Background: Up-frameshift (Upf) factors eliminate aberrant mRNAs that contain a premature termination codon (PTC).
Results: The Upf complex was recruited to a PTC product and promoted the degradation of a model unfolded protein.
Conclusion: Upf factors facilitate the ubiquitin-dependent degradation of products derived from mRNA containing specific PTCs.
Significance: The findings reveal a mechanism of quality control that prevents the production of aberrant products derived from mRNAs containing specific PTCs.
Keywords: Gene Regulation, mRNA Decay, Protein Folding, Protein Turnover, Ribosomes
Abstract
Up-frameshift (Upf) factors eliminate aberrant mRNAs containing a specific premature termination codon (PTC). Here, we show that Upf complex facilitates the ubiquitin-dependent degradation of products derived from mRNA containing specific PTCs in Saccharomyces cerevisiae. The efficiency of recruitment of the Upf complex to a PTC product was correlated with the decay of the PTC product. Upf factors promoted the degradation of the human von Hippel-Lindau (VHL) protein, which is an unfolded protein in yeast cells, in a manner that depends on the presence of a faux 3′-UTR. Mass spectrometric analysis and Western blot analysis revealed that Hsp70 was associated with the PTC product. These findings suggest that the Upf complex may be recruited to ribosomes in a faux 3′-UTR-dependent manner and then associates with aberrant products to facilitate their degradation by the proteasome.
Introduction
To ensure the fidelity of gene expression, prokaryotic and eukaryotic cells have evolved several mRNA surveillance mechanisms that degrade aberrant mRNA (1, 2). The mRNA surveillance systems not only contribute to the maintenance of cellular homeostasis, but are also implicated in certain diseases (3, 4). One of the best characterized mRNA surveillance pathways is nonsense-mediated mRNA decay (NMD),3 in which the Up-frameshift (Upf) complex composed of Upf protein 1 (Upf1), Upf2, and Upf3 recognizes and eliminates mRNAs containing a PTC. Translation termination at a PTC leads to the assembly of the Upf complex, which couples premature translation termination to mRNA decay by interacting with both eukaryotic translation termination factors (i.e. eRF1 and eRF3) and mRNA degradation enzymes (1, 2, 5, 6).
In mammalian cells, translation termination codons and exon-exon junctions are cis-acting elements that allow the recognition of PTCs (1, 2). The exon junction complex, which is assembled 20–24 nucleotides upstream of a splice junction (7, 8), marks upstream stop codons as premature. In yeast, flies, worms, and plants, PTC recognition can occur independently of a downstream exon boundary, and exon junction complex protein components are not required for NMD (9–13). Therefore, other determinants must present the downstream signal required for NMD substrate recognition. The “faux 3′-UTR” model proposes that premature translation termination is intrinsically aberrant because the stop codon is not in the appropriate context (14, 15). In this model, ribosomes terminating at a PTC cannot interact with a specific set of proteins, and consequently, the termination reaction is impaired or too slow. Impaired translation termination at a PTC results in the assembly of the Upf complex and the rapid degradation of the mRNA (14, 15). The faux 3′-UTR model is supported by the observation that NMD targets mRNAs with extended 3′-UTRs in yeast, flies, worms, plants, and mammalian cells (11, 12, 16–18). These findings indicate that in eukaryotes, the distance between a termination codon and its downstream messenger ribonucleoprotein context provides important positional information that facilitates the recognition of aberrant translation termination events.
Translation repression and co-translational protein degradation by the proteasome also play crucial roles as quality control systems that target aberrant mRNAs (19–24). Various products derived from PTC-containing mRNAs show the same decay rates in wild-type and upf1Δ cells (17). However, we have shown that the Upf complex facilitates the degradation of an aberrant protein derived from mRNAs containing a PTC at specific positions in a manner consistent with the faux 3′-UTR model (25). How the Upf complex recognizes the specific PTC products and stimulates their degradation by the proteasome remains to be elucidated.
Upf1 interacts with E2 ubiquitin ligase Ubc3 with its the Cys-His-rich repeated N terminus (CH domain) and serves as substrate for the in vitro self-ubiquitination activity (26). The modification of Upf1 is required for the binding to Upf3 and NMD in vivo (26). These findings suggest that Upf1 may act as an E3 ubiquitin ligase on its association with Upf3 and play a crucial role in signaling to the NMD pathway. The role of ubiquitination activity of Upf1 in the degradation of the PTC product remains to be elucidated.
In this study, we investigated the mechanism of the recognition and degradation of the specific PTC product Pgk1-300 by the Upf complex. The efficiency of ubiquitination of Pgk1-300 was not affected by the deletion of Upf1, suggesting that Upf1 may play a minor role in the ubiquitination of Pgk1-300. Upf1 did not facilitate the degradation of Pgk1-300(K0), which contains no Lys residue, suggesting that Upf1 stimulates degradation of the PTC products by the proteasome in a ubiquitination-dependent manner. Immunoprecipitation experiments showed that the Upf complex associates with this PTC product and that the efficiency of co-purification of the PTC product with Upf1 correlated with the rate of decay of the PTC product. Upf1 also promoted the degradation of the human von Hippel-Lindau (VHL) protein, a model unfolded protein in yeast cells, in a manner that depended on a faux 3′-UTR, but not when the VHL chaperone human elongin BC was co-expressed in yeast. Immunoprecipitation and mass spectrometric analysis revealed that the Pgk1-300 protein interacts with Hsp70 that plays a crucial role in the degradation of misfolded protein (27, 28). These results indicate that the Upf factors may form the complex containing Hsp70 with the PTC products and stimulate their ubiquitination-dependent proteasomal degradation.
EXPERIMENTAL PROCEDURES
Strains and Other Methods
Yeast strains and plasmids are listed in Table 1. Information about the oligonucleotides used in this study is listed in Table 2. Polysome analysis was performed as described (20).
TABLE 1.
Yeast strains and plasmids used in this study
TABLE 2.
List of oligonucleotides primers used for plasmid construction
Determination of the Half-life of the Reporter Proteins by Western Blotting
To determine the stabilities of the reporter proteins, cells were harvested at the indicated times after the addition of cycloheximide (0.1 mg/ml). Proteins encoded by the reporter genes were detected by Western blotting and visualized with horseradish peroxidase-conjugated secondary antibody (Amersham Biosciences). Band intensities were quantitated on an LAS3000 mini using MultiGauge version 3.0 (Fujifilm Co. Ltd.), and the relative levels of products were determined based on comparison with a standard curve. When the intensity of a band was outside the range of the standard curve, a series of dilutions of the sample was prepared. The intensities of the bands of the diluted samples were compared with those of the standard curve, and the relative level of the product in the sample as compared with the control product was determined.
