Background: Hydroquinone is a benzene metabolite shown to lead to decreased DNA methylation.
Results: Hydroquinone exposure increases Ten Eleven Translocation 1 methylcytosine dioxygenase activity and 5-hydroxymethylcytosine levels and decreases DNA methylation.
Conclusion: Hydroquinone leads to DNA demethylation through a Ten Eleven Translocation 1-dependent mechanism.
Significance: This mechanism may explain observations of decreased DNA methylation and cytotoxicity following exposure to benzene and hydroquinone.
Keywords: Dioxygenase, DNA Methylation, Epigenetics, Quinones, Reactive Oxygen Species (ROS), Benzene, DNA Demethylation, Hydroquinone, Ten Eleven Translocation 1, TET1
Abstract
DNA methylation regulates gene expression throughout development and in a wide range of pathologies such as cancer and neurological disorders. Pathways controlling the dynamic levels and targets of methylation are known to be disrupted by chemicals and are therefore of great interest in both prevention and clinical contexts. Benzene and its metabolite hydroquinone have been shown to lead to decreased levels of DNA methylation, although the mechanism is not known. This study employs a cell culture model to investigate the mechanism of hydroquinone-mediated changes in DNA methylation. Exposures that do not affect HEK293 cell viability led to genomic and methylated reporter DNA demethylation. Hydroquinone caused reactivation of a methylated reporter plasmid that was prevented by the addition of N-acetylcysteine. Hydroquinone also caused an increase in Ten Eleven Translocation 1 activity and global levels of 5-hydroxymethylcytosine. 5-Hydroxymethylcytosine was found enriched at LINE-1 prior to a decrease in both 5-hydroxymethylcytosine and 5-methylcytosine. Ten Eleven Translocation-1 knockdown decreased 5-hydroxymethylcytosine formation following hydroquinone exposure as well as the induction of glutamate-cysteine ligase catalytic subunit and 14-3-3σ. Finally, Ten Eleven Translocation 1 knockdown decreased the percentage of cells accumulating in G2+M following hydroquinone exposure, indicating that it may have a role in cell cycle changes in response to toxicants. This work demonstrates that hydroquinone exposure leads to active and functional DNA demethylation in HEK293 cells in a mechanism involving reactive oxygen species and Ten Eleven Translocation 1 5-methylcytosine dioxygenase.
Introduction
DNA methylation is dynamic, and the mechanisms involved in methylation and demethylation represent an interface between the environment and gene expression (1). Changes in DNA methylation have been implicated in a wide range of pathologies including cancer (2), neurodevelopmental disorders (3), and chronic inflammation (4). Efforts to understand mechanisms involved in DNA methylation and demethylation therefore have enormous translational potential. DNA methylation is established and maintained by the family of DNA methyltransferases (DNMTs),3 although mechanisms underlying demethylation have been more difficult to elucidate. Whereas Arabidopsis DNA glycosylases DME and ROS1 participate in exchanging methylcytosines with cytosines, no analogous pathway has been discovered in mammalian cells (5). A number of chemicals, such as 5-aza-2′-deoxycytidine, decrease DNA methylation passively by inhibiting maintenance methylation as DNA is synthesized (6). However, this mechanism depends on cell division and may not explain active demethylation in post-mitotic cells such as neurons (7) and in the early stages of embryogenesis.
Pharmaceutical, industrial, and environmental chemicals have also been shown to decrease DNA methylation, but the mechanisms are unclear (8, 9). Several chemicals have been shown to have a passive effect on methylation by modifying DNMT activity (10, 11). An interesting target that could explain active demethylation involves the Ten Eleven Translocation (TET) family of Fe2+ and α-ketoglutarate-dependent 5mC dioxygenases (12, 13). In a mechanism proposed by Guo et al., TET proteins catalyze the oxidation of 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC), followed by deamination to 5-hydroxymethyluracil by cytidine deaminases. Subsequent base excision repair machinery would then replace 5-hydroxymethyluracil with unmethylated cytosine (14).
In this study, we focused on changes in DNA methylation by the benzene metabolite hydroquinone (HQ). Benzene is a ubiquitous environmental toxicant found in petroleum products and cigarette smoke (15) that has been associated with aplastic anemia and acute myelogenous leukemia (16). HQ is one of the most abundant metabolites of benzene (17, 18) and has been shown to increase levels of reactive oxygen species (ROS) (19, 20), induce mitotic arrest (21), and promote apoptosis (20). Although benzene and benzene metabolite exposures have also been shown to be associated with loss of genomic methylation (9, 22), no mechanisms have been described to explain the observed decreases.
In our study, we investigated a mechanism of DNA demethylation induced by exposure to HQ in a cell culture model. Evidence will be presented showing that HQ mediates DNA demethylation in a mechanism involving ROS and the TET1 pathway in HEK293 cells.
EXPERIMENTAL PROCEDURES
Chemicals and Reagents
Unless otherwise stated, all chemicals and reagents were obtained from Sigma-Aldrich. Nit-GFP, human APOBEC2 (AP2), and TET1 catalytic domain plasmids were obtained from Dr. Hongjun Song's laboratory (JHU) and used according the published methods (14). Silencer Select Pre-designed siRNA against Tet1(4392420; ID s37194) and control (4390846) were obtained from Ambion and used at 10 nm.
Cell Culture
HEK293 cells were obtained from ATCC and grown in DMEM supplemented with 10% FBS. Hydroquinone, menadione, buthionine sulfoximine, and N-acetylcysteine (NAC) stocks were freshly prepared in H2O and sterile-filtered.
MTT Assay
Cytotoxicity was measured using standard 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) protocol. In brief, 1.5 × 104 cells were seeded in each well of a 96-well plate. Cells were exposed to the indicated concentrations of HQ for 24 h, washed with PBS, and incubated with 10 mg/ml MTT reagent in PBS at 37 °C for 1 h. Cells were lysed, and formazan crystals were dissolved in 100 μl of dimethyl sulfoxide. Absorbance was read at 570 nm using a microplate reader, and data were expressed as a percentage of nonexposed cell MTT reduction.
