Background: CXCR4/CXCL12 axis provides directional cues for breast cancer cells to metastasize to specific organs.
Results: LIP regulates the expression of CXCR4, promoting CXCR4/CXCL12-mediated breast cancer cell migration.
Conclusion: LIP is a previously unrecognized transcriptional regulator of CXCR4.
Significance: Our results revealed a potential link between heregulin, LIP, and CXCR4 and may have important therapeutic implications for metastatic breast cancer.
Keywords: Breast Cancer, C/EBP Transcription Factor, Chemotaxis, Cxcr4, Metastasis
Abstract
Metastasis is the primary cause of death in cancer patients. CXCR4/CXCL12 chemokine axis provides directional cues for breast cancer cells to metastasize to specific organs. Despite their potential clinical importance, how CXCR4 expression in breast cancer cells is regulated at the molecular level is not well understood. We identified an isoform of C/EBPβ, liver-enriched inhibitory protein (LIP), as a previously unrecognized transcriptional regulator of CXCR4 in breast cancer cells. LIP up-regulated the transcription of CXCR4 through direct interaction with the CXCR4 promoter. The increase in CXCR4 mRNA was paralleled by an increased cell surface expression of the CXCR4, which in turn promoted CXCR4-mediated breast cancer cell migration. A significant positive correlation between LIP and CXCR4 expression was observed in stage III and IV human breast carcinoma specimens. Neuregulin 1 (or NRG1, hereafter referred to as heregulin) increased CXCR4 expression in breast cancer cells, and this coincided with increased LIP binding on the CXCR4 promoter. These findings may have important implications for understanding the molecular basis of CXCR4-mediated breast cancer cell metastasis and could potentially allow us to develop novel strategies to reduce morbidity and mortality in patients with metastatic breast cancer.
Introduction
Earlier diagnosis and development of new drugs significantly improved survival for breast cancer patients. However, the metastatic migration of breast cancer cells is almost never completely curable and remains the actual cause of morbidity and mortality. The development of metastasis is a complicated, multistep process that includes loss of cellular adhesion, extravasation, chemoattraction, infiltration, and colonization as well as angiogenesis (1). Therefore, understanding molecular events underlying various stages of metastasis is obviously a daunting task but one that is of utmost importance to develop new therapeutic targets for breast cancer malignancy.
The CXCR4/CXCL12 (SDF-1α) axis is a major driving force behind the metastatic migration of breast cancer cells (2, 3) and has been proven to play a role in all steps of metastasis (4). Müller et al. (3) showed that the level of CXCR4 is higher in malignant breast tumors than in their normal healthy counterparts, suggesting that its expression level correlates with increased metastasis-associated mortality. Neutralizing the interaction of CXCR4/CXCL12 in vivo significantly impaired the metastasis of breast cancer cells and cell migration (3). Kato et al. (5) have shown that the expression of CXCR4 in surgically resected invasive ductal carcinomas is significantly correlated with the degree of lymph node metastasis. Another study has also described that breast cancer cells metastasized to the lungs express very high levels of CXCR4 as compared with the parental cells (6). These results are further substantiated by the fact that CXCR4 is one of the few genes that is up-regulated in bone-metastasized breast cancer cells (7). Consistent with these studies, knockdown of endogenous CXCR4 gene expression in breast cancer cells resulted in significant inhibition of breast cancer cell migration in vitro (8). Furthermore, our previous results showed that activation of CXCR4/CXCL12 signaling induces blood vessel instability, resulting in the penetration of breast tumor cells through the human brain microvascular endothelial cells (9). All of these data provide compelling evidence that CXCR4/CXCL12 axis plays a pivotal role in spreading breast cancer cells to different organs. However, there is only a limited understanding of how CXCR4 is regulated at the molecular level in the context of breast cancer metastasis.
C/EBP is a member of the basic leucine zipper family of transcription regulators and consists of at least six isotypes. Among isoforms, C/EBPβ (also known as liver-enriched activator protein (LAP)2 or Cebpβ in rodents) is indispensable for ductal morphogenesis and functional differentiation of mammary epithelial cells (10). C/EBPβ exists as three isoforms (LAP1, LAP2, and LIP) that are generated by in-frame alternative translation initiation (11). Among these isoforms, LIP displays an increased affinity for DNA, but it lacks a portion of its trans-activating domain, rendering it able to antagonize the transcriptional activation of LAP or other C/EBPs and leucine zipper proteins in substoichiometric ratios (12). The ratio of these isoforms varies during mammary development and tumorigenesis (13–15). The dominant-negative LIP isoform is predominantly expressed during proliferative cellular responses and is associated with aggressive breast cancer cells (11). In line with this, most of human breast-infiltrating ductal carcinomas that express high levels of LIP are estrogen receptor- and progesterone receptor-negative and aneuploid (12). Importantly, C/EBPβ is overexpressed at late stages in carcinogenesis of breast cancer (16), suggesting that it could potentially contribute to metastatic progression of breast cancer. C/EBPβ also plays an important role in the evasion of metastatic breast cancer cells from the cytostatic effects of TGF-β (17).
Although LIP expression appears to be closely implicated in regulation of invasive and metastatic properties of breast cancer cells, whether LIP is directly involved in breast cancer metastasis and, if so, how LIP regulates breast cancer cell migration at the molecular level remain elusive. Here, we demonstrate that LIP functions as a regulator of CXCR4 and modulates breast cancer cell migration and invasion.
Interestingly, heregulin (HRG), which is known to promote invasion and metastasis of breast cancer cells (18), up-regulated the LIP level, and this coincided with increased surface expression of CXCR4. Thus, our results also provide new insights into the potential mechanistic link between HRG, LIP, and CXCR4 in breast cancer cell metastasis.
EXPERIMENTAL PROCEDURES
Cell Culture
Breast cancer cells were maintained in medium and supplements as recommended by ATCC. Platinum-GP retroviral packaging cell line (Cell Biolabs, Inc.) were grown in DMEM supplemented with 10% FBS. Cells were grown to 90–95% confluence and treated with heregulin (100 ng/ml) (Peprotech).
Human Tissue Analysis and Animal Studies
All tissue specimens were collected after each patient provided written informed consent and were used under a protocol reviewed and approved by the University of Pittsburgh Institutional Review Board. All experiments using animals were done in accordance with a protocol approved by the Institutional Animal Care and Use Committee (IACUC) of University of Pittsburgh. LIP- and control vector-transduced breast cancer cells were transplanted intravenously into 6–8-week-old NOD/SCID-interleukin-2 receptor-γc-null mice (The Jackson Laboratory). Recipient animals found in moribund condition were euthanized according to the approved protocol.
Luciferase Reporter Gene Assay
CXCR4 reporter plasmids were kindly gifted from Dr. Yasukawa, Ehime University, Japan. Mutations were introduced in the putative C/EBP binding site using the site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions. The original sequence 5′-CTCAAACTTAGGAAATGCCTCTGG-3′ was changed to CTCAAACATAGGGAATGCCTCTGG by introducing two nucleotide substitutions as underlined. A nine-base pair deletion was introduced in the putative C/EBP binding site at −230 to −222 using PCR such that the sequence 5′-CTCAAACTTAGGAAATGCCTCTGG-3′ was changed to 5′-CTC-(deletion)-AAATGCCTCTGG 3′. A luciferase reporter gene assay was performed by the dual-luciferase reporter assay kit (Promega) on Veritas microplate luminometer (Tuner Biosystems). The average ratios of luciferase activity to Renilla activity was measured by Veritas software (Tuner Biosystems).
Electrophoretic Mobility Shift Assay (EMSA)
DNA binding assays were performed with nuclear extracts prepared from LIP- and control vector-transduced breast cancer cells, as described previously (19). The double-stranded oligonucleotide corresponding to the C/EBP binding element within the CXCR4 promoter (5′-TTCCCTCAAACTTAGGAAATGCCTCTGG-3′) and to YY1 (5′-TAGCAAGGATGGACGCGCCACAGAGAGAC-3′) were end-labeled with [γ-32P]ATP by using T4 polynucleotide kinase according to the manufacturer's instructions (New England Biolabs). For supershift analysis, 3 μg of nuclear protein extract was incubated with antibodies at room temperature for 30 min before the binding reaction. Each nuclear extract was incubated with 2 μg of poly(dI-dC), 10× binding buffer (100 mm Tris-Cl, pH 7.5, 500 mm NaCl, 10 mm EDTA, 10 mm dithiothreitol, 50% glycerol), 1.5 μl of 50% glycerol, and 10,000 cpm of radiolabeled oligonucleotide in a final volume of 20 μl at room temperature for 30 min. For the competition reaction, the C/EBP binding site-mutated oligonucleotide (5′-TTCCCTCAAACAGCTCTCTAGCCTCTGG-3′) or C/EBP binding site-deleted oligonucleotide (5′-TTCCCTCAAACGCCTCTGGG-3′) or nonspecific oligonucleotide (AP1 consensus oligonucleotide, 5′-CGGCTCCGGACTCACTACCGAACCA-3′) was used. The reaction mixtures were resolved on a non-denaturing 6% polyacrylamide gel. Antibodies used for the supershift assays were anti-C/EBPβ antibody (c-19, Santa Cruz Biotechnology), anti-YY-1 (H-414, Santa Cruz Biotechnology), or anti-FLAG (M2, Sigma) or anti-HA (F-7, Santa Cruz Biotechnology) antibody.
