Background: The Lewis X carbohydrate antigen is abundantly expressed in several stem cell populations.
Results: Fut10 is responsible for the synthesis of unique types of Lewis X on N-glycans and alters stem cell functions.
Conclusion: Fut10 are required for the maintenance of stem cells and neural development.
Significance: Learning Fut10 function is crucial for understanding how stemness is maintained.
Keywords: Embryonic Stem Cell, Glycobiology, Glycosyltransferases, HPLC, Neural Stem Cell, Bisecting N-Glycans, Stemness
Abstract
Lewis X (LeX, Galβ1–4(Fucα1–3)GlcNAc) is a carbohydrate epitope that is present at the nonreducing terminus of sugar chains of glycoproteins and glycolipids, and is abundantly expressed in several stem cell populations. LeX antigen can be used in conjunction with fluorescence-activated cell sorting to isolate neurosphere-forming neural stem cells (NSCs) from embryonic mouse brains. However, its function in the maintenance and differentiation of stem cells remains largely unknown. In this study, we examined mice deficient for fucosyltransferase 9 (Fut9), which is thought to synthesize most, if not all, of the LeX moieties in the brain. We found that the number of NSCs was increased in the brain of Fut9−/− embryos, suggesting that Fut9-synthesized LeX is dispensable for the maintenance of NSCs. Another α1,3-fucosyltransferase gene, fucosyltransferase 10 (Fut10), is expressed in the ventricular zone of the embryonic brain. Overexpression of Fut10 enhanced the self-renewal of NSCs. Conversely, suppression of Fut10 expression induced the differentiation of NSCs and embryonic stem cells. In addition, knockdown of Fut10 expression in the cortical ventricular zone of the embryonic brain by in utero electroporation of Fut10-miRNAs impaired the radial migration of neural precursor cells. Our data suggest that Fut10 is involved in a unique α1,3-fucosyltransferase activity with stringent substrate specificity, and that this activity is required to maintain stem cells in an undifferentiated state.
Introduction
The Lewis X (LeX)2 (Galβ1–4(Fucα1–3)GlcNAc) carbohydrate epitope, which is present in antigens recognized by antibodies for stage-specific embryonic antigen-1 or leukocyte cluster of differentiation 15 (CD15), is a marker of the inner cell mass of the mouse blastocyst and its derivative, embryonic stem (ES) cells. LeX expression is progressively down-regulated in the early mouse embryo as development proceeds, and in differentiating ES cells in vitro (1–4). LeX is also expressed by neural precursor cells in the germinal zone of the embryonic mouse telencephalon (5–7). Neural stem cells (NSCs) and their progeny, neural progenitor cells, are collectively referred to be neural precursor cells and are present not only in embryonic brains but also in the subependyma of adult brains. When cultured in a colony-forming neurosphere assay, LeX-positive cells from the adult mouse subependyma generated more neurospheres than LeX-negative cells (8), suggesting that neurosphere-forming NSCs in the adult brain abundantly express LeX. Although the expression of LeX is considered to be tightly associated with the stemness of various types of stem cells, the functional roles of LeX in neural development and the maintenance of stem cell pluripotency remain to be investigated.
LeX is synthesized by adding fucose with an α1,3-linkage to the N-acetylglucosamine (GlcNAc) of the N-acetyllactosamine backbone at the nonreducing terminus of sugar chains (9). This reaction is catalyzed by α1,3-fucosyltransferases. Three active α1,3-fucosyltransferases, Fut4, Fut7, and Fut9, exist in rodents and are well characterized. Fut4 and Fut9 are expressed in the brain. Fut9 is thought to synthesize most, if not all, of the LeX moieties on glycoproteins and glycolipids in the brain (10). Despite the expected roles of Fut9-synthesized LeX in neural development, Fut9-deficient mice exhibit minor alterations in brain morphology and subtle behavioral abnormalities (11, 12). Recently, two novel putative α1,3-fucosyltransferases, Fut10 and Fut11, have been identified in mice and humans on the basis of structural homology to other α1,3-fucosyltransferases (13). Neither Fut10 nor Fut11 increased the amount of LeX when overexpressed in cultured cells or exhibited enzymatic activities with conventional oligosaccharide substrates (14, 15). However, Fut10 was reported to transfer fucose to GlcNAc at the innermost core position of N-glycans (16), in contrast to the conventional α1,3-fucosyltransferases that add fucose to GlcNAc at the nonreducing end. In this study, we present evidence that Fut10 increases the amount of LeX structures on bisecting N-glycans of glycoproteins and plays an important role in the self-renewal of stem cells.
