accumulation of the phosphorylated form of the intracellular domain of Notch1 (NICD1), another target of Fbxw7, but not that of BBF2H7 (Fig. 8C). Notch suppresses the differentiation and proliferation of early chondrogenic cells (46), suggesting that the accumulation of Notch1 induced by depletion of Fbxw7 might inhibit chondrocyte differentiation.
To eliminate any effect of Notch1 accumulation induced by Fbxw7 ablation during early chondrogenesis, we depleted Fbxw7 2 days after the onset of chondrocyte differentiation and exposed the cells from day 0 to DAPT, an inhibitor of γ-secretase activity that is necessary for Notch activation. Under these conditions, ablation of Fbxw7 resulted in a slight enhancement of chondrogenesis compared with that apparent in control cells (Fig. 8D), and it triggered a substantial increase in the abundance of BBF2H7(N) (E). We next examined the effect of Fbxw7α overexpression on chondrogenesis in murine chondrogenic ATDC5 cells. Forced expression of Fbxw7α resulted in marked inhibition of chondrogenesis in ATDC5 cells (Fig. 8, F and G), an effect that was accompanied by down-regulation of BBF2H7(N) (H). Expression of Fbxw7α(ΔF) had no such effects, suggesting that they were attributable to the ubiquitin ligase activity of Fbxw7α. Together, these results indicate that Fbxw7 regulates chondrogenesis both positively and negatively by promoting the degradation of Notch and BBF2H7, respectively.
DISCUSSION
With the use of DiPIUS, an unbiased approach to the identification of substrates for specific ubiquitin ligases, we found that OASIS and BBF2H7 are substrates of Fbxw7. Validation analyses confirmed that OASIS and BBF2H7 are bona fide substrates of Fbxw7, and we further found that Fbxw7 plays key roles in bone and cartilage formation by controlling the levels of OASIS and BBF2H7, respectively.
Our analysis of changes in the abundance of Fbxw7 mRNA during osteogenic and chondrocytic differentiation suggested that Fbxw7 might antagonize osteogenesis and chondrogenesis by promoting the degradation of OASIS and BBF2H7, respectively, predominantly during the early phase of such differentiation. Subsequent down-regulation of Fbxw7 during the later stages of differentiation might allow the up-regulation of Col1A1 and Sec23a and, thereby, promote the production of bone and cartilage matrix proteins by osteoblasts and chondrocytes, respectively. The accumulation of these matrix proteins in the ER may induce ER stress and, thereby, trigger the cleavage and activation of OASIS and BBF2H7. Such a sequence of events would constitute a positive feedback loop, suppression of which by Fbxw7 might impede the premature activation of the Col1A1 and Sec23a genes and, thereby, prevent excessive accumulation of matrix proteins before cell maturation.
In addition to osteogenesis and chondrogenesis, Fbxw7 controls adipogenesis by promoting the degradation of KLF5, SREBPs, and c/EBPα (29–32). Inactivation of Fbxw7 in mouse preadipocytes or adult human mesenchymal stem cells promotes their differentiation into mature adipocytes (31, 32). Genetic ablation or RNAi-mediated depletion of Fbxw7 in mouse liver induced fatty liver, reminiscent of that associated with nonalcoholic steatohepatitis in humans (29, 30). Fbxw7 also targets peroxisome proliferator-activated receptor γ (PPARγ) coactivator 1α (PGC-1α) for degradation (47), although the relationship between Fbxw7 and PGC-1α in adipogenesis or lipogenesis remains to be determined. Therefore, we therefore propose that Fbxw7 inhibits mesenchymal differentiation and maturation by concomitant targeting of multiple transcriptional factors for degradation and that it may, therefore, contribute to the maintenance of mesenchymal stem cells (Fig. 9). The biological relevance of Fbxw7 in each differentiation pathway appears to be determined by the expression of corresponding substrates and by the expression and activation of protein kinases that phosphorylate the CPDs of these proteins.
FIGURE 9.