Immunoprecipitation
Yeast strains were grown in 1 liter of selective medium containing 2% glucose (w/v) to an A600 of 0.8 and harvested by centrifugation. The cell pellet was snap-frozen in liquid nitrogen and then ground in liquid nitrogen using a mortar and a pestle. The resulting cell powder was then resuspended on ice in 3 ml of lysis buffer (20 mm HEPES-KOH, pH 7.4, 2 mm, Mg(OAc)2, 100 mm KOAc, 1 mm DTT, 1 mm PMSF) supplemented with a protease inhibitor mixture (Complete tablets; Roche Applied Science). Extracts were then transferred to 1.5-ml tubes and clarified by centrifugation at 4 °C for 10 min at 10,000 rpm. Supernatant was recovered and centrifuged at 4 °C for 10 min at 10,000 rpm to remove membranes and insoluble material. Extracts were kept in aliquots at −80 °C until ready to use. Cell extracts were incubated with anti-DTKDDDDK tag antibody beads (Wako) in IXA-100 buffer (29) and then washed three times and eluted with 0.5 mg/ml FLAG peptide.
Plasmid Construction
Plasmids expressing VHL proteins were constructed as follows. To construct pKK244 (pCRR4Blunt-TOPOR-VHL), total RNA was extracted from HEK293 cells with TRIzol (Invitrogen) based on the manufacturer's protocol. First-strand cDNA synthesized from the HEK293 total RNA was PCR-amplified using the primers OIT1859 (5′-ATGCCCCGGAGGGCGGAGAACTGGGACGAGGCCGAGGTAGGCGCGGAGGAGGCAGGCGTCGAAGAGTACGGCCCTGAAGAAGACGGCGGGGAGGAGTCGG-3′) and OIT1811 (5′-GCTACGGGATCCTCAATCTCCCATCCGTTGATGTGCAATGCGCTCCTGTGTC-3′). Cloning of PCR products in vector pCR4Blunt-TOPO was performed using Zero Blunt TOPO PCR cloning kit for sequencing (Invitrogen) and carried out according to the manufacturer's instructions. To construct pKK246 (pGPDp-FLAG-VHL-WUTR), SpeI-BamHI fragment of FLAG-VHL was amplified in a PCR using two primers OIT1810 (5′-GCTACGACTAGTATGGACTACAAGGACGACGATGACAAGCCCCGGAGGGCGGAGAACTGGGACGAGGCCGAGGT-3′) and OIT1811 with pKK244 as the template, and was replaced with a corresponding fragment of pIT2044 (Kuroha et al. (25)). pKK247 (pGPDp-FLAG-VHL-LUTR) was constructed by replacing the pKK246 BamHI-XhoI fragment with a fragment amplified by PCR using primers OIT1021 (5′-ACTTCGGTAAGGCTTTGGAGTAAGGATCCCAACCAGACCATTCTTGGCC-3′) and OIT1017 (5′-CCGCTCGAGATTGACCAATATATGTCTCTGAATGCC-3′) with pIT2044 as the template. pKK276 (pGPDp-VHL-LUTR) was constructed by replacing the pKK247 SpeI-BamHI fragment with a fragment amplified by PCR using primers OIT2180 (5′-GCTACGACTAGTATGCCCCGGAGGGCGGAGAACTGGGACGAGGCCGAG-3′) and OIT1811 with pKK244 as the template. To construct pKK273 (pGPDp-HA-pgk1-300 (K0)), lysine codons in PGK1 gene were replaced by arginine codons by site-directed mutagenesis with the primers shown in Table 1. To construct pKK278 (p414GPDp-Myc-elongin B) and pKK279 (p415GPDp-Myc-elongin C), SpeI-BamHI fragment of elongin B or C were amplified in a PCR using the primer sets, OIT2371 (5′-GCTACGACTAGTATGGAACAGAAGTTGATTTCCGAAGAAGACCTCGACGTGTTCCTCATGATCCGGCGCCACAAGACCAC-3′) and OIT2372 (5′-GCTACGGGATCCTCACTGCACGGCTTGTTCATTGGCACTGCTTCCCG-3′), or OIT2373 (5′-GCTACGACTAGTATGGAACAGAAGTTGATTTCCGAAGAAGACCTCGATGGAGAGGAGAAAACCTATGGTGGCTGTGAAGG-3′) and OIT2374 (5′-GCTACGGGATCCTTAACAATCTAAGAAGTTCGCAGCCATCAGCAGTT-3′), respectively, with elongin B cDNA (provided by Open Biosystems) and elongin C cDNA (provided by Kazusa DNA Research Institute) as the template, and were cloned into p414GPD or p415GPD respectively.
RESULTS
Upf1 Stimulates Degradation of the PTC Products by the Proteasome in a Ubiquitination-dependent Manner
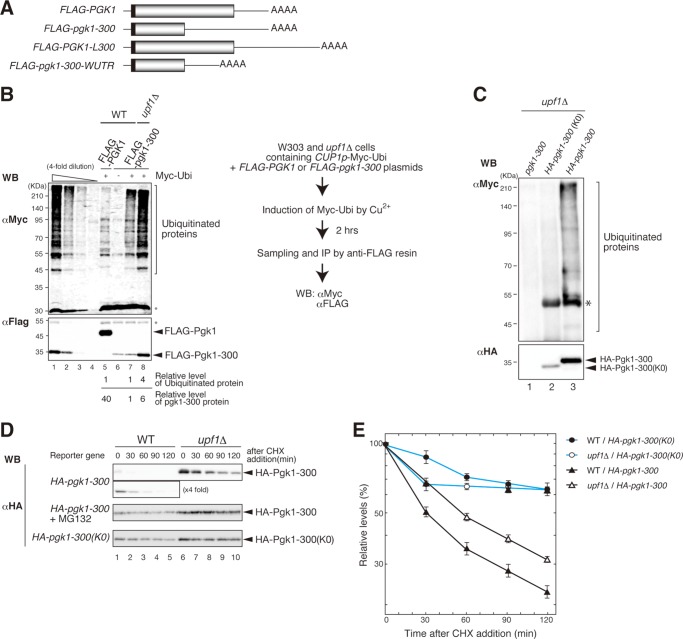
We previously reported that truncated protein derived from FLAG-pgk1-300 or FLAG-his3-100 mRNA is rapidly degraded in a Upf complex-dependent manner (25). Another study showed that Upf1 may act as an E3 ubiquitin ligase through its association with Upf3 in yeast, and mutations that affect this activity affect RNA degradation by NMD (26). Therefore, Upf1 may function as an E3 ubiquitin ligase that targets PTC products for rapid degradation by the proteasome. To test this possibility, we examined the role of Upf1 in the ubiquitination of PTC products derived from the reporter genes listed in Fig. 1A. Myc-tagged ubiquitin (Myc-Ubi) was transiently overproduced in wild-type or upf1Δ mutant cells expressing the PTC product, FLAG-Pgk1-300. Affinity-purified FLAG-Pgk1-300 protein was analyzed by SDS-PAGE followed by immunoblotting with anti-Myc antibodies to detect ubiquitinated proteins (Fig. 1B, upper panel). The signals corresponding to higher molecular weight PTC products were detected only in