Background: Precise spatiotemporal rearrangement of the actin cytoskeleton is essential for cell migration and adhesion.
Results: SWAP-70 bundles actin filaments and interacts with cofilin in vitro and in murine mast cells.
Conclusion: SWAP-70 is a parallel and anti-parallel bundling protein that participates in the organization of F-actin networks.
Significance: Identification and characterization of a regulator of F-actin dynamics improve our understanding on the spatiotemporal control of cytoskeletal processes.
Keywords: Actin, Cell Biology, Cofilin, Cytoskeleton, Mast Cell, F-actin Bundling, SWAP-70
Abstract
Coordinated assembly and disassembly of actin into filaments and higher order structures such as stress fibers and lamellipodia are fundamental for cell migration and adhesion. However, the precise spatiotemporal regulation of F-actin structures is not completely understood. SWAP-70, a phosphatidylinositol 3,4,5-trisphosphate-interacting, F-actin-binding protein, participates in actin rearrangements through yet unknown mechanisms. Here, we show that SWAP-70 is an F-actin-bundling protein that oligomerizes through a Gln/Glu-rich stretch within a coiled-coil region. SWAP-70 bundles filaments in parallel and anti-parallel fashion through its C-terminal F-actin binding domain and delays dilution-induced F-actin depolymerization. We further demonstrate that SWAP-70 co-localizes and directly interacts with cofilin, an F-actin severing and depolymerization factor, and contributes to the regulation of cofilin activity in vivo. In line with these activities, upon stem cell factor stimulation, murine bone marrow-derived mast cells lacking SWAP-70 display aberrant regulation of F-actin and actin free barbed ends dynamics. Moreover, proper stem cell factor-dependent cofilin activation via dephosphorylation and subcellular redistribution into a detergent-resistant cytoskeletal compartment also require SWAP-70. Together, these findings reveal an important role of SWAP-70 in the dynamic spatiotemporal regulation of F-actin networks.
Introduction
The dynamic reorganization of the actin cytoskeleton is essential for many cellular processes, including endocytosis, migration, polarization, and adhesion. Monomeric actin assembles into filaments that are incorporated into higher order cellular structures such as stress fibers, ruffles, lamellipodia, and filopodia. These processes are dynamically controlled by a range of actin regulatory molecules under tight spatiotemporal regulation that maintain appropriate dynamic actin turnover (1–3). Cross-linking and bundling proteins, for instance, are essential to arrange filaments into bundles and networks providing mechanical stability to F-actin structures (4, 5). Defects in F-actin bundling impair cell migration and adhesion (6–8). Bundling proteins act in concert with filament nucleators, elongators, and capping and severing proteins. A central molecule regulating F-actin is cofilin (actin depolymerizing factor/cofilin), a filament-severing and depolymerization factor involved in F-actin assembly and disassembly (9, 10). Tight spatiotemporal regulation of cofilin activity by pH, by direct binding to several proteins and phosphatidylinositol 4,5-bisphosphate, by phosphorylation, and by subcellular localization (11–13) is critical for proper actin dynamics (14). Disturbance of cofilin activity results in impaired chemotaxis and is observed in invasive metastatic cancers (15, 16). Recently, the complex and coordinated assembly and disassembly of specific F-actin structures have been viewed as a function of multidomain scaffold proteins that integrate actin binding and regulatory activities with other protein/protein interactions, membrane binding, and/or signaling domains (17). However, neither these multidomain scaffold proteins nor their underlying mechanisms and the regulatory networks that control F-actin dynamics are fully understood.
The switch-associated protein 70 (SWAP-70), initially identified in activated murine B cells (18), is mainly expressed in hematopoietic cells (19, 20). SWAP-70's structure consists of an N-terminal putative EF-hand domain, a pleckstrin homology domain that binds phosphatidylinositol 3,4,5-trisphosphate and is responsible for membrane localization, a tri-partite coiled-coil region, and a C-terminal F-actin-binding site (21, 22), through which SWAP-70 specifically binds nonmuscle but not muscle actin. Although Swap-70−/− mice are generally healthy (23), SWAP-70 is involved in migration, polarization, adhesion, immune functions, and differentiation of B cells, dendritic cells, mast cells, and/or erythroid progenitors (23–28). SWAP-70 interacts with the Rho GTPases Rac1 and RhoA, localizes to F-actin-rich structures such as lamellipodia, membrane ruffles, and stress fibers, and was proposed to control F-actin rearrangements in different cell types (20, 21, 26, 28). However, SWAP-70's specific functions and mechanisms in these processes remained largely unknown.
Here, we investigated the role of SWAP-70 in regulating F-actin rearrangements, and we report a novel function of SWAP-70 in the stabilization of actin filaments in vitro and in the contribution to F-actin dynamics in mast cells.
EXPERIMENTAL PROCEDURES
DNA Constructs
Mouse SWAP-70 and mouse cofilin-1 cDNAs were PCR-amplified from pMIG-SWAP-70 (24) and a cofilin-1 cDNA clone (number 3110001F06, ImaGenes, Source BioScience), respectively, cloned into pENTR-D-TOPO (Invitrogen), and subcloned into pDEST-15 and pDEST-17 (Invitrogen) for bacterial expression and pDN-Cerulean and pDN-Venus (pDEST-31 derivatives, Dr. Attila Toth, Technische Universität Dresden, Germany) for mammalian expression using the Gateway system (Invitrogen). SWAP-70 C-term (SWAP-70 aa2 444–585) was described earlier (29). SWAP-70w/oCC (SWAP-70Δ312–529; SWAP-70 without the aa 312–525 corresponding to the three coiled-coil regions) was generated by mutagenesis of pENTR-D-TOPO-SWAP-70 using specific primers and subsequently subcloned into pDEST-17.
Mice, Cells, and Transfection
Swap-70−/− mice backcrossed onto a 129/SvEMS background have been previously described (23). Mice were bred and maintained in the animal facility of the Medical Faculty, Technische Universität Dresden (Dresden, Germany), according to institutional guidelines. NIH 3T3 cells were maintained in DMEM (Invitrogen) with 10% FCS (Invitrogen), 1% penicillin/streptomycin at 37 °C in 5% CO2. Recombinant murine SCF and IL-3 were either collected from supernatants of IL-3-producing X63 cells and SCF-secreting baby hamster kidney cells, respectively, or purchased from PeproTech. SCF and IL-3 concentrations were measured by ELISA (PeproTech). BMMCs were obtained from 6- to 9-week-old WT and Swap-70−/− mice by flushing femur marrow and culturing cells in the presence of IL-3 (60 ng/ml) in Hybridoma medium (Invitrogen) with 2% FCS and 1% penicillin/streptomycin. After 3 weeks of culture, BMMCs, defined as c-kitR+ FcϵR1+, represent >98% of the cells. BMMCs used for experiments were 3–6 weeks old. Transfection of NIH 3T3 was performed with Lipofectamine 2000 (Invitrogen) according to the manufacturer's recommendations.
Antibodies, Protein Expression, and Purification
Polyclonal anti-SWAP-70 and anti-cofilin-1 antibodies were raised in rabbits and purified on His-SWAP-70 and GST-cofilin-1 columns, respectively, by affinity chromatography. Anti-actin, anti-cofilin, anti-GST, and anti-GAPDH were from Santa Cruz Biotechnology; anti-phospho-cofilin S3 was from Cell Signaling; anti-His was from Dianova; and anti-Arp2 was from Sigma. His- and GST-tagged proteins were expressed in Escherichia coli BL21(DE3) and purified according to standard protocols (Qiagen). Cells were lysed by sonication in His buffer (50 mm NaH2PO4, pH 8.0, 300 mm NaCl, 10 mm imidazole) or GST buffer (50 mm Tris, pH 8.0, 1 mm Na2S2O5, 1 mm EDTA) supplemented with 1 mg/ml lysozyme, 10 μg/ml RNase A (Roche Applied Science), 5 μg/ml DNase I (Roche Applied Science), and protease inhibitors (Roche Applied Science). Tagged proteins were purified from cleared lysates with either nickel-nitrilotriacetic acid-agarose (Qiagen) or glutathione-agarose (Sigma) columns for His- and GST-tagged proteins, respectively, followed by gel filtration or dialysis in SWAP-70's dialysis buffer (10 mm Tris, pH 7.4, 150 mm NaCl, 1 mm DTT, 0.1 mm EDTA, 10% glycerol) or cofilin-1 dialysis buffer (20 mm Tris, pH 7.4, 100 mm NaCl, 1 mm DTT, 0.1 mm EDTA, 10% glycerol). SWAP-70 was gel-filtered on a Superdex 75 HPLC column (Amersham Biosciences). Proteins were snap-frozen and kept at −80 °C. Human fascin was purified as described previously (30). Proteins were precleared by centrifugation at 40,000 rpm for 20 min at 4 °C before all experiments.
Cell Extracts, Fractionation, and Immunoprecipitation
BMMCs were IL-3-starved overnight and stimulated with 20 ng/ml SCF before cell lysis. NIH 3T3 were serum-starved for 2 h and stimulated with 15 nm EGF. Total cell extracts were prepared with RIPA buffer (50 mm Tris, pH 8.0, 150 mm NaCl, 0.5% sodium deoxycholate, 1% Nonidet P-40, 0.1% SDS, 5 mm Na3VO4, 10 mm NaF, protease inhibitors (Roche Applied Science)). For cofilin localization in detergent-resistant and -soluble fractions, stimulated BMMCs were lysed in 250 mm Tris, pH 7.4, 2.5 mm MgCl2, 250 mm NaCl, 1 mm ATP, 1 mm EGTA, 1% Triton X-100, 5 mm Na3VO4, 10 mm NaF, protease inhibitors (Roche Applied Science) and cleared by centrifugation for 30 min at 13,000 rpm (4 °C) before centrifuging the supernatants for 2 h at 4 °C and 40,000 rpm (Beckman MAX-XP). Supernatants containing the detergent-soluble or cytoplasmic fraction were collected, and pellets, containing the detergent-resistant or cytoskeletal fraction, were washed with lysis buffer before being resuspended in SDS-PAGE loading buffer (29). For actin partitioning, SCF-stimulated BMMCs were lysed in ice-cold lysis buffer (10 mm imidazole, pH 7.0, 10 mm EGTA, 40 mm KCl, 1% Triton X-100, 5 mm Na3VO4, 10 mm NaF, protease inhibitors (Roche Applied Science)). Samples were centrifuged at 13,000 rpm for 2 min to isolate the Triton X-100-insoluble fraction, containing higher order networks/bundled F-actin (low speed pellet, LSP). Supernatants were further ultracentrifuged at 45,000 rpm for 20 min at 4 °C to obtain the noncross-linked F-actin (high speed pellet, HSP) and the Triton X-100-soluble fraction, containing soluble G-actin (high speed supernatant, HSS) (31, 32). Samples were resuspended in SDS-PAGE loading buffer.
Cell extracts for immunoprecipitation were prepared with Nonidet P-40 lysis buffer (20 mm Tris/HCl, pH 7.4, 20 mm MgCl2, 150 mm NaCl, 1% Nonidet P-40, 5 mm Na3VO4, 10 mm NaF, protease inhibitors (Roche Applied Science)). Extracts were precleared with protein G-agarose beads (Invitrogen) and subjected to immunoprecipitation with 3 μg of antibody. Samples were incubated for 2 h at 4 °C with gentle rocking before the addition of 30 μl of protein G-agarose beads followed by 1 h of incubation. Beads were washed three times and resuspended in SDS-PAGE loading buffer. Protein concentration was determined by the Bradford method using Roti-Quant (Carl Roth, Germany).