Chromatin Immunoprecipitation (ChIP) Assay
Cells were fixed by the addition of 37% formaldehyde to a final concentration of 1% formaldehyde and incubation at room temperature for 10 min. Cross-linking was stopped by the addition of glycine to a final concentration of 0.125 m. Nuclei were pelleted and lysed by incubation in nuclear lysis buffer (50 mm Tris-HCl, pH 8.1, 10 mm EDTA, 1% SDS in the presence of proteinase inhibitors). The chromatin fractions were immunoprecipitated with anti-C/EBPβ or anti-FLAG (M2, Sigma) or anti-HA (F-7, Santa Cruz Biotechnology) or anti-YY1 (H-414, Santa Cruz Biotechnology) antibody. DNA-protein complexes were immunoprecipitated with salmon sperm DNA prebound protein A/G-agarose. DNA was extracted with phenol/chloroform, precipitated, redissolved, and used as templates for PCR. Different PCR cycles (ranging from 24 to 32) were used to evaluate each assay, and the lowest possible cycle was chosen for the presentation. PCR for input controls were performed at the same number of PCR cycles as the immunoprecipitated complexes. The primers used for the PCR correspond to regions flanking the C/EBP binding site within the CXCR4 gene promoter are: sense, 5′-CAGAGAGACGCGTTCCTAGC-3′; antisense, 5′-CGGGTGGTCGGTAGTGAGT-3′. Primers used in PCR to amplify the YY1-binding site of the CXCR4 promoter are: 5′-TTCCATCCACTTTAGCAAGGA-3′; antisense, 5′-CTCCCAGAGGCATTTCCTAA-3′.
Chemotaxis Assay and Matrigel Invasion Assay
The modified Boyden chamber (48-well) (Neuroprobe) was used for both chemotaxis and invasion assay. Serum-starved LIP- and control vector-transduced breast cancer cells were detached in DMEM media. Lower compartments of the Boyden chamber were filled with CXCL12 (125 ng/ml or indicated concentrations; Peprotech) in DMEM and then covered with a 10-μm-pore polycarbonate membrane. For chemotaxis assay, the membrane was precoated with human collagen IV (Sigma) (25 μg/ml in DMEM) for 2 h at 37 °C. To verify the specificity of the cell migration, cells were preincubated with anti-CXCR4 antibody (25 μg/ml, clone 12G5) (R&D Systems) for 1 h. For an invasion assay, 10-μm-pore polycarbonate membrane was coated with Matrigel according to the manufacturer's instructions (BD Biosciences). 200 μl of cells at a density of 4 × 106 cells/ml were loaded into the upper compartments, and the chamber was incubated at 37 °C, 5% CO2 for 16 h. The membrane was stained by Diff-quick fixative (Dade Diagnostics). Cells that had migrated across the membrane were counted under microscope. Five fields were counted for each sample in duplicate or triplicate.
Flow Cytometry
Cells were removed from flasks with a non-enzymatic cell dissociation solution (Cell Stripper; Mediatech). Cells were incubated with biotin-conjugated mouse monoclonal anti-human CXCR4 (clone 12G5; R&D Systems, MN) followed by streptavidin-conjugated phycoerythrin (eBioscience). Analysis was done using a Coulter Epics cytometer instrument and Expo 32 ADC software (Beckman Coulter).
Expression Vectors and Generation of Stable Cell Lines
The coding sequence of LIP isoform was PCR-amplified and subcloned into XhoI and EcoRI sites of retroviral vector MSCV-IRES-GFP. The forward PCR primer for LIP was 5′-CCGCTCGAGATGGCGGCGGGCTT-3′. The reverse primer was 5′-GCGAATTCCTAGCAGTGGCCGGA-3′. pCMV-FLAG LAP2 (#15738) (17), pCMV-HA LIP (#15739) (17), pLKO.1 puro CXCR4 siRNA-1 (#12271) (20), Scramble shRNA (#1864) (21), and pLKO.1-TRC control (#10879) (22) constructs were obtained from Addgene. C/EBPβ MISSION shRNA constructs were from Sigma Aldrich. To establish retrovirus-producing cell line, Platinum-GP retroviral packaging cell line (Cell Biolabs) was transfected with human LIP MSCV-GFP vector along with pVSV-G (purchased from Stratagene) by Lipofectamine 2000 (Invitrogen). Two days after transfection, culture medium containing high-titer virus was harvested and used to infect breast cancer cells by ViraDuctin retrovirus transduction kit (Cell Biolabs). Lentivirus particles are produced from 293T cells and used to infect cells using ViraDuctin lentivirus transduction kit (Cell Biolabs).
Tartrate-resistant Acid Phosphatase (TRAP) Staining and Immunohistochemistry
Femurs from transplanted mice were fixed in 4% paraformaldehyde, decalcified in 10% EDTA, and then embedded in paraffin. For identification of osteoclasts, the sections were deparaffinzed, dehydrated, and stained using the TRAP staining kit (Sigma) according to the manufacturer's instructions. For identification of GFP-expressing cells, the sections were immunolabeled with goat polyclonal anti-GFP antibody (Novus Biologicals; 1:500) for 2 h at room temperature and subsequently incubated with biotinylated goat-specific secondary antibody (Vector Laboratories) followed by 3,3′-diaminobenzidine staining according to the manufacturer's instructions (Vector Laboratories).
RESULTS
LIP Has the Ability to Directly Bind to the CXCR4 Promoter and Regulates the Transcription of CXCR4 in Breast Cancer Cells
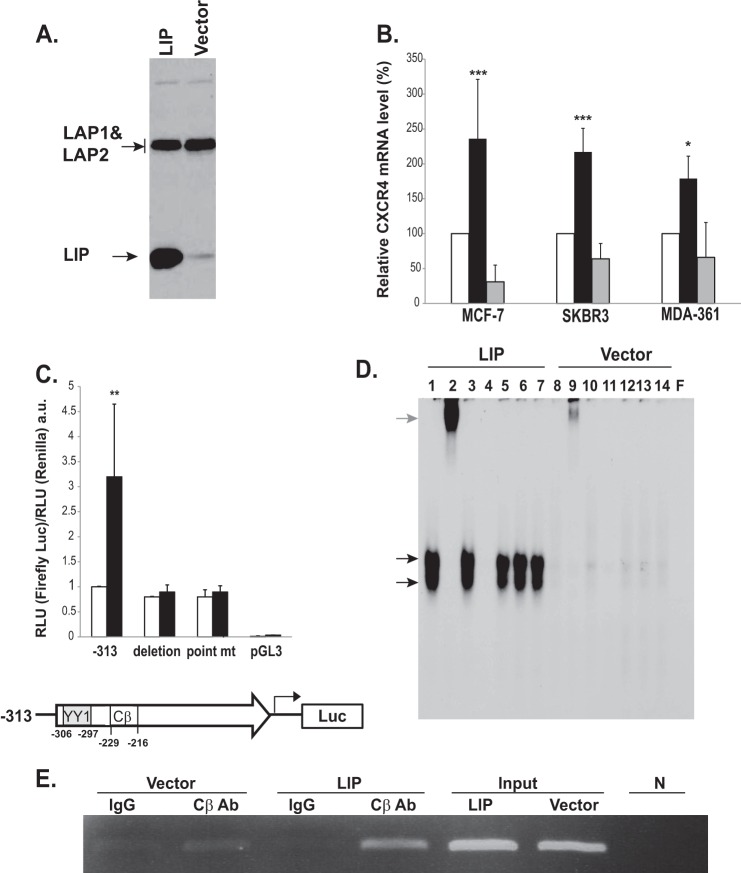
LIP has been known to be overexpressed in several human cancers including breast cancer. Dysregulated expression of LIP has been associated with aggressive and metastatic properties of breast cancer cells (12, 17, 23). Thus, we considered it a reasonable possibility that LIP can function as a potential regulator of metastasis-related molecules such as CXCR4. The presence of putative C/EBP binding sites (TKNNGNAAY) (−226 to −218) in the human CXCR4 promoter further substantiates this speculation. We first examined the ability of LIP to regulate the transcription of CXCR4. A bicistronic retroviral vector encoding LIP and GFP was constructed and used to transduce breast cancer cells. LIP expression level was elevated in cell lysates extracted from the LIP-transduced MCF-7 breast cancer cells (Fig. 1A). Control vector-transduced MCF-7 cells also exhibited a low endogenous level of LIP expression (Fig. 1A). Ectopic expression of LIP was found to increase CXCR4 mRNA levels in breast cancer cells, as evidenced by real-time quantitative PCR analysis (Fig. 1B). Treatment of LIP-transduced cells with actinomycin D was sufficient to block the increase in CXCR4 mRNA (see the gray bars in Fig. 1B), suggesting that the increase in CXCR4 mRNA occurred at the transcriptional level. Among the breast cancer cell lines tested, LIP-transduced MCF-7 and SKBR3 cells exhibited relatively high levels of CXCR4 transcripts and were thus used for further investigation. We next performed an in vitro reporter assay to evaluate whether CXCR4 promoter activity is modulated by LIP-dependent transactivation. The increased expression of LIP led to a 3–4-fold increase in CXCR4 promoter activity, suggesting that LIP transcriptionally regulates CXCR4 expression (Fig. 1C). To examine whether the increase in CXCR4 promoter activity is dependent on recognition of the specific C/EBPβ binding site, we generated CXCR4 promoter mutant constructs; one with a 9-bp deletion encompassing the putative C/EBPβ binding site and the other with two substitution mutations in the putative binding site. As shown in Fig. 1C, the mutant promoters containing either the substitution or deletion significantly diminished LIP-induced CXCR4 promoter activity.