the presence of Myc-Ubi, but not in the presence of untagged ubiquitin (Ubi) (Fig. 1B, lanes 5–8). The intensity of signals corresponding to higher molecular weight PTC products derived from affinity-purified wild-type FLAG-Pgk1 in wild type was slightly decreased as compared with those of FLAG-Pgk1-300 (Fig. 1B, upper panel, lanes 5 and 7). The levels of affinity-purified FLAG-Pgk1 are 40-fold higher than those of FLAG-pgk1-300 (Fig. 1B, lower panel, lanes 5 and 8). These results indicate that FLAG-Pgk1-300 was more efficiently ubiquitinated than wild-type FLAG-Pgk1. The amount of ubiquitinated FLAG-Pgk1-300 in upf1Δ mutant cells was higher as compared with that in wild-type cells (Fig. 1B, upper panel, lanes 5 and 8). The level of immunoprecipitated FLAG-Pgk1-300 protein was also increased 4-fold in upf1Δ mutant cells as compared with that in wild-type cells (Fig. 1B, lower panel, lanes 5 and 8). We estimated the efficiency of ubiquitination of FALG-Pgk1-300 protein in wild type and upf1 mutant as a ratio between the levels of affinity-purified protein and ubiquitinated protein that are recognized by anti-Myc antibody. The efficiency of ubiquitination of FLAG-Pgk1-300 products was almost the same in wild-type and upf1Δ cells (Fig. 1B, compare lanes 7 and 8). These results strongly suggest that Upf1 plays only a minor role in the ubiquitination of the FLAG-Pgk1-300 protein. Similar results were obtained when ubiquitination of FLAG-His3-100 was analyzed in wild-type and upf1Δ mutant cells (data not shown).
FIGURE 1.
Upf1 stimulates degradation of the PTC products by the proteasome in a ubiquitination-dependent manner. A, schematic drawing of the mRNA derived from FLAG-PGK1 reporter genes. The open boxes indicate the open reading frame, filled boxes indicate FLAG tag, lines represent UTRs, and AAAA denotes the poly(A) tail. B, the Pgk1-300 PTC product was ubiquitinated even in the absence of Upf1. W303 and W303upf1Δ cells with pGPDp-FLAG-PGK1 or pGPDp-FLAG-pgk1-300 containing pCUP1p-Myc-Ubi or pCUP1p-Ubi were grown until A600 = 0.6. For the expression of Myc-Ubi from the CUP1 promoter, cells were incubated with 0.1 mm CuSO4 for 2 h before the addition of cycloheximide (0.1 mg/ml). W303 cells and W303upf1Δ cells harboring an indicated plasmid were harvested at the indicated times after the addition of cycloheximide. Cell extracts were prepared by Y-PER reagent (Pierce) and subjected to anti-FLAG immunoprecipitation followed by Western blotting (WB) with anti-Myc (top panel) or anti-FLAG antibodies (bottom panel). The band intensities of the samples were compared with a standard curve by using a series of dilutions of samples in lane 8. Asterisks indicate the endogenous products that are recognized with antibodies. C, lysine-less Pgk1-300 was not ubiquitinated. W303upf1Δ cells with the reporters indicated above the panel were transformed with pCUP1p-Myc-Ubi. Samples were prepared and subjected to anti-HA immunoprecipitation followed by Western blotting with anti-Myc (top panel) or anti-HA antibodies (bottom panel). The asterisk indicates the antibody heavy chain band. D, Upf1 did not stimulate the degradation of HA-Pgk1-300 containing no lysine residues. W303 and W303upf1Δ cells with pGPDp-HA-pgk1-300 or pGPDp-HA-pgk1-300 (K0) were grown in selective medium. After the addition of cycloheximide (CHX) as described under “Experimental Procedures,” cells were harvested at the indicated times, and samples were analyzed by Western blotting with anti-HA antibodies. For Western blot of W303 cells with pGPDp-HA-pgk1-300, 5-fold samples were also used. When indicated, cells were grown in the presence of 0.2 mm MG132. E, the relative levels of remaining HA-Pgk1-300 and HA-Pgk1-300(K0) protein after inhibition of translation are plotted. Error bars indicate S.D.
To evaluate the role of ubiquitination in the Upf1-dependent degradation of PTC products, we constructed the HA-pgk1-300(K0) reporter gene, which contains no lysine residues. Because FLAG tag sequence contains Lys residues, we utilized HA tag to detect the products. We observed no ubiquitination when HA-Pgk1-300(K0) was co-expressed with Myc-Ubi (Fig. 1C), and HA-Pgk1-300(K0) was significantly stabilized both in wild type and in upf1Δ cells (Fig. 1D, middle panel, and Fig. 1E). We also confirmed that the proteasome inhibitor MG132 drastically stabilized HA-Pgk1-300 both in wild type and in upf1Δ cells (Fig. 1D, bottom panel). These results indicate that Upf1 facilitates the degradation of the ubiquitinated forms of the PTC products.