EXPERIMENTAL PROCEDURES
Mice and Genotyping
CD1 (ICR) mice (SLC, Japan) were used to isolate NSCs and for in utero electroporation except when specifically described. Noon of the plugged date was considered as embryonic day (E) 0.5. Mice heterozygous for Fut9 were maintained in the C57/BL6 background. PCR primers used for genotyping Fut9 mutants were: fut9 F300 (5′-ACAACAAATCCCATGCGGTC-3′), CB341R (5′-CATGTGATTCCCAAAACCG-3′), and fut9 KO R3 (5′-GCCATGATGGATACTTTCT-3′). DNA was obtained from the tip of the tail and DNA was amplified for 35 cycles in a thermal cycler (PerkinElmer Life Sciences) with denaturation at 95 °C for 30 s, annealing at 58 °C for 30 s, and extension at 72 °C for 45 s. All experiments were carried out with the permission of the institutional Animal Research Committee.
Cell Culture and Fluorescence-activated Cell Sorting
The protocol used to generate neurospheres in vitro has been described (17, 18). Briefly, cells from the embryonic cortex or medial ganglionic eminence (GE) were mechanically triturated in serum-free media and cultured in the presence of 10 ng/ml of FGF-2 and 2 μg/ml of heparin (Sigma). After 7 days in vitro, the number of floating sphere colonies (neurospheres) possessing a diameter >0.08 mm was counted. Stem cell self-renewal was assessed by counting the number of new neurospheres that arose after primary single neurospheres were mechanically dissociated into single cells, and cultured for 6 days in 0.2 ml of serum-free media containing 20 ng/ml of EGF, FGF2, and heparin.
Neuro2a and COS1 cells were maintained in 10% FBS/DMEM culture medium. R1 ES cells (19) were grown on gelatin-coated plates and maintained in 15% FBS/DMEM medium containing 1000 units/ml of LIF (Millipore), 0.8 μm of the MEK inhibitor PD0325901, and 1.0 μm of the GSK3 inhibitor CT99021 (both from Axon Medchem BV). Cells were transfected with expression plasmids by using Lipofectamine 2000 transfection reagent (Invitrogen). A flow cytometer (FACS Aria Diva; BD Biosciences) equipped with an argon ion laser (488 nm, 200 milliwatt) was used to collect GFP+ cells.
Plasmids and Retrovirus Preparation
Full-length Fut10 gene was amplified by RT-PCR from E14.5 mouse brain cDNA, sequenced, and subcloned into the pCX expression vector or pMXIE retroviral vector to generate retrovirus, which expresses Fut10 and GFP as described (20). The BLOCK-iTTM PolII miR RNAi Expression Vector Kit (Invitrogen) was used for constructing miRNA expression vector. Three 21-nucleotide sequences, two (#1, 5′-GAGAGCTGGCGAGCTTCATTA-3′ and #2, 5′-CGGCTTCTGACAGCTCTCAAT-3′) from the open reading frame and one (#3, 5′-TACTGGCTGTCTTCTGTGGAA-3′) from the 3′ untranslated region of Fut10, were selected. DNA fragments containing each pair of sense and antisense together with loop sequences were subcloned into the pcDNA6.2-GW/EmGFP or pMXIE vectors to generate Fut10 miRNA expression or retrovirus plasmids. A packaging cell line, ΦNX-Eco (ATCC 3443), was transiently transfected with the retrovirus plasmids using FuGENE 6 (Roche Applied Science) and cultured for 3 days at 30 °C. The supernatant was filtered, centrifuged to yield a titer more than 2 × 107 cfu/ml, and aliquoted at −80 °C until use. Cells were spin-infected at 1,000 rpm with 2 × 105 of retrovirus in the presence of 5 μg/ml of Polybrene (Sigma) at room temperature for 1.5 h.
Histochemistry and in Situ Hybridization
Brains were sectioned along the coronal plane into 14-μm thick sections, and then incubated with biotinylated lotus lectin (1:200; Vector Laboratories) at 4 °C overnight. Alternatively, cryosections were incubated with anti-LeX mouse monoclonal antibody (Clone MMA, Lab Vision Corporation), followed by a biotinylated goat anti-mouse IgM antibody (Vector Laboratories) at 4 °C overnight. The sections were subjected to a Tyramide Signal Amplification (TSATM) biotin system (PerkinElmer Life Sciences). LeX antigens were detected and visualized by incubating the sections with an avidin and biotinylated peroxidase complex (Vectastain Elite ABC kit, Vector Laboratories) and a peroxidase substrate (3,3′-diaminobenzidine; Dojindo).
Digoxygenin (DIG)-labeled single-stranded riboprobes directed against the entire coding region of the Fut4, Fut7, Fut9, or Fut10 genes were synthesized using the DIG RNA labeling mixture (Roche Applied Science). In situ hybridization was performed as described previously (21). Briefly, cryosections were treated with proteinase K (1 μg/ml, 60–90 min, room temperature), and then hybridized at 65 °C overnight in hybridization buffer with sense or antisense DIG-labeled RNA probes. DIG-labeled RNA probes were detected and visualized by incubating the sections with alkaline phosphatase-conjugated anti-DIG antibody (1:2000; Roche) and an alkaline phosphatase substrate (nitro blue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate).
RT-PCR
Total RNA isolation, cDNA synthesis, and RT-PCR analysis were carried out as described (20). The sense and antisense primers used were summarized in Table 1.