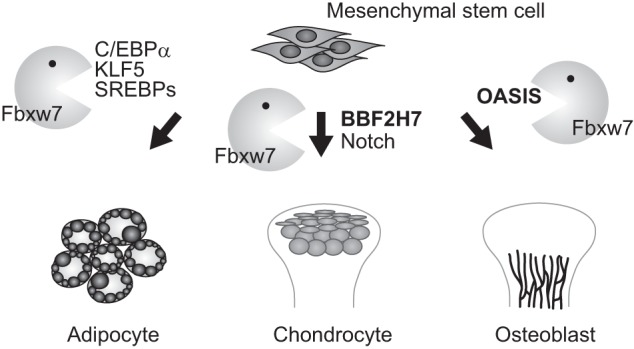
Fbxw7 controls mesenchymal differentiation. Fbxw7 mediates ubiquitin-dependent degradation of substrates that contribute to adipogenesis, chondrogenesis, and osteogenesis. Thus, it inhibits adipogenesis by targeting C/EBPα, KLF5, and SREBPs for degradation. We show that Fbxw7 also regulates chondrogenesis and osteogenesis by promoting the degradation of Notch, BBF2H7, and OASIS, respectively.
The regulation of certain transcriptional factors in response to specific conditions depends on their localization. Under normoxic conditions, hypoxia-inducible factor 1α (HIF1α) is localized to the cytoplasm, where it undergoes oxygen-dependent proline hydroxylation followed by von Hippel-Lindau protein-dependent proteasomal degradation. During hypoxia, however, the proline hydroxylases are inactive, and HIF1α is able to translocate to the nucleus and induce the transcription of genes related to oxygen homeostasis and metabolism (48). Notch proteins are transcriptional factors that contain both EGF-like extracellular domains and cytoplasmic regions. The binding of a Notch ligand to the EGF-like extracellular domain triggers Notch cleavage both outside and within the transmembrane domain, resulting in the nuclear translocation of the cleaved Notch intracellular domain and its association with the constitutive DNA-binding protein RBP-Jκ (49). Finally, SREBPs are localized at the ER membrane under normal conditions. In response to a reduction in the intracellular level of cholesterol, however, they move to the Golgi complex, where they are cleaved sequentially by two proteases. These cleavage steps release the mature form of SREBPs, which enters the nucleus and activates the transcription of genes related to cholesterol and fatty acid metabolism (50). Fbxw7 targets nuclear forms of all of these transcriptional factors (HIF1α, Notch, and SREBPs) as well as those of OASIS and BBF2H7 for degradation (5, 12, 13, 31, 51, 52). Fbxw7 might, therefore, function to turn off the transactivation activity of these factors in the nucleus, thereby contributing to the strict regulation of transcription in response to various stimuli or conditions.
Acknowledgments
We thank T. Kitamura for pMX-puro; J. M. Cunningham and K. Hanada for the mCAT-1 plasmid; M. Oda, E. Koba, K. Oyamada, T. Takami, N. Nishimura, K. Tsunematsu, and other laboratory members for technical assistance; and A. Ohta for help with preparation of the manuscript.
This work was supported in part by a grant from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.

This article contains supplemental Table S1.
- SCF
- Skp1-Cul1-F-box protein
- SREBP
- sterol response element-binding protein
- OASIS
- old astrocyte specifically induced substance
- BBF2H7
- BBF2 human homolog on chromosome 7
- ER
- endoplasmic reticulum
- DAPT
- N-[N-(3,5-difluorophenacetyl)-l-alanyl]-S-phenylglycine t-butyl ester
- DiPIUS
- differential proteomics-based identification of ubiquitylation substrates
- DiPIUS-NL
- differential proteomics-based identification of ubiquitylation substrates nonlabeling
- SILAC
- stable isotope labeling with amino acids in cell culture
- IPI
- International Protein Index
- CPD
- Cdc4 phosphodegron
- FL
- full-length
- N
- nuclear.