PTC Product Associates with Upf Complex in RNA-independent Manner
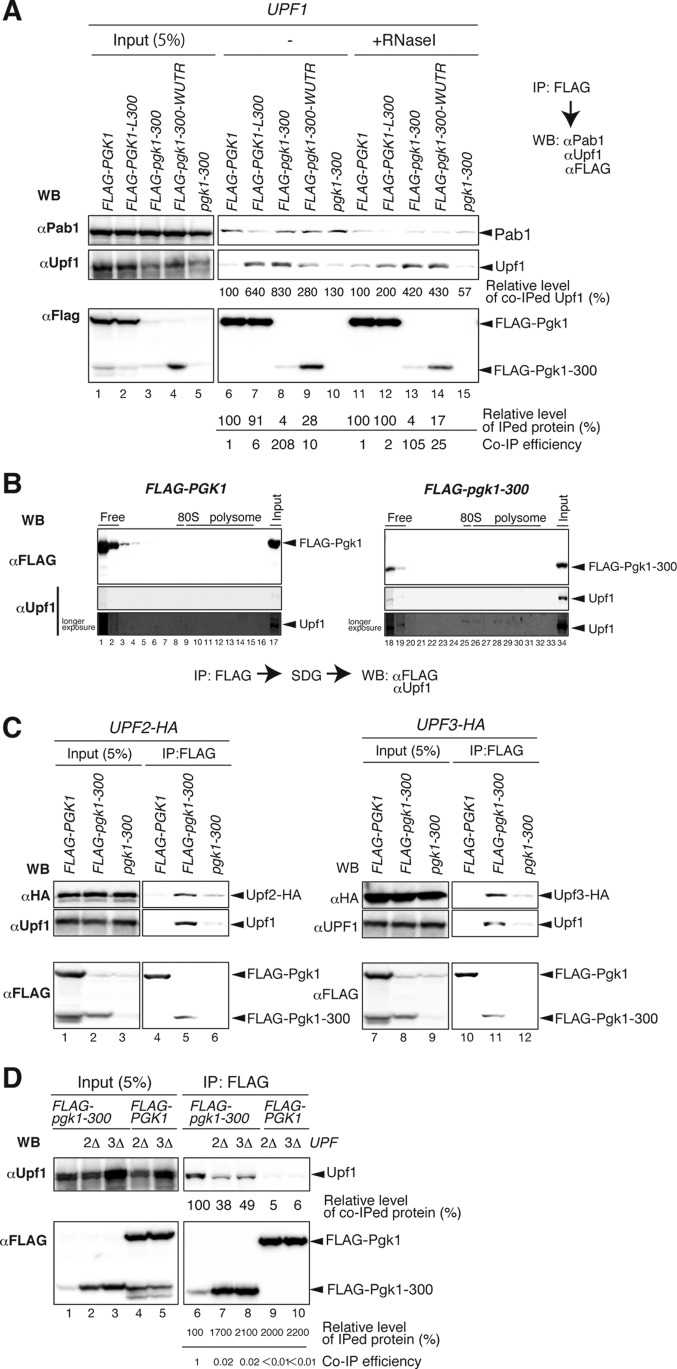
The mechanisms underlying the recognition and degradation of PTC products mediated by Upf factors remain unclear. We therefore examined the association between Upf factors and the PTC product by co-immunoprecipitation (co-IP) analysis. Extracts from wild-type cells expressing FLAG-Pgk1, FLAG-Pgk1-300, or Pgk1-300 were immunoprecipitated using an anti-FLAG resin, and the precipitates were analyzed by Western blotting with anti-FLAG or anti-Upf1 antibodies. Upf1 was more efficiently co-immunoprecipitated with FLAG-Pgk1-300 protein than with FLAG-Pgk1 protein (Fig. 2A, lanes 6 and 8). FLAG-pgk1-300-WUTR contains a short ORF flanked by the wild-type 3′-UTR, and mRNA derived from this reporter gene was not subjected to NMD (25). Although this reporter gene encodes a protein that is identical to the one produced from FLAG-pgk1-300-LUTR, Upf1 does not significantly promote the proteasomal degradation of the FLAG-Pgk1-300 protein (25). Here, Upf1 was co-immunoprecipitated with FLAG-Pgk1-300 derived from FLAG-pgk1-300-WUTR, but less efficiently than with the same protein derived from FLAG-pgk1-300 (Fig. 2A, lanes 8, 9, 13, and 14). FLAG-PGK1-L300 mRNA contains an intact ORF flanked by a long 3′-UTR and is degraded by NMD in accordance with the faux 3′-UTR model (25). However, Upf1 does not stimulate the degradation of FLAG-Pgk1 protein derived from FLAG-PGK1-L300 (25). Here, the presence of a long 3′-UTR promoted the interaction between Upf1 and the product (Fig. 2A, lanes 6 and 7), but this interaction was largely dependent on RNA (Fig. 2A, lanes 11 and 12). These results indicate that both a faux 3′-UTR and a truncated ORF are required for the RNA-independent interaction of Upf1 with a PTC product and that the co-IP efficiency of Upf1 and the FLAG-Pgk1-300 protein is highly correlated with the decay rates of the PTC product.
FIGURE 2.
A PTC product associates with Upf complex in an RNA-independent manner. A, Upf1 was more efficiently co-immunoprecipitated with FLAG-Pgk1-300 than with FLAG-Pgk1. W303 cells were transformed with pGPDp-FLAG-PGK1, pGPDp-FLAG-PGK1-L300, pGPDp-FLAG-pgk1-300, pGPDp-FLAG-pgk1-300-WUTR, or pGPDp-pgk1-300. FLAG-Pgk1 and FLAG-pgk1-300 proteins were affinity-purified from cell extracts. Where indicated (+RNase I), cell lysates were immunoprecipitated with anti-FLAG resin under conditions of RNase I treatment. Co-IP efficiency was calculated by dividing the relative level of co-immunoprecipitated (CoIPed) protein (%) by the relative level of immunoprecipitated (IPed) protein (%). WB, Western blotting. B, Upf1 was associated with the PTC product in ribosome-free fractions. Extracts of wild-type cells containing pGPDp-FLAG-PGK1 and pGPDp-FLAG-pgk1-300 were fractionated by sucrose density gradient (SDG) centrifugation, immunoprecipitated using anti-FLAG resin, and analyzed by Western blotting. The positions of ribosomes free (free), 80 S ribosomes (80S), and polysomes are indicated. C, Upf2 and Upf3 were also efficiently co-immunoprecipitated with Pgk1-300. W303UPF2-HA or W303UPF3-HA cells were transformed with pGPDp-FLAG-PGK1, pGPDp-FLAG-pgk1-300, or pGPDp-pgk1-300. FLAG-Pgk1 and FLAG-pgk1-300 proteins were affinity-purified from cell extracts. Total extracts (Input; left panels) and immunoprecipitated samples (IP; right panels) were analyzed by Western blotting with anti-HA (top panel), anti-Upf1 (middle panel), or anti-FLAG antibodies (bottom panel). D, both Upf2 and Upf3 were required for the association of Upf1 with the Pgk1-300 PTC product. W303, W303upf2Δ, or W303upf3Δ cells were transformed with the indicated reporters, and FLAG-pgk1-300 was affinity-purified. Total extracts (Input; left panels) and immunoprecipitated samples (IP; right panels) were analyzed by Western blotting as in C.