TABLE 1.
PCR primers
| Gene | Primer | Sequence |
|---|---|---|
| Fut4 | Sense | 5′-GACCTGGTGAAGGAACTCCA-3′ |
| Antisense | 5′-AGGCACGAAGCGCTCATAGT-3′ | |
| Fut7 | Sense | 5′-TGTGTTCGGTCGCGCCAGCG-3′ |
| Antisense | 5′-TCAAGCCTGGAACCAGCTTT-3′ | |
| Fut9 | Sense | 5′-ACAACAAATCCCATGCGGTC-3′ |
| Antisense | 5′-GTGGGAATCAGATTTTTATC-3′ | |
| Fut10 | Sense | 5′-CAGGGTGTGGGCAAACAGTAG-3′ |
| Antisense | 5′-GAATTAAGCTTACTCTGTCAGT-3′ | |
| Oct3/4 | Sense | 5′-CGTTCTCTTTGGAAAGGTGTTC-3′ |
| Antisense | 5′-TACTCGAACCACATCCTTCTCT-3′ | |
| Nanog | Sense | 5′-CACTGACATGAGTGTGGGTCTTC-3′ |
| Antisense | 5′-GGAGGAGAGTTCTTGCATCTGCT-3′ | |
| Cdx2 | Sense | 5′-CAAGGAGAGGAAAATCAAGAAGAAG-3′ |
| Antisense | 5′-CAAGGAGGTCACAGGACTCAAG-3′ | |
| Fgf5 | Sense | 5′-AAAGTCAATGGCTCCCACGAA-3′ |
| Antisense | 5′-GAACAGTGACGGTGAAGGAAA-3′ | |
| Sox2 | Sense | 5′-TCTGTGGTCAAGTCCGAGGCCA-3′ |
| Antisense | 5′-TGCGAAGCGCCTAACGTACCAC-3′ | |
| Brachyury | Sense | 5′-AGTATGAACCTCGGATTCAC-3′ |
| Antisense | 5′-CCGGTTGTTACAAGTCTCAG-3′ | |
| Gata4 | Sense | 5′-AGCCTACATGGCCGACGTGG-3′ |
| Antisense | 5′-TCAGCCAGGACCAGGCTGTT-3′ | |
| Hnf4 | Sense | 5′-CCATGGTGTTAAAGGACGTGC-3′ |
| Antisense | 5′-TAGGATTCAGATCCCGAGCC-3′ | |
| β-Actin | Sense | 5′-AGGCCAACCGTGAAAAGATGAC-3′ |
| Antisense | 5′-GTACATGGTGGTACCACCAGAC-3′ |
N-Glycan Analysis
N-Glycans of glycoproteins from cells or tissues were analyzed as described previously (22, 23). Briefly, samples were precipitated with acetone, lyophilized, and hydrazinolized to release N-glycans. For purification and in-column N-acetylation, the hydrazinolized sample solution was loaded onto a graphite carbon column (GL-Pak Carbograph, GL Science), washed with 50 mm ammonium acetate buffer (pH 7.0), and then eluted with 50 mm triethylamine acetate buffer (pH 7.0) in 60% acetonitrile. The reducing ends of liberated N-glycans were tagged with the fluorophore 2-aminopyridine (PA) and the PA-tagged N-glycans were purified using a cellulose column (Takara Bio).
PA-sugar chains were separated by high-performance liquid chromatography (HPLC) as reported previously (22, 23). Briefly, PA-tagged N-glycans were applied onto a Microgranular DE52-packed column (GE Healthcare) to obtain neutral N-glycans. The neutral N-glycans were size fractionated by normal phase HPLC with a Shodex Asahipak NH2P-50 4E column (Showa Denko), and each fraction was further separated by reverse-phase HPLC with a Develosil C30-UG-5 column (Nomura Chemical). PA-sugar chains were detected at excitation and emission wavelengths of 320 and 400 nm, respectively. N-Glycans were identified by calculating the mannose unit value from normal phase HPLC and the glucose unit value from reverse-phase HPLC.
In Vitro Fucosyltransferase Assay
COS1 cells transfected with plasmids that express Fut9 (pCX-Fut9) or Fut10 (pCX-Fut10) were washed in 25 mm MES buffer (pH 7.4) containing 20 mm MnCl2 for 10 min, and then harvested by scraping. Cells were pelleted by centrifugation and resuspended in 100 μl of cold 1% Triton X-100. The samples were sonicated briefly, then centrifuged at 17,800 × g for 10 min at 4 °C. The supernatant, which was used as a source of Fut9 and Fut10, was incubated with substrate (PA-sugar chains or Neuro2a cell extracts) in 50 mm cacodylate buffer (pH 6.8), 5 mm ATP, 75 μm GDP-l-fucose (Calbiochem), and 25 mm MnCl2 at 37 °C for 2 h or overnight. The enzyme reaction was terminated by incubating the reaction mixtures at 98 °C for 3 min. The reaction mixtures were centrifuged at 17,800 × g for 5 min at 4 °C. Each supernatant was subjected to normal phase HPLC analysis. When Neuro2a cell extracts, prepared as described above, were used as a substrate, the N-glycans of glycoproteins in the reaction mixture were released by hydrazinolysis, and then tagged with PA. The PA-tagged N-glycans were analyzed by normal and reverse-phase HPLC.