Pulldown Assays
For in vitro pulldown assays, 0.4 μm of purified GST-SWAP-70, GST-Rac1, GST-cofilin, or GST as control and 0.2–0.5 μm His-SWAP-70 were mixed in 20 mm Tris, pH 8.0, 100 mm NaCl, 10% glycerol, 1 mm EDTA, 1 mm MgCl2, 0.1% Nonidet P-40, 1 mm DTT, 0.2 mm PMSF (SWAP-70-cofilin) or 20 mm Tris, pH 7.5, 100 mm NaCl, 10% glycerol, 1 mm EDTA, 25 mm CaCl2, 0.1% Nonidet P-40, 1 mm DTT (SWAP-70/SWAP-70 interaction) and rocked for 1 h at 4 °C. Glutathione-agarose beads were then added and incubated with the samples for 30 min, washed, and resuspended in SDS-PAGE loading buffer. Samples were then subjected to SDS-PAGE and immunoblotting. Equal loading of GST proteins was determined by anti-GST blots or Coomassie staining.
Analytical Gel Filtration and Velocity Sedimentation
For oligomerization studies, recombinant purified SWAP-70, SWAP-70w/oCC, and SWAP-70 C-term, as well as protein standards (gel filtration markers kit for protein molecular mass of 29–700 kDa, Sigma), were analyzed individually by gel filtration at 60 ml/h flow on a Superdex 200 HPLC column (Amersham Biosciences). The elution volumes of the protein standards were fit to a linear regression with respect to their known Stokes radius (RS). Protein standards and their RS were as follows: carbonic anhydrase (2.01 nm), BSA (3.6 nm), alcohol dehydrogenase (4.6 nm), β-amylase (5.4 nm), and apoferritin (6.1 nm). The resulting straight-line equation was used to determine the RS for SWAP-70 and derivatives.
For velocity sedimentation studies, 30 μg of SWAP-70, BSA (4.6 S), β-amylase (9.2 S), and apoferritin (16.3 S) were loaded onto a 10-ml linear 15–40% glycerol gradients made in 20 mm Tris, pH 7.5, 100 mm NaCl, 1 mm EDTA, 25 mm CaCl2, 1 mm DTT. Gradients were centrifuged in a Beckman Coulter Optima LE-80K ultracentrifuge using an SW 40Ti, swinging bucket rotor at 32,000 rpm for 34 h. Fractions (0.4 ml) were collected from the bottom of the gradient, acetone-precipitated, and resolved in SDS-PAGE. Signals were quantified by densitometry using the Fiji software. Intensities were plotted by fraction and compared with protein standards to determine the relative sedimentation coefficient (s20,w referred here as S) of SWAP-70 according to Ref. 33. Smax was calculated with the formula Smax = 0.00361 M2/3 (33), where M is mass in daltons and Smax is expressed in Svedberg units (S). The Smax/S ratio to estimate protein globularity of proteins has been described elsewhere (33, 34). Native molecular weight was determined using the equation M = 4.205 (SRS) (33).
Pyrene-Actin Polymerization and Depolymerization
Protocols and buffers for in vitro actin studies have been described before (35, 36). Human platelet nonmuscle actin (APHL99; Cytoskeleton) was diluted in G buffer (2 mm Tris/HCl, pH 8.0, 0.5 mm DTT, 0.2 mm ATP, 0.1 mm CaCl2), allowed to depolymerize for 1 h on ice, spun for 30 min at 13,000 rpm (4 °C) to remove residual nondepolymerized filaments, and used for experiments. Polymerization and depolymerization reactions contained 2 μm nonmuscle actin doped with 10% rabbit muscle pyrene-actin (AKL99, Cytoskeleton). Prior to polymerization, calcium-actin was exchanged for magnesium-actin by incubating actin in 5 mm MgCl2, 20 μm EGTA for 2 min. Polymerization was induced by addition of volume of 10× KMEI (500 mm KCl, 10 mm MgCl2, 10 mm EGTA, 100 mm imidazole, pH 7.0) with or without buffer control or proteins to be tested and mixed immediately prior to the assay.
For depolymerization experiments, 2–3 μm actin (10% pyrene-actin) was polymerized for 3 h at 25 °C in the dark. To start depolymerization, F-actin was diluted to 0.5 or 0.1 μm with 1× KMEI (made in G-Mg buffer, G buffer with MgCl2 instead CaCl2) and test components and homogenized by gently pipetting two times. Time between mixing components and data collection was 20–25 s. Pyrene fluorescence was measured using a FLUORstar OPTIMA plate reader (BMG LABTECH) with excitation of 355 nm and emission of 405 nm at 25 °C. Fluorescence bleaching during the course of experiments was determined to be insignificant. Raw polymerization data were normalized subtracting the offset from zero and then dividing by the plateau value of actin alone. For the time to half-maximal polymerization (t½, max) determination, reactions were normalized by dividing their own plateau and solving for the time at half-maximal fluorescence (0.5 arbitrary units) in the linear range of the reactions (36). Raw disassembly data were normalized by dividing all data points by each reaction's offset. To visualize actin filaments, samples were fixed with Alexa Fluor 488/phalloidin (Invitrogen) 1:1 molar ration with F-actin, immediately diluted to 10 nm in fluorescence buffer (25 mm imidazole, pH 7.0, 25 mm KCl, 4 mm MgCl2, 1 mm EGTA, 100 mm DTT) (37), and spotted onto 0.1 mg/ml poly-l-lysine (Sigma)-coated coverslips using cut pipette tips to minimize shearing. Filaments were visualized using an epifluorescence microscope (Axiophot, Carl Zeiss, Germany) and a PL APO ×100 1.4 NA oil objective. Images were acquired at 25 °C with an AxioCam MRm camera using AxioVision software (version 4.4) and processed with the Fiji software.
Actin Filament Bundling
Low speed pelleting assays to assess F-actin bundling were performed as described (35). Briefly, actin (3 μm) was polymerized as for depolymerization assays; samples were mixed with test proteins, incubated for 30 min at room temperature, and pelleted at 13,000 rpm for 30 min at 4 °C. Supernatants were collected and precipitated with ice-cold acetone. Pellets, containing networked/bundled F-actin, were washed and resuspended in 1× SDS-PAGE loading buffer. Samples were analyzed on Coomassie-stained gels. Protein bands were quantified by densitometry using the Fiji software.
Actin Filament Binding and Fluorescence Quenching
High speed co-sedimentation assays were performed as described previously (35). F-actin was prepared at 2 μm final concentration as described above. Samples were mixed with test proteins, incubated for 30 min at room temperature, and pelleted at 45,000 rpm (125,000 × g) for 20 min at 4 °C in a Beckman MAX-XP ultracentrifuge. Supernatants were collected and precipitated with ice-cold acetone. Pellets were washed and resuspended in SDS-PAGE loading buffer. Samples were analyzed on Coomassie-stained gels. Control assays determining the amount of pelleted SWAP-70 in the absence of F-actin were performed, and the values were subtracted at each point before calculating dissociation equilibrium constants (Kd). For fluorescence quenching, 2 μm F-actin (10% pyrene-actin) was mixed with test components and carefully homogenized by pipetting twice with cut tips, and fluorescence was measured immediately as for polymerization assays. The delay between mixing of components and the first readout was 20–25 s. The effect of SWAP-70 on pyrene-actin and pyrene-labeled F-actin fluorescence was determined according to Ref. 36 by mixing a range of F-actin (10% pyrene-actin) concentrations with 0.5 μm SWAP-70 or SWAP-70's dialysis buffer, allowing it to equilibrate 24 h, and measuring fluorescence.
Nonmuscle Actin Preparation
Nonmuscle actin (β and γ) as complex with profilin was purified from pig spleen using poly-l-proline affinity chromatography as described before (38). One kg of pig spleen was homogenized using an immersion blender and suspended in H buffer (50 mm Tris, 5 mm EGTA, 2 mm DTT, 5 mm benzamidine, 0.5 mm ATP, 0.1 mm PMSF, 200 mg/ml sucrose, pH 8.0). After readjusting the pH, the suspension was centrifuged in an Avanti J-30I (JLA 16.250 rotor, Beckman Coulter, Fullerton, CA) for 30 min at 38,000 × g. The supernatant was centrifuged again in an Optima XPN-90 ultracentrifuge (Ti45 rotor, Beckman Coulter) for 30 min at 100,000 × g. The resulting supernatant was decanted carefully, filtered, and loaded overnight onto a chromatography column containing poly-l-proline peptides coupled to CNBr-Sepharose. The profilin-actin complexes were eluted from the column in P buffer (30 mm Tris, 100 mm KCl, 100 mm glycine, 1 mm DTT, 5 mm benzamidine, 30% DMSO, pH 7.4); the organic solvent was then separated by exhaustive dialysis against G buffer (5 mm Tris, 0.2 mm ATP, 0.5 mm DTT, 0.2 mm CaCl2, 0.1 mg/ml NaN3, pH 8.0) and gel filtration on a HiLoad 26/60 Superdex 75 HPLC column (GE Healthcare).
Total Internal Reflection Fluorescence Microscopy (TIRFM)
TIRFM with ATTO488-labeled nonmuscle actin was performed as described before (39). Images from an Olympus IX-81 inverted microscope equipped with an Apo N ×60 TIRF objective were captured every 2 s with 100–150-ms exposure times by a Hamamatsu Orca-R2 CCD camera (Hamamatsu Corp., Bridgewater, NJ). With a 2 × 2-binning mode, the pixel size corresponded to 0.22 μm. Right before coating, precleaned coverslips were extensively rinsed with 70 °C double distilled water and dried with N2. For coating, 2 mg/ml mPEG-silane MW 2,000 (Laysan Bio, Arab, AL) were dissolved in 80% ethanol at pH 2.0, adjusted with HCl. 300–400 μl of this solution were added on the coverslips, which were then dried at 65 °C for at least 12 h. Coverslips were rinsed carefully with filtered double distilled water and dried with N2 immediately prior the flow chambers preparation.
Elongation rates of filaments were calculated by manual tracking of growing barbed ends with ImageJ software after post-processing the time-lapse movies by background subtraction (30-pixel rolling ball radius). At least 15 filaments from three movies per condition were measured. Parallel and anti-parallel filament bundling was determined by following the time-resolved fusion of single filaments from at least three movies.
Electron Microscopy of Negatively Stained Actin Filament Bundles
F-actin was prepared and mixed with test proteins as described for bundling assays. Samples containing 2.1 μm F-actin and 1 μm SWAP-70 or 2 μm SWAP-70 C-term or buffer control were diluted 1:3 prior to experiments. Formvar-coated copper slot grids were placed onto 5-μl samples for 1 min, quickly rinsed in double-distilled water, stained with 2% uranyl acetate, and air-dried before analysis with a TECNAI12 transmission electron microscope (FEI) operated at 120 kV.
Immunofluorescence and Flow Cytometry
For BMMC immunostaining, cells were IL-3 starved overnight, plated on 20 μg/ml fibronectin-coated coverslips, allowed to adhere for 1 h at 37 °C and treated with 25 ng/ml SCF (conditioned medium) for 15 min. For intracellular staining, cells were fixed and permeabilized using a cytofix/cytoperm kit (BD Biosciences) according to the manufacturer's indications. Coverslips were mounted in Fluoromount-G (SouthernBiotech). Anti-rabbit Alexa Fluor 633 and anti-goat Alexa Fluor 594 secondary antibodies were from Invitrogen. Co-localization was determined with the Co-localization Threshold plugin (Fiji). This plugin calculates Pearson's R values that indicate co-localization significance (1 corresponding to a perfect co-localization and 0 to a complete exclusion).
For flow cytometry staining, cells were IL-3-starved overnight, then serum-starved for an additional 2 h, SCF-stimulated, and fixed with paraformaldehyde. Next, cells were permeabilized with 0.2% Triton X-100 in FACS buffer (2% FCS, 2 mm EDTA, PBS), stained with 22 nm rhodamine/phalloidin (Invitrogen), and analyzed in a LSRII (BD Biosciences). Data were analyzed using FlowJo software.
Actin Free Barbed-end Quantification
Actin free barbed ends were stained as follows. BMMCs were IL-3-starved overnight, plated on 20 μg/ml fibronectin-coated coverslips, allowed to adhere for 1 h at 37 °C, and treated with 25 ng/ml SCF (conditioned medium) for 15 min. Coverslips were quickly rinsed in warm PBS, permeabilized, and labeled with 0.4 μm Alexa Fluor 568-labeled actin in permeabilization buffer (20 mm HEPES, 138 mm KCl, 4 mm MgCl2, 3 mm EGTA, 0.2 mg/ml saponin, 1% BSA, 1 mm ATP, 3 μm phalloidin) for 45 s at 37 °C and immediately fixed with 4% paraformaldehyde in Kreb's S buffer (20 mm HEPES, 145 mm NaCl, 5 mm KCl, 1.2 mm CaCl2, 1.3 MgCl2, 1.2 NaH2PO4, 10 mm glucose, 0.4 m sucrose) for 10 min. F-actin was then stained with Alexa Fluor 488-phalloidin as described previously (40, 41). Images were acquired by confocal microscopy as described below. For quantification of actin free barbed ends and F-actin, background was subtracted first, and Alexa Fluor 568 (labeled barbed ends) or Alexa Fluor 488 (F-actin) pixel intensity was quantified (Fiji) on entire cells and normalized to each cell's area.
Microscopy and Image Analysis
Unless otherwise stated, samples were analyzed at room temperature using a Leica TCS SP5 confocal laser scanning microscope (Leica, Germany) and an HCX PL APO ×63 1.4 NA oil objective. Images were acquired in the linear range of the detector's response preventing pixel saturation with LAS AF software (Leica) and analyzed with the Fiji software. Figures were prepared with Adobe Photoshop and Illustrator. For acceptor photobleaching FRET, transfected NIH 3T3 cells were grown on poly-l-lysine-coated (100 μg/ml) coverslips, serum-starved for 2 h, stimulated with 15 nm EGF in DMEM, and immediately fixed with 4% paraformaldehyde, PBS for 10 min. Acceptor photobleaching FRET macro, LAS software (Leica), was employed to acquire images. FRET efficiency was determined using the pbFRET version 1 plugin (Dr. Mike Lorenz, MPI-CBG, Germany) on the ImageJ (National Institutes of Health) or Fiji software. This plugin calculates FRET efficiency (% FRET) by subtracting pixel-by-pixel images of donor pre-bleach from donor post-bleach, divided by donor post-bleach ((post-pre)/post), and normalizing in this way for expression levels.
Statistical Analysis
Statistical analysis and graphing were carried out with Prism 5 (GraphPad) and Numbers (Apple). Statistical significance was determined by paired two-tailed Student's t test (confidence interval 95%).
RESULTS
Dilution-induced F-actin Depolymerization Is Delayed by SWAP-70
SWAP-70 binds to nonmuscle F-actin (21, 22), yet the role of this interaction was previously unknown. In vitro His6-SWAP-70 (hereafter SWAP-70) binds to nonmuscle F-actin with an estimated Kd of 1.1 μm (Fig. 1A). From binding experiments we calculated that SWAP-70 binds F-actin at ∼2:1 (SWAP-70/actin) stoichiometry at saturation (Fig. 1A). To better understand the role of SWAP-70 in regulating the actin cytoskeleton, we first examined its effects on the rate of nonmuscle actin polymerization and depolymerization in vitro. SWAP-70 had no effects on the rate of actin polymerization in pyrene-actin assays (Fig. 1B). Compared with the curve's plateau of control reactions, a slight increase in fluorescence was observed in samples containing SWAP-70 likely because of the known association of SWAP-70 to F-actin and/or filament bundling/cross-linking. In another approach using TIRFM of freely diffusing and growing actin filaments, SWAP-70 also showed no effects on filament elongation and nucleation (Fig. 1, C and D; supplemental Movie S1). Interestingly, SWAP-70 caused actin filaments to firmly adhere to non-PEG-coated coverslips even at concentrations as low as 50 nm without changing the filament elongation and nucleation rates as visualized by TIRFM (supplemental Movie S1 and data not shown). Of note, PEG coating of glass surfaces, which was used for all subsequent TIRFM assays and prevented SWAP-70-mediated adhesion of F-actin to the slides, did not affect elongation rates of either control or test samples. In contrast to actin polymerization, when we evaluated dilution-induced F-actin disassembly in pyrene-actin assays, SWAP-70 markedly attenuated depolymerization in a concentration-dependent manner (Fig. 1E). Dilution of F-actin to a final concentration of 0.5 μm, which is the critical concentration of actin at the pointed ends (42), did not alter this effect (data not shown). SWAP-70 alone neither significantly quenched nor increased the fluorescence of pyrene-labeled G- or F-actin (data not shown). To confirm these results, F-actin induced to depolymerize in the presence or absence of SWAP-70 was stained with labeled phalloidin, spotted on coverslips, and observed by light microscopy. As expected, after F-actin depolymerization more long filaments were observed in samples containing SWAP-70 compared with the control (Fig. 1, F and G). Interestingly, in the presence of SWAP-70 filaments were frequently intertwined; thus, the filament length measured for these samples could also be regarded as the length of filament bundles or cross-linked filaments. Together, these results suggest that SWAP-70 attenuates F-actin depolymerization induced by dilution.
FIGURE 1.
SWAP-70 binds F-actin, delays F-actin depolymerization induced by dilution, and does not affect filament polymerization and elongation. A, binding of SWAP-70 to nonmuscle F-actin. Increasing concentrations of SWAP-70 (0.25, 0.5, 1, 2, and 4 μm) were incubated with 0.5 μm F-actin followed by a high speed pelleting assay. SWAP-70 bound to F-actin was quantified by densitometry of Coomassie-stained gels. The actual amount of F-actin (actin in pellet) per reaction was measured as 0.21 ± 0.017 μm. The average of four independent experiments is shown. Curve shows the best fit for the data. Error bars indicate ± S.E. B, SWAP-70 has no effects on in vitro pyrene-actin polymerization. 2 μm nonmuscle G-actin (10% pyrene-labeled) was polymerized in the presence or absence of SWAP-70. a.u., arbitrary units. C, spontaneous assembly of nonmuscle actin filaments (1.3 μm, 23% ATTO488-labeled) in the presence or absence of 0.5 μm SWAP-70 visualized by TIRFM. Red circles mark the barbed end of a growing filament. Time is in seconds (upper right corner). D, elongation rate of actin filaments is not affected by SWAP-70. At least 15 filaments from three movies per condition were measured. Error bars indicate ± S.D. E, time course of F-actin depolymerization induced by dilution at 0.1 μm in the presence of variable concentrations of SWAP-70. 2 μm F-actin was first mixed with SWAP-70 or buffer (negative control), incubated for 2 min, and then diluted 20-fold. F, visualization of F-actin from depolymerization experiments as in E. Representative images before and after F-actin depolymerization. Time is in minutes (upper right corner) after depolymerization. G, filament length was measured 25 min after F-actin depolymerization from experiments as in F. At least 50 filaments per condition were analyzed. Representative data of at least three independent experiments are shown. Scale bar, 10 μm. See also supplemental Movie S1.
SWAP-70 Bundles Actin Filaments through Its C-terminal Region
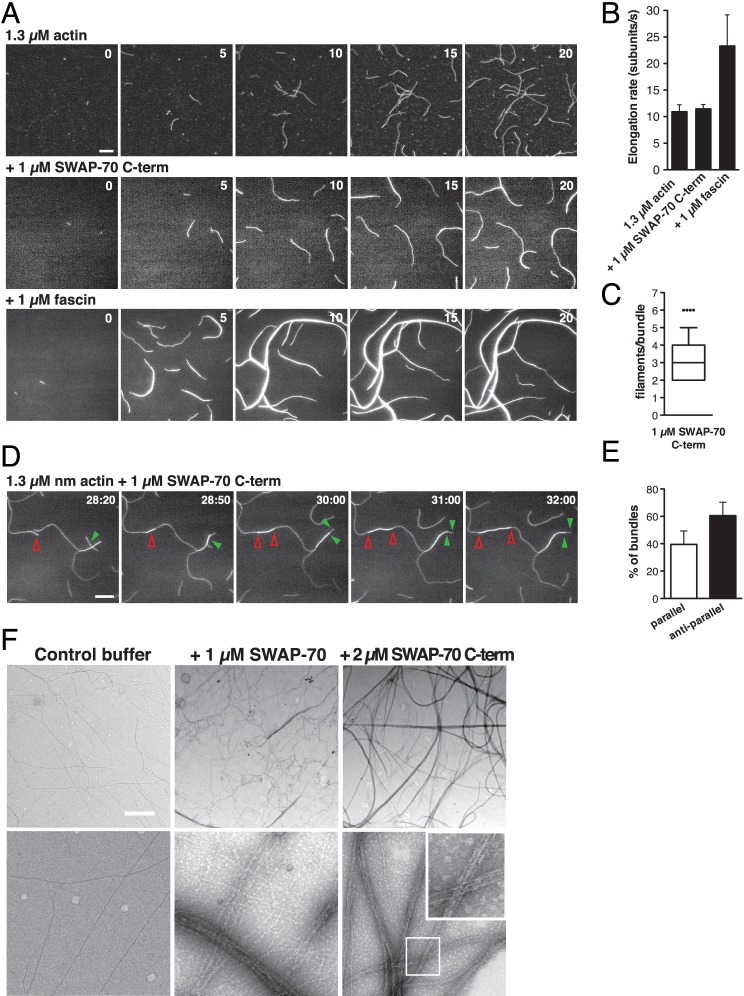
The fact that SWAP-70 binds F-actin together with our initial observations (see above) led us to ask whether SWAP-70 bundles/cross-links actin filaments. Low speed pelleting assays revealed that SWAP-70 significantly increased the amount of bundled/cross-linked actin filaments (Fig. 2, A–C). Previous studies identified a prominent F-actin-binding site near the C terminus of SWAP-70. This site was proposed to be masked in a folded, auto-inhibitory conformation of the full-length protein (20, 21). A truncated SWAP-70 carrying this F-actin-binding site and part of the coiled-coil region (His6-SWAP-70(444–585), hereafter SWAP-70 C-term) was sufficient and more efficient to bundle/cross-link actin filaments (Fig. 2, A–C), consistent with the higher binding affinity of SWAP-70 C-term for F-actin compared with the full-length protein (21). The increase in bundled/cross-linked F-actin was concomitant with increasing concentrations of SWAP-70 and SWAP-70 C-term (Fig. 2C). In these assays, an approximate ratio of 2:1 (SWAP-70 C-term/F-actin) was needed to bundle/cross-link all available F-actin, which in our assays corresponded to 87 ± 4% of the total actin. A maximum of 47.5 ± 3% of bundled/cross-linked F-actin was observed in the presence of SWAP-70 (Fig. 2C). The effects of increasing concentrations of F-actin on a constant amount of SWAP-70 or SWAP-70 C-term were comparable (data not shown). These observations were confirmed by light microscopy (Fig. 2D). Addition of SWAP-70 and SWAP-70 C-term caused the formation of thick and entangled filaments. Next, we performed TIRFM of growing filaments in the presence or absence of SWAP-70 C-term to observe F-actin bundling/cross-linking dynamics by real time imaging. Actin filaments that grew in the presence of SWAP-70 C-term elongated as control filaments and were frequently fused into bundles when they came in contact with an adjacent filament or bundle (Fig. 3, A and B and supplemental Movie S2), although not as efficiently as with the bundling protein fascin (30). Bundles formed in the presence of SWAP-70 C-term contained on average 3.3 ± 1.1 filaments after 30 min of actin polymerization (Fig. 3C). Analysis of the directionality of single filaments that incorporated into bundles revealed that SWAP-70 C-term generated bundles in a parallel and anti-parallel fashion, with a slight preference for the anti-parallel orientation (Fig. 3, D and E, and supplemental Movie S3). Finally, to further characterize the structure of the F-actin bundles generated by SWAP-70, electron micrographs were obtained from negatively stained F-actin with SWAP-70 or SWAP-70 C-term. Detailed observation revealed organized bundles consisting on average of four filaments per bundle (Fig. 3F), in line with our TIRFM data. These results establish SWAP-70 as an F-actin bundling protein and reveal that SWAP-70's C-terminal 144 residues are sufficient for F-actin bundling.