Western Blotting
Whole cell lysates were prepared using radioimmunoprecipitation assay buffer and included addition of protease inhibitors. Protein concentrations were measured using the Bradford assay, and 30 μg of whole cell lysates was separated on a 4–20% Tris-glycine gradient gel (Invitrogen). Nuclear lysates were prepared using Panomics Nuclear Extract kit (Fremont, CA) and measured using the Bradford assay. 15 μg of nuclear lysates was separated on a 4–12% Tris-glycine gradient gel (Invitrogen). Protein was transferred to a nitrocellulose membrane, blocked with 0.5% casein, and incubated overnight at 4 °C with antibodies against V5©(Invitrogen; 1:5000), FLAG-M2©(Sigma; 1:2000), actin (Sigma; 1:5000), TET1 (Abnova; 1:1000), or TATA-binding protein (Millipore; 1:1000). Blots were incubated with secondary antibody (1:10,000) for 1 h before visualization on the Licor Odyssey Imager.
qRT-PCR
RNA was isolated using TRIzol and quantified using NanoDrop. 1 μg was used to generate cDNA (SuperscriptIII; Invitrogen). qPCR was performed using a Bio-Rad iCycler and iQ SYBR Green supermix according to the manufacturer's instructions.
Primers used were as follows: 14-3-3σ forward, GGCCATGGACATCAGCAAGAA and reverse, CGAAAGTGGTCTTGGCCAGAG; GAPDH forward, ACATCGCTCAGACACCATG and reverse, TGTAGTTGAGGTCAATGAAGGG; GCLC forward, AAAAGTCCGGTTGGTCCTG and reverse, CCTGGTGTCCCTTCAATCATG; TET1 forward, TTCGTCACTGCCAACCTTAG and reverse, ATGCCTCTTTCACTGGGTG; TET2 forward, CACTGCATGTTTGGACTTCTG and reverse, TGCTCATCCTCAGGTTTTCC; TET3 forward, TCCGGATTGAGAAGGTCATC and reverse, CCAGGCCAGGATCAAGATAA.
Immunodotblotting
Genomic DNA was isolated using GenElute Mammalian Genomic DNA kit (Sigma) and measured using a NanoDrop (Thermo Scientific). 250 ng was denatured with 0.4 m NaOH at 95 °C for 10 min. DNA was immobilized on nitrocellulose membrane, dried, and fixed by vacuum baking at 80 °C. The membrane was then blocked with 0.5% casein and probed with antibodies against 8-hydroxy-2′-deoxyguanosine (8-oxo-dG) (Trevigen; 1:2500), 5-mC (Active Motif; 1:1000), or 5-hmC (Active Motif; 1:8000). Mouse anti-single-stranded DNA (Abcam; 1:1500) antibody was used simultaneously for normalizing 5hmC. Antibody binding was measured using the Licor Odyssey. Methylene blue staining of DNA (0.04% with 0.3 m sodium acetate) was used for normalization of 8-oxo-dG and 5mC antibodies. Intensity of methylene blue was measured using ImageJ software (National Institutes of Health). Linear range of detection of DNA was determined by loading increasing amounts of single-stranded DNA and probing with increasing dilutions of anti-single-stranded DNA antibody. 5hmC Ab specificity was confirmed by blocking antibody with 2 μg/ml 5hmC or 5mC at room temperature for 2 h and blotting DNA from mock-transfected and TET1-overexpressing HEK293 cells.
Methylation-sensitive Reporter Assay
Nit-GFP plasmid was methylated using 4 units of CpG methyltransferase and 0.6 mm S-adenosylmethionine (New England Biolabs) at 30 °C for 12 h and purified using Qiagen gel extraction columns. 2.5 × 105 HEK293 cells were transfected with 0.5 μg of methylated or unmethylated Nit-GFP plasmid, and co-transfections were used at 1:4 GFP:TET1 (+ control), AP2, or βgal for 48 h using Lipofectamine. Cells were exposed to 60 μm HQ for 24 h, beginning 24 h after transfection. Cells were harvested using 0.25% trypsin-EDTA, fixed using ice-cold methanol:acetone (1:1), and stored at −20 °C. Cells were washed three times and resuspended in 4 °C PBS. GFP fluorescence was measured using a FACSCalibur flow cytometer (BD Biosciences) and analyzed using CellQuest software. Dead cell and doublet discrimination were accomplished using forward scatter versus side scatter gating, and at least 10,000 events were analyzed in each sample for all experiments.
Bisulfite Sequencing
Bisulfite sequencing was performed as reported by Guo et al. (14). At the end of the 24-h exposure to 60 μm HQ, DNA from cells transfected with methylated Nit-GFP plasmid was isolated and bisulfite modified using the EpiTect bisulfite kit (Qiagen). The Nit-GFP promoter was PCR-amplified using primers recognizing the bisulfite-modified sequence and gel-purified. Bands of the predicted size were cloned into TOPO TA vector (Invitrogen) for sequencing (Johns Hopkins University DNA Analysis Facility). 6 CpGs within the promoter were read in 23 clones. Primers used for bisulfite-sequencing of Nit-GFP promoter were forward, TTTTTTATTAGTGATAGAGAAAAGTGAAA and reverse, CAAATAAACTTCAAAATCAACTTACC (14).
ROS Measurement
Cells were grown in black 96-well plates and incubated with 50 μm (final concentration) 2′,7′-dichlorodihydrofluorescein diacetate (DCF-DA) in DMEM lacking phenol red for 30 min. DCF-DA was removed by washing twice with PBS, and cells were exposed to HQ, NAC (5 mm), or in combination. Fluorescence was measured at 485em 522ex beginning at 30 min and followed through indicated time points.
Hydroxymethylated/Methylated DNA Immunoprecipitation
Genomic DNA was isolated from cells exposed to hydroquinone or control conditions. 8 μg of DNA was sheared using intermittent sonication and validated to be 100–500 bp by gel electrophoresis. 2 μg of DNA was heat-denatured for 10 min and placed immediately in an ice bath. DNA was incubated with antibody against 5mC, 5hmC, or matched isotype control at 4 °C overnight. Protein G magnetic beads (Invitrogen) were added for 1 h at 4 °C, and bead-antibody-DNA complexes were magnetically separated, followed by three washes in immunoprecipitation buffer (4 °C PBS with 0.05%Triton X-100). Protein was digested using proteinase K digestion buffer (50 mm Tris, pH 8.0, 10 mm EDTA) overnight at 50 °C. Proteinase K was inactivated by incubation at 80 °C for 30 min. DNA was purified using nucleic acid columns (Sigma). Immunoprecipitated DNA was used as a template for qPCR, and -fold enrichment was calculated using input DNA.