FIGURE 1.
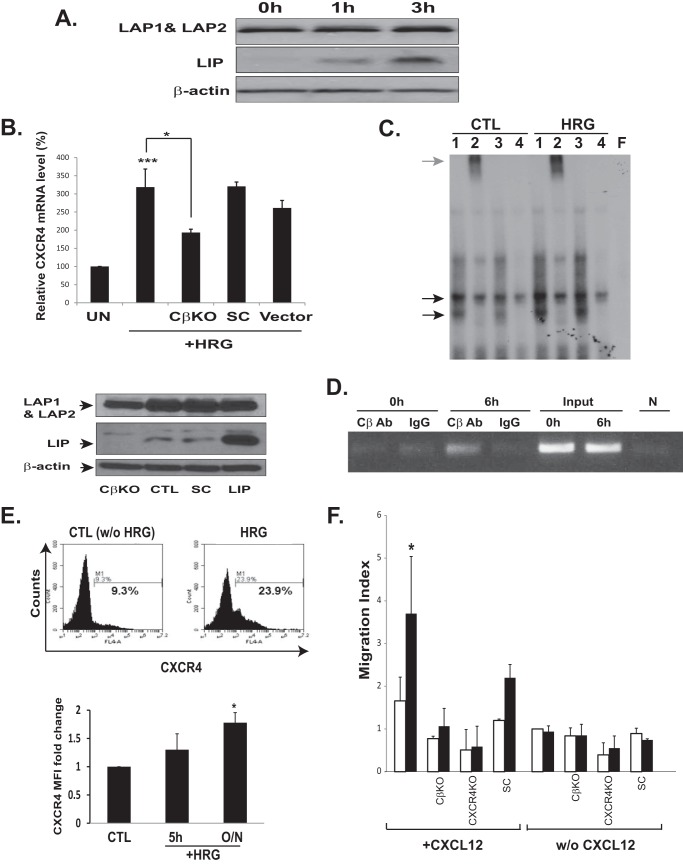
LIP increases CXCR4 transcription in breast cancer cells. A, Western blot analyses of LIP-transduced breast cancer cells. MCF-7 cells were retrovirally transduced with either control MSCV-GFP vector (Vector) or MSCV-LIP-GFP vector (LIP). The anti-C/EBPβ antibody was raised against the C termini of C/EBPβ and thus recognized all isoforms. The three different isoforms of C/EBPβ, LAP1, LAP2, and LIP, were detected in whole-cell extracts. Note that control vector-transduced cells (Vector) express a low level of endogenous LIP. A stronger LIP band was clearly detectable in an over-exposed blot (see supplemental Fig. S1). B, quantitative PCR analysis of CXCR4 in LIP- and control vector-transduced breast cancer cell lines. MCF-7, SKBR3, and MDA-MB361 cells were transduced with either control MSCV-GFP vector (white bars) or MSCV-LIP-GFP vector (black bars). Actinomycin D treatment (4 μg/ml) (gray bars) blocked the increase in CXCR4 mRNA induced by LIP. CXCR4 expression was normalized against either 18 S RNA or GAPDH. The levels of CXCR4 mRNA in the control cells were arbitrarily set at 100%. Results represent the mean ± S.D. of at least three independent experiments, each done in duplicate. *, p < 0.05; ***, p < 0.005. C, effects of LIP expression on the transcriptional activity of the CXCR4. MCF-7 cells were transiently co-transfected with firefly luciferase reporter vectors harboring CXCR4 promoter region (−313) and Renilla luciferase vectors followed by LIP (black bars) or control vector infection (white bars). Deletion or substitution mutations of the putative C/EBP binding site within the CXCR4 promoter abolished LIP-induced CXCR4 promoter activity. At 2–3 days after transduction, cells were lysed, and luciferase signals were measured. The results were presented by the relative luciferase units (RLU; a.u., absorbance units) divided by the activity of cells transfected with the basic reporter construct alone. The y axis is the ratio of firefly to Renilla RLU values. **, p < 0.01 The lower diagram shows the location of putative YY1 and C/EBP binding site. D, equal amounts of nuclear extract from LIP- and control vector-transduced MCF-7 cells were preincubated for 30 min in the presence (lane 2) or absence (lane 1) of anti-C/EBPβ antibody. Lane 1 shows binding between probe and nuclear extracts. Lane 2 shows the supershifted complex formed between the oligonucleotide, the nuclear extract, and the C/EBPβ antibody. Normal IgG was used as a supershift control (lane 3). For competition experiments, a 100-fold excess of unlabeled, cold wild type (lane 4) or C/EBP binding site-mutated (lane 5) or C/EBP binding site-deleted (lane 6) or unrelated AP1 consensus site (lane 7) oligonucleotides were premixed with radiolabeled C/EBP probe. The last lane (F) shows free probe. The black arrows indicate the C/EBPβ-DNA complex. The gray arrow indicates the supershifted C/EBPβ-DNA complex. The free probe has been allowed to run off the gel to better resolve the multiple complexes. E, ChIP analysis. Chromatin extracts were prepared from LIP- or control vector (Vector)-transduced MCF-7 cells. The fragmented chromatin was subjected to immunoprecipitation with control antibodies (IgG) or anti-C/EBPβ antibody (CβAb). Eluted DNA was amplified by the PCR primers designed to specifically amplify the proximal CXCR4 promoter regions containing the binding sites for C/EBP. The expected size of the PCR product is 120 base pairs. N, no template controls.
To further investigate if LIP transactivates CXCR4 promoter activity via its direct binding to the putative C/EBP binding site, we performed an EMSA using 32P-labeled oligonucleotide probes containing the consensus C/EBP binding site. DNA binding activity was clearly detected in nuclear extracts from LIP-transduced cells (lane 1, black arrows) (Fig. 1D). This DNA binding activity disappeared in the presence of unlabeled (cold) wild type competitor (lane 4) but was unaffected by oligonucleotides containing mutated (lane 5) or deleted C/EBP consensus binding sequence (lane 6) or unrelated oligonucleotides (AP1) (lane 7), indicating that the observed signals are specific. The addition of anti-C/EBPβ antibody resulted in supershifted DNA-protein complexes confirming the identity of the protein causing the shift (lane 2, gray arrow). Of note, despite the relatively high levels of endogenous LAPs (LAP1 and LAP2) in vector-transduced control cells (see Fig. 1A), no apparent DNA binding activity was observed. This suggests that LIP may have a major and indispensable role in modulating transcriptional activity of CXCR4 in breast cancer cells. To corroborate the above findings in living cells, we performed ChIP assay. Cell lysates were immunoprecipitated with antibody against C/EBPβ. Subsequent PCR amplification using primers specific to the CXCR4 promoter produced a band of the expected size (120 base pair) in LIP-transduced cells (Fig. 1E). Low or negligible levels of PCR-amplified bands were detected in the control vector-transduced cells.