To examine the complex containing FLAG-Pgk1-300 and Upf factors, we analyzed polysomes isolated by sucrose density gradient centrifugation followed by Western blotting. Affinity-purified FLAG-Pgk1-300 and co-immunoprecipitated Upf1 were distributed in the ribosome-free fractions (Fig. 2B, lanes 1, 2, 18, and 19), indicating that Upf1 associates with the PTC product that is released from the ribosome. Because Upf1 stably interacts with PTC product expressed from the reporter mRNA that contain a 3′-UTR, we investigated the association of Upf2 and Upf3 with the PTC products by immunoprecipitation of FLAG-Pgk1 or FLAG-Pgk1-300 from extracts of UPF2-HA and UPF3-HA cells. Upf1, Upf2-HA, and Upf3-HA were more efficiently co-immunoprecipitated with FLAG-Pgk1-300 than with FLAG-Pgk1 (Fig. 2C). Moreover, the levels of Upf1 co-purified with FLAG-Pgk1-300 were greatly reduced in upf2Δ and upf3Δ mutant cells (Fig. 2D, lanes 6–8). These findings suggested that the FLAG-Pgk1-300 protein interacts with the Upf1-Upf2-Upf3 complex, and both Upf2 and Upf3 are required for the efficient interaction between Upf1 and the PTC product. Taken together, our results suggest that Upf factors may transiently form a complex with the nascent FLAG-Pgk1-300 polypeptide on the ribosome at a PTC, and this complex may remain intact after the release of polypeptide from the ribosome (see Fig. 5).
FIGURE 5.
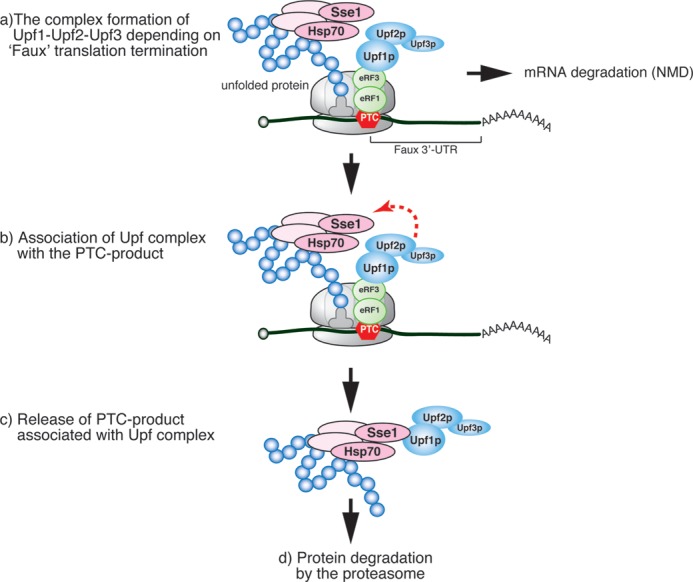
Model for rapid degradation of truncated aberrant protein derived from aberrant mRNA containing a specific premature termination codon. a, premature translation termination leads to recruitment of Upf1. b, when truncated polypeptide is unfolded, Upf complex forms a complex with truncated polypeptide. c, the PTC product is released from ribosome but still associates with Upf complex. d, finally, Upf complex-truncated polypeptide complex is rapidly degraded by the proteasome in a ubiquitination-independent manner.
Upf1 Facilitates the Degradation of Unfolded VHL Protein
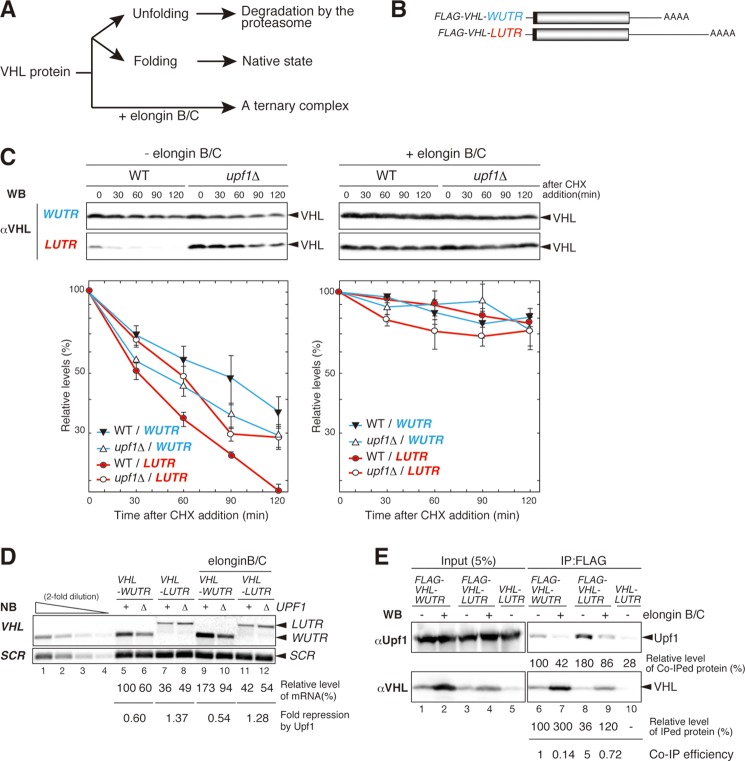
We previously showed that Upf1 stimulates the degradation of truncated proteins in a manner that depends on the presence of a faux 3′-UTR (25). In this study, we found that the truncated PTC product Pgk1-300 could interact with Upf factors, whereas intact Pgk1 protein could not (Fig. 2C). These results strongly suggest that truncated Pgk1-300 proteins have specific molecular characteristics that promote proteolysis. In particular, PTC products that are truncated may not fold properly. To determine whether the unfolded conformation of a PTC product is important for its Upf factor-mediated proteasomal degradation, we investigated the degradation of human VHL tumor suppressor protein expressed in yeast, which is a typical unfolded protein in yeast (28, 30, 31). In the presence of cofactor elongin B/C protein, VHL folding is coupled to its assembly into a ternary complex. In the absence of elongin B/C, two distinct chaperon pathways mediate VHL folding and quality control (28). Newly synthesized VHL is folded by Hsp70 and TRiC, whereas misfolded VHL is associated with the Hsp90 complex containing Hsp70, Sti1, and See1 and is degraded by the ubiquitin-proteasome pathway (28). We used VHL protein as a model unfolded protein to determine whether protein folding is crucial for the stimulation of proteasomal degradation by Upf1.
To investigate whether the Upf complex promoted the degradation of folded and unfolded VHL proteins in a faux 3′-UTR-dependent manner, we constructed two reporter genes, FLAG-VHL-WUTR (WUTR) and FLAG-VHL-LUTR (LUTR) (Fig. 3B), and examined the stability of the corresponding protein products after inhibition of translation. We found that VHL protein derived from the LUTR reporter gene was more labile in wild-type cells (50% of the protein remaining after cycloheximide addition, t½ = 30 min) than in upf1Δ cells (t½ = 85 min) in the absence of elongin B/C (Fig. 3C, left panels). VHL protein derived from the WUTR reporter gene was more stable in wild-type cells (t½ = 75 min) than in upf1Δ cells (t½ = 45 min) in the absence of elongin B/C (Fig. 3C, left panels). These results indicate that Upf1 facilitates the degradation of VHL protein derived from the LUTR reporter gene but not from the WUTR reporter gene. In addition, VHL proteins derived from all reporter genes were significantly stabilized in both strains when elongin B/C was co-expressed (Fig. 3C right panels). Because the LUTR reporter was subjected to NMD even in the presence of elongin B/C (Fig. 3D), these results suggest that VHL protein degradation induced by Upf1 is dependent on both a faux 3′-UTR and an unfolded conformation of VHL. Furthermore, Upf1 efficiently formed a complex with VHL protein derived from the LUTR reporter gene, and this interaction was suppressed by elongin B/C (Fig. 3E). These results strongly suggest that Upf1 recognizes the unfolded conformation of the VHL protein, in a manner dependent upon the presence of a faux 3′-UTR, and promotes the degradation of VHL by the proteasome.