TET Activity
Total TET activity was measured using the Epigentek Epigenase 5mC hydroxylase kit (Farmingdale, NY) according to the supplier's protocol. Nuclear lysates were prepared using Panomics Nuclear Extraction kit (Fremont, CA) and measured using the Bradford assay. Hydroxymethylated product formed was measured within a standard curve (0–2 ng), and activity was calculated as ng/min/mg. Data are expressed as -fold change over unexposed nuclear lysates. Overexpression of the TET1 catalytic domain was used as a positive control. Primers used in hydroxymethylated/methylated DNA immunoprecipitation were as follows: LINE1 forward, TGCGGAGAAATAGGAACACTTTT and reverse, TGAGGAATCGCCACACTGACT (23); 14-3-3σ forward, CATTTAGGCAGTCTGATTCC and reverse, GCTCACGCCTGTCATCTC (24); glutamate-cysteine ligase catalytic subunit (GCLC) forward, CGTCCCAAGTCTCACAGTCA and reverse, CTTTACGCAAACGCGACATA (25).
DNA Content
Cells were harvested using 0.25% trypsin-EDTA, resuspended in 300 μl of PBS at 4 °C, and fixed overnight at −20 °C in 1:1 methanol:acetone, added dropwise with intermittent vortexing. Cells were washed with cold PBS and incubated with 0.25 ml of 5 μg/ml RNase, 15 min at 37 °C. 0.25 ml of 100 μg/ml propidium iodide was added for 60 min at room temperature. DNA content was measured using a FACSCalibur flow cytometer and analyzed using Cell Quest software (BD Biosciences). Doublet discrimination was accomplished by gating on forward scatter-H v. FL3A and FL3A v. FL3-H. At least 10,000 events were analyzed in each sample.
Statistical Evaluation
GraphPad Prism version 5 was used for statistical analyses. All data are summarized as mean ± S.E. Student's t test was performed for all single analyses. Multiple comparisons were made using one-way analysis of variance (ANOVA) and Tukey's, Dunnett's, or Newman-Keul post hoc tests. p values = 0.05 were considered significant.
RESULTS
Low Concentrations of Hydroquinone Decrease DNA Methylation in a Mechanism Involving ROS
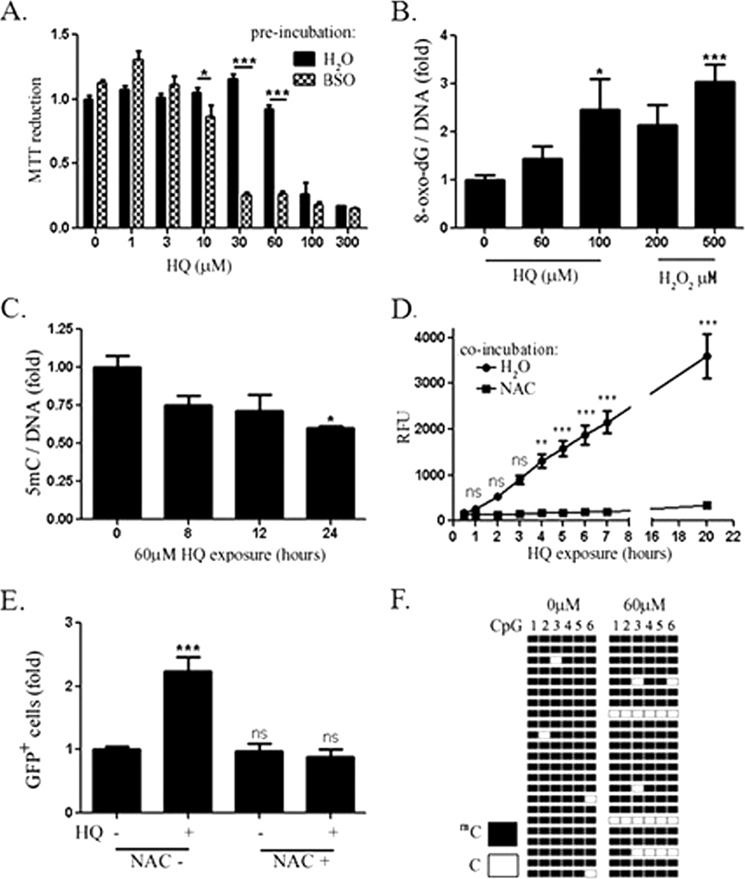
The MTT assay was used to determine concentrations of HQ which did not affect viability of HEK293 cells. Significant changes in MTT reduction were observed following exposure for 24 h to concentrations of 100 μm and higher (Fig. 1A). Preincubation of cells with buthionine sulfoximine, an inhibitor of glutathione synthesis, increased the sensitivity of cells to HQ, indicating the involvement of glutathione and that toxicity is mediated in part by ROS. 8-oxo-dG was also measured by immunodotblotting genomic DNA. Significant increases in 8-oxo-dG staining were observed at 100 μm HQ and 500 μm H2O2 (Fig. 1B) but not at 60 μm HQ, although a 40% decrease in genomic 5mC was observed (Fig. 1C). Increases in ROS were observed as early at 4 h following exposure to 60 μm HQ and levels of ROS were lower in cells incubated with the anti-oxidant (NAC) (Fig. 1D).
FIGURE 1.
A, MTT assay results following HQ exposure for 24 h are shown. Cells were incubated for 14 h with 0.5 mm buthionine sulfoximine or an equivalent volume of vehicle prior to HQ exposure. B, cells were exposed to indicated concentrations of HQ for 20 h or H2O2 for 0.5 h . C, 250 ng of genomic DNA was then denatured, vacuum-baked to a nitrocellulose membrane, and probed with antibody against 8-oxo-dG or 5mC and normalized to methylene blue staining. D, ROS was measured by DCF-DA oxidation following 60 μm HQ exposure with and without NAC. E, reactivation of methylated reporter plasmid by HQ alone or in combination with NAC is shown. GFP+ cells were quantified by flow cytometry. n = 4, Values are mean ± S.E. (error bars). F, bisulfite sequencing of methylated methylation-sensitive reporter plasmid promoter in control and HQ-exposed HEK293 cells. Six CpGs were measured in 23 clones.
A possible mechanism involved in decreased methylation in the absence of oxidative DNA damage could involve inhibition of DNMT1, as many of the cells would have passed through S phase within 24 h. A reporter assay was established to examine this possibility. The Nit-GFP plasmid is replication-incompetent and would therefore be less likely to undergo demethylation passively (16). Moreover, the functionality of CpG methylation could be ascertained by measuring GFP expression. A nearly complete repression of GFP was observed in cells transfected with the reporter plasmid that was methylated with CpG methyltransferase and S-adenosylmethionine (data not shown). At 24 h following exposure to 60 μm HQ, a >2-fold increase in the number of GFP+ cells was observed (Fig. 1E). Moreover, the increase in GFP+ cells was prevented by co-incubating HQ-exposed cells with NAC, suggesting that ROS was involved in reactivation of the methylated plasmid by HQ. Bisulfite sequencing of the promoter confirmed both methylation of the plasmid promoter and an increase in unmethylated CpGs following 24-h exposure to HQ (Fig. 1F).
Hydroquinone Increases Genomic 5-Hydroxymethylcytosine Levels through TET1 5mC Dioxygenase
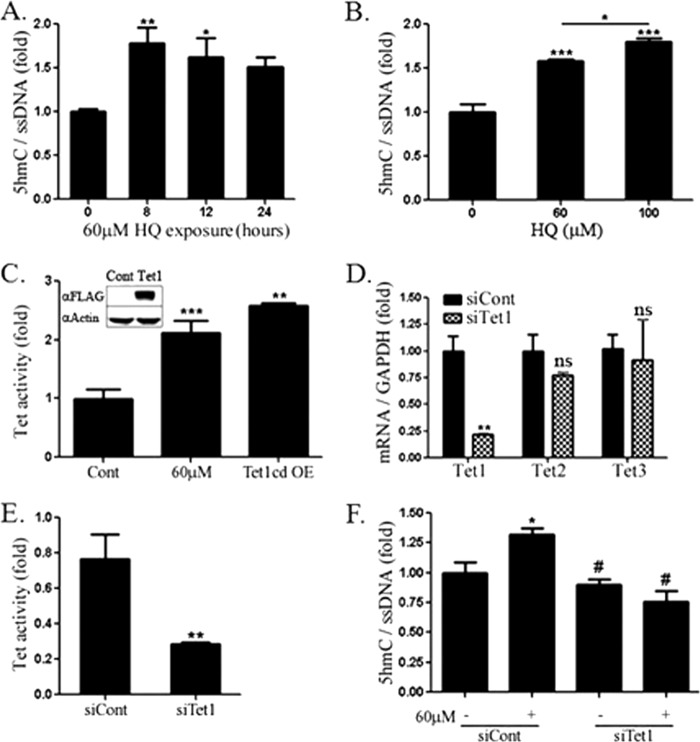
Due to the recent interest in active demethylation pathways involving TET proteins, immunodotblotting was used to measure 5hmC in genomic HEK293 DNA following HQ exposure. Noting a decrease in 5mC following 24 h of HQ exposure, a time course of 5hmC was conducted to examine the potential for 5hmC as a demethylation intermediate. 5hmC levels increased at 8 and 12 h of HQ exposure, but were not significantly higher at 24 h (Fig. 2A). A dose response was observed for 5hmC levels 18 h following exposure to HQ (Fig. 2B) and menadione (data not shown), which is chemically similar to HQ and other ROS-inducing quinones.
FIGURE 2.
A, 5hmC in HEK293 cells following 8-, 12-, and 24-h exposure to HQ measured using immunodotblotting. DNA was immobilized onto nitrocellulose paper and probed with antibodies against 5hmC and ssDNA for normalization. IR dye-conjugated IgG antibodies (mouse: 600, rabbit: 800) were used for simultaneous visualization by Licor Odyssey Imager. B, 5hmC dose response following 18-h exposure to HQ. C, 5hmC formed from nuclear extracts of HEK293 cells exposed to 0 or 60 μm HQ or overexpressing (OE) FLAG-tagged Tet1 catalytic domain. D, Tet1, 2, and 3 mRNA measured by qRT-PCR following transfection with Tet1-targeted or control siRNA. Tet expression was normalized to GAPDH. E, 5hmC formed from HEK293 nuclear extracts following control or Tet1 knockdown. F, immunodotblotting of genomic DNA from cells exposed to 60 μm HQ following control or Tet1-targeted knockdown. Values are mean ± S.E. (error bars), n = 3. *, significance compared with control by t test (D and E) and one-way ANOVA and Newman-Keuls (B) or Tukey post-test (C and F). #, significance compared with siCont + HQ.
TET activity was higher in HEK293 cells exposed to HQ to levels similar in cells overexpressing TET1 catalytic domain (Fig. 2C). Although there are three distinct family members of TET proteins, we found that siRNA-mediated knockdown of TET1 led to an approximate 75% decrease in both Tet1 mRNA (Fig. 2D) and total TET activity (Fig. 2E). siRNA against Tet1 did not affect Tet2 or Tet3 mRNA levels (Fig. 2D), but completely abrogated HQ-induced increases in 5hmC (Fig. 2F), indicating that the increase in 5hmC in HEK293 cells exposed to HQ requires TET1.
Hydroquinone Exposure Leads to Increased Nuclear TET1 Levels in HEK293 Cells
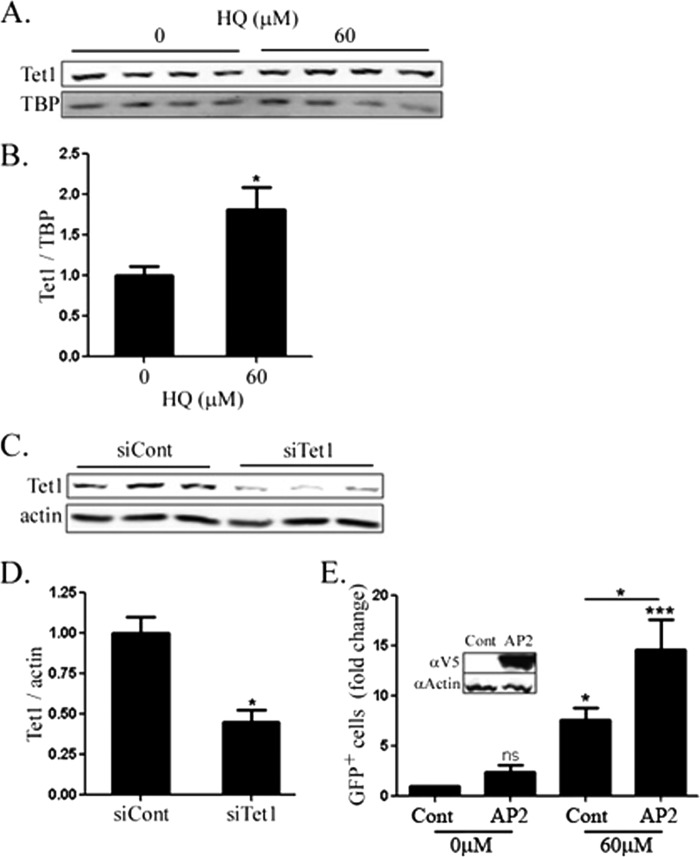
To determine the relationship between TET1 protein and activity, levels of TET1 protein were measured in nuclei by Western blotting. A 75% increase was observed in TET1 protein following exposure to 60 μm HQ (Fig. 3, A and B). A 60% decrease in the band was also observed in cells transfected with TET1 siRNA, thereby confirming the identity of the band as TET1 (Fig. 3, C and D).