YY1 is one of well known transcriptional repressors of CXCR4 (24). YY1 binding site (−306 to −297) is located near the putative C/EBP binding site (see a diagram in Fig. 1C). Therefore, it is conceivable that the binding of LIP might interfere with the binding of YY1 to the CXCR4 promoter, thus relieving the repression effects of YY1. To address this issue, we performed EMSA experiments. The intensity of the shifted bands (Fig. 2A, black arrows) was clearly reduced in LIP-transduced cells as compared with that of the control cells (lane 7 versus 1), suggesting that the ectopic LIP expression affected the binding of YY1 to the CXCR4 promoter. Specificity of the binding was verified by a YY1 antibody supershift assay (lanes 8 and 9). ChIP assays were then performed using YY1 antibody to assess how LIP modulates in vivo binding of YY1 to the CXCR4 promoter. The YY1 binding was greatly decreased in LIP-expressing cells as compared with control cells (Fig. 2B). These findings are consistent with the EMSA results and indicate that the increase in LIP abundance led to the decreased YY1 binding to CXCR4 promoter in breast cancer cells.
FIGURE 2.
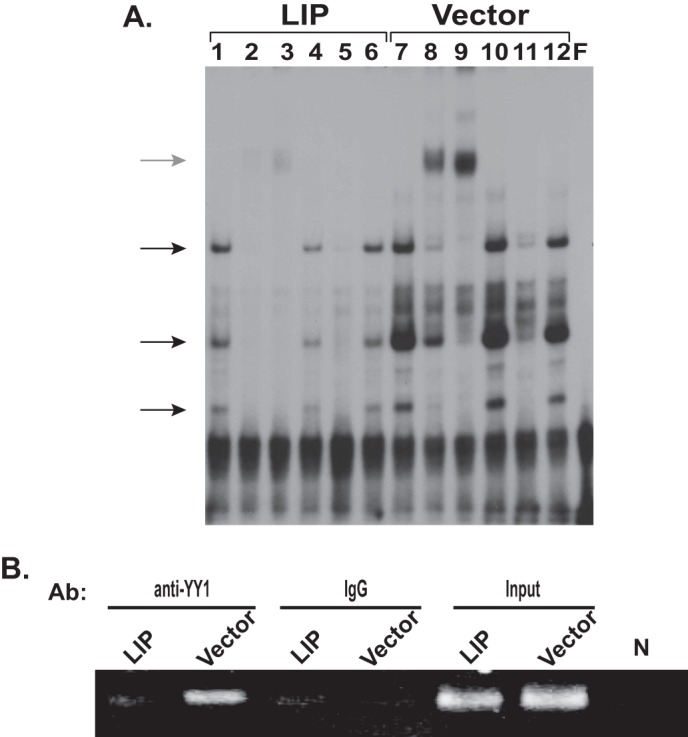
LIP modulates YY1 binding to the CXCR4 promoter. A, gel-shift assays were basically performed as described in Fig. 1D using nuclear extracts from LIP- or control vector (Vector)-transduced MCF-7 cells. The black arrows indicate the YY1-DNA complex. The gray arrow indicates the supershifted YY1-DNA complex. Lanes 1 and 7, probe with nuclear extracts; lanes 2 and 3 and lanes 8 and 9, probe with nuclear extract preincubated with anti-YY1 antibody (0.4 μg for lanes 2 and 8 and 1 μg for lanes 3 and 9); lanes 4 and 10, supershift control with normal IgG; lanes 5 and 11, probe with nuclear extract in the presence of excess unlabeled wild type probe; lanes 6 and 12, probe with nuclear extract in the presence of excess unlabeled AP1 consensus site oligonucleotide. The last lane (F) shows free probe. F, free probe. B, ChIP analysis of YY1 binding to the CXCR4 promoter in the LIP-transduced cells. The enrichment of CXCR4 promoter in the ChIP DNA pulled by the YY1 antibody (Ab) was assayed. As a negative control, a non-immune IgG was used. N, no template controls.
LIP Binds to the CXCR4 Promoter as Hetero- and Homodimers
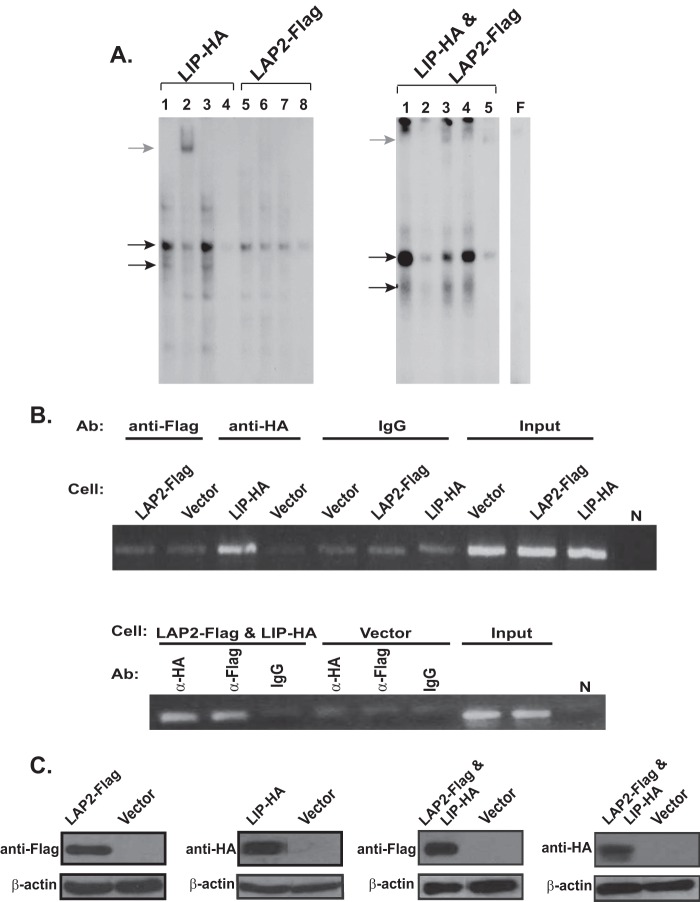
Homo- or heterodimer formation is required for the function of C/EBPs. This raises the question of whether the LAP isoforms have similar regulatory roles in modulating CXCR4 expression in breast cancer cells. However, unlike LIP, the ectopic expression of LAP1 or LAP2 did not exert any noticeable effects on CXCR4 expression as demonstrated by flow cytometry analysis (see supplemental Fig. S2). Although this finding indicates that the increased levels of LAP do not appear to play much of a role in regulating CXCR4 in breast cancer cells, LAP can still serve as a heterodimeric partner for LIP to transcriptionally activate CXCR4. To address this issue, we utilized FLAG-tagged LAP2 and HA-tagged LIP cDNA constructs as C/EBPβ antibodies that selectively recognize each isoform are not commercially available. EMSA analysis yielded two major shifted bands in the cells expressing HA-tagged LIP (LIP-HA cells) (Fig. 3A, left panel). The addition of anti-HA antibody reduced the intensity of both bands (lane 2), suggesting that the upper band is a LAP/LIP heterodimer and the lower band is a LIP/LIP homodimer. Similarly, anti-HA antibody caused the disruption of the two retarded bands in the MCF-7 cells co-expressing FLAG-tagged LAP2 and HA-tagged LIP (LIP-HA and LAP2-FLAG cells) (Fig. 3A, right panel, lane 2). In the same cells, anti-FLAG antibody preferentially reduced the intensity of the upper band but not that of the lower band (Fig. 3A, right panel, lane 3), confirming the identity of the retarded complexes. Meanwhile, much fainter retarded complexes were detected in the cells expressing FLAG-tagged LAP2 alone (LAP2-FLAG cells) as compared with those detected in LIP-HA cells (Fig. 3A, left panel, lane 5 versus lane 1). This result further supports the notion that ectopic LAP2 expression alone does not potentiate the binding of the CEBPβ complexes to the CXCR4 promoter as much as does LIP. ChIP analysis clearly demonstrated the preferential recruitment of LIP to the CXCR4 promoter as compared with LAP2 (Fig. 3B, upper panel). Consistent with the EMSA analysis, recruitment of LAP2 to the CXCR4 promoter was markedly increased when LIP was co-expressed (Fig. 3B, lower panel). Although LAP isoforms are abundantly expressed, endogenous LIP was expressed at a much lower level and was barely detectable as shown in Fig. 1A. Therefore, although both LAP/LIP and LIP/LIP bind to CXCR4 promoter, it appears that the dimerization and subsequent DNA binding are mainly dependent on the levels of LIP rather than LAPs.
FIGURE 3.