FIGURE 3.
Upf1 facilitates the degradation of unfolded VHL protein depending on the presence of a faux 3′-UTR. A, the folding or unfolding pathways for VHL protein expressed in yeast. In the presence of elongin BC, VHL folding is coupled to its assembly into a ternary complex, and VHL is stable. In the absence of elongin BC, two distinct chaperon pathways mediate VHL folding and quality control. Newly synthesized VHL is folded by Hsp70 and TRiC, but misfolded VHL is associated with Hsp90 in a complex containing Hsp70, Sti1, and Sse1 and is ultimately degraded by the ubiquitin-proteasome pathway. B, schematic representation of the FLAG-VHL reporter mRNAs. The open boxes indicate the open reading frame, filled boxes indicate FLAG tag, lines represent UTRs, and AAAA denotes the poly(A) tail. C, Upf1 facilitates the proteasomal degradation of FLAG-VHL protein, dependent on the presence of a faux 3′-UTR. Samples were prepared of W303 and W303upf1Δ cells with the indicated plasmids in the presence or absence of elongin B/C. To express elongin B/C in yeast, we transformed strains with pKK278 (p414GPDp-Myc-elongin B) and pKK279 (p415GPDp-Myc-elongin C). The stabilities of VHL proteins derived from FLAG-VHL-LUTR or FLAG-VHL-WUTR were determined by Western blotting with anti-VHL antibodies (top and middle panels). CHX, cycloheximide. The relative levels of FLAG-VHL protein remaining after inhibition of translation are plotted (bottom panels). Error bars indicate S.D. D, Upf1 facilitates the degradation of FLAG-VHL-LUTR mRNA. Northern blotting (NB) with a digoxigenin-labeled VHL probe was performed. The relative levels for each mRNA were normalized to the FLAG-VHL-WUTR mRNA level in W303 cells in absence of elongin B/C, which was assigned a value of 100, and SCR1 mRNA levels were used as a loading control of RNA samples. E, Upf1 was more efficiently co-immunoprecipitated with unfolded VHL protein derived from LUTR reporter than with folded VHL protein. W303 cells were transformed with pGPDp-FLAG-VHL-LUTR, pGPDp-FLAG-VHL-WUTR, or pGPDp-VHL-LUTR. FLAG-VHL proteins were affinity-purified from cell extracts. Total extracts (Input; left panels) and immunoprecipitated samples (IP; right panels) were analyzed by Western blotting with anti-Upf1 (top panel) or anti-FLAG antibodies (bottom panel).
Pgk1-300 Protein Is Associated with Hsp70
Correct VHL folding is mediated by TRiC/CCT and Hsp70, but misfolded VHL is associated with the Hsp90 complex consisting of Hsp70, Sti1, and Sse1 (28). Thus, both folding and degradation of VHL requires Hsp70 in yeast. Because VHL protein derived from the LUTR reporter gene was more labile in wild-type cells than in upf1Δ cells (Fig. 3C), we suspected the Hsp70 might associate with Pgk1-300. We identified the 70-kDa protein that is co-immunoprecipitated with FLAG-Pgk1-300 but not with Pgk1 (Fig. 4A). Mass spectrometric analysis revealed that this protein corresponds to Ssa1 and Ssa2. Furthermore, Western blot analysis of affinity-purified FLAG-Pgk1-300 and FLAG-Pgk1 using an anti-Hsp70 antibody (3A3) that recognizes yeast Ssa1, Ssa2, Ssa3, and Ssa4 (32) revealed that Hsp70 proteins were specifically associated with FLAG-Pgk1-300 (Fig. 4B). These results indicate that Hsp70 interacts with Pgk1-300 and may be involved in the stimulation of proteasomal degradation of truncated proteins derived from aberrant mRNAs.
FIGURE 4.
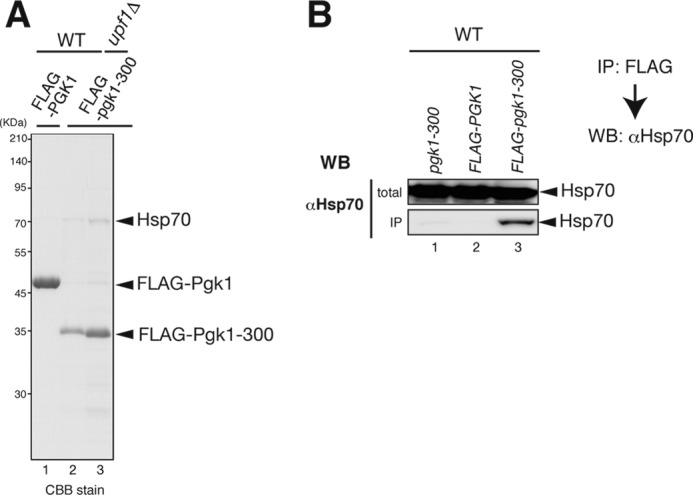
FLAG-pgk1-300 protein is associated with Hsp70. A, identification of the protein associated with FLAG-Pgk1-300. FLAG-Pgk1-300 proteins were affinity-purified from cell extracts, and co-purified 70-kDa protein was analyzed by mass spectrometry as shown previously (34). CBB stain, Coomassie Brilliant Blue stain. B, immunoblots demonstrating that Hsp70 was co-immunoprecipitated with FLAG-Pgk1-300. The total extracts (Input; top panels) and immunoprecipitated samples (IP; bottom panels) were analyzed by Western blotting (WB) with anti-Hsp70 monoclonal 3A3 antibodies (Abcam) which recognizes yeast Ssa1, Ssa2, Ssa3, and Ssa4 (32).