REFERENCES
- 1. Frescas D., Pagano M. (2008) Deregulated proteolysis by the F-box proteins SKP2 and β-TrCP. Tipping the scales of cancer. Nat. Rev. Cancer 8, 438–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nakayama K. I., Nakayama K. (2006) Ubiquitin ligases. Cell-cycle control and cancer. Nat. Rev. Cancer 6, 369–381 [DOI] [PubMed] [Google Scholar]
- 3. Welcker M., Clurman B. E. (2008) FBW7 ubiquitin ligase. A tumour suppressor at the crossroads of cell division, growth and differentiation. Nat. Rev. Cancer 8, 83–93 [DOI] [PubMed] [Google Scholar]
- 4. Jin J., Cardozo T., Lovering R. C., Elledge S. J., Pagano M., Harper J. W. (2004) Systematic analysis and nomenclature of mammalian F-box proteins. Genes Dev. 18, 2573–2580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hubbard E. J., Wu G., Kitajewski J., Greenwald I. (1997) sel-10, a negative regulator of lin-12 activity in Caenorhabditis elegans, encodes a member of the CDC4 family of proteins. Genes Dev. 11, 3182–3193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sundaram M., Greenwald I. (1993) Suppressors of a lin-12 hypomorph define genes that interact with both lin-12 and glp-1 in Caenorhabditis elegans. Genetics 135, 765–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Koepp D. M., Schaefer L. K., Ye X., Keyomarsi K., Chu C., Harper J. W., Elledge S. J. (2001) Phosphorylation-dependent ubiquitination of cyclin E by the SCFFbw7 ubiquitin ligase. Science 294, 173–177 [DOI] [PubMed] [Google Scholar]
- 8. Moberg K. H., Bell D. W., Wahrer D. C., Haber D. A., Hariharan I. K. (2001) Archipelago regulates Cyclin E levels in Drosophila and is mutated in human cancer cell lines. Nature 413, 311–316 [DOI] [PubMed] [Google Scholar]
- 9. Strohmaier H., Spruck C. H., Kaiser P., Won K. A., Sangfelt O., Reed S. I. (2001) Human F-box protein hCdc4 targets cyclin E for proteolysis and is mutated in a breast cancer cell line. Nature 413, 316–322 [DOI] [PubMed] [Google Scholar]
- 10. Yada M., Hatakeyama S., Kamura T., Nishiyama M., Tsunematsu R., Imaki H., Ishida N., Okumura F., Nakayama K., Nakayama K. I. (2004) Phosphorylation-dependent degradation of c-Myc is mediated by the F-box protein Fbw7. EMBO J. 23, 2116–2125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Welcker M., Orian A., Jin J., Grim J. E., Grim J. A., Harper J. W., Eisenman R. N., Clurman B. E. (2004) The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proc. Natl. Acad. Sci. U.S.A. 101, 9085–9090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gupta-Rossi N., Le Bail O., Gonen H., Brou C., Logeat F., Six E., Ciechanover A., Israël A. (2001) Functional interaction between SEL-10, an F-box protein, and the nuclear form of activated Notch1 receptor. J. Biol. Chem. 276, 34371–34378 [DOI] [PubMed] [Google Scholar]
- 13. Oberg C., Li J., Pauley A., Wolf E., Gurney M., Lendahl U. (2001) The Notch intracellular domain is ubiquitinated and negatively regulated by the mammalian Sel-10 homolog. J. Biol. Chem. 276, 35847–35853 [DOI] [PubMed] [Google Scholar]
- 14. Nateri A. S., Riera-Sans L., Da Costa C., Behrens A. (2004) The ubiquitin ligase SCFFbw7 antagonizes apoptotic JNK signaling. Science 303, 1374–1378 [DOI] [PubMed] [Google Scholar]
- 15. Wei W., Jin J., Schlisio S., Harper J. W., Kaelin W. G., Jr. (2005) The v-Jun point mutation allows c-Jun to escape GSK3-dependent recognition and destruction by the Fbw7 ubiquitin ligase. Cancer Cell 8, 25–33 [DOI] [PubMed] [Google Scholar]