LIP binds to the CXCR4 promoter as LAP/LIP hetero- and LIP/LIP homodimers. A, cells were transfected with HA-tagged LIP (LIP-HA) and FLAG-tagged LAP2 (LAP2-FLAG) either alone (left panel) or together (right panel: LIP-HA & LAP2-FLAG). Left panel: lanes 2 and 6 were preincubated with anti-HA and anti-FLAG antibody, respectively. Lanes 1 and 5, probe with nuclear extracts; lanes 3 and 7, supershift control with IgG; lanes 4 and 8, probe with nuclear extract in the presence of excess unlabeled wild type probe. Right panel: lane 1, probe with nuclear extracts; lane 2, supershift with anti-HA antibody; lane 3, supershift with anti-FLAG antibody; lane 4, supershift control with normal IgG; lane 5, probe with nuclear extract in the presence of excess unlabeled wild type probe. Black arrows designate shifted bands whose intensities were diminished by adding each antibody. The last lane (F) shows free probe. B, chromatin samples were prepared as described above. ChIP analysis was performed using anti-FLAG or anti-HA antibodies (Ab). Immunoprecipitated DNA was assayed for the enrichment of CXCR4 promoter. Consistent with the EMSA results above, the ectopic expression of LIP (LIP-HA) but not LAP-2 (LAP2-FLAG) showed a significant enrichment of CXCR4 promoter. As a negative control, a non-immune IgG was used. Lower panel, ChIP analysis with cells co-transfected with HA-tagged LIP (LIP-HA) and FLAG-tagged LAP2 (LAP2-FLAG). N, no template controls. C, the expression of HA-tagged LIP and FLAG-tagged LAP2 was assessed by Western blot analysis using respective antibodies.
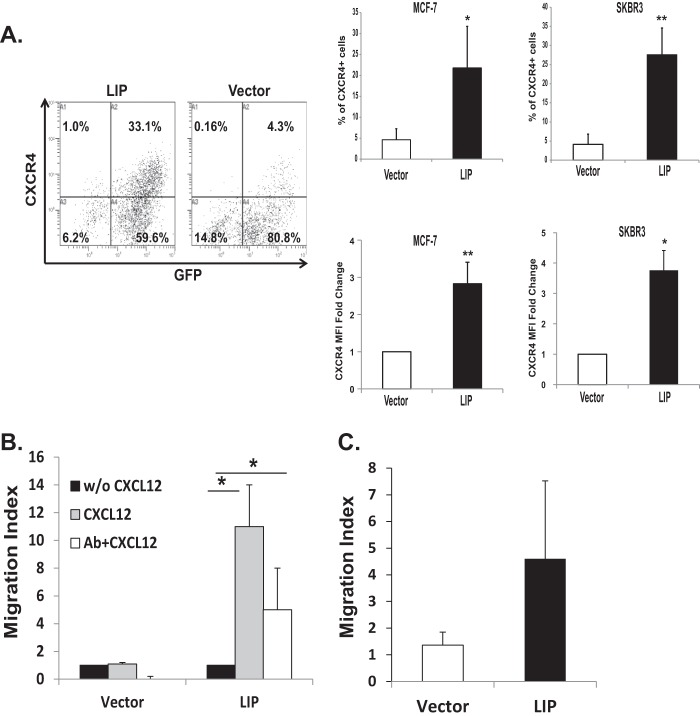
LIP Increases the Expression of Functional Cell Surface CXCR4
Next, we examined whether the increased CXCR4 mRNA levels induced by LIP expression is parallel to increases in its protein level. G-protein-coupled receptors (GPCR) including CXCR4 are regulated in a dynamic and complex manner by endocytosis or internalization. As CXCL12-induced cell migration requires the cell surface expression of CXCR4, we assessed the levels of CXCR4 at the cell surface. Percentage of transduced cells (GFP+) was similar between LIP- and control vector-transduced cells (∼85–95%) (Fig. 4A, left panel). However, among the transduced cells (GFP+), LIP-transduced cells showed a significantly elevated level of CXCR4 compared with control vector-transduced cells (33 versus 4.3%) (Fig. 4A, left panel, upper right quadrant). A similar increase in cell surface expression of CXCR4 was also observed in the LIP-expressing SKBR3 cells (Fig. 4A, right panel).
FIGURE 4.
LIP induces increases in functional cell surface CXCR4 receptor in breast cancer cells. A, flow cytometry analysis of cells transduced with either MSCV control (Vector) or MSCV-LIP (LIP) vectors. Transduced MCF-7 and SKBR3 cells were harvested by non-enzymatic cell dissociation. Cell surface expression of CXCR4 was analyzed by flow cytometry using biotinylated CXCR4 antibody and allophycocyanin (or phycoerythrin)-conjugated streptavidin. Left, transduced MCF-7 cells were assessed by GFP expression. Note that MCF-7 cells expressing MSCV-LIP increased the percentage of CXCR4 expressing cells. Numbers indicate the percentage in each quadrant. Results are presented as the percentage (%) of CXCR4-positive (CXCR4+, upper right panels) or shown as -fold change in MFI relative to vector-transduced control cells (lower right panels). Data are representative of at least 10 independent experiments. *, p < 0.05; **, p < 0.01. B, the transduced cells were subjected to the transwell chemotaxis migration assay in the presence (gray bars) or absence (black bars) of CXCL12 (125 ng/ml). Cells were preincubated (white bars) with CXCR4 neutralizing antibody, as described under “Experimental Procedures.” The chemotaxis was evaluated by counting the number of cells that had migrated through the membrane in three-four representative fields. Chemotactic activity was expressed as a migration index, which is the ratio of the number of cells migrating toward CXCL12 to the number of cells migrating toward medium control. *. p < 0.05. C, for the invasion assay, transduced cells were plated in a modified Boyden chamber coated with Matrigel. The bottom chamber was loaded with CXCL12 (125 ng/ml). The number of migrated cells on the lower surface of the membrane was counted (5 random 100× fields per well). Results are expressed as the migration index calculated as above.
We then determined whether the increase in cell surface CXCR4 expression is functionally relevant for breast cancer cell migration. LIP- and control vector (Vector)-transduced cells were assessed in transwell assays scoring chemotaxis toward CXCL12. The chemotactic activity was ∼10-fold higher in LIP-transduced cells compared with control vector-transduced cells (Fig. 4B). The migratory response of the LIP-transduced cells to CXCL12 was significantly attenuated by neutralizing anti-CXCR4 antibody. Furthermore, LIP enhanced the ability of MCF-7 cells to invade through the extracellular matrix barriers, as evidenced by invasion assays (Fig. 4C).
To investigate how LIP affects the metastatic potential of breast cancer cells in vivo, LIP-transduced SKBR3 and MCF-7 cells were intravenously injected into 6–8-week-old NOD/SCID/IL2Rγ null mice. Recipient mice that were transplanted with LIP-transduced cells displayed typical signs of cancer burdens, including weight loss, spiky coat, kyphosis, and decreased mobility. In particular, the mice transplanted with LIP-transduced SKBR3 cells (n = 7) exhibited severe signs of morbidity for unknown reasons and died within 3 weeks after transplantation so that we were not able to examine metastasis on these recipient animals. In contrast, the recipient animals transplanted with control-vector-transduced SKBR3 cells (n = 6) survived beyond 5–6 weeks post-transplant.
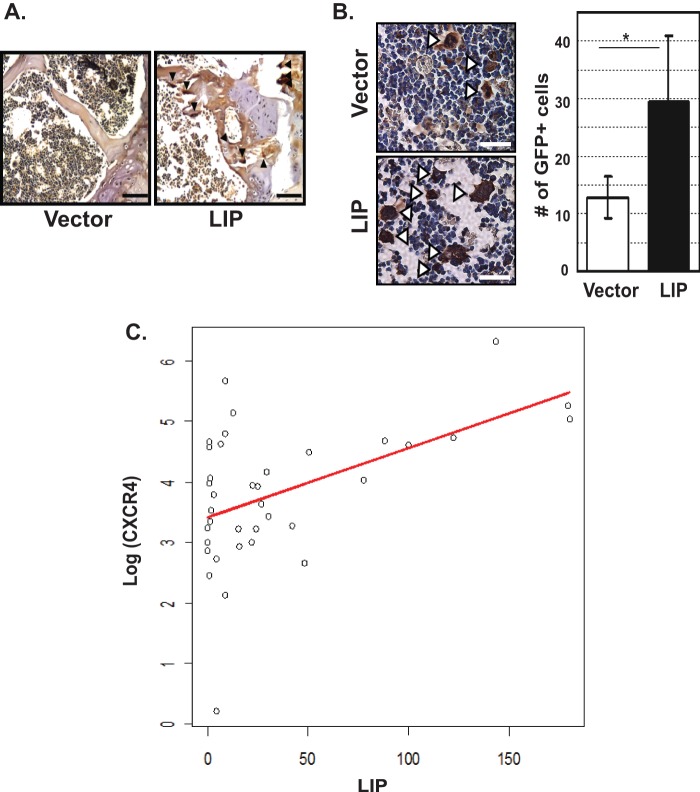
Although animals transplanted with LIP-transduced MCF-7 cells showed significantly better post-transplant survival rates, no statistically significant difference was found in the number of lung metastatic foci between control- and LIP cell-transplanted animals (data not shown). Because the most frequent site of breast cancer metastasis is the bone (25) and this commonly causes osteolytic lesions (26), the bones from the transplanted animals were carefully examined. The recipient animals transplanted with LIP-transduced MCF-7 cells (n = 5) had a higher frequency of osteoclast cells in their bones as compared with the animals transplanted with control cells (n = 2), as evidenced by TRAP staining (Fig. 5A). To quantitatively measure metastasized cells, recipient bones were immunohistochemically stained with anti-GFP antibody. A significantly higher frequency of GFP-positive cells was detected in the bones from animals transplanted with LIP-transduced cells compared with animals transplanted control vector-transduced cells (Fig. 5B).