In Utero Electroporation
In utero electroporation was performed as described previously (21). Three μg of plasmids in 1 μl of a solution containing 0.05% fast green was microinjected through the uterus into the lateral ventricle of E14.5 fetal brains. After injecting the plasmids, the embryos were placed between 2 electrodes (CUY650-P5, NEPA GENE). An electroporator (CUY21, NEPA GENE) was used to deliver six 50-ms pulses of 35 V at 75-ms intervals.
RESULTS
Increased Numbers of Neural Stem Cells in the Fut9−/− Embryonic Brain
To investigate the effects of Fut9 gene disruption on the NSC pool size in embryonic mouse brains, we utilized a clonal colony-forming neurosphere assay (17, 18). Contrary to the expectation that Fut9-synthesized LeX plays a significant role in the maintenance of NSCs, the number of NSCs in the brain of Fut9−/− embryos was greater than that in brains of their littermate controls (Fig. 1A). The difference between the number of NSCs in the dorsal forebrain of Fut9−/− embryos and in brains of control embryos was statistically significant (t(25) = 5.559, p < 0.001), but the difference in the number of NSCs in the ventral forebrain of brains of Fut9−/− embryos and brains of control embryos was not statistically significant (t(18) = 0.705, p = 0.490).
FIGURE 1.
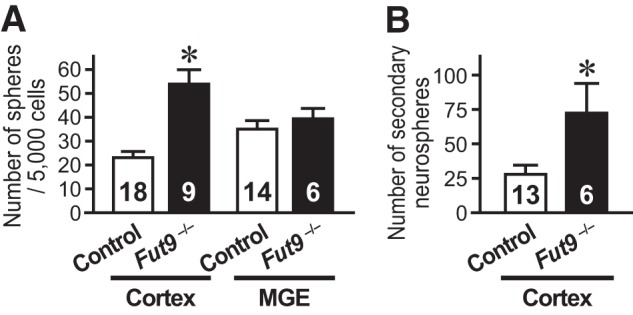
Cortical neural stem cell numbers are increased in the E15.5 Fut9−/− brain. A, NSCs isolated from the cortex or medial GE of E15.5 Fut9−/− embryos or littermate control embryos are shown. B, to assess the self-renewal capacity of primary sphere-forming NSCs, single primary neurosphere colonies of similar size were manually dissociated and recultured. More secondary spheres were formed from each primary Fut9−/− sphere than from each primary control sphere. Error bars indicate S.E. and n values are shown within the columns. *, p < 0.05 by Student's t test.
We assessed the self-renewal capacity of NSCs isolated from the embryonic brain by passaging single primary neurospheres and counting the resultant secondary neurospheres, because the number of secondary neurospheres reflects how many times an NSC undergoes symmetric expansive divisions during primary neurosphere formation. A greater number of secondary neurospheres was generated from Fut9−/− primary neurospheres than from control primary neurospheres (t(17) = 2.556, p = 0.021; Fig. 1B). These results suggest that Fut9-synthesized LeX is dispensable for the NSC maintenance in the dorsal forebrain of mouse embryos. If LeX is required to maintain NSCs in an undifferentiated state, a specific LeX could be synthesized by one of the other α1,3-fucosyltransferases.
Fut10 Is Expressed by Lex-positive Neural Precursor Cells in the Embryonic Brain
The fucose-binding lectin Lotus tetragonolobus agglutinin (LTA) binds specifically to the LeX sugar chain Galβ1–4(Fucα1–3)GlcNAcβ1–3-R. LTA does not bind glycans containing either sialyl LeX or VIM-2 determinants, nor does it bind the isomeric Lea, Galβ1–3(Fucα1–4)GlcNAc-R (24). Thus, biotinylated lotus lectin was used to examine the expression pattern of LeX in the embryonic brain. Lectin staining of E15.5 brains indicates that the LeX epitope is present in the ventricular zone (VZ) and the subventricular zone (SVZ) around the lateral ventricles, the upper cortical layers, and the mantle zone of the GE (Fig. 2A). To examine which glycoconjugates, glycoproteins or glycolipids, convey the lectin-binding LeX moieties, we pretreated the sections with acetone/chloroform or peptide:N-glycosidase F before the LTA staining. We observed positive signals in the sections pretreated with acetone/chloroform, which were comparable with those in the non-treated one (Fig. 2B), but no signal in the peptide:N-glycosidase F-pretreated sections (Fig. 2C). These results suggest that LeX epitopes attach to N-glycans of glycoproteins.
FIGURE 2.