FIGURE 2.

SWAP-70 bundles actin filaments through its C-terminal region. A, SWAP-70 bundles actin filaments in low speed pelleting assays. 2 μm F-actin was mixed with 2 μm SWAP-70, 2 μm SWAP-70 C-term, or buffer (control). Supernatants (S) contain nonbundled F-actin, and pellets (P) bundled F-actin. Samples were analyzed on Coomassie-stained gels. B, quantification by densitometry of bundled actin (actin in pellet) from at least three experiments as in A. Error bars indicate ± S.D. C, quantification by densitometry of bundled F-actin in the presence of the indicated proteins after low speed pelleting assay. 1 μm F-actin was mixed with SWAP-70 or SWAP-70 C-term (0.25, 0.5, 1, 2, 4, and 8 μm) and processed as in A. The average of three independent experiments is shown. Curves show the best fit for the data. Error bars indicate ± S.E. D, visualization of bundled F-actin by fluorescent microscopy in the presence of the indicated proteins. Images are displayed using a 6_shades look-up table (LUT) in Fiji. Color calibration bar is shown on the right. Scale bar, 10 μm.
FIGURE 3.
SWAP-70-induced bundles contain parallel and anti-parallel actin filaments. A, dynamics of actin filament bundling by SWAP-70 C-term. Spontaneous polymerization of nonmuscle actin filaments (1.3 μm, 23% ATTO488-labeled) on mPEG-coated coverslips in the presence or absence of SWAP-70 C-term or fascin at the indicated concentrations. Time is in minutes. Scale bar, 10 μm. B, SWAP-70 C-term does not alter the elongation rate of growing actin filaments. At least 15 single, not bundled, filaments from three movies per condition were measured. Error bars indicate ± S.D. C, number of filaments per bundle in the presence of SWAP-70 C-term 30 min after polymerization. Filaments from at least 100 bundles from four movies were visually counted. Data are shown using a Box-and-Whisker plot as follows: box indicates 25th and 75th percentiles; error bars represent 10th and 90th percentiles; middle line is the median; dots are outlier values. D, SWAP-70 C-term bundles actin filaments in parallel and anti-parallel fashion visualized by TIRFM. Conditions are as in A. Arrowheads mark the bundling events and track barbed ends of filaments. Red open arrowheads mark anti-parallel and green solid arrowheads parallel growing filaments. Time in min/s. Scale bar, 10 μm. E, orientation of actin filaments in SWAP-70 C-term-induced bundles. At least 100 bundles from four movies were visually analyzed. F, electron microscopy of SWAP-70-induced F-actin bundles. F-actin was negatively stained with uranyl acetate. Samples contained 2.1 μm F-actin. Upper panels show low magnification; lower panels show high magnification. Inset shows an enlarged view of the white box. Scale bar, 100 nm. Representations of at least three independent experiments are shown. See also supplemental Movies S2 and S3.
SWAP-70 Oligomerization Mediates F-actin Bundling
To mediate F-actin bundling, a protein must contain at least two F-actin-binding domains or a single binding site and self-association capacity (43). The C-terminal region of SWAP-70 is sufficient to bind and bundle F-actin suggesting that SWAP-70 oligomerizes to bundle filaments. Thus, SWAP-70's capacity to oligomerize was examined. Pulldown assays with purified recombinant GST-SWAP-70 and SWAP-70 demonstrated interaction between these two proteins (Fig. 4A). Similarly, immunoprecipitation (IP) of Venus-SWAP-70 from transfected NIH 3T3 cells revealed its association with endogenous SWAP-70 (Fig. 4B). As an additional approach to study SWAP-70 oligomerization, we evaluated its hydrodynamic properties employing analytical gel filtration (size exclusion chromatography) and glycerol gradient velocity sedimentation with purified proteins (Fig. 4, C–E), and we calculated its native molecular weight. In gel filtration, SWAP-70 was eluted as a single peak with an experimentally calculated Stokes radius (RS) of 4.8 ± 0.14 nm (Fig. 4D). Determination of SWAP-70's sedimentation coefficient (S also known as s20,w), using velocity sedimentation on 15–40% glycerol gradients, revealed the presence of at least three distinct populations of SWAP-70 with peaks in fractions 9.7, 13.6, and 16.7 (Fig. 4E). Some protein was also observed in the last two fractions likely because of remaining protein that did not migrate into the gradient or stuck to the tube. With respect to known protein standards, the three prominent peaks had approximate S values of 14.2 S, 10.4 S, and 7.4 S, respectively. Compared with gel filtration, the identification of more protein populations in velocity sedimentation assays might reflect different stability of SWAP-70 oligomers under various experimental conditions, e.g. less stable under greater physical constraints in gel filtration. The native molecular masses (M = 4.205 (SRS) (33)) for the three SWAP-70 populations were determined to be 286 kDa (14.2 S), 209 kDa (10.4 S), and 148 kDa (7.4 S). Considering that the molecular mass of SWAP-70 obtained from the aa sequence is 71.6 kDa, these findings suggest the presence of tetramers, trimers, and dimers. The Smax/S ratios for the these three populations were 1.1, 1.2, and 1.4, respectively, indicating that SWAP-70 is a fairly globular protein (33).
FIGURE 4.
SWAP-70 forms oligomers. A, His-SWAP-70 binds GST-SWAP-70 in pulldown assays. 0.5 μm of His- and GST-SWAP-70 or GST control were mixed and incubated prior isolation of GST proteins. B, co-immunoprecipitation of endogenous SWAP-70 with Venus-SWAP-70. Protein extracts from NIH 3T3 cells expressing Venus-SWAP-70 were immunoprecipitated (IP) with anti-GFP or anti-IgG isotype control. Samples were resolved by SDS-PAGE and immunoblotted (IB). C, analytical gel filtration analysis of SWAP-70, SWAP-70 C-term, and SWAP-70w/oCC. Vertical offset chromatogram of the indicated recombinant purified proteins is shown. D, determination of SWAP-70's RS by gel filtration. Protein standards used to calibrate the column are shown. Experimentally determined RS values of the indicated proteins are shown in the table below. ± indicates S.D. E, determination of the sedimentation coefficient (S) of SWAP-70. 15–40% glycerol linear gradients were used in velocity sedimentation assays. Protein standards BSA (4.6 S), β-amylase (9.2 S), and apoferritin (16.3 S) were used to calculate SWAP-70's S value. Samples were collected from the bottom (fraction 1) to the top (fraction 25) of the gradient, precipitated, run in SDS-polyacrylamide gels, and immunoblotted (IB). Note that SWAP-70 shows at least three main peaks on fractions 9.7, 13.6, and 16.7 (from left to right in the chart), which correspond to estimated S values of 14.2 S, 10.4 S, and 7.4 S, respectively, suggesting the presence of tetramers, trimers, and dimers. The experimentally determined native molecular masses (see under “Experimental Procedures”) for SWAP-70 tetramers, trimers, and dimers are 286 kDa (14.2 S), 209 kDa (10.4 S), and 148 kDa (7.4 S), respectively. Theoretical molecular mass of SWAP-70 is 71.6 kDa. Svedberg units (S). Representatives of four independent experiments are shown. F, native gel electrophoresis of recombinant SWAP-70 reveals oligomers. 1 μg of SWAP-70 and 0.4 μg of SWAP-70 C-term were resolved and immunoblotted with anti-SWAP-70. Proteins were precleared by ultracentrifugation at 40,000 rpm for 30 min at 4 °C to remove possible protein aggregates. Note that the buffer conditions used for the gel filtration experiments vary from the ones used in these assays. Representatives of at least four independent experiments are shown. G, SWAP-70w/oCC binds F-actin similar to full-length SWAP-70. 2 μm F-actin was incubated with 2 μm SWAP-70 or 2 μm SWAP-70w/oCC followed by a high speed pelleting assay (co-sedimentation assay). Samples were analyzed on Coomassie-stained gels. Supernatants (S) contain G-actin, and pellets (P) contain F-actin together with F-actin-bound proteins. H, quantification by densitometry of proteins bound (protein in pellets) or not bound (protein in supernatants) to F-actin from at least four independent experiments as in G. Error bars indicate ± S.D. a.u., arbitrary units. I, SWAP-70w/oCC fails to bundle actin filaments in low speed pelleting assays. Conditions are as in Fig. 2A. J, quantification by densitometry of bundled actin (actin in pellet) from four independent experiments as in I. Error bars indicate ± S.D. Representatives of at least three independent experiments are shown.
The RS of SWAP-70 C-term determined by gel filtration was 3.8 ± 0.05 nm (Fig. 4D). This value is higher than the RS of the 66-kDa protein BSA (3.6 nm), suggesting that the size and mass of SWAP-70 C-term are reminiscent of a tetramer because SWAP-70 C-term's theoretical mass is 19 kDa. These findings reinforce the notion of the autoinhibitory state of the full-length protein, suggesting that other domains of SWAP-70, absent on SWAP-70 C-term, participate to regulate oligomerization of the protein. Further evidence of SWAP-70 oligomerization was obtained from immunoblots after native gel electrophoresis of recombinant SWAP-70 and SWAP-70 C-term, where at least two extra protein bands of molecular mass two to four times larger than the monomeric proteins were observed for SWAP-70 and SWAP-70 C-term (Fig. 4F). Together, these four independent observations suggest that SWAP-70 oligomerizes most likely into dimers and tetramers.
We hypothesized that the predicted C-terminal coiled-coil regions, spanning residues 312–529, are responsible for SWAP-70 oligomerization. To corroborate that oligomerization is responsible for F-actin bundling, we generated a deletion mutant lacking these coiled-coil regions (SWAP-70Δ312–529, hereafter SWAP-70w/oCC). Purified SWAP-70w/oCC had an estimated RS of 3.3 ± 0.06 nm as assessed by gel filtration (Fig. 4, C and D). Considering SWAP-70 as a globular protein, this value, lower than BSA's RS of 3.6 nm, is congruent with a protein of mass very close to the theoretical molecular mass of SWAP-70w/oCC (46 kDa). This observation posits SWAP-70w/oCC as a monomeric protein and suggests that the coiled-coil regions of SWAP-70 are responsible for oligomerization. Because SWAP-70 C-term spans aa 444–585 and presumably tetramerizes, we conclude that the region between aa 444 and 529 is critical for oligomerization. Next, we tested the F-actin binding and bundling activity of SWAP-70w/oCC in high and low speed pelleting assays, respectively. As expected, although this oligomerization-deficient mutant retained a similar F-actin binding capacity as full-length SWAP-70 (Fig. 4, G and H), it did not induce F-actin bundling (Fig. 4, I and J). Together, these results suggest that SWAP-70 bundles actin filaments by self-oligomerization, and we hypothesize that SWAP-70 exerts similar functions in vivo.
Altered Actin Cytoskeleton Dynamics of BMMCs in Absence of SWAP-70
Upon SCF stimulation, Swap-70−/− BMMCs show enhanced membrane protrusion activity and defects in homotypic adhesion and migration (28). To validate the in vivo functions of SWAP-70 in a cell type that is physiologically relevant for SWAP-70 and to explore the relevance of SWAP-70's cytoskeletal modulatory activity, we used murine BMMCs. Mast cells are known to utilize a highly organized actin cytoskeleton to achieve their surveillance and protective functions, which are fundamental for innate and adaptive immunity (44). We analyzed the amounts of bundled and free actin filaments in Swap-70+/+ (WT) and Swap-70−/− BMMCs using a previously described cell fractionation method (32). Compared with WT BMMCs, the amount of bundled F-actin (LSP) was significantly lower in Swap-70−/− cells (Fig. 5, A and B). Concomitant significant increases in G-actin (HSS) and noncross-linked F-actin (HSP) were observed in mutant cells. SCF stimulation for 15 min did not markedly change the amount of actin in any of the three protein fractions (LSP, HSS, and HSP) prepared under these conditions. As an independent assay to measure the amount of F-actin, in this case total F-actin (bundled and nonbundled), we performed flow cytometry of rhodamine/phalloidin-labeled BMMCs. Compared with WT, Swap-70−/− cells have significantly less F-actin revealed by the lower mean fluorescence intensity levels (Fig. 5, C and D). These results are in agreement with the in vitro F-actin bundling activity of SWAP-70 suggesting a corresponding function in vivo.
FIGURE 5.
SWAP-70 deficiency alters actin dynamics in SCF-stimulated BMMCs. A, SWAP-70 deficiency results in decreased bundled F-actin in BMMCs. Partitioning of actin into Triton X-100-insoluble LSP (higher order bundled F-actin), HSP (noncross-linked F-actin), and Triton X-100-soluble HSS (monomeric actin) is shown. Proteins were resolved by SDS-PAGE and immunoblotted. A representative experiment is shown. B, actin levels in each fraction were quantified by densitometry from four independent experiments as in A. Values were normalized to total actin (LSP+HSS+HSP) in WT 0 min SCF (referred as control) and expressed as percentage. The star indicates a p value <0.05 comparing the same fractions in WT and Swap-70−/−. Error bars indicate ± S.E. C, SWAP-70 deficiency leads to lower total F-actin levels in BMMCs. Flow cytometry histogram of cells stained for F-actin with rhodamine-phalloidin. Gray curve represents unstained cells. At least 10,000 cells per condition were recorded. D, F-actin mean fluorescence intensity (MFI) values from four independent experiments as in C were normalized to WT 0 min SCF and expressed in arbitrary units (a.u.). Error bars indicate ± S.D. E, visualization of actin free barbed ends in BMMCs. Cells were permeabilized and labeled for barbed ends (Alexa Fluor 568-actin). Samples were analyzed by confocal microscopy. Optical section thickness ∼1 μm. Scale bar, 10 μm. Representative cells are shown. F, quantification of actin free barbed ends from experiments as in E. 35–45 cells were analyzed per condition. Data are shown using a Box-and-Whisker plot as in Fig. 2E. WT (+/+) or Swap-70−/− (−/−). a.u., arbitrary units. Representative of at least three independent experiments are shown.
Next, we quantified actin free barbed ends by microscopy in WT and Swap-70−/− BMMCs as readout of active actin dynamics (45). Barbed-end numbers determined by confocal microscopy were similar in resting WT and Swap-70−/− BMMCs (Fig. 5, E and F). However, the number of barbed ends significantly decreased upon SCF stimulation in WT, whereas in mutant cells this reduction was considerably compromised. Collectively, these data reveal the in vivo relevance of SWAP-70 for maintaining balanced actin dynamics.
Cofilin Associates Directly with SWAP-70
We identified cofilin-1 (hereafter referred to as cofilin) as a putative interacting partner of SWAP-70 by mass spectrometry analyses of anti-SWAP-70 co-IPs from SCF-stimulated BMMCs (data not shown). This putative interaction became particularly interesting considering the defects of Swap-70−/− BMMCs on actin dynamics (Fig. 5) and the known functions of cofilin in the generation of barbed ends in vivo, along with its involvement in actin dynamics and directed cell migration (46, 47). To confirm this interaction, we performed IPs of endogenous cofilin from epidermal growth factor (EGF)-stimulated NIH 3T3 cells transfected with Venus-tagged SWAP-70, and we detected physical interaction (Fig. 6A). EGF stimulation was used because the interaction was initially identified in SCF-activated BMMCs, and the co-localization of SWAP-70 and cofilin in BMMCs significantly increases upon SCF stimulation (Fig. 6B). To further explore the interaction between SWAP-70 and cofilin, and to determine whether it is stimulus-dependent, acceptor photobleaching FRET assays were performed with NIH 3T3 cells transfected with Venus-SWAP-70 and Cerulean-cofilin. In starved cells, cofilin and SWAP-70 showed positive FRET signals, and this interaction increased over time after EGF stimulation (Fig. 6, C and D). Control cells transfected with only Venus and Cerulean showed negative FRET efficiencies (Fig. 6D). FRET between cofilin and SWAP-70 preferentially localized to areas close to the plasma membrane (Fig. 6E), which are rich in actin-remodeling proteins, including cofilin (48, 49). This suggests a regulated and functional interaction of both proteins.
FIGURE 6.
Cofilin associates directly with SWAP-70. A, co-immunoprecipitation of cofilin and Venus-SWAP-70. Protein extracts from NIH 3T3 cells expressing Venus-SWAP-70 were subjected to Co-IP with anti-cofilin or anti-IgG isotype control. Immunocomplexes were resolved by SDS-PAGE and immunoblotted (IB). B, co-localization of SWAP-70 and cofilin increases upon SCF stimulation in BMMCs. Immunofluorescence of WT BMMCs stimulated or not with SCF. Images were acquired by confocal microscopy. Pearson's R values were calculated as described under “Experimental Procedures” (a value of 1 indicates perfect co-localization). 50–65 cells were analyzed per condition. Representative cells are shown. ± indicates S.D. Scale bar, 10 μm. Representative of two independent experiments is shown. C–E, FRET interaction between Venus-SWAP-70 and Cerulean-cofilin increases after cytokine stimulation. NIH 3T3 cells expressing Venus-SWAP-70 and Cerulean-cofilin or controls were stimulated with 15 nm EGF and analyzed by acceptor photobleaching FRET. Fluorescence intensities were acquired before and after bleaching a region of interest (white box) with a 510 nm laser (100% power). FRET efficiency was determined as described under “Experimental Procedures.” Representative images for one cell 3 min after stimulation are shown. Scale bar, 10 μm. 20–30 cells were analyzed per condition. D, FRET efficiency quantification from experiments in C. Data were normalized for protein expression levels per cell. Error bars indicate ± S.D. E, Venus-SWAP-70 and Cerulean-cofilin interact more efficiently close to the plasma membrane. FRET efficiency is displayed as a three-dimensional surface plot. Inset shows the image used. A representative cell is shown. F, purified cofilin directly binds SWAP-70 in in vitro pulldown assays. GST-Rac1 was used as a positive control. G, quantification by densitometry of at least three independent experiments as in F. Error bars indicate ± S.D. H, binding affinity of SWAP-70 and cofilin in in vitro pulldown assays. Increasing concentrations of GST-cofilin (0.1–1.6 μm) were incubated with 0.4 μm SWAP-70. Samples were resolved and immunoblotted with anti-SWAP-70. Quantification by densitometry of three independent experiments. Curve shows the best fit for the data. Error bars indicate ± S.D. a.u., arbitrary units. Representative of at least three independent experiments are shown.
To investigate whether cofilin and SWAP-70 bind directly, we performed in vitro pulldown assays with purified proteins, and we observed that GST-cofilin and SWAP-70 directly associated (Fig. 6, F and G). Purified SWAP-70 has been reported to interact with Rac1 in in vitro pulldowns (50) and was used as a positive control in our assays. The estimated Kd value for the SWAP-70/cofilin interaction was calculated to be 0.26 μm (Fig. 6H). Together, our results provide evidence for a direct and functional interaction between SWAP-70 and cofilin, and they suggest that both proteins cooperate in regulating the actin cytoskeleton, particularly in response to cytokine stimuli.
SWAP-70 Hinders Cofilin Severing Activity on Actin Filaments
To understand the role of the interaction between SWAP-70 and cofilin, we tested whether SWAP-70 affects the binding of cofilin to F-actin in co-sedimentation assays. Addition of SWAP-70 caused a slight but statistically significant concentration-dependent increase in the amount of cofilin in the supernatant, i.e. not bound to F-actin (Fig. 7A). To corroborate these results, we determined whether addition of SWAP-70 counteracts the cofilin-induced fluorescence quenching that exists upon binding of cofilin to pyrene-labeled F-actin (12, 51). We observed that the cofilin-mediated fluorescence quenching was partially reverted by increasing concentrations of SWAP-70 (Fig. 7A, lower chart), as predicted from a displacement of cofilin from F-actin. SWAP-70 alone did not significantly affect pyrene-actin fluorescence (data not shown).
FIGURE 7.
Cofilin activity is moderately diminished by SWAP-70 in vitro. A, SWAP-70 displaces cofilin from F-actin. 2 μm F-actin (10% pyrene-actin) was mixed with SWAP-70 (0.1, 0.25, 0.5, and 1 μm), incubated for 10 min, and mixed with 2 μm cofilin followed by a high speed pelleting assay. Supernatants (S) and pellets (P) were analyzed on Coomassie-stained gels and quantified by densitometry. Soluble cofilin (not bound to F-actin) in the control without SWAP-70 was subtracted from every data point and plotted as percentage of total cofilin. One star indicates a p value <0.05; two stars indicate a p value <0.01. Error bars indicate ± S.E. Data from three independent experiments are shown. Below, SWAP-70 counteracts the pyrene fluorescence quenching caused by cofilin. Conditions as in A, but after cofilin addition pyrene fluorescence was immediately measured. The fluorescence value after cofilin addition was subtracted from the value before cofilin addition and normalized to the control without SWAP-70 (arbitrarily set to 0 for plotting purposes). Representative of three independent experiments is shown. B and C, SWAP-70 partially prevents the effects of cofilin on F-actin depolymerization. F-actin depolymerization induced by dilution in the presence of SWAP-70 and/or cofilin. B, F-actin was mixed with SWAP-70, incubated 10 min, and diluted 4-fold with or without cofilin. C, magnesium-actin was polymerized in the presence of SWAP-70 or buffer for 3 h and then diluted as in B. D, visualization of F-actin from the experiment in C. At least 10 random images per condition were acquired. Representative images are shown. Time is in minutes. Scale bar, 10 μm. E, modest decrease on cofilin severing activity under actin polymerization conditions in the presence of SWAP-70. Time course of actin polymerization in the presence of cofilin and SWAP-70. a.u., arbitrary units. F, SWAP-70 C-term slightly decreases cofilin severing activity under TIRFM polymerization conditions. Proteins were preincubated for 1 min before the addition of nonmuscle G-actin. Fascin was used as control. Time is in minutes. Scale bar, 10 μm. G, quantification of the number of free filaments 15 min after polymerization began. Shown are the fold increases between two conditions. All single, free filaments in the field of view (0.01 × 0.01 cm) from three movies per condition were counted. Error bars indicate ± S.D. See also supplemental Movie S4.
Based on these results, we hypothesized that SWAP-70 affects cofilin activity by interfering with its binding to F-actin. To test this hypothesis, we analyzed the effect of cofilin on dilution-induced F-actin depolymerization in the presence of SWAP-70. We considered the results of cofilin-mediated quenching (described above) to discern between the loss of fluorescence due to quenching and disassembly in our depolymerization experiments. The concentration of cofilin used for depolymerization (0.1–0.2 μm) only caused minimal quenching (∼10%), consistent with other reports (12). As expected, cofilin increased the rate of F-actin depolymerization (Fig. 7B), interpreted as the drop in fluorescence. This increase in depolymerization was partially prevented by the presence of SWAP-70. In a second approach, actin was polymerized in the presence or absence of SWAP-70 and then induced to depolymerize by dilution with or without cofilin. In agreement with our previous results, in the presence of SWAP-70 F-actin showed little depolymerization upon addition of cofilin, whereas in the absence of SWAP-70, cofilin enhanced F-actin depolymerization (Fig. 7C). Samples from these reactions were stained with labeled phalloidin and were analyzed by microscopy. Addition of cofilin to F-actin yielded abundant and very short filaments, as expected from cofilin activities, whereas addition of cofilin to SWAP-70-bundled F-actin mainly resulted in long filaments with the sporadic presence of short ones (Fig. 7D). The effects of SWAP-70 in these experiments might be complex as SWAP-70 interferes with F-actin depolymerization through bundling, affects cofilin binding to F-actin, and directly interacts with cofilin. In an attempt to discern these possibilities, we first used pyrene-actin polymerization assays in the presence of cofilin to address whether the observed effects of SWAP-70 on cofilin activity reflect reduced filament severing. This approach is exclusively sensitive to severing, as barbed ends newly generated by cofilin serve as seeds for new filaments to grow, thus greatly increasing the rate of filament assembly (10, 52). Similar to yeast cofilin-1 (12), cofilin increased the rate of actin polymerization in a concentration-dependent manner (data not shown). However, SWAP-70 modestly reduced cofilin severing under these conditions (Fig. 7E). We then used TIRFM as an independent method to visualize actin filament severing in the presence of cofilin and SWAP-70. In our TIRFM setup cofilin at 100 nm efficiently severed actin filaments. Under TIRFM polymerization conditions, SWAP-70 had no significant effect on filament severing (data not shown), suggesting that SWAP-70 cannot significantly alter cofilin severing at least under these experimental conditions.
To further investigate whether newly formed SWAP-70-bundled actin filaments are permissive to cofilin severing, we polymerized actin in the presence of cofilin and SWAP-70 C-term under TIRFM conditions. Bundle formation by SWAP-70 C-term was not considerably altered in the presence of cofilin, similar to fascin-induced bundles (Fig. 7F and supplemental Movie S4). To determine filament severing, we counted the number of free filaments as a marker for cofilin activity (53) 15 min after polymerization began. In the presence of cofilin, the number of free filaments increased 3.7-fold compared with the actin control (Fig. 7G). A higher increase of 4.4-fold was observed when samples contained fascin, which is in line with the fascin-mediated enhancement of cofilin severing (30). Interestingly, the increase in the number of free filaments in the presence of SWAP-70 C-term was only 1.7-fold, which is two times lower than the control (Fig. 7G). This suggests reduced severing by cofilin or an increased filament/bundle re-annealing rate. In light of the weaker bundling activity of SWAP-70 C-term compared with fascin, we favor reduced severing to cause this effect, contrasting fascin. Of note, however, as with the fascin control, SWAP-70 C-term-induced bundles thickened faster in the presence of cofilin indicating residual severing within bundles (30). These results show a partial reduction of cofilin severing activity, which might be more pronounced in steady state SWAP-70-induced F-actin bundles, similar to the effects observed in our depolymerization experiments (Fig. 7, B–D). Together, our data suggest bundling to be the most likely cause of SWAP-70's effects on cofilin activity, similar to spontaneously formed bundles (54) and other bundling proteins (53), and reveal a complex functional interplay among F-actin, cofilin, and SWAP-70. Based on our findings, we speculate that SWAP-70 also exerts additional function(s) on cofilin in vivo, particularly upon cytokine stimuli, by mechanisms that we cannot fully recapitulate in vitro.
Lack of SWAP-70 in BMMCs Results in Deregulation of Cofilin Activity
To assess the in vivo impact of SWAP-70 deficiency on cofilin activity, we first determined the levels of phosphorylated cofilin (Ser(P)-3) using total cell extracts of SCF-stimulated WT or Swap-70−/− BMMCs. Phosphorylation of cofilin in WT cells decreased after 5 min of SCF stimulation, remained low until 15 min, and then rose by 30 min to the levels in resting cells (Fig. 8, A and B). However, in Swap-70−/− BMMCs cofilin phosphorylation did not change upon stimulation and was significantly higher at 15 min, while in resting cells the levels were similar to WT. Cofilin protein and mRNA levels were similar in WT and Swap-70−/− BMMCs (Fig. 8A, middle blot, and data not shown). A similar difference in cofilin phosphorylation levels was independently observed when cofilin was immunoprecipitated from Nonidet P-40 lysates of BMMCs (Fig. 8C). In these lysates, mostly detergent-soluble proteins are isolated, whereas in total cell extracts (Fig. 8A), detergent-soluble along with detergent-insoluble membrane- and cytoskeleton-associated proteins are obtained; thus, Nonidet P-40 lysates are enriched in the pool of detergent-soluble cofilin. SCF stimulation caused significant cofilin dephosphorylation (∼3.3-fold) in WT but not in Swap-70−/− BMMCs, which instead showed a ∼1.5-fold increase (Fig. 8C). A similar aberrant cofilin dephosphorylation was observed in Swap-70−/− mouse embryonic fibroblasts stimulated with EGF (data not shown). These data suggest that SWAP-70 is involved in the cytokine-dependent regulation of cofilin activity.
FIGURE 8.
Cofilin activity is misregulated in BMMCs in the absence of SWAP-70. A, reduced cofilin dephosphorylation in response to SCF in Swap-70−/− BMMCs. Immunoblots (IB) of total cell extracts resolved by SDS-PAGE. p-cofilin (phospho-cofilin). B, quantification by densitometry of p-cofilin levels from 2 to 4 independent experiments as in A. p-cofilin was plotted relative to the control WT 0 min SCF set arbitrarily as 1. a.u., arbitrary units. C, reduced cofilin dephosphorylation in Swap-70−/− BMMCs. Immunoprecipitation of soluble cofilin from Nonidet P-40 lysates of SCF-stimulated BMMCs. Immunocomplexes were analyzed by immunoblot with anti-p-cofilin and then re-probed with anti-cofilin to determine total cofilin amounts. p-cofilin/cofilin (p-cof/cof) ratio is shown and was determined by densitometry. A representative of two independent experiments is shown. D, lack of SWAP-70 causes cofilin to aberrantly distribute into Triton X-100-insoluble, cytoskeletal protein fractions. Cell fractionation into Triton X-100-soluble (cytoplasmic) and Triton X-100-insoluble (cytoskeletal/membrane) fractions. Proteins resolved by SDS-PAGE were immunoblotted. A representative experiment is shown. Protein levels are shown in bar charts below the immunoblots. Cofilin/tubulin and SWAP-70/tubulin ratios from four independent experiments were quantified by densitometry and were plotted relative to cytoplasmic WT 0 min SCF set arbitrarily as 1. E, localization of cofilin to F-actin rich regions is increased in Swap-70−/− BMMCs. Cells were immunostained and analyzed by confocal microscopy. Representative cells are shown. Scale bar, 10 μm. F, Pearson's R values (co-localization index) were calculated for cofilin/F-actin and Arp2/F-actin as described under “Experimental Procedures.” 85–100 cells were analyzed per condition. WT (+/+) or Swap-70−/− (−/−) are shown. Error bars indicate ± S.D. Representative of at least three independent experiments are shown.
The subcellular localization of SWAP-70 changes upon cell stimulation in some cell types (20, 21, 29). Based on our biochemical evidence, we hypothesized that SWAP-70 deficiency alters cofilin subcellular distribution upon cytokine stimulation in BMMCs. In WT cells, SCF stimulation triggered enrichment of detergent-soluble, cytoplasmic cofilin concomitant with a similar increase of detergent-soluble SWAP-70 (Fig. 8D). Conversely, SCF stimulation of Swap-70−/− cells induced a significant decrease in the amount of soluble cofilin, concomitant with a slight increase in the amount of detergent-insoluble, cytoskeletal/membrane-associated cofilin. This enrichment of insoluble cofilin occurred only in Swap-70−/− cells. WT BMMCs also showed reduction of cytoskeletal/membrane-associated SWAP-70 after SCF stimulation. Both proteins showed comparable SCF-mediated redistribution into detergent-soluble and -insoluble fractions, which was abrogated in the absence of SWAP-70. These observations suggest similar subcellular dynamics of cofilin and SWAP-70 in response to SCF stimulation, which could involve a direct protein/protein interaction because their co-localization is cytokine-dependent and preferentially occurs at the cell periphery (Fig. 6, B, D, and E). In line with this hypothesis, the co-localization between cofilin and F-actin in resting and SCF-stimulated BMMCs was significantly higher in Swap-70−/− cells as assessed by immunofluorescence (Fig. 8, E and F). Of note, this co-localization was mostly confined to the cell periphery. As reference, the co-localization of F-actin with Arp2, a subunit of the Arp2/3 complex, was not significantly affected in the absence of SWAP-70 (Fig. 8F). These observations suggest that regardless of the stimulation SWAP-70 deficiency causes increased association of cofilin with the cytoskeletal and/or membrane compartments.
Cofilin activity is regulated in a local activation/global inhibition fashion (15, 48). Thus, a commonly used surrogate marker for cofilin activity in vivo is the generation of actin free barbed ends (46, 55), although barbed ends might also arise from other mechanisms (45). Compared with WT cells, Swap-70−/− BMMCs are impaired in reducing the number of actin free barbed ends upon SCF stimulation (Fig. 5F), consistent with an aberrant regulation of cofilin activity in the absence of SWAP-70. Collectively, our results suggest that SWAP-70 is involved in regulating the activity and subcellular distribution of cofilin upon SCF stimulation in BMMCs.
DISCUSSION
Actin dynamics is a multifactorial process involving the interplay of numerous actin-binding proteins and protein complexes that provide the precise spatiotemporal control of the actin cytoskeleton necessary for cell motility, adhesion, and polarization. The complex regulation of these activities and how they dynamically organize and maintain actin filaments into specialized higher order structures are not completely understood. Although previous studies have proposed SWAP-70 to regulate cytoskeletal F-actin rearrangements (20, 21, 26, 28), the mechanism of this control remained unknown. In this study, we describe how SWAP-70 contributes to the control of actin dynamics. We demonstrate the following: (i) SWAP-70 bundles actin filaments and delays their depolymerization, and (ii) it is involved in modulating cofilin activity upon cytokine stimulation in BMMCs likely by fine-tuning cofilin association with the cytoskeletal/membrane compartment.