FIGURE 5.
LIP promotes osteoclast formation and the development of bone metastasis. A, representative TRAP-stained bone sections from recipient animals. Transduced MCF-7 cells were sorted for GFP expression and expanded. Cells were non-enzymatically detached and were transplanted into NOD-scid IL2rγnull mice (2 × 106 cells/mouse). Thirty days after transplantation, the bones from recipient animals were examined. Osteoclasts were visualized by TRAP staining and counterstained with hematoxylin. Arrowheads indicate osteoclasts. Scale bars, 100 μm. B, bone sections from the above recipient animals were immunostained using anti-GFP antibody. Left, representative bone sections stained with anti-GFP antibody. Open arrowheads indicate GFP+ cells. Scale bars, 50 μm. Right, quantification of total number of GFP+ cells per field of analysis. Y axis shows the total number of GFP expressing cells per field. At least 200 cells per field in a minimum of two randomly selected fields were counted. Data were shown as the mean ± S.D. *, p < 0.05. C, a positive correlation between LIP and CXCR4 protein expression in advanced breast cancer patients. Protein lysates from breast cancer tissues (stages III and IV, n = 37) were analyzed by Western blot for the expressions of LIP and CXCR4. Protein expression levels were quantitatively estimated by densitometric analysis and normalized to β-actin. To standardize and correct the density among films, the standard sample was loaded on each gel. Data points for each tissue specimen were plotted as a scatter plot. A simple linear regression analysis was performed to assess a linear association between LIP and CXCR4, where LIP was a predictor and CXCR4 was the response variable. A log-transform was applied to the values of CXCR4 to satisfy the normality assumption. The results show that the LIP values predict the level of CXCR4 well. The estimated correlation coefficient between LIP and CXCR4 on a raw scale was 0.54. p = 0.001 (two-sided).
LIP Expression Is Correlated with the Increased Expression of CXCR4 in Advanced-stage Breast Cancer Patients
Having observed a relationship between LIP and CXCR4 in breast cancer cell cells, we next determined if the same relationship exists in patients with advanced breast cancer (stages III and IV). To this end, we examined the protein levels of LIP and CXCR4 in surgically resected snap-frozen breast cancer specimens (n = 37). The results showed that the LIP values predict the level of CXCR4 well (Fig. 5C). More specifically, the fitted regression line is log (CXCR4) = 3.4 + 0.01 × LIP, and the results indicated that the unit increase of the CXCR4 level on a log scale is strongly positively associated with the unit increase of the LIP values with the two-sided p value of 0.001. Meanwhile, no correlation was found between LIP and CXCR4 in normal breast specimens (n = 5, data not shown).
Heregulin Up-regulates the Expression of CXCR4 in Breast Cancer Cells
HRG promotes metastatic migration of breast cancer cells (27, 28), and blockage of HRG expression inhibits metastasis of breast cancer (18). Interestingly, HRG treatment increased the levels of LIP but not the other C/EBPβ isoforms (Fig. 6A). Because LIP expression led to increased levels of CXCR4 mRNA, we examined whether the observed increase in LIP protein expression by HRG is sufficient to increase the transcription of CXCR4. Indeed, HRG treatment significantly increased CXCR4 mRNA levels (Fig. 6B). The selective knockdown of LIP can provide a clue as to whether HRG-induced CXCR4 mRNA expression is mediated via LIP. However, the selective knockdown of LIP without targeting other C/EBPβ (for example, LAP1 and LAP2) is not technically feasible, as C/EBPβ is translated into three different isoforms from the same mRNA via a leaky ribosomal scanning mechanism (17). Nevertheless, our attempt to knockdown C/EBPβ via shRNA-mediated silencing permitted almost complete knockdown of LIP (Fig. 6B, lower panels). Although the down-regulation of LAPs was also observed (∼50–60% reduction in protein level), a further down-regulation was not achievable by any other combinations of siRNA oligonucleotides or vector-based shRNAs. This is most likely due to the fact that LIP is present at considerably lower levels compared with LAPs. Knockdown of LIP significantly reduced, albeit not completely abrogated, the ability of HRG to induce CXCR4 mRNA (Fig. 6B, upper panel), suggesting that the increase in CXCR4 mRNA in HRG-treated breast cancer cells was at least in part dependent on LIP. EMSA analysis further demonstrated that HRG treatment induced the binding of endogenous LAP/LIP and LIP/LIP complexes to CXCR4 promoter (Fig. 6C). ChIP analysis also revealed that HRG treatment enhanced the binding of LIP to the CXCR4 promoter in vivo (Fig. 6D). The increase of LIP binding by HRG treatment was concomitant with a significant increase (p < 0.05) in the cell surface expression of CXCR4 (Fig. 6E), and this coincided with the enhanced ability for HRG-treated cells to migrate toward CXCL12 (Fig. 6F). shRNA-mediated down-regulation of either C/EBPβ or CXCR4 significantly reduced the migration of MCF-7 cells toward CXCL12, demonstrating that HRG-induced chemotactic migration is both LIP- and CXCR4-dependent (Fig. 6F).
FIGURE 6.
Heregulin up-regulates the expression of CXCR4 in breast cancer cells. A, HRG preferentially up-regulated the LIP isoform in breast cancer cells. MCF-7 cells were serum-starved overnight followed by HRG treatment (100 ng/ml) for the indicated time points. Total proteins were probed with antibodies as indicated. A representative Western blot of at least three independent experiments is shown. B, HRG was sufficient to induce the transcription of CXCR4 in breast cancer cells. MCF-7 cells were starved overnight in serum-free medium and then either left untreated (UN) or treated with 100 ng/ml HRG for 2–3 h. CXCR4 mRNA levels were determined using quantitative PCR as detailed in Fig. 1. shRNA targeting C/EBPβ (CβKO) and non-silencing scrambled control shRNA (SC) were also transduced. The mRNA levels of untreated cells were arbitrarily set at 100%. Results represent the mean ± S.D. of at least three independent experiments, each performed in duplicate. *, p < 0.05; ***, p < 0.005. Lower, Western blot analysis of C/EBPβ knockdown cells; knockdown of C/EBPβ resulted in reduced expression of all three isoforms of C/EBPβ. Densitometry analysis indicated the almost complete knockdown of LIP and ∼50% reduction of LAP. CTL, parental MCF-7 cells; LIP, LIP-transduced cells. C, nuclear extracts prepared from HRG-treated (HRG) and untreated (CTL) MCF-7 cells were subjected to EMSA analysis. The black arrows indicate the C/EBPβ-DNA complex. The gray arrow indicates the supershifted C/EBPβ-DNA complex. The last lane (F) shows free probe. Lanes 1, probe with nuclear extracts; lanes 2, probe with nuclear extract pre-incubated with anti-C/EBPβ antibody; lanes 3, normal IgG; lanes 4, cold wild type probe. D, MCF-7 cells were harvested at the indicated time after HRG (100 ng/ml) treatment. ChIP analysis was performed using anti-C/EBPβ antibody as described in Fig. 1. IgG antibody (Ab) was used as a control. N, no template controls. E, flow cytometric analysis of CXCR4 expression in HRG-treated MCF-7 cells. The surface expression of CXCR4 was variable depending on cell confluency (38). To quantify CXCR4 cell surface expression, MCF-7 Cells were grown to 90–95% confluence before HRG treatment. Cells were left untreated (CTL) or were treated with 100 ng/ml HRG for the time points indicated. CXCR4 cell surface expression was analyzed as described above. Upper, representative flow cytometric histograms showing cell surface CXCR4 expression in MCF-7 cells that were either untreated (CTL) or treated with HRG for overnight. Numbers indicate the percentage of gated cells expressing CXCR4. Lower, the data plotted show -fold change in mean fluorescence intensity. The mean fluorescence intensity of untreated cells was set at 1.0. The graph shows mean values of at least five independent experiments. *, p < 0.05. F, the chemotactic migration of cells was assessed in the presence (black bars) or absence (white bars) of HRG as described in Fig. 4B. C/EBPβ (CβKO) and CXCR4 (CXCR4 KO) knockdown cells were plated in the upper well of a transwell chamber. SC, scrambled siRNA-transduced MCF-7 cells. CXCL12 (125 ng/ml) was placed in the lower chamber. Chemotactic activity was expressed as a migration index, which is the ratio of the number of cell migrating toward CXCL12 to the number of cells migrating toward medium control (in the absence of CXCL12 and HRG).
DISCUSSION
Current therapies still fail to eliminate migrating (metastasizing) breast cancer cells. The complicated multistep nature of metastasis makes it difficult to dissect the molecular mechanisms involved in the metastatic cascade.
CXCR4 is the G protein-coupled chemokine receptor that mediates entry of T cell tropic HIV virus (29). It also plays a critical role in mediating stem cell homing to the niche (30). Furthermore, CXCR4 signaling axis endows breast cancer cells with metastatic ability and thus emerged as one of the most relevant targets for modulating breast cancer cell metastasis (3). Even though the CXCR4 signaling axis is one of the promising “druggable” targets for intervening metastasis, CXCR4 is expressed on several tissues, and there have been some concerns about using drugs to target CXCR4. In fact, AMD-3100, a CXCR4 receptor blocker, exerted unfavorable side effects in the clinical trial developing gastrointestinal side effects, thrombocytopenia, and atrial and ventricular arrhythmias (31). Very little is known about the molecular mechanisms that regulate CXCR4 receptor in breast cancer cells.
In this study we have identified the C/EBPβ isoform, LIP, as a previously unrecognized transcriptional regulator of CXCR4. Retroviral-mediated ectopic expression of LIP in breast cancer cells resulted in up-regulation of CXCR4 mRNA. EMSA and ChIP analyses demonstrated that the ectopic expression of LIP, but not LAP, enhanced CXCR4 transcription. LIP binds to the CXCR4 promoter by forming LAP/LIP hetero- and LIP/LIP homodimer. Thus LAPs are necessary as a dimerization partner for LIP. Nevertheless, ectopic expression of LAPs had only a limited effect on CXCR4 expression. The most proficiently translated isoform of C/EBPβ is LAP2 followed by LAP1, and thus these isoforms are abundantly expressed in breast cancer cells (32). In contrast, LIP is expressed at significantly lower levels and is barely detectable. Therefore, it is likely that LAP/LIP hetero- and LIP/LIP homodimers are formed at a rate dictated by the availability of LIP. This may explain why the expression of CXCR4 is largely dependent on the availability of LIP but not LAP1/2.
LIP is generally recognized as being a repressor of transcription due to its lack of activation domains (but it retains the DNA binding capability) (33). Thus, it was somewhat surprising that LIP enhanced transcription activation of CXCR4. Our results showed that LIP interfered with the binding of YY1, a strong CXCR4 repressor, and this might lead to increased CXCR4 expression. It is also conceivable that the LAP/LIP or LIP/LIP complex interacts with other transcriptional regulatory proteins to activate CXCR4 transcription since the C/EBP-family members often form complexes with other transcription factors.
LIP expression increased cell surface levels of the CXCR4 in breast cancer cells. The LIP-induced CXCR4 expression was functionally effective, as its expression level is closely correlated with the ability of breast cancer cells to migrate in in vitro invasion and chemotaxis assay in response to CXCL12. The recipient animals transplanted with LIP-transduced cells developed many signs of illness including wasting syndrome (cachexia) and exhibited high osteoclastogenic activity. We observed an increased trend for metastasis in the mice transplanted with LIP-transduced cells. Some recipient animals (especially the mice transplanted with LIP-transduced SKBR3 cells) became moribund shortly after transplantation and had to be euthanized. One of the potential causes of this early post-transplant death is that LIP may not only increase the metastatic capacity but could also enhance the aggressiveness of breast cancer cells. In fact, ectopic expression of LIP in TM3 breast cancer cells induces epithelial proliferation and the formation of mammary hyperplasias (34), substantiating such a possibility.
In advanced-stage human breast carcinoma specimens (n = 37), but not in normal specimen (n = 5), we observed a strong correlation between CXCR4 and LIP protein expression, lending further support for a potential role for LIP in regulating CXCR4 expression in advanced breast cancer patients.
HRG has been shown to play a relevant role for the aggressiveness and metastatic capacity of breast cancer cells (27, 28) and is associated with gefitinib resistance (35). Our study revealed that HRG regulates CXCR4 expression. HRG treatment increased the level of LIP without affecting the expression of the other C/EBPβ isoforms (Fig. 6A). This concomitantly enhanced LAP/LIP and LIP/LIP binding activity to CXCR4 promoter, resulting in a significant increase of the cell surface expression of CXCR4.
HRG binds to the ErbB3 and ErbB4 and leads to the formation of heterodimers with ErbB2, which subsequently activates ErbB2 downstream signaling pathways (36). Because ErbB2 activation is known to induce the expression of CXCR4 in breast cancer cells (37), one can hypothesize that HRG may induce CXCR4 expression through the activation of the ErbB2 signaling pathway. However, whereas ErbB2 enhances the CXCR4 expression by inhibition of proteasome-mediated degradation (rather than mRNA accumulation) (37), our study clearly demonstrates that HRG increases the CXCR4 gene at a transcriptional level as evidenced by Chip and Q-RT-PCR analysis. However, it is still an open possibility that HRG may regulate CXCR4 expression through both transcriptional and post-transcriptional mechanisms.
Taken together, our study showed that LIP serves a previously unappreciated role as a transcriptional regulator of CXCR4 gene in breast cancer cells. Furthermore, our study demonstrates that HRG regulates CXCR4 expression in breast cancer cells at least partially through LIP, thus providing potential new targets for the treatment and prevention of CXCR4-mediated breast cancer metastasis.
Acknowledgments
This project used the UPCI flow cytometry and animal facility, which was supported in part by the P30CA047904 award from the National Institutes of Health.
This work was supported in part by Department of Defense Grant W81XWH-09-1-0350 (to B.-C. L.).

This article contains supplemental Figs. S1 and S2.
- LAP
- liver-enriched activator protein
- LIP
- liver-enriched inhibitory protein
- HRG
- heregulin
- TRAP
- tartrate-resistant acid phosphatase.
REFERENCES
- 1. Tang X., Sun Z., Runne C., Madsen J., Domann F., Henry M., Lin F., Chen S. (2011) A critical role of Gβγ in tumorigenesis and metastasis of breast cancer. J. Biol. Chem. 286, 13244–13254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chiang A. C., Massagué J. (2008) Molecular basis of metastasis. N. Engl. J. Med. 359, 2814–2823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Müller A., Homey B., Soto H., Ge N., Catron D., Buchanan M. E., McClanahan T., Murphy E., Yuan W., Wagner S. N., Barrera J. L., Mohar A., Verástegui E., Zlotnik A. (2001) Involvement of chemokine receptors in breast cancer metastasis. Nature 410, 50–56 [DOI] [PubMed] [Google Scholar]
- 4. Vaday G. G., Hua S. B., Peehl D. M., Pauling M. H., Lin Y. H., Zhu L., Lawrence D. M., Foda H. D., Zucker S. (2004) CXCR4 and CXCL12 (SDF-1) in prostate cancer. Inhibitory effects of human single chain Fv antibodies. Clin. Cancer Res. 10, 5630–5639 [DOI] [PubMed] [Google Scholar]
- 5. Kato M., Kitayama J., Kazama S., Nagawa H. (2003) Expression pattern of CXC chemokine receptor-4 is correlated with lymph node metastasis in human invasive ductal carcinoma. Breast Cancer Res. 5, R144–R150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Helbig G., Christopherson K. W., 2nd, Bhat-Nakshatri P., Kumar S., Kishimoto H., Miller K. D., Broxmeyer H. E., Nakshatri H. (2003) NF-κB promotes breast cancer cell migration and metastasis by inducing the expression of the chemokine receptor CXCR4. J. Biol. Chem. 278, 21631–21638 [DOI] [PubMed] [Google Scholar]
- 7. Kang Y., Siegel P. M., Shu W., Drobnjak M., Kakonen S. M., Cordón-Cardo C., Guise T. A., Massagué J. (2003) A multigenic program mediating breast cancer metastasis to bone. Cancer Cell 3, 537–549 [DOI] [PubMed] [Google Scholar]
- 8. Chen Y., Stamatoyannopoulos G., Song C. Z. (2003) Down-regulation of CXCR4 by inducible small interfering RNA inhibits breast cancer cell invasion in vitro. Cancer Res. 63, 4801–4804 [PubMed] [Google Scholar]
- 9. Lee B. C., Lee T. H., Avraham S., Avraham H. K. (2004) Involvement of the chemokine receptor CXCR4 and its ligand stromal cell-derived factor 1α in breast cancer cell migration through human brain microvascular endothelial cells. Mol. Cancer Res. 2, 327–338 [PubMed] [Google Scholar]
- 10. Seagroves T. N., Krnacik S., Raught B., Gay J., Burgess-Beusse B., Darlington G. J., Rosen J. M. (1998) C/EBPβ, but not C/EBPα, is essential for ductal morphogenesis, lobuloalveolar proliferation, and functional differentiation in the mouse mammary gland. Genes Dev. 12, 1917–1928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Baldwin B. R., Timchenko N. A., Zahnow C. A. (2004) Epidermal growth factor receptor stimulation activates the RNA binding protein CUG-BP1 and increases expression of C/EBPβ-LIP in mammary epithelial cells. Mol. Cell. Biol. 24, 3682–3691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zahnow C. A., Younes P., Laucirica R., Rosen J. M. (1997) Overexpression of C/EBPβ-LIP, a naturally occurring, dominant-negative transcription factor, in human breast cancer. J. Natl. Cancer Inst. 89, 1887–1891 [DOI] [PubMed] [Google Scholar]
- 13. Robinson G. W., Johnson P. F., Hennighausen L., Sterneck E. (1998) The C/EBPβ transcription factor regulates epithelial cell proliferation and differentiation in the mammary gland. Genes Dev. 12, 1907–1916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li H., Baldwin B. R., Zahnow C. A. (2011) LIP expression is regulated by IGF-1R signaling and participates in suppression of anoikis. Mol. Cancer 10, 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Arnal-Estapé A., Tarragona M., Morales M., Guiu M., Nadal C., Massagué J., Gomis R. R. (2010) HER2 silences tumor suppression in breast cancer cells by switching expression of C/EBPss isoforms. Cancer Res. 70, 9927–9936 [DOI] [PubMed] [Google Scholar]
- 16. Zahnow C. A. (2002) CCAAT/enhancer binding proteins in normal mammary development and breast cancer. Breast Cancer Res. 4, 113–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gomis R. R., Alarcón C., Nadal C., Van Poznak C., Massagué J. (2006) C/EBPβ at the core of the TGFβ cytostatic response and its evasion in metastatic breast cancer cells. Cancer Cell 10, 203–214 [DOI] [PubMed] [Google Scholar]
- 18. Tsai M. S., Shamon-Taylor L. A., Mehmi I., Tang C. K., Lupu R. (2003) Blockage of heregulin expression inhibits tumorigenicity and metastasis of breast cancer. Oncogene 22, 761–768 [DOI] [PubMed] [Google Scholar]
- 19. Lee B. C., Lee T. H., Zagozdzon R., Avraham S., Usheva A., Avraham H. K. (2005) Carboxyl-terminal Src kinase homologous kinase negatively regulates the chemokine receptor CXCR4 through YY1 and impairs CXCR4/CXCL12 (SDF-1α)-mediated breast cancer cell migration. Cancer Res. 65, 2840–2845 [DOI] [PubMed] [Google Scholar]
- 20. Orimo A., Gupta P. B., Sgroi D. C., Arenzana-Seisdedos F., Delaunay T., Naeem R., Carey V. J., Richardson A. L., Weinberg R. A. (2005) Stromal fibroblasts present in invasive human breast carcinomas promote tumor growth and angiogenesis through elevated SDF-1/CXCL12 secretion. Cell 121, 335–348 [DOI] [PubMed] [Google Scholar]
- 21. Sarbassov D. D., Guertin D. A., Ali S. M., Sabatini D. M. (2005) Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 307, 1098–1101 [DOI] [PubMed] [Google Scholar]
- 22. Moffat J., Grueneberg D. A., Yang X., Kim S. Y., Kloepfer A. M., Hinkle G., Piqani B., Eisenhaure T. M., Luo B., Grenier J. K., Carpenter A. E., Foo S. Y., Stewart S. A., Stockwell B. R., Hacohen N., Hahn W. C., Lander E. S., Sabatini D. M., Root D. E. (2006) A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell 124, 1283–1298 [DOI] [PubMed] [Google Scholar]
- 23. Wethmar K., Smink J. J., Leutz A. (2010) Upstream open reading frames. Molecular switches in (patho)physiology. Bioessays 32, 885–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Moriuchi M., Moriuchi H., Margolis D. M., Fauci A. S. (1999) USF/c-Myc enhances, while Yin-Yang 1 suppresses, the promoter activity of CXCR4, a coreceptor for HIV-1 entry. J. Immunol. 162, 5986–5992 [PubMed] [Google Scholar]
- 25. Kominsky S. L., Davidson N. E. (2006) A “bone” fide predictor of metastasis? Predicting breast cancer metastasis to bone. J. Clin. Oncol. 24, 2227–2229 [DOI] [PubMed] [Google Scholar]
- 26. Kozlow W., Guise T. A. (2005) Breast cancer metastasis to bone. Mechanisms of osteolysis and implications for therapy. J. Mammary Gland Biol. Neoplasia 10, 169–180 [DOI] [PubMed] [Google Scholar]
- 27. Cheng L., Zha Z., Lang B., Liu J., Yao X. (2009) Heregulin-β1 promotes metastasis of breast cancer cell line SKBR3 through upregulation of Snail and induction of epithelial-mesenchymal transition. Cancer Lett. 280, 50–60 [DOI] [PubMed] [Google Scholar]
- 28. Mazumdar A., Adam L., Boyd D., Kumar R. (2001) Heregulin regulation of urokinase plasminogen activator and its receptor. Human breast epithelial cell invasion. Cancer Res. 61, 400–405 [PubMed] [Google Scholar]
- 29. Dimitrov D. S. (1997) How do viruses enter cells? The HIV coreceptors teach us a lesson of complexity. Cell 91, 721–730 [DOI] [PubMed] [Google Scholar]
- 30. Peled A., Petit I., Kollet O., Magid M., Ponomaryov T., Byk T., Nagler A., Ben-Hur H., Many A., Shultz L., Lider O., Alon R., Zipori D., Lapidot T. (1999) Dependence of human stem cell engraftment and repopulation of NOD/SCID mice on CXCR4. Science 283, 845–848 [DOI] [PubMed] [Google Scholar]
- 31. Hendrix C. W., Collier A. C., Lederman M. M., Schols D., Pollard R. B., Brown S., Jackson J. B., Coombs R. W., Glesby M. J., Flexner C. W., Bridger G. J., Badel K., MacFarland R. T., Henson G. W., Calandra G. (2004) Safety, pharmacokinetics, and antiviral activity of AMD3100, a selective CXCR4 receptor inhibitor, in HIV-1 infection. J. Acquir. Immune Defic. Syndr. 37, 1253–1262 [DOI] [PubMed] [Google Scholar]
- 32. Uematsu S., Kaisho T., Tanaka T., Matsumoto M., Yamakami M., Omori H., Yamamoto M., Yoshimori T., Akira S. (2007) The C/EBP β isoform 34-kDa LAP is responsible for NF-IL-6-mediated gene induction in activated macrophages, but is not essential for intracellular bacteria killing. J. Immunol. 179, 5378–5386 [DOI] [PubMed] [Google Scholar]
- 33. Kowenz-Leutz E., Leutz A. (1999) A C/EBPβ isoform recruits the SWI/SNF complex to activate myeloid genes. Mol. Cell 4, 735–743 [DOI] [PubMed] [Google Scholar]
- 34. Zahnow C. A., Cardiff R. D., Laucirica R., Medina D., Rosen J. M. (2001) A role for CCAAT/enhancer binding protein β-liver-enriched inhibitory protein in mammary epithelial cell proliferation. Cancer Res. 61, 261–269 [PubMed] [Google Scholar]
- 35. Hutcheson I. R., Knowlden J. M., Hiscox S. E., Barrow D., Gee J. M., Robertson J. F., Ellis I. O., Nicholson R. I. (2007) Heregulin β1 drives gefitinib-resistant growth and invasion in tamoxifen-resistant MCF-7 breast cancer cells. Breast Cancer Res. 9, R50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fitzpatrick V. D., Pisacane P. I., Vandlen R. L., Sliwkowski M. X. (1998) Formation of a high affinity heregulin binding site using the soluble extracellular domains of ErbB2 with ErbB3 or ErbB4. FEBS Lett. 431, 102–106 [DOI] [PubMed] [Google Scholar]
- 37. Li Y. M., Pan Y., Wei Y., Cheng X., Zhou B. P., Tan M., Zhou X., Xia W., Hortobagyi G. N., Yu D., Hung M. C. (2004) Upregulation of CXCR4 is essential for HER2-mediated tumor metastasis. Cancer Cell 6, 459–469 [DOI] [PubMed] [Google Scholar]
- 38. Carlisle A. J., Lyttle C. A., Carlisle R. Y., Maris J. M. (2009) CXCR4 expression heterogeneity in neuroblastoma cells due to ligand-independent regulation. Mol. Cancer 8, 126. [DOI] [PMC free article] [PubMed] [Google Scholar]