- 16. Liu N., Li H., Li S., Shen M., Xiao N., Chen Y., Wang Y., Wang W., Wang R., Wang Q., Sun J., Wang P. (2010) The Fbw7/human CDC4 tumor suppressor targets proproliferative factor KLF5 for ubiquitination and degradation through multiple phosphodegron motifs. J. Biol. Chem. 285, 18858–18867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhao D., Zheng H. Q., Zhou Z., Chen C. (2010) The Fbw7 tumor suppressor targets KLF5 for ubiquitin-mediated degradation and suppresses breast cell proliferation. Cancer Res. 70, 4728–4738 [DOI] [PubMed] [Google Scholar]
- 18. Akhoondi S., Sun D., von der Lehr N., Apostolidou S., Klotz K., Maljukova A., Cepeda D., Fiegl H., Dafou D., Dofou D., Marth C., Mueller-Holzner E., Corcoran M., Dagnell M., Nejad S. Z., Nayer B. N., Zali M. R., Hansson J., Egyhazi S., Petersson F., Sangfelt P., Nordgren H., Grander D., Reed S. I., Widschwendter M., Sangfelt O., Spruck C. (2007) FBXW7/hCDC4 is a general tumor suppressor in human cancer. Cancer Res. 67, 9006–9012 [DOI] [PubMed] [Google Scholar]
- 19. Matsuoka S., Oike Y., Onoyama I., Iwama A., Arai F., Takubo K., Mashimo Y., Oguro H., Nitta E., Ito K., Miyamoto K., Yoshiwara H., Hosokawa K., Nakamura Y., Gomei Y., Iwasaki H., Hayashi Y., Matsuzaki Y., Nakayama K., Ikeda Y., Hata A., Chiba S., Nakayama K. I., Suda T. (2008) Fbxw7 acts as a critical fail-safe against premature loss of hematopoietic stem cells and development of T-ALL. Genes Dev. 22, 986–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Thompson B. J., Jankovic V., Gao J., Buonamici S., Vest A., Lee J. M., Zavadil J., Nimer S. D., Aifantis I. (2008) Control of hematopoietic stem cell quiescence by the E3 ubiquitin ligase Fbw7. J. Exp. Med. 205, 1395–1408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Reavie L., Della Gatta G., Crusio K., Aranda-Orgilles B., Buckley S. M., Thompson B., Lee E., Gao J., Bredemeyer A. L., Helmink B. A., Zavadil J., Sleckman B. P., Palomero T., Ferrando A., Aifantis I. (2010) Regulation of hematopoietic stem cell differentiation by a single ubiquitin ligase-substrate complex. Nat. Immunol. 11, 207–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hoeck J. D., Jandke A., Blake S. M., Nye E., Spencer-Dene B., Brandner S., Behrens A. (2010) Fbw7 controls neural stem cell differentiation and progenitor apoptosis via Notch and c-Jun. Nat. Neurosci. 13, 1365–1372 [DOI] [PubMed] [Google Scholar]
- 23. Matsumoto A., Onoyama I., Sunabori T., Kageyama R., Okano H., Nakayama K. I. (2011) Fbxw7-dependent degradation of Notch is required for control of “stemness” and neuronal-glial differentiation in neural stem cells. J. Biol. Chem. 286, 13754–13764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. King B., Trimarchi T., Reavie L., Xu L., Mullenders J., Ntziachristos P., Aranda-Orgilles B., Perez-Garcia A., Shi J., Vakoc C., Sandy P., Shen S. S., Ferrando A., Aifantis I. (2013) The ubiquitin ligase FBXW7 modulates leukemia-initiating cell activity by regulating MYC stability. Cell 153, 1552–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Reavie L., Buckley S. M., Loizou E., Takeishi S., Aranda-Orgilles B., Ndiaye-Lobry D., Abdel-Wahab O., Ibrahim S., Nakayama K. I., Aifantis I. (2013) Regulation of c-Myc ubiquitination controls chronic myelogenous leukemia initiation and progression. Cancer Cell 23, 362–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Takeishi S., Matsumoto A., Onoyama I., Naka K., Hirao A., Nakayama K. I. (2013) Ablation of Fbxw7 eliminates leukemia-initiating cells by preventing quiescence. Cancer Cell 23, 347–361 [DOI] [PubMed] [Google Scholar]
- 27. Sancho R., Jandke A., Davis H., Diefenbacher M. E., Tomlinson I., Behrens A. (2010) F-box and WD repeat domain-containing 7 regulates intestinal cell lineage commitment and is a haploinsufficient tumor suppressor. Gastroenterology 139, 929–941 [DOI] [PubMed] [Google Scholar]
- 28. Babaei-Jadidi R., Li N., Saadeddin A., Spencer-Dene B., Jandke A., Muhammad B., Ibrahim E. E., Muraleedharan R., Abuzinadah M., Davis H., Lewis A., Watson S., Behrens A., Tomlinson I., Nateri A. S. (2011) FBXW7 influences murine intestinal homeostasis and cancer, targeting Notch, Jun, and DEK for degradation. J. Exp. Med. 208, 295–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Onoyama I., Suzuki A., Matsumoto A., Tomita K., Katagiri H., Oike Y., Nakayama K., Nakayama K. I. (2011) Fbxw7 regulates lipid metabolism and cell fate decisions in the mouse liver. J. Clin. Invest. 121, 342–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kumadaki S., Karasawa T., Matsuzaka T., Ema M., Nakagawa Y., Nakakuki M., Saito R., Yahagi N., Iwasaki H., Sone H., Takekoshi K., Yatoh S., Kobayashi K., Takahashi A., Suzuki H., Takahashi S., Yamada N., Shimano H. (2011) Inhibition of ubiquitin ligase F-box and WD repeat domain-containing 7α (Fbw7α) causes hepatosteatosis through Krüppel-like factor 5 (KLF5)/peroxisome proliferator-activated receptor γ2 (PPARγ2) pathway but not SREBP-1c protein in mice. J. Biol. Chem. 286, 40835–40846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sundqvist A., Bengoechea-Alonso M. T., Ye X., Lukiyanchuk V., Jin J., Harper J. W., Ericsson J. (2005) Control of lipid metabolism by phosphorylation-dependent degradation of the SREBP family of transcription factors by SCF(Fbw7). Cell Metab. 1, 379–391 [DOI] [PubMed] [Google Scholar]
- 32. Bengoechea-Alonso M. T., Ericsson J. (2010) The ubiquitin ligase Fbxw7 controls adipocyte differentiation by targeting C/EBPα for degradation. Proc. Natl. Acad. Sci. U.S.A. 107, 11817–11822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Murakami T., Kondo S., Ogata M., Kanemoto S., Saito A., Wanaka A., Imaizumi K. (2006) Cleavage of the membrane-bound transcription factor OASIS in response to endoplasmic reticulum stress. J. Neurochem. 96, 1090–1100 [DOI] [PubMed] [Google Scholar]
- 34. Kondo S., Saito A., Hino S., Murakami T., Ogata M., Kanemoto S., Nara S., Yamashita A., Yoshinaga K., Hara H., Imaizumi K. (2007) BBF2H7, a novel transmembrane bZIP transcription factor, is a new type of endoplasmic reticulum stress transducer. Mol. Cell. Biol. 27, 1716–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Honma Y., Kanazawa K., Mori T., Tanno Y., Tojo M., Kiyosawa H., Takeda J., Nikaido T., Tsukamoto T., Yokoya S., Wanaka A. (1999) Identification of a novel gene, OASIS, which encodes for a putative CREB/ATF family transcription factor in the long-term cultured astrocytes and gliotic tissue. Brain Res. Mol. Brain Res. 69, 93–103 [DOI] [PubMed] [Google Scholar]
- 36. Nikaido T., Yokoya S., Mori T., Hagino S., Iseki K., Zhang Y., Takeuchi M., Takaki H., Kikuchi S., Wanaka A. (2001) Expression of the novel transcription factor OASIS, which belongs to the CREB/ATF family, in mouse embryo with special reference to bone development. Histochem. Cell Biol. 116, 141–148 [DOI] [PubMed] [Google Scholar]
- 37. Murakami T., Saito A., Hino S., Kondo S., Kanemoto S., Chihara K., Sekiya H., Tsumagari K., Ochiai K., Yoshinaga K., Saitoh M., Nishimura R., Yoneda T., Kou I., Furuichi T., Ikegawa S., Ikawa M., Okabe M., Wanaka A., Imaizumi K. (2009) Signalling mediated by the endoplasmic reticulum stress transducer OASIS is involved in bone formation. Nat. Cell Biol. 11, 1205–1211 [DOI] [PubMed] [Google Scholar]
- 38. Saito A., Hino S., Murakami T., Kanemoto S., Kondo S., Saitoh M., Nishimura R., Yoneda T., Furuichi T., Ikegawa S., Ikawa M., Okabe M., Imaizumi K. (2009) Regulation of endoplasmic reticulum stress response by a BBF2H7-mediated Sec23a pathway is essential for chondrogenesis. Nat. Cell Biol. 11, 1197–1204 [DOI] [PubMed] [Google Scholar]
- 39. Yumimoto K., Matsumoto M., Oyamada K., Moroishi T., Nakayama K. I. (2012) Comprehensive identification of substrates for F-box proteins by differential proteomics analysis. J. Proteome Res. 11, 3175–3185 [DOI] [PubMed] [Google Scholar]
- 40. Matsumoto M., Hatakeyama S., Oyamada K., Oda Y., Nishimura T., Nakayama K. I. (2005) Large-scale analysis of the human ubiquitin-related proteome. Proteomics 5, 4145–4151 [DOI] [PubMed] [Google Scholar]
- 41. Kamura T., Hara T., Matsumoto M., Ishida N., Okumura F., Hatakeyama S., Yoshida M., Nakayama K., Nakayama K. I. (2004) Cytoplasmic ubiquitin ligase KPC regulates proteolysis of p27(Kip1) at G1 phase. Nat. Cell Biol. 6, 1229–1235 [DOI] [PubMed] [Google Scholar]
- 42. Kamura T., Hara T., Kotoshiba S., Yada M., Ishida N., Imaki H., Hatakeyama S., Nakayama K., Nakayama K. I. (2003) Degradation of p57Kip2 mediated by SCFSkp2-dependent ubiquitylation. Proc. Natl. Acad. Sci. U.S.A. 100, 10231–10236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kamura T., Sato S., Iwai K., Czyzyk-Krzeska M., Conaway R. C., Conaway J. W. (2000) Activation of HIF1α ubiquitination by a reconstituted von Hippel-Lindau (VHL) tumor suppressor complex. Proc. Natl. Acad. Sci. U.S.A. 97, 10430–10435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hayashi S., McMahon A. P. (2002) Efficient recombination in diverse tissues by a tamoxifen-inducible form of Cre. A tool for temporally regulated gene activation/inactivation in the mouse. Dev. Biol. 244, 305–318 [DOI] [PubMed] [Google Scholar]
- 45. Onoyama I., Tsunematsu R., Matsumoto A., Kimura T., de Alborán I. M., Nakayama K., Nakayama K. I. (2007) Conditional inactivation of Fbxw7 impairs cell-cycle exit during T cell differentiation and results in lymphomatogenesis. J. Exp. Med. 204, 2875–2888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zanotti S., Canalis E. (2010) Notch and the skeleton. Mol. Cell. Biol. 30, 886–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Olson B. L., Hock M. B., Ekholm-Reed S., Wohlschlegel J. A., Dev K. K., Kralli A., Reed S. I. (2008) SCFCdc4 acts antagonistically to the PGC-1α transcriptional coactivator by targeting it for ubiquitin-mediated proteolysis. Genes Dev. 22, 252–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ke Q., Costa M. (2006) Hypoxia-inducible factor-1 (HIF-1). Mol. Pharmacol. 70, 1469–1480 [DOI] [PubMed] [Google Scholar]
- 49. Bray S. J. (2006) Notch signalling. A simple pathway becomes complex. Nat. Rev. Mol. Cell Biol. 7, 678–689 [DOI] [PubMed] [Google Scholar]
- 50. Brown M. S., Goldstein J. L. (1997) The SREBP pathway. Regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell 89, 331–340 [DOI] [PubMed] [Google Scholar]
- 51. Cassavaugh J. M., Hale S. A., Wellman T. L., Howe A. K., Wong C., Lounsbury K. M. (2011) Negative regulation of HIF-1α by an FBW7-mediated degradation pathway during hypoxia. J. Cell. Biochem. 112, 3882–3890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Flügel D., Görlach A., Kietzmann T. (2012) GSK-3β regulates cell growth, migration, and angiogenesis via Fbw7 and USP28-dependent degradation of HIF-1α. Blood 119, 1292–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]