SWAP-70 Oligomerizes to Bundle Actin Filaments
Our data show that SWAP-70 restricts F-actin depolymerization. Several lines of evidence reveal that SWAP-70 bundles actin filaments via its C-terminal region, which contains the F-actin-binding site. Using TIRFM, we showed that SWAP-70-induced bundles contain on average four filaments that are assembled in parallel and/or anti-parallel fashion. The single F-actin-binding site on SWAP-70's C terminus and the presence of a C-terminal coiled-coil region suggested oligomerization as the mechanism for bundling, similar to α-actinin (56) and EPLIN (57). This hypothesis is further supported by the identification of SWAP-70 and SWAP-70 C-term oligomers in gel filtration and velocity sedimentation experiments in near-physiological buffer conditions. In addition, SWAP-70 oligomers were isolated from cells and by pulldown experiments in vitro (Fig. 4). An oligomerization-deficient mutant of SWAP-70 lacking the acidic C-terminal stretch of the tri-partite coiled-coil region (SWAP-70w/oCC) fails to bundle actin filaments while still retaining F-actin binding, confirming that oligomerization of SWAP-70 mediates F-actin bundling. The region of SWAP-70 we suggest to be critical for oligomerization locates between aa 444 and 525. This stretch has a pI of 4.8 and is rich in glutamine and glutamic acid, which together account for 41% of the 81 aa. Glutamine-rich regions were described as modules involved in multimerization of proteins even if present in coiled-coil regions (58), and the only protein closely related to SWAP-70, DEF6, also harbors a Gln/Glu-rich (31%) stretch suggested to be involved in its multimerization, which, however, is functionally different and not related to F-actin dynamics (59).
In vivo, SWAP-70 deficiency results in less bundled and total F-actin levels in BMMCs (Fig. 5), providing evidence for SWAP-70's bundling activity in cells. In addition, SWAP-70 shows stimulation-dependent localization to membrane ruffles, lamellipodia, and loose actin filament arrays (20, 21). Likewise, an overexpressed SWAP-70 C-terminal fragment (aa 448–585) localized to cortical actin and stress fibers in COS7 cells (21). These subcellular localizations are consistent with the role of SWAP-70 in bundling and are also observed for the bundling protein α-actinin (56). Thus, we conclude that SWAP-70 oligomerizes, likely forming dimers and/or tetramers, and thereby bundles actin filaments in vitro. Dimerization and tetramerization, as observed in vitro, may also occur in vivo and is subject to future studies. We suggest that post-translational modifications and/or other regulatory mechanisms enable SWAP-70 to oligomerize possibly by changing its tertiary structure and thus control its functions. Binding of SWAP-70 to phosphatidylinositol 3,4,5-trisphosphate, Rac1, or other protein(s) or phosphorylation may thus regulate the bundling activity by exposing the auto-inhibited C-terminal F-actin-binding site as well as the oligomerization domain. In B cells, phosphorylation of SWAP-70 at Tyr-517 diminishes but does not abolish its association with F-actin (29), seemingly regulating, at least in part, the dissociation rate of SWAP-70 from F-actin. SWAP-70's bundling activity might be regulated by similar mechanisms in BMMCs.
SWAP-70 delays dilution-induced F-actin depolymerization in a concentration-dependent manner. Similar results were obtained with SWAP-70 C-term (data not shown), which is sufficient to bundle filaments. We favor filament stabilization through bundling to be the main mechanism because SWAP-70 does not bind barbed ends, as revealed by the unaffected actin polymerization and filament elongation rates in the presence of the protein (Fig. 1). However, pointed end binding cannot be completely excluded. SWAP-70-induced filament bundling most likely reduces monomer release from filament ends due to inter-filament cross-linking, hence slowing down F-actin depolymerization similar to EPLIN (57), fascin (30), and other bundling proteins (53).
SWAP-70 Contributes to Cofilin Regulation in BMMCs
We demonstrate here a previously unknown interaction between SWAP-70 and cofilin. Our data suggest that this is a direct interaction that requires cytokine stimulation to occur in cells, where the complex preferentially localizes close to the plasma membrane. Our biochemical studies on the role of the interaction provide evidence that SWAP-70 diminishes cofilin activity on F-actin under our polymerization and depolymerization conditions (Fig. 7). SWAP-70 causes displacement of cofilin from F-actin (Fig. 7A), similar to the competition of yeast Coronin-1B with cofilin (12). This may provide a mechanistic explanation for the inhibitory effects of SWAP-70 on cofilin. Considering our in vitro and in vivo data, the effects we observed on cofilin activity may be due to filament bundling by SWAP-70. In support of this, although SWAP-70 C-term reduces cofilin activity (Fig. 7, F and G), it seems not to bind cofilin directly in vitro in pulldown assays (data not shown). Alternatively, and not necessarily mutually exclusive, SWAP-70 might displace cofilin from actin filaments through a direct interaction rather than by competition with cofilin for F-actin binding.
Our in vivo data show a cytokine-dependent interaction between cofilin and SWAP-70. This suggests that the interaction might be more complex than we can possibly recreate in vitro, and it may involve regulatory events that promote the proteins to interact in certain cellular compartments, such as the membrane/proximal interactions we observe. We hypothesize that as for F-actin bundling, cofilin binding is regulated through a conformational change of SWAP-70 triggered via signaling pathways downstream of cytokine stimuli. SWAP-70 deficiency in BMMCs causes abnormal regulation of cofilin activity evidenced by impaired spatial distribution of cofilin, altered free barbed end dynamics, and decreased cofilin dephosphorylation upon SCF stimulation. Although other mechanisms may generate free barbed ends as well (45), considering our results showing a direct, functional interaction between cofilin and SWAP-70, we favor cofilin activity's misregulation in the absence of SWAP-70 to be the cause for the free barbed-end phenotype. The aberrant subcellular distribution of cofilin in Swap-70−/− BMMCs may trigger abnormal regulation of cofilin activity by altering cofilin's precise spatial and temporal regulation cycle. Coronin 1B regulates cofilin directly by controlling cofilin's spatial distribution and function on actin filaments according to their nucleotide state (12). SWAP-70 may participate in the control of cofilin activity following a similar type of spatial regulation. Direct binding of SWAP-70 to cofilin could, to some extent, regulate cofilin localization in cells, similar to tropomyosin (60). In the absence of SWAP-70, the stimulus-dependent SWAP-70-mediated spatial targeting of cofilin is likely impaired, exposing the abnormally distributed cofilin to aberrant regulation. Likewise, the F-actin bundling activity of SWAP-70 may not be ruled out as participating directly or indirectly in these processes, for example, by shaping the F-actin network architecture and thus controlling accessibility of cofilin to such structures. Other proteins that interact with SWAP-70, such as RhoGTPases, which regulate kinases and phosphatases that in turn control cofilin activity, may also affect these processes. The extensive in vitro evidence presented in this study, however, supports the notion that SWAP-70 regulates cofilin's spatial distribution primarily through direct binding and SWAP-70-mediated F-actin bundling.
SWAP-70 is involved in polarization, adhesion, and migration (28, 29, 61), which heavily depend on F-actin reorganization. The functions of SWAP-70 described here may explain the respective deficiencies observed in Swap-70−/− cells, because many F-actin-stabilizing proteins play fundamental roles in migration, polarization, and adhesion (7, 8, 62, 63). Supporting this concept, the importance of anti-parallel bundling proteins, of which SWAP-70 may be considered a new member, has recently been highlighted by the finding that myosin activity on actin networks, which is vital for cell migration, depends on filament orientation; anti-parallel filaments are the preferred substrate for contraction and disassembly (64).
Acknowledgments
We thank Dr. Attila Toth (Technische Universität Dresden, Dresden, Germany) for providing the gateway system's plasmids and reagents; Dr. Michaël Chopin for fruitful discussions; Dr. Tatsiana Ripich and Dr. François McNicoll for critical reading of the manuscript; and the Jessberger laboratory members for constructive discussions and criticism. We are thankful to the reviewers for comments and input, which notably improved this manuscript.
This work was supported by Deutsche Forschungsgemeinschaft Grants SPP1394 (to R. J.) and SPP1464 (to J. F.).

This article contains supplemental Movies 1–4.
- aa
- amino acid
- SCF
- stem cell factor
- BMMC
- bone marrow-derived mast cell
- IP
- immunoprecipitation
- TIRFM
- total internal reflection fluorescence microscopy
- LSP
- low speed pellet
- HSP
- high speed pellet
- HSS
- high speed supernatant
- C-term
- C terminus.
REFERENCES
- 1. Pollard T. D., Borisy G. G. (2003) Cellular motility driven by assembly and disassembly of actin filaments. Cell 112, 453–465 [DOI] [PubMed] [Google Scholar]
- 2. Pollard T. D., Cooper J. A. (2009) Actin, a central player in cell shape and movement. Science 326, 1208–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Le Clainche C., Carlier M. F. (2008) Regulation of actin assembly associated with protrusion and adhesion in cell migration. Physiol. Rev. 88, 489–513 [DOI] [PubMed] [Google Scholar]
- 4. Vignjevic D., Yarar D., Welch M. D., Peloquin J., Svitkina T., Borisy G. G. (2003) Formation of filopodia-like bundles in vitro from a dendritic network. J. Cell Biol. 160, 951–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Stevenson R. P., Veltman D., Machesky L. M. (2012) Actin-bundling proteins in cancer progression at a glance. J. Cell Sci. 125, 1073–1079 [DOI] [PubMed] [Google Scholar]
- 6. Adams J. C. (2004) Roles of fascin in cell adhesion and motility. Curr. Opin. Cell Biol. 16, 590–596 [DOI] [PubMed] [Google Scholar]
- 7. Yamakita Y., Matsumura F., Lipscomb M. W., Chou P. C., Werlen G., Burkhardt J. K., Yamashiro S. (2011) Fascin1 promotes cell migration of mature dendritic cells. J. Immunol. 186, 2850–2859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hamill K. J., Hopkinson S. B., Skalli O., Jones J. C. (2012) Actinin-4 in keratinocytes regulates motility via an effect on lamellipodia stability and matrix adhesions. FASEB J. 27, 546–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bamburg J. R., McGough A., Ono S. (1999) Putting a new twist on actin: ADF/cofilins modulate actin dynamics. Trends Cell Biol. 9, 364–370 [DOI] [PubMed] [Google Scholar]
- 10. Andrianantoandro E., Pollard T. D. (2006) Mechanism of actin filament turnover by severing and nucleation at different concentrations of ADF/cofilin. Mol. Cell 24, 13–23 [DOI] [PubMed] [Google Scholar]
- 11. Frantz C., Barreiro G., Dominguez L., Chen X., Eddy R., Condeelis J., Kelly M. J., Jacobson M. P., Barber D. L. (2008) Cofilin is a pH sensor for actin free barbed end formation: role of phosphoinositide binding. J. Cell Biol. 183, 865–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gandhi M., Achard V., Blanchoin L., Goode B. L. (2009) Coronin switches roles in actin disassembly depending on the nucleotide state of actin. Mol. Cell 34, 364–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. van Rheenen J., Song X., van Roosmalen W., Cammer M., Chen X., Desmarais V., Yip S. C., Backer J. M., Eddy R. J., Condeelis J. S. (2007) EGF-induced PIP2 hydrolysis releases and activates cofilin locally in carcinoma cells. J. Cell Biol. 179, 1247–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Condeelis J. (2001) How is actin polymerization nucleated in vivo? Trends Cell Biol. 11, 288–293 [DOI] [PubMed] [Google Scholar]
- 15. van Rheenen J., Condeelis J., Glogauer M. (2009) A common cofilin activity cycle in invasive tumor cells and inflammatory cells. J. Cell Sci. 122, 305–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sun C. X., Magalhães M. A., Glogauer M. (2007) Rac1 and Rac2 differentially regulate actin free barbed end formation downstream of the fMLP receptor. J. Cell Biol. 179, 239–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chesarone M. A., Goode B. L. (2009) Actin nucleation and elongation factors: mechanisms and interplay. Curr. Opin. Cell Biol. 21, 28–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Borggrefe T., Wabl M., Akhmedov A. T., Jessberger R. (1998) A B-cell-specific DNA recombination complex. J. Biol. Chem. 273, 17025–17035 [DOI] [PubMed] [Google Scholar]
- 19. Borggrefe T., Masat L., Wabl M., Riwar B., Cattoretti G., Jessberger R. (1999) Cellular, intracellular, and developmental expression patterns of murine SWAP-70. Eur. J. Immunol. 29, 1812–1822 [DOI] [PubMed] [Google Scholar]
- 20. Hilpelä P., Oberbanscheidt P., Hahne P., Hund M., Kalhammer G., Small J. V., Bähler M. (2003) SWAP-70 identifies a transitional subset of actin filaments in motile cells. Mol. Biol. Cell 14, 3242–3253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ihara S., Oka T., Fukui Y. (2006) Direct binding of SWAP-70 to nonmuscle actin is required for membrane ruffling. J. Cell Sci. 119, 500–507 [DOI] [PubMed] [Google Scholar]
- 22. Wakamatsu I., Ihara S., Fukui Y. (2006) Mutational analysis on the function of the SWAP-70 PH domain. Mol. Cell. Biochem. 293, 137–145 [DOI] [PubMed] [Google Scholar]
- 23. Borggrefe T., Keshavarzi S., Gross B., Wabl M., Jessberger R. (2001) Impaired IgE response in SWAP-70-deficient mice. Eur. J. Immunol. 31, 2467–2475 [DOI] [PubMed] [Google Scholar]
- 24. Chopin M., Chacón-Martínez C. A., Jessberger R. (2011) Fine tuning of IRF-4 expression by SWAP-70 controls the initiation of plasma cell development. Eur. J. Immunol. 41, 3063–3074 [DOI] [PubMed] [Google Scholar]
- 25. Ocana-Morgner C., Wahren C., Jessberger R. (2009) SWAP-70 regulates RhoA/RhoB-dependent MHCII surface localization in dendritic cells. Blood 113, 1474–1482 [DOI] [PubMed] [Google Scholar]
- 26. Pearce G., Angeli V., Randolph G. J., Junt T., von Andrian U., Schnittler H. J., Jessberger R. (2006) Signaling protein SWAP-70 is required for efficient B cell homing to lymphoid organs. Nat. Immunol. 7, 827–834 [DOI] [PubMed] [Google Scholar]
- 27. Ripich T., Jessberger R. (2011) SWAP-70 regulates erythropoiesis by controlling α-4 integrin. Haematologica 96, 1743–1752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sivalenka R. R., Jessberger R. (2004) SWAP-70 regulates c-kit-induced mast cell activation, cell-cell adhesion, and migration. Mol. Cell. Biol. 24, 10277–10288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pearce G., Audzevich T., Jessberger R. (2011) SYK regulates B-cell migration by phosphorylation of the F-actin interacting protein SWAP-70. Blood 117, 1574–1584 [DOI] [PubMed] [Google Scholar]
- 30. Breitsprecher D., Koestler S. A., Chizhov I., Nemethova M., Mueller J., Goode B. L., Small J. V., Rottner K., Faix J. (2011) Cofilin cooperates with fascin to disassemble filopodial actin filaments. J. Cell Sci. 124, 3305–3318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Watts R. G., Howard T. H. (1992) Evidence for a gelsolin-rich, labile F-actin pool in human polymorphonuclear leukocytes. Cell Motil. Cytoskeleton 21, 25–37 [DOI] [PubMed] [Google Scholar]
- 32. Gu C., Yaddanapudi S., Weins A., Osborn T., Reiser J., Pollak M., Hartwig J., Sever S. (2010) Direct dynamin-actin interactions regulate the actin cytoskeleton. EMBO J. 29, 3593–3606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Erickson H. P. (2009) Size and shape of protein molecules at the nanometer level determined by sedimentation, gel filtration, and electron microscopy. Biol. Proced. Online 11, 32–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schürmann G., Haspel J., Grumet M., Erickson H. P. (2001) Cell adhesion molecule L1 in folded (horseshoe) and extended conformations. Mol. Biol. Cell 12, 1765–1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Harris E. S., Higgs H. N. (2006) Biochemical analysis of mammalian formin effects on actin dynamics. Methods Enzymol. 406, 190–214 [DOI] [PubMed] [Google Scholar]
- 36. Mullins R. D., Machesky L. M. (2000) Actin assembly mediated by Arp2/3 complex and WASP family proteins. Methods Enzymol. 325, 214–237 [DOI] [PubMed] [Google Scholar]
- 37. Harris E. S., Rouiller I., Hanein D., Higgs H. N. (2006) Mechanistic differences in actin bundling activity of two mammalian formins, FRL1 and mDia2. J. Biol. Chem. 281, 14383–14392 [DOI] [PubMed] [Google Scholar]
- 38. Lindberg U., Schutt C. E., Hellsten E., Tjäder A. C., Hult T. (1988) The use of poly(l-proline)-Sepharose in the isolation of profilin and profilactin complexes. Biochim. Biophys. Acta 967, 391–400 [DOI] [PubMed] [Google Scholar]
- 39. Breitsprecher D., Kiesewetter A. K., Linkner J., Vinzenz M., Stradal T. E., Small J. V., Curth U., Dickinson R. B., Faix J. (2011) Molecular mechanism of Ena/VASP-mediated actin-filament elongation. EMBO J. 30, 456–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cai L., Marshall T. W., Uetrecht A. C., Schafer D. A., Bear J. E. (2007) Coronin 1B coordinates Arp2/3 complex and cofilin activities at the leading edge. Cell 128, 915–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bryce N. S., Clark E. S., Leysath J. L., Currie J. D., Webb D. J., Weaver A. M. (2005) Cortactin promotes cell motility by enhancing lamellipodial persistence. Curr. Biol. 15, 1276–1285 [DOI] [PubMed] [Google Scholar]
- 42. Pollard T. D., Blanchoin L., Mullins R. D. (2000) Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annu. Rev. Biophys. Biomol. Struct. 29, 545–576 [DOI] [PubMed] [Google Scholar]
- 43. Bartles J. R. (2000) Parallel actin bundles and their multiple actin-bundling proteins. Curr. Opin. Cell Biol. 12, 72–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Galli S. J., Nakae S., Tsai M. (2005) Mast cells in the development of adaptive immune responses. Nat. Immunol. 6, 135–142 [DOI] [PubMed] [Google Scholar]
- 45. Lorenz M., DesMarais V., Macaluso F., Singer R. H., Condeelis J. (2004) Measurement of barbed ends, actin polymerization, and motility in live carcinoma cells after growth factor stimulation. Cell Motil. Cytoskeleton 57, 207–217 [DOI] [PubMed] [Google Scholar]
- 46. Ghosh M., Song X., Mouneimne G., Sidani M., Lawrence D. S., Condeelis J. S. (2004) Cofilin promotes actin polymerization and defines the direction of cell motility. Science 304, 743–746 [DOI] [PubMed] [Google Scholar]
- 47. Bernstein B. W., Bamburg J. R. (2010) ADF/cofilin: a functional node in cell biology. Trends Cell Biol. 20, 187–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mouneimne G., DesMarais V., Sidani M., Scemes E., Wang W., Song X., Eddy R., Condeelis J. (2006) Spatial and temporal control of cofilin activity is required for directional sensing during chemotaxis. Curr. Biol. 16, 2193–2205 [DOI] [PubMed] [Google Scholar]
- 49. Lai F. P., Szczodrak M., Block J., Faix J., Breitsprecher D., Mannherz H. G., Stradal T. E., Dunn G. A., Small J. V., Rottner K. (2008) Arp2/3 complex interactions and actin network turnover in lamellipodia. EMBO J. 27, 982–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Shinohara M., Terada Y., Iwamatsu A., Shinohara A., Mochizuki N., Higuchi M., Gotoh Y., Ihara S., Nagata S., Itoh H., Fukui Y., Jessberger R. (2002) SWAP-70 is a guanine-nucleotide-exchange factor that mediates signalling of membrane ruffling. Nature 416, 759–763 [DOI] [PubMed] [Google Scholar]
- 51. Blanchoin L., Pollard T. D. (1999) Mechanism of interaction of Acanthamoeba actophorin (ADF/Cofilin) with actin filaments. J. Biol. Chem. 274, 15538–15546 [DOI] [PubMed] [Google Scholar]
- 52. Carlier M. F., Laurent V., Santolini J., Melki R., Didry D., Xia G. X., Hong Y., Chua N. H., Pantaloni D. (1997) Actin depolymerizing factor (ADF/cofilin) enhances the rate of filament turnover: implication in actin-based motility. J. Cell Biol. 136, 1307–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Schmoller K. M., Semmrich C., Bausch A. R. (2011) Slow down of actin depolymerization by cross-linking molecules. J. Struct. Biol. 173, 350–357 [DOI] [PubMed] [Google Scholar]
- 54. Michelot A., Berro J., Guérin C., Boujemaa-Paterski R., Staiger C. J., Martiel J. L., Blanchoin L. (2007) Actin-filament stochastic dynamics mediated by ADF/cofilin. Curr. Biol. 17, 825–833 [DOI] [PubMed] [Google Scholar]
- 55. Marshall T. W., Aloor H. L., Bear J. E. (2009) Coronin 2A regulates a subset of focal-adhesion-turnover events through the cofilin pathway. J. Cell Sci. 122, 3061–3069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Otey C. A., Carpen O. (2004) α-Actinin revisited: a fresh look at an old player. Cell Motil. Cytoskeleton 58, 104–111 [DOI] [PubMed] [Google Scholar]
- 57. Maul R. S., Song Y., Amann K. J., Gerbin S. C., Pollard T. D., Chang D. D. (2003) EPLIN regulates actin dynamics by cross-linking and stabilizing filaments. J. Cell Biol. 160, 399–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Fiumara F., Fioriti L., Kandel E. R., Hendrickson W. A. (2010) Essential role of coiled coils for aggregation and activity of Q/N-rich prions and polyQ proteins. Cell 143, 1121–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hey F., Czyzewicz N., Jones P., Sablitzky F. (2012) DEF6, a novel substrate for the Tec kinase ITK, contains a glutamine-rich aggregation-prone region and forms cytoplasmic granules that co-localize with P-bodies. J. Biol. Chem. 287, 31073–31084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ono S., Ono K. (2002) Tropomyosin inhibits ADF/cofilin-dependent actin filament dynamics. J. Cell Biol. 156, 1065–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ocaña-Morgner C., Reichardt P., Chopin M., Braungart S., Wahren C., Gunzer M., Jessberger R. (2011) Sphingosine 1-phosphate-induced motility and endocytosis of dendritic cells is regulated by SWAP-70 through RhoA. J. Immunol. 186, 5345–5355 [DOI] [PubMed] [Google Scholar]
- 62. Chin Y. R., Toker A. (2010) The actin-bundling protein palladin is an Akt1-specific substrate that regulates breast cancer cell migration. Mol. Cell 38, 333–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Skau C. T., Courson D. S., Bestul A. J., Winkelman J. D., Rock R. S., Sirotkin V., Kovar D. R. (2011) Actin filament bundling by fimbrin is important for endocytosis, cytokinesis, and polarization in fission yeast. J. Biol. Chem. 286, 26964–26977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Reymann A. C., Boujemaa-Paterski R., Martiel J. L., Guérin C., Cao W., Chin H. F., De La Cruz E. M., Théry M., Blanchoin L. (2012) Actin network architecture can determine myosin motor activity. Science 336, 1310–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]