FIGURE 3.
A, nuclear Tet1 measured in HEK293 nuclear lysates following 20-h exposure to HQ. B, quantification of A. Tata-binding protein was used as a loading control for nuclear lysates. C, Tet1 levels measured by Western blotting HEK293 whole cell lysates following control or Tet1-targeted siRNA. D, quantification of C. Actin was used as a loading control. E, reactivation of methylation-sensitive reporter by HQ alone or with overexpression of human V5-tagged cytidine deaminase AP2. GFP+ cells were quantified by flow cytometry. Values are mean ± S.E. (error bars), n = 3. *, significance compared with control by t test (B and D) or one-way ANOVA and Tukey post-test (E).
Because HQ exposure led to functional demethylation, 5hmC formation, and an increase in total Tet activity and nuclear TET1 levels, we reasoned that overexpression of deaminase proteins should enhance reactivation of the methylated GFP reporter. Indeed, overexpression of V5-tagged human AP2 increased the GFP+ population ∼2-fold following 60 μm HQ exposure (Fig. 3E).
TET1 Mediates Gene Expression Induced by Hydroquinone
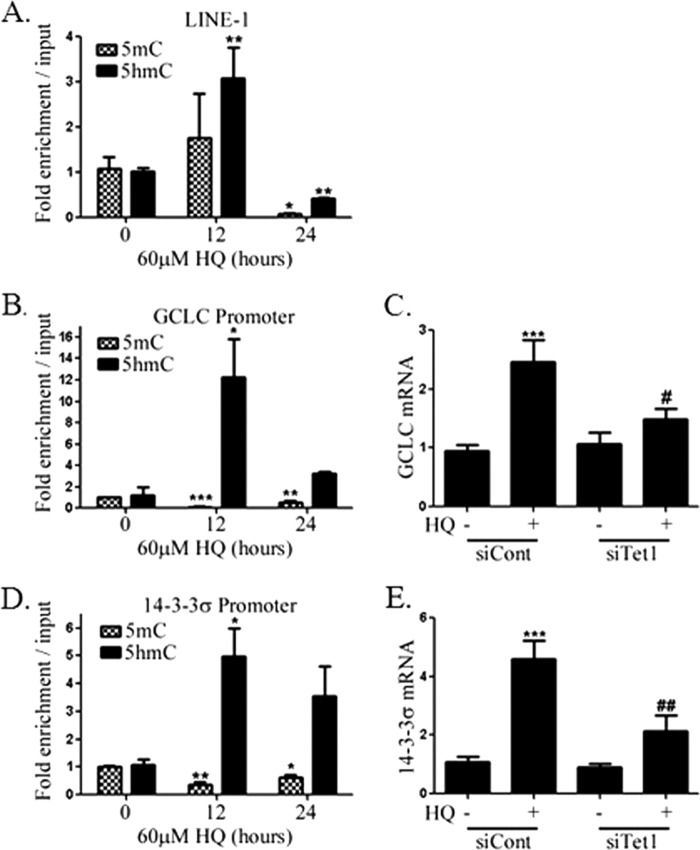
The results from the experiments involving the reporter plasmid suggest that HQ increases 5hmC and the involvement of TET1. Genomic regions enriched for 5hmC and 5mC were identified by immunoprecipitation and qPCR. A 3-fold enrichment of the open reading frame 2 (ORF2) of LINE-1 was observed in cells exposed to 60 μm HQ for 12 h without a significant change in 5mC (Fig. 4A). At 24 h, the enrichment of 5mC and 5hmC was decreased by 10- and 2-fold, respectively. The data support a role for HQ-induced 5hmC formation prior to demethylation, consistent with recently described Tet-mediated oxidation-deamination demethylation pathways. In addition, the effects of HQ are widespread, considering the frequency of LINE-1 in the human genome (26).
FIGURE 4.
Genomic DNA was sonicated and immunoprecipitated using antibodies against 5hmC or 5mC and used as templates for qPCR to measure enrichment of LINE-1 (A), GCLC promoter (B), and 14-3-3σ promoter (D) in HEK293 cells following 0-, 12-, and 24-h exposure to 60 μm HQ. ΔΔCt values were normalized to input values. mRNA was isolated from cells exposed to 60 μm HQ for 24 h following knockdown with control siRNA or siRNA targeting Tet1, and qRT-PCR was used to measure levels of GCLC (C) and 14-3-3σ (E). GAPDH was used as a control in all qRT-PCRs. Values are mean ± S.E. (error bars), n = 3. *, significance compared with control by one-way ANOVA with Dunnett's post-test. #, indicate significance compared with siCont + 60 μm HQ (C and E).
We next investigated 5hmC at promoters of genes that respond to HQ. GCLC is a component of the glutamate-cysteine ligase protein, the rate-limiting enzyme in glutathione synthesis. ROS disrupts Keap1-dependent sequestration of the transcription factor Nrf2, resulting in the transcription of numerous antioxidant response element-containing genes, including GCLC. 5hmC was enriched ∼12-fold at the promoter of GCLC following a 12-h exposure to HQ, whereas 5mC levels were significantly lower at 12 and 24 h (Fig. 4B). Whereas GCLC mRNA was induced by ∼2.5-fold in response to HQ in HEK293 cells, induction was attenuated in cells following siRNA-mediated TET1 knockdown (Fig. 4C). No changes in oxidized glutathione were observed following exposure to HQ with or without TET1 knockdown (data not shown).
Because previous studies reported accumulation of cells in G2+M phase after exposure to HQ, we investigated the effect of HQ on methylation of the14-3-3σ promoter. 14-3-3σ is a p53-regulated inhibitor of cdc2-cyclinB1 nuclear translocation and has been shown to be regulated by promoter methylation (27–30). We observed an enrichment of 5hmC (Fig. 4D) at the 14-3-3σ promoter following a 12-h exposure to HQ and decreased 5mC at 12 and 24 h. Induction of 14-3-3σ by HQ was also blocked by TET1 knockdown (Fig. 4E). Interestingly, 14-3-3σ promoter hypermethylation has been reported in numerous cancers and correlates strongly with a decrease in expression (31). In addition, 14-3-3σ expression can be increased by exposure to 5-aza-2′-deoxycytidine (32), and its overexpression leads to a mitotic arrest in different cell types (29). Further experiments were directed to examine whether TET1 plays a role in the changes to cell cycle that result from exposure to HQ.
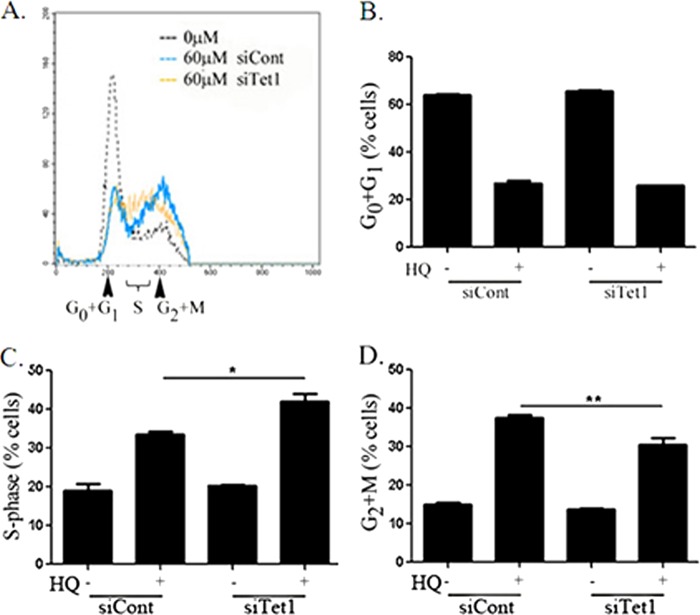
TET1 Mediates Cell Cycle Changes following Hydroquinone Exposure
HQ led to an increase in the percentage of cells in G2+M from 15% in unexposed cells to 37.5% measured by DNA content analysis using flow cytometry (Fig. 5A). There was no change in the sub-G0 population by HQ exposure. The percentage of cells in G0+G1 phases decreased following HQ exposure, but TET1 knockdown did not affect this decrease (Fig. 5B). The percentage of cells in S phase increased following TET1 knockdown in HQ-exposed cells, from 33 to 42% (Fig. 5C). TET1 knockdown decreased the number of cells in G2+M following HQ exposure to ∼30% (Fig. 5D). Neither TET1 nor control knockdown had any effect on unexposed cells, and control knockdown did not affect the cell cycle compared with untransfected cells. The data suggest that TET1 may play a role in promoting the cellular response to ROS-inducing agents, such as promoting arrest of the cell cycle to allow for efficient DNA repair.
FIGURE 5.
A, representative cell cycle profiles of HEK293 cells exposed to HQ for 24 h following control or Tet1 knockdown are shown. Cells were fixed using methanol:acetone (1:1) and stained with propidium iodide. B–D, 10,000 events were collected in each sample following control or Tet1 knockdown and exposure to 0 or 60 μm HQ (+) and percentage of cells in G0+G1 (B), S (C), and G2+M (D) phases were quantified. n = 3, * p < 0.05 by one-way ANOVA and Tukey post-test. Error bars, S.E.
DISCUSSION
Epidemiologic and laboratory data suggest a negative correlation between DNA methylation and benzene exposures (9, 22). We investigated the mechanism to account for the decrease by examining the effects of the major benzene metabolite, HQ, in HEK293 cells. Here, we demonstrate that concentrations of HQ that did not affect viability led to decreased DNA methylation in HEK293 cells in a mechanism involving ROS. Using an immunodotblotting technique, we show that genome-wide DNA methylation levels are decreased following a 24-h exposure to HQ. A GFP reporter plasmid revealed that HQ-mediated demethylation results in an increase in gene expression. Because the plasmid is replication-incompetent (14), it is likely that the demethylation is active in nature and does not involve inhibiting the activity of DNMTs. HQ is a well known inducer of ROS and oxidative stress through the depletion of glutathione (19, 20). The addition of NAC abrogated the reactivation of methylated reporter plasmid, suggesting that HQ caused demethylation in a mechanism involving ROS. ROS and oxidative stress have been shown to lead to both increases (31) and decreases (32, 33) in DNA methylation. Interestingly, the low concentrations of HQ used did not significantly increase oxidized glutathione, lipid peroxidation (data not shown), or 8-oxo-dG formation, indicating that DNA demethylation was induced by relatively low levels of ROS that do not result in oxidative stress. This may be particularly relevant in cells and tissues that are sensitive to changes in redox status or in cases of signaling events that may be mediated by ROS.
Whereas mechanisms explaining DNA demethylation have been largely focused on inhibition of the DNMT family of DNA methyltransferases, recent evidence suggests a role for the TET family of 5mC dioxygenases in active DNA demethylation. The proposed mechanism entails the conversion of 5mC to 5hmC catalyzed by TET1, followed by deamination to 5hmU and subsequent base excision repair, resulting in an unmethylated cytosine (12–14). Our studies provide support that this mechanism is responsible for DNA demethylation in cells exposed to HQ. Immunodotblots of genomic DNA from HEK293 cells exposed to 60 or 100 μm HQ reveal significantly higher levels of 5hmC than unexposed controls. 5hmC was also enriched at ORF2 of the retrotransposon LINE-1 12 h following HQ exposure, followed by a decrease in both 5mC and 5hmC at 24 h. As LINE-1 is estimated to comprise 15–19% of human genomic DNA (26), these results support the data from immunoblots of genome-wide increases in 5hmC and also show that the increase in 5hmC is followed by a decrease in genomic methylation, which suggests that 5hmC is an intermediate in DNA demethylation in HQ-exposed cells. The increase and decrease in 5hmC and 5mC, respectively, are also associated with gene expression. Enrichment of 5hmC at the promoters of GCLC and 14-3-3σ was associated with increased expression of their mRNAs after exposure to HQ. Because 5mC and 5hmC have different binding proteins, suggesting very different biological roles in epigenetic regulation, it is not clear whether demethylation is necessary for gene expression or if 5hmC formation alone is sufficient.
Previous studies involving chemical- and radiation-induced changes to cytosine suggest that direct oxidation of 5mC can lead to 5hmC (34, 35). The lack of 8-oxo-dG formation suggests that ROS resulting from 60 μm HQ does not directly oxidize 5mC. Interestingly, exposure to HQ exposure resulted in an increase in nuclear Tet activity. Moreover, siRNA-mediated knockdown of TET1 significantly decreased total TET activity and prevented HQ-mediated 5hmC formation. We also detected higher levels of nuclear TET1 by Western blotting with no change in mRNA levels (data not shown). Although it is unclear how protein levels are affected by HQ, TET1 has been shown to form multimers via oxidation (12), raising the possibility that its nuclear transport import or export was altered. It is also possible that the increase in activity was due to post-translational modifications that were caused by ROS. Importantly, the increase in activity is likely due to changes in protein levels or post-translational modifications and not in cofactors, such as α-ketoglutarate, because an excess of cofactors is included in the TET activity assay. Finally, the involvement of a TET1-mediated DNA demethylation mechanism was supported by the increase in GFP expression in cells transfected with the cytidine deaminase AP2.
HQ exposure has long been known to increase ROS and affect the cell cycle, an effect attributed to disruption of microtubule assembly through sulfhydryl reactivity (18, 21). Here we demonstrate that TET1 mediates the expression of 14-3-3σ, a p53 target gene, in response to HQ exposure. Moreover, TET1 partially mediates the HQ-induced accumulation of cells in G2+M phases of the cell cycle, which could be due to its influence over 14-3-3σ. It is possible that TET1 is involved in preventing cells from progressing through the cell cycle when ROS levels are elevated. It is not clear whether TET proteins play a role in carcinogenesis or cell stress responses, although the role of TET1 in HQ-mediated expression of genes involved in cell cycle regulation and antioxidant defense warrants further study. Tumor tissues are reportedly low in 5hmC levels, and loss of TET expression has been demonstrated in malignant glioblastoma (36) and hepatocellular carcinoma samples (37), suggesting that loss of TET activity may contribute to tumorigenesis. To our knowledge, this is the first study to demonstrate that responses to ROS-mediated perturbations in the cell cycle are modulated by TET1.
In summary, we have shown that a ROS-inducing chemical increases genome-wide and gene-specific 5hmC formation through TET1 methylcytosine dioxygenase. We suggest that TET1 represents a link between cellular redox status and maintenance of the epigenome. Future studies will examine whether the observed effects are unique to HQ or shared by other xenobiotics that increase ROS.
Acknowledgments
We thank Drs. Hongjun Song (Johns Hopkins University) and Junjie U. Guo (Massachusetts Institute of Technology) for providing reagents.
This work was supported, in whole or in part, by National Institutes of Health Grant 1U18TR000547. This work was also supported by Johns Hopkins University Center in Urban and Environmental Health Grant 30 ES003819.
- DNMT
- DNA methyltransferase
- ANOVA
- analysis of variance
- AP2
- APOBEC2
- DCF-DA
- 2′,7′-dichlorodihydrofluorescein diacetate
- GCLC
- glutamate-cysteine ligase catalytic subunit
- 5hmC
- 5-hydroxymethylcytosine
- HQ
- hydroquinone
- 5mC
- 5-methylcytosine
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium
- NAC
- N-acetylcysteine
- 8-oxo-dG
- 8-hydroxy-2′-deoxyguanosine
- qRT-PCR
- quantitative RT-PCR
- ROS
- reactive oxygen species
- TET1
- Ten Eleven Translocation 1.
REFERENCES
- 1. Jaenisch R., Bird A. (2003) Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat. Genet. 33, 245–254 [DOI] [PubMed] [Google Scholar]
- 2. Tsai H. C., Baylin S. B. (2011) Cancer epigenetics: linking basic biology to clinical medicine. Cell Res. 21, 502–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Millan M. J. (2013) An epigenetic framework for neurodevelopmental disorders: from pathogenesis to potential therapy. Neuropharmacology 68, 2–82 [DOI] [PubMed] [Google Scholar]
- 4. Ishida K., Kobayashi T., Ito S., Komatsu Y., Yokoyama T., Okada M., Abe A., Murasawa A., Yoshie H. (2012) Interleukin-6 gene promoter methylation in rheumatoid arthritis and chronic periodontitis. J. Periodontol. 83, 917–925 [DOI] [PubMed] [Google Scholar]
- 5. Ponferrada-Marín M. I., Martínez-Macías M. I., Morales-Ruiz T., Roldán-Arjona T., Ariza R. R. (2010) Methylation-independent DNA binding modulates specificity of repressor of silencing 1 (ROS1) and facilitates demethylation in long substrates. J. Biol. Chem. 285, 23032–23039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Christman J. K. (2002) 5-Azacytidine and 5-aza-2′-deoxycytidine as inhibitors of DNA methylation: mechanistic studies and their implications for cancer therapy. Oncogene 21, 5483–5495 [DOI] [PubMed] [Google Scholar]
- 7. Guo J. U., Ma D. K., Mo H., Ball M. P., Jang M. H., Bonaguidi M. A., Balazer J. A., Eaves H. L., Xie B., Ford E., Zhang K., Ming G. L., Gao Y., Song H. (2011) Neuronal activity modifies the DNA methylation landscape in the adult brain. Nat. Neurosci. 14, 1345–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. O'Driscoll C. M., Coulter J. B., Bressler J. P. (2013) Induction of a trophoblast-like phenotype by hydralazine in the P19 embryonic carcinoma cell line. Biochim. Biophys. Acta 1833, 460–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bollati V., Baccarelli A., Hou L., Bonzini M., Fustinoni S., Cavallo D., Byun H. M., Jiang J., Marinelli B., Pesatori A. C., Bertazzi P. A., Yang A. S. (2007) Changes in DNA methylation patterns in subjects exposed to low-dose benzene. Cancer Res. 67, 876–880 [DOI] [PubMed] [Google Scholar]
- 10. Lee B. H., Yegnasubramanian S., Lin X., Nelson W. G. (2005) Procainamide is a specific inhibitor of DNA methyltransferase 1. J. Biol. Chem. 280, 40749–40756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lee W. J., Zhu B. T. (2006) Inhibition of DNA methylation by caffeic acid and chlorogenic acid, two common catechol-containing coffee polyphenols. Carcinogenesis 27, 269–277 [DOI] [PubMed] [Google Scholar]
- 12. Tahiliani M., Koh K. P., Shen Y., Pastor W. A., Bandukwala H., Brudno Y., Agarwal S., Iyer L. M., Liu D. R., Aravind L., Rao A. (2009) Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324, 930–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ito S., D'Alessio A. C., Taranova O. V., Hong K., Sowers L. C., Zhang Y. (2010) Role of TET proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 466, 1129–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Guo J. U., Su Y., Zhong C., Ming G. L., Song H. (2011) Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell 145, 423–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ong C. N., Lee B. L., Shi C. Y., Ong H. Y., Lee H. P. (1994) Elevated levels of benzene-related compounds in the urine of cigarette smokers. Int. J. Cancer 59, 177–180 [DOI] [PubMed] [Google Scholar]
- 16. International Agency for Research on Cancer (1987) Overall evaluations of carcinogenicity: an updating of IARC monographs volumes 1 to 42. IARC Monogr. Eval. Carcinog. Risks Hum. Suppl. 7, 1–440 [PubMed] [Google Scholar]
- 17. Parke D. V., Williams R. T. (1953) The metabolism of benzene containing C14 benzene. Biochem. J. 54, 231–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Snyder R., Hedli C. C. (1996) An overview of benzene metabolism. Environ. Health Perspect. 104, 1165–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kolachana P., Subrahmanyam V. V., Meyer K. B., Zhang L., Smith M. T. (1993) Benzene and its phenolic metabolites produce oxidative DNA damage in HL60 cells in vitro and in the bone marrow in vivo. Cancer Res. 53, 1023–1026 [PubMed] [Google Scholar]
- 20. Shen D. X., Shi X., Fu J. L., Zhang Y. M., Zhou Z. C. (2003) The role of thiol reduction in hydroquinone-induced apoptosis in HEK293 cells. Chem. Biol. Interact. 145, 225–233 [DOI] [PubMed] [Google Scholar]
- 21. Parmentier R., Dustin P., Jr. (1948) Early effects of hydroquinone on mitosis. Nature 161, 527. [DOI] [PubMed] [Google Scholar]
- 22. Ji Z., Zhang L., Peng V., Ren X., McHale C. M., Smith M. T. (2010) A comparison of the cytogenetic alterations and global DNA hypomethylation induced by the benzene metabolite, hydroquinone, with those induced by melphalan and etoposide. Leukemia 24, 986–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Muotri A. R., Marchetto M. C., Coufal N. G., Oefner R., Yeo G., Nakashima K., Gage F. H. (2010) L1 retrotransposition in neurons is modulated by MeCP2. Nature 468, 443–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Westfall M. D., Mays D. J., Sniezek J. C., Pietenpol J. A. (2003) The ΔNp63α phosphoprotein binds the p21 and 14-3-3σ promoters in vivo and has transcriptional repressor activity that is reduced by Hay-Wells syndrome-derived mutations. Mol. Cell. Biol. 23, 2264–2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Du W., Rani R., Sipple J., Schick J., Myers K. C., Mehta P., Andreassen P. R., Davies S. M., Pang Q. (2012) The FA pathway counteracts oxidative stress through protection of antioxidant defense gene promoters. Blood 119, 4142–4151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lander E. S., Linton L. M., Birren B., Nusbaum C., Zody M. C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Funke R., Gage D., Harris K., Heaford A., Howland J., Kann L., Lehoczky J., LeVine R., McEwan P., McKernan K., Meldrim J., Mesirov J. P., Miranda C., Morris W., Naylor J., Raymond C., Rosetti M., Santos R., Sheridan A., Sougnez C., Stange-Thomann N., Stojanovic N., Subramanian A., Wyman D., Rogers J., Sulston J., Ainscough R., Beck S., Bentley D., Burton J., Clee C., Carter N., Coulson A., Deadman R., Deloukas P., Dunham A., Dunham I., Durbin R., French L., Grafham D., Gregory S. (2001) Initial sequencing and analysis of the human genome. Nature 409, 860–921 [DOI] [PubMed] [Google Scholar]
- 27. Hermeking H., Lengauer C., Polyak K., He T. C., Zhang L., Thiagalingam S., Kinzler K. W., Vogelstein B. (1997) 14-3-3σ is a p53-regulated inhibitor of G2/M progression. Mol. Cell 1, 3–11 [DOI] [PubMed] [Google Scholar]
- 28. Lodygin D., Hermeking H. (2005) The role of epigenetic inactivation of 14-3-3σ in human cancer. Cell Res. 15, 237–246 [DOI] [PubMed] [Google Scholar]
- 29. Ferguson A. T., Evron E., Umbricht C. B., Pandita T. K., Chan T. A., Hermeking H., Marks J. R., Lambers A. R., Futreal P. A., Stampfer M. R., Sukumar S. (2000) High frequency of hypermethylation at the 14-3-3σ locus leads to gene silencing in breast cancer. Proc. Natl. Acad. Sci. U.S.A. 97, 6049–6054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yi B., Tan S. X., Tang C. E., Huang W. G., Cheng A. L., Li C., Zhang P. F., Li M. Y., Li J. L, Yi H., Peng F., Chen Z. C., Xiao Z. Q. (2009) Inactivation of 14-3-3σ by promoter methylation correlates with metastasis in nasopharyngeal carcinoma. J. Cell. Biochem. 106, 858–866 [DOI] [PubMed] [Google Scholar]
- 31. O'Hagan H. M., Wang W., Sen S., Destefano Shields C., Lee S. S., Zhang Y. W., Clements E. G., Cai Y., Van Neste L., Easwaran H., Casero R. A., Sears C. L., Baylin S. B. (2011) Oxidative damage targets complexes containing DNA methyltransferases, SIRT1, and polycomb members to promoter CpG islands. Cancer Cell 20, 606–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lertratanangkoon K., Wu C. J., Savaraj N., Thomas M. L. (1997) Alterations of DNA methylation by glutathione depletion. Cancer Lett. 120, 149–156 [DOI] [PubMed] [Google Scholar]
- 33. Cerda S., Weitzman S. A. (1997) Influence of oxygen radical injury on DNA methylation. Mutat. Res. 386, 141–152 [DOI] [PubMed] [Google Scholar]
- 34. Castro G. D., Díaz Gómez M. I., Castro J. A. (1996) 5-Methylcytosine attack by hydroxyl free radicals and during carbon tetrachloride promoted liver microsomal lipid peroxidation: structure of reaction products. Chem. Biol. Interact. 99, 289–299 [DOI] [PubMed] [Google Scholar]
- 35. Wagner J. R., Cadet J. (2010) Oxidation reactions of cytosine DNA components by hydroxyl radical and one-electron oxidants in aerated aqueous solutions. Acc. Chem. Res. 43, 564–571 [DOI] [PubMed] [Google Scholar]
- 36. Orr B. A., Haffner M. C., Nelson W. G., Yegnasubramanian S., Eberhart C. G. (2012) Decreased 5-hydroxymethylcytosine is associated with neural progenitor phenotype in normal brain and shorter survival in malignant glioma. PLoS One 7, e41036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liu C., Liu L., Chen X., Shen J., Shan J., Xu Y., Yang Z., Wu L., Xia F., Bie P., Cui Y., Bian X. W., Qian C. (2013) Decrease of 5-hydroxymethylcytosine is associated with progression of hepatocellular carcinoma through down-regulation of TET1. PLoS One 8, e62828. [DOI] [PMC free article] [PubMed] [Google Scholar]