DISCUSSION
Our previous study demonstrated that the Upf complex plays critical roles not only in NMD, but also in the rapid degradation of the PTC product by the proteasome (25). In this study, we demonstrated that the ubiquitination is required for the Upf1-dependent degradation of the PTC product (Fig. 1). However, the overall ubiquitination of FLAG-Pgk1-300 or FLAG-His3-100 was not significantly stimulated by Upf1 (Fig. 1B and data not shown), suggesting that Upf1 may not be involved in the ubiquitination of Pgk1-300 (Fig. 1B). Alternatively, specific ubiquitination by Upf1 may be crucial for rapid degradation of a truncated protein by the proteasome. Upf1 has an E3 ubiquitin ligase activity, and self-ubiquitination of Upf1 is required for the interaction with Upf3 and NMD (26). The mutations in CH domain of Upf1 cause defects in self-ubiquitination and interaction with Upf3 (26). One possible explanation for these findings is that Upf1 may ubiquitinate Pgk1-300, thereby stimulating the degradation of the truncated protein. To test this possibility, the Lys residue that is crucial for rapid degradation by the proteasome must be identified.
We have also provided evidence that the Upf complex selectively recognizes PTC products derived from mRNAs containing a specific premature termination codon (25). Our results demonstrated that Upf1 stimulates the degradation of VHL protein in the absence of elongin B/C (Fig. 3). An important question is how the Upf complex mediates the recognition of the PTC products by the proteasome. Although certain chaperones specifically recognize unfolded proteins and facilitate the proper folding of nascent peptides in eukaryotes, the specific chaperon facilitates the degradation of associated protein. For example, the Hsp90 complex containing Sse1, Hsp70, Sti1, and Hsp90 facilitates the degradation of VHL protein by the proteasome in the absence of cofactor elongin B/C (28). Interestingly, comprehensive analysis of interactions between yeast proteins revealed that Upf1 could interact with Sse1 protein that is an ATP exchange factor of Hsp70 (33). Therefore, one possibility is that the Upf1 complex may interact with Sse1, a component of the Hsp90 complex containing Sse1, Hsp70, Sti1, and Hsp90. Co-immunoprecipitation experiments revealed that Upf complex directly or indirectly interacts with the truncated proteins Pgk1-300 (Fig. 2) and the unfolded model protein human VHL (Fig. 3). Because Hsp70 interacts with truncated Pgk1-300 protein (Fig. 4) and VHL (28), we suspect that the truncated aberrant protein associates with a putative higher order complex composed of Upf factors and the Hsp90 complex. Because the folding of VHL by TRiC is inhibited by the Hsp90 complex, the interaction between Upf1 and Hsp90 complex may facilitate the recognition and degradation of VHL by the proteasome. To investigate this possibility, the roles of components of Hsp90 complex in the Upf-dependent degradation of proteins by the proteasome should be examined.
Alternatively, the Upf complex may recruit the proteasome and facilitate the degradation of associated PTC product. Consistent with this, a comprehensive two-hybrid analysis showed that Upf1p interacts with Rpn11p, a 19 S subunit of the proteasome (33); however, there is no further evidence that Upf1 interacts directly with the proteasome. Further experiments will be necessary to investigate the possible interactions between Upf1 and the proteasome.
Our results also demonstrated that Upf factors stimulate the degradation of Pgk1-300 (Fig. 1) (25), as well as that of VHL protein in the absence of elongin B/C (Fig. 3), only when these proteins were expressed from aberrant mRNAs with long 3′-UTRs. In yeast, formation of the Upf complex has been proposed to be dependent on the presence of a long 3′-UTR. Upf complex formation might be crucial for rapid degradation of aberrant mRNAs and the proteins they encode. We propose a model in which yeast Upf1 stimulates the proteasomal degradation of a PTC product derived from an aberrant mRNA containing a faux 3′UTR (Fig. 5). In this model, the Upf complex is recruited to ribosomes at a PTC in a faux 3′-UTR-dependent manner and then interacts with the unfolded PTC product via the Hsp90 complex. The PTC product in the complex is maintained in the unfolded state and can therefore be efficiently recognized and degraded by the proteasome. One important question is how the Upf complex associates with the PTC product during translation. Future studies will determine the importance of Upf complex-mediated rapid degradation of PTC products in the processing of aberrant proteins.
Acknowledgments
We thank Dr. Allan Jacobson and Dr. Roy Bijoyita for valuable discussion and helpful suggestions. We thank Dr. Mutsuhito Ohno, Dr. Makoto Kitabatake, and Dr. Yasushi Saeki for materials. We also thank all members of the laboratory, especially Dr. Isao Kashima, for technical support.
This work was supported by a grant-in-aid for young scientists (to K. K.) and Grant-in-aid for Scientific Research on Innovative Areas “RNA regulation” 20112006 (to T. I.) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
- NMD
- nonsense-mediated mRNA decay
- PTC
- premature termination codon
- Upf
- Up-frameshift
- IP
- immunoprecipitation
- His3
- imidazoleglycerol-phosphate dehydratase
- Pgk1
- 3-phosphoglycerate kinase 1
- VHL
- von Hippel-Lindau
- Ubi
- ubiquitin.
REFERENCES
- 1. Kervestin S., Jacobson A. (2012) NMD: a multifaceted response to premature translational termination. Nat. Rev. Mol. Cell Biol. 13, 700–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lejeune F., Maquat L. E. (2005) Mechanistic links between nonsense-mediated mRNA decay and pre-mRNA splicing in mammalian cells. Curr. Opin. Cell Biol. 17, 309–315 [DOI] [PubMed] [Google Scholar]
- 3. Holbrook W. P., Oskarsdóttir A., Fridjónsson T., Einarsson H., Hauksson A., Geirsson R. T. (2004) No link between low-grade periodontal disease and preterm birth: a pilot study in a healthy Caucasian population. Acta Odontol. Scand. 62, 177–179 [DOI] [PubMed] [Google Scholar]
- 4. Kuzmiak H. A., Maquat L. E. (2006) Applying nonsense-mediated mRNA decay research to the clinic: progress and challenges. Trends Mol. Med. 12, 306–316 [DOI] [PubMed] [Google Scholar]
- 5. Conti E., Izaurralde E. (2005) Nonsense-mediated mRNA decay: molecular insights and mechanistic variations across species. Curr. Opin. Cell Biol. 17, 316–325 [DOI] [PubMed] [Google Scholar]
- 6. Amrani N., Sachs M. S., Jacobson A. (2006) Early nonsense: mRNA decay solves a translational problem. Nat. Rev. Mol. Cell Biol. 7, 415–425 [DOI] [PubMed] [Google Scholar]
- 7. Le Hir H., Izaurralde E., Maquat L. E., Moore M. J. (2000) The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon-exon junctions. EMBO J. 19, 6860–6869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Singh G., Kucukural A., Cenik C., Leszyk J. D., Shaffer S. A., Weng Z., Moore M. J. (2012) The cellular EJC interactome reveals higher-order mRNP structure and an EJC-SR protein nexus. Cell 151, 750–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Culbertson M. R., Leeds P. F. (2003) Looking at mRNA decay pathways through the window of molecular evolution. Curr. Opin. Genet. Dev. 13, 207–214 [DOI] [PubMed] [Google Scholar]
- 10. Gatfield D., Unterholzner L., Ciccarelli F. D., Bork P., Izaurralde E. (2003) Nonsense-mediated mRNA decay in Drosophila: at the intersection of the yeast and mammalian pathways. EMBO J. 22, 3960–3970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Longman D., Plasterk R. H., Johnstone I. L., Cáceres J. F. (2007) Mechanistic insights and identification of two novel factors in the C. elegans NMD pathway. Genes Dev. 21, 1075–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kertész S., Kerényi Z., Mérai Z., Bartos I., Pálfy T., Barta E., Silhavy D. (2006) Both introns and long 3′-UTRs operate as cis-acting elements to trigger nonsense-mediated decay in plants. Nucleic Acids Res. 34, 6147–6157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Muhlrad D., Parker R. (1994) Premature translational termination triggers mRNA decapping. Nature 370, 578–581 [DOI] [PubMed] [Google Scholar]
- 14. Amrani N., Dong S., He F., Ganesan R., Ghosh S., Kervestin S., Li C., Mangus D. A., Spatrick P., Jacobson A. (2006) Aberrant termination triggers nonsense-mediated mRNA decay. Biochem. Soc. Trans. 34, 39–42 [DOI] [PubMed] [Google Scholar]
- 15. Amrani N., Ganesan R., Kervestin S., Mangus D. A., Ghosh S., Jacobson A. (2004) A faux 3′-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature 432, 112–118 [DOI] [PubMed] [Google Scholar]
- 16. Behm-Ansmant I., Kashima I., Rehwinkel J., Saulière J., Wittkopp N., Izaurralde E. (2007) mRNA quality control: an ancient machinery recognizes and degrades mRNAs with nonsense codons. FEBS Lett. 581, 2845–2853 [DOI] [PubMed] [Google Scholar]
- 17. Muhlrad D., Parker R. (1999) Aberrant mRNAs with extended 3′ UTRs are substrates for rapid degradation by mRNA surveillance. RNA 5, 1299–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bühler M., Steiner S., Mohn F., Paillusson A., Mühlemann O. (2006) EJC-independent degradation of nonsense immunoglobulin-μ mRNA depends on 3′ UTR length. Nat. Struct. Mol. Biol. 13, 462–464 [DOI] [PubMed] [Google Scholar]
- 19. Ito-Harashima S., Kuroha K., Tatematsu T., Inada T. (2007) Translation of the poly(A) tail plays crucial roles in nonstop mRNA surveillance via translation repression and protein destabilization by proteasome in yeast. Genes Dev. 21, 519–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Inada T., Aiba H. (2005) Translation of aberrant mRNAs lacking a termination codon or with a shortened 3′-UTR is repressed after initiation in yeast. EMBO J. 24, 1584–1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dimitrova L. N., Kuroha K., Tatematsu T., Inada T. (2009) Nascent peptide-dependent translation arrest leads to Not4p-mediated protein degradation by the proteasome. J. Biol. Chem. 284, 10343–10352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bengtson M. H., Joazeiro C. A. (2010) Role of a ribosome-associated E3 ubiquitin ligase in protein quality control. Nature 467, 470–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tsuboi T., Kuroha K., Kudo K., Makino S., Inoue E., Kashima I., Inada T. (2012) Dom34:hbs1 plays a general role in quality-control systems by dissociation of a stalled ribosome at the 3′ end of aberrant mRNA. Mol. Cell 46, 518–529 [DOI] [PubMed] [Google Scholar]
- 24. Brandman O., Stewart-Ornstein J., Wong D., Larson A., Williams C. C., Li G. W., Zhou S., King D., Shen P. S., Weibezahn J., Dunn J. G., Rouskin S., Inada T., Frost A., Weissman J. S. (2012) A ribosome-bound quality control complex triggers degradation of nascent peptides and signals translation stress. Cell 151, 1042–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kuroha K., Tatematsu T., Inada T. (2009) Upf1 stimulates degradation of the product derived from aberrant messenger RNA containing a specific nonsense mutation by the proteasome. EMBO Rep. 10, 1265–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Takahashi S., Araki Y., Ohya Y., Sakuno T., Hoshino S., Kontani K., Nishina H., Katada T. (2008) Upf1 potentially serves as a RING-related E3 ubiquitin ligase via its association with Upf3 in yeast. RNA 14, 1950–1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bercovich B., Stancovski I., Mayer A., Blumenfeld N., Laszlo A., Schwartz A. L., Ciechanover A. (1997) Ubiquitin-dependent degradation of certain protein substrates in vitro requires the molecular chaperone Hsc70. J. Biol. Chem. 272, 9002–9010 [DOI] [PubMed] [Google Scholar]
- 28. McClellan A. J., Scott M. D., Frydman J. (2005) Folding and quality control of the VHL tumor suppressor proceed through distinct chaperone pathways. Cell 121, 739–748 [DOI] [PubMed] [Google Scholar]
- 29. Inada T., Winstall E., Tarun S. Z., Jr., Yates J. R., 3rd, Schieltz D., Sachs A. B. (2002) One-step affinity purification of the yeast ribosome and its associated proteins and mRNAs. RNA 8, 948–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Spiess C., Miller E. J., McClellan A. J., Frydman J. (2006) Identification of the TRiC/CCT substrate binding sites uncovers the function of subunit diversity in eukaryotic chaperonins. Mol. Cell 24, 25–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Melville M. W., McClellan A. J., Meyer A. S., Darveau A., Frydman J. (2003) The Hsp70 and TRiC/CCT chaperone systems cooperate in vivo to assemble the von Hippel-Lindau tumor suppressor complex. Mol. Cell. Biol. 23, 3141–3151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Arnold C. E., Wittrup K. D. (1994) The stress response to loss of signal recognition particle function in Saccharomyces cerevisiae. J. Biol. Chem. 269, 30412–30418 [PubMed] [Google Scholar]
- 33. Kaake R. M., Milenković T., Przulj N., Kaiser P., Huang L. (2010) Characterization of cell cycle specific protein interaction networks of the yeast 26S proteasome complex by the QTAX strategy. J. Proteome Res. 9, 2016–2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. McCloskey A., Taniguchi I., Shinmyozu K., Ohno M. (2012) hnRNP C tetramer measures RNA length to classify RNA polymerase II transcripts for export. Science 335, 1643–1646 [DOI] [PubMed] [Google Scholar]
- 35. Fujii K., Kitabatake M., Sakata T., Miyata A., Ohno M. (2009) A role for ubiquitin in the clearance of nonfunctional rRNAs. Genes Dev. 23, 963–974 [DOI] [PMC free article] [PubMed] [Google Scholar]