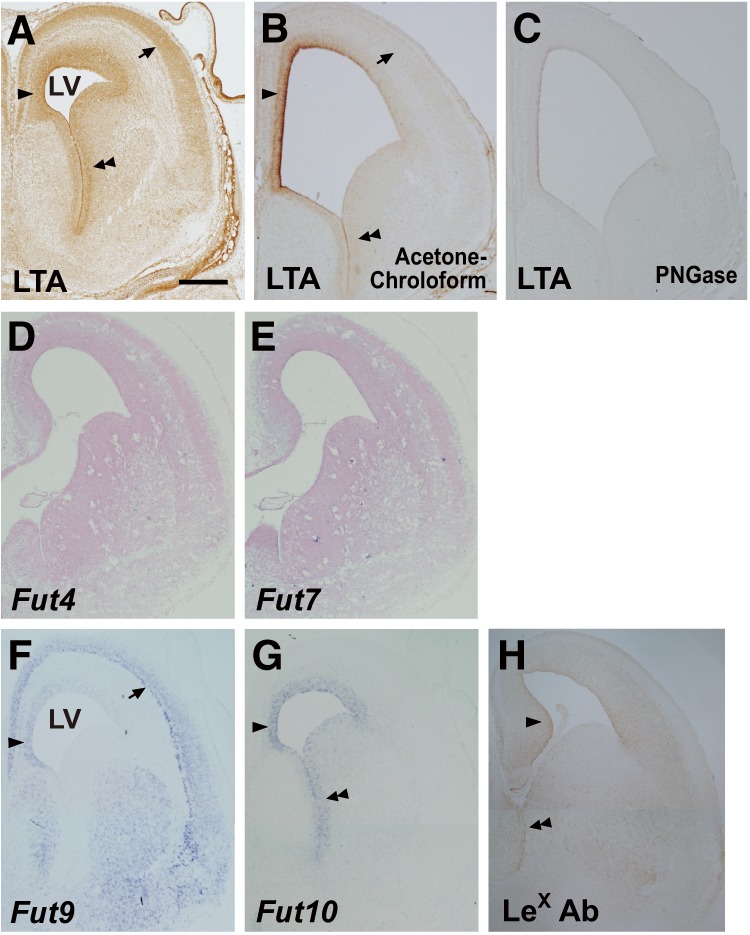
Expression of LeX and α1,3-fucosyltransferase genes. A–C, coronal cryosections of E15.5 brains were stained with LTA lectin, which were non-treated (A) or pretreated with acetone/chloroform to release glycolipids (B) or peptide:N-glycosidase F (PNGase) to release N-glycans (C). D–G, the cryosections were subjected to in situ hybridization with probes directed against Fut4 (D), Fut7 (E), Fut9 (F), or Fut10 (G). LTA staining and an in situ hybridization signal for Fut9 are visible in the upper cortical layers (arrows in A and F) and the dorsomedial portion of the VZ/SVZ (single arrowheads in A and F), whereas LTA staining and the in situ hybridization signal for Fut10 overlap in the VZ/SVZ of the cortex and GE (single and double arrowheads in A and G). H, coronal cryosections of E15.5 Fut9−/− brains were immunostained with anti-LeX antibody. Positive signal was detected in the VZ/SVZ (single and double arrowheads). Scale bar = 0.5 mm.
To determine which fucosyltransferases could synthesize LeX in the embryonic brain, we performed in situ hybridization with probes directed against Fut4, Fut7, Fut9, and Fut10. Fut4 and Fut7 were barely expressed in the VZ/SVZ (Fig. 2, D and E). In contrast, Fut9 was expressed in the dorsomedial aspect, but not the ventrolateral aspect, of the VZ/SVZ (Fig. 2F). This aspect of Fut9 expression explains the above findings that the brains of Fut9−/− embryos have an increased number of NSCs in the cortical VZ/SVZ but not in the subcortical VZ/SVZ. The expression patterns of LeX and Fut9 overlapped completely in the upper cortical layers and the GE mantle zone, suggesting that migrating neuroblasts from the VZ/SVZ up-regulate Fut9 expression and increase LeX synthesis. We next examined the expression of Fut10, and discovered that mRNA for this gene was abundant throughout the entire VZ/SVZ of the embryonic brain (Fig. 2G). Finally, we investigated the expression of LeX antigens in E15.5 Fut9−/− brains by immunostaining using an anti-LeX antibody and detected positive signals in the VZ/SVZ (Fig. 2H). These results suggest that Fut10 could be an active α1,3-fucosyltransferase, although its enzymatic activities to synthesize LeX antigens have not been provided.
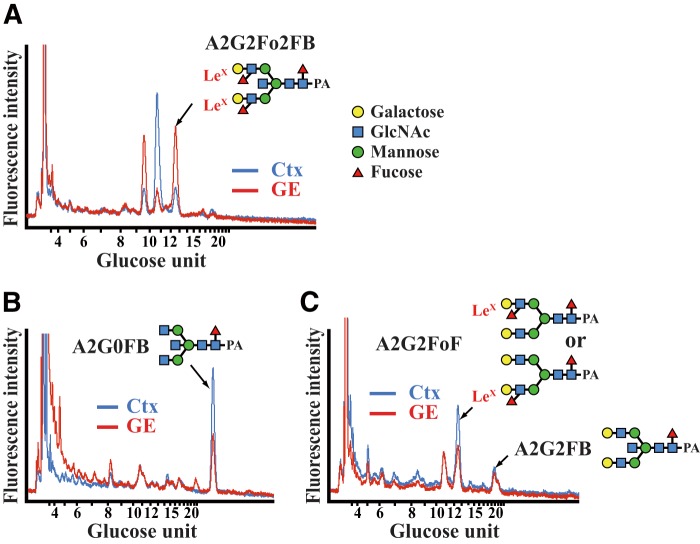
To verify the presence of LeX antigens in the embryonic brain, we used HPLC to analyze the N-glycans of the dorsomedial cortex and the VZ/SVZ of the GE of E15.5 brains. LeX antigens were detected on the N-glycans of both tissues. The LeX-containing bisecting N-glycan, A2G2Fo2FB, was more abundant in the VZ/SVZ of the GE than in the cortex (Fig. 3A), despite the fact that its substrate, A2G0FB, was expressed more strongly in the cortex than in the VZ/SVZ of the GE and that A2G2FB was comparable in these tissues (Fig. 3, B and C). The LeX-containing biantennary N-glycan, A2G2FoF, was also expressed in the cortex (Fig. 3C). These findings suggest that Fut10 increases the amount of LeX by preferentially attaching fucose to bisecting N-glycans. We cannot exclude the possibilities that a scarce amount of Fut9 is expressed and synthesizes A2G2Fo2FB in the VZ/SVZ of the GE or that the sample was contaminated with cells from the underlying mantle zone, which express Fut9 and could synthesize A2G2Fo2FB. However, these possibilities are unlikely because the amount of A2G2Fo2FB was much less in the dorsomedial cortex, where Fut9 is strongly expressed, than in the VZ/SVZ of the GE. Thus, these results suggest that Fut10 could be or be related to a unique α1,3-fucosyltransferase with stringent substrate specificity.
FIGURE 3.
Identification of LeX-containing N-glycans. A–C, N-glycans were isolated from the dorsomedial cortex (Ctx) and the VZ/SVZ of the GE of E15.5 brains, and then analyzed by HPLC. Reverse-phase HPLC charts of fraction 8.2 (mannose unit 8.50–8.83, A), fraction 5.2 (mannose unit 5.50–5.83, B), and fraction 7.2 (mannose unit 7.50–7.83, C) are shown.
Fut10 Is Responsible for the Synthesis of Unique Types of Lex on N-Glycans
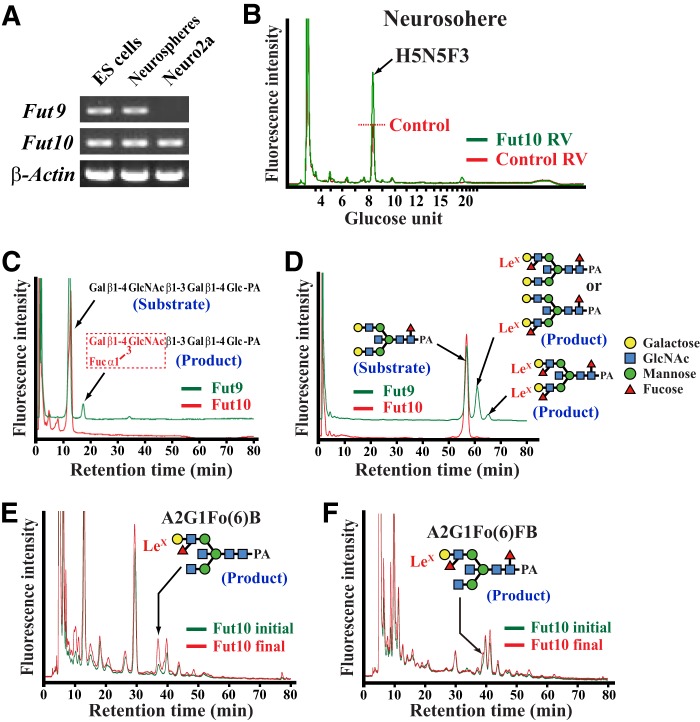
To investigate whether Fut10 exhibits α1,3-fucosyltransferase activity and synthesizes LeX antigens on N-glycans, we overexpressed Fut10 in neurospheres derived from the GE of the E15.5 brain, which expresses both Fut9 and Fut10 (Fig. 4A). HPLC analysis of neurospheres infected with Fut10-expressing retroviruses revealed that a peak in fraction 8.2 (mannose unit 8.50–8.83) increased. We used mass spectrometry to determine that the peak consists of 5 hexoses, 5 N-acetylhexosamines, and 3 fucoses (Fig. 4B). Although we could not identify its structure, this N-glycan likely contains 2 or 3 outer fucoses (e.g. stereoisomers of A2G2Fo2FB).
FIGURE 4.
Fut10-related α1,3-fucosyltransferase activities. A, RT-PCR for Fut9, Fut10, and β-actin expression in ES cells, Neuro2a cells, and neurospheres derived from the E15.5 ganglionic eminence. B, reverse-phase HPLC elution patterns of PA-tagged N-glycans from neurospheres infected with Fut10 (green) or control (red) retroviruses (RV) are shown. Reverse-phase HPLC was conducted on normal phase HPLC fraction 8.2 (mannose unit 8.50–8.83). C and D, sugar chain substrates were mixed with COS1 cell extracts, which served as a source of Fut9 or Fut10 enzyme activity, and then the reaction mixture was analyzed by normal phase HPLC. Whereas Fut9-mediated Lex synthesis generated lacto-N-fuconeopentaose from lacto-N-neotetraose (C) and A2G2Fo(3 or 6)FB and A2G2Fo2FB from the bisecting N-glycan substrate A2G2FB (D), no Fut10 activity was detected on these substrates. E and F, Neuro2a cell extracts were used as a source of substrates and mixed with the Fut10 enzyme source. N-Glycans were extracted from the reaction mixture and analyzed by HPLC. Normal phase HPLC fractions that contain N-glycans with mannose unit 7–8 (E) and 8–9 (F) were further separated by reverse-phase HPLC. Incubation with Fut10 increased the peak of LeX-containing A2G1Fo(6)B (E) and A2G1Fo(6)FB (F).
We performed in vitro α1,3-fucosyltransferase assays to confirm the enzymatic activity of Fut10 and determine its substrate specificity (25). Cell extracts of COS1 cells that overexpress Fut9 or Fut10 were used as a source of enzymes because naive untransfected cells exhibit no α1,3-fucosyltransferase activities. The first sugar substrate we tested in the assay was PA-tagged lacto-N-neotetraose. Although Fut9 produced LeX by attaching fucose to this substrate, Fut10 showed no activity (Fig. 4C). We then tested the bisecting N-glycan A2G2FB. Although Fut9 synthesized LeX at the nonreducing terminal to generate A2G2Fo(3 or 6)FB and A2G2Fo2FB, Fut10 did not (Fig. 4D). Finally, we tested if Fut10 exhibited α1,3-fucosyltransferase activity with glycoprotein substrates present in Neuro2a neuroblastoma cell extracts, because Neuro2a cells express very little Fut9 (Fig. 4A). After incubation, a reaction mixture of Neuro2a cell extracts and control COS1 cell extracts showed no apparent increase of LeX-containing N-glycans. However, levels of both A2G1Fo(6)FB and A2G1Fo(6)B increased after incubating a mixture of Neuro2a cell extracts and Fut10-overexpressing COS1 cell extracts (Fig. 4, E and F). We did not detect increased levels of A2G1Fo(6)FB or A2G1Fo(6)B in a reaction consisting only of Fut10-overexpressing COS1 cell extracts, indicating that Fut10 increases the amount of LeX on the bisecting N-glycans of glycoproteins that are specifically expressed on Neuro2a neuroblastoma cells. Although it remains to be investigated whether or not Fut10 itself is an α1,3-fucosyltransferase, these results suggest that Fut10 is involved in an α1,3-fucosyltransferase activity with strict substrate specificity and that this activity synthesizes LeX only on restricted glycoprotein substrates.
Fut10 Is Required for the Maintenance of ES Cells and Neural Stem Cells
To investigate the function of Fut10 and the LeX that it synthesizes, we used miRNAs to knockdown gene expression. Three miRNA-expressing vectors were constructed: two target sequences within the coding region and one targets a sequence in the 3′ UTR (Fig. 5A). All miRNAs also expressed enhanced green fluorescent protein. Individually or in combination, the miRNAs efficiently down-regulated endogenous Fut10 expression in Neuro2a cells (Fig. 5, B and C). Next, we transfected ES cells with the Fut10-miRNA vectors, and collected the GFP+ cells 48 h later by using fluorescence-activated cell sorting. We confirmed that Fut10 expression was knocked down in the transfected ES cells (t(8) = 2.852, p = 0.021; Fig. 6A). The expression of pluripotent stem cell-specific genes was significantly down-regulated in the Fut10-miRNA transfectants (t(8) = 4.758, p = 0.001 for Oct3/4 and t(8) = 4.072, p = 0.004 for Nanog; Fig. 6B), suggesting that Fut10 expression is required to maintain ES cells in an undifferentiated state. We also examined the expression of Sox2, an early neural marker, and Gata4, a primitive endodermal marker. Both genes were selectively up-regulated in Fut10-miRNA overexpressing ES cells (t(6) = 3.366, p = 0.015 for Sox2 and t(4) = 3.500, p = 0.025 for Gata4; Fig. 6C). It is possible that knockdown of Fut10 caused the ES cells to differentiate into cells that are equivalent to those of the cylinder stage epiblast, which expresses both Sox2 and Gata4 but not Brachyury, a mesodermal marker.
FIGURE 5.
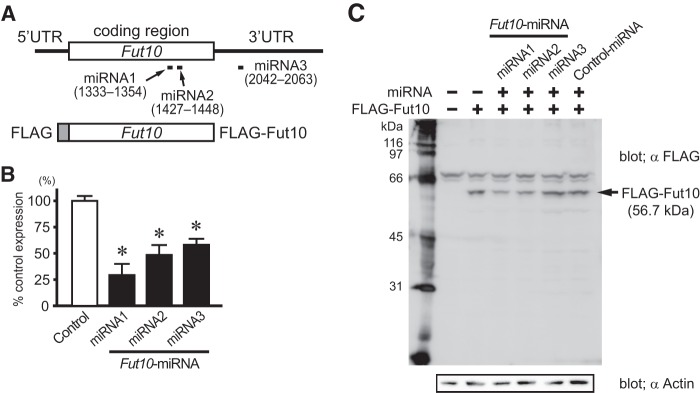
Fut10-miRNAs knockdown the expression of Fut10. A, full-length Fut10, FLAG-tagged Fut10, and the target sequences of the miRNAs are shown. B, Fut10-miRNAs were transfected into Neuro2a cells and Fut10 mRNA was quantified by quantitative RT-PCR 3 days later. The expression level of Fut10 relative to that of β-actin is shown. Values are mean ± S.E. (n = 3). *, p < 0.05. C, Fut10-miRNAs were transfected into Neuro2a cells and the cells were subjected to Western blotting. Fut10-miRNA1 and miRNA2, but not miRNA3, knocked down the expression of FLAG-tagged Fut10.
FIGURE 6.
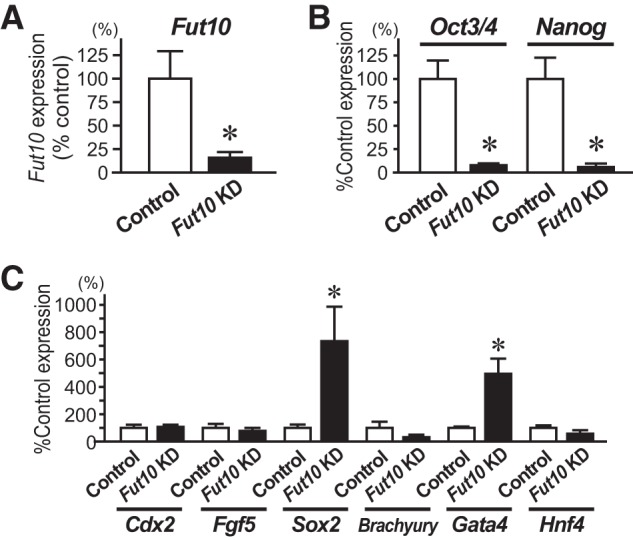
Knockdown of Fut10 enhances ES cell differentiation. A–C, ES cells were transfected with Fut10-miRNA expression plasmids and transfectants were collected 48 h later by cell sorting for GFP fluorescence. Quantitative RT-PCR was performed for Fut10 (A), undifferentiated cell markers (B), and differentiated cell markers (C) and the expression levels of those genes relative to that of β-actin are shown. Error bars indicate S.E. from three or more independent experiments. *, p < 0.05 by Student's t test.
To investigate the function of Fut10 in NSCs, we infected primary neurospheres with retrovirus expressing Fut10, Fut10-miRNA, or Fut9. First, we examined expression levels of active α1,3-fucosyltransferase genes and found that overexpression or knockdown of Fut10 did not induce the expression of Fut4, Fut7, or Fut9 (Fig. 7A). We then assessed the function of Fut10 expression in the maintenance of NSCs by passaging single retrovirus-infected neurospheres derived from the embryonic medial GE, and then counting the resultant secondary neurospheres. Although overexpressing Fut9 in primary neurospheres showed little effects on the formation of secondary neurospheres, overexpressing Fut10 increased the number of secondary neurospheres compared with that of neurospheres infected with control retroviruses (F(3,11) = 11.40, p = 0.001; Fig. 7B). These findings suggest that Fut10 enhances the self-renewal of NSCs. To exclude the possibility that Fut10 overexpression increases Fut9 activity, we utilized neurospheres derived from the GE of E15.5 Fut9−/− embryos. The difference between the number of secondary neurospheres obtained from Fut9−/− primary neurospheres infected with either control retroviruses or Fut10-expressing retroviruses was similar to the difference between the number obtained from Fut9+/− primary neurospheres infected with either control retroviruses or Fut10-expressing retroviruses (F(2,6) = 18.06, p = 0.003 for Fut9+/− and F(2,12) = 17.33, p < 0.001 for Fut9−/−; Fig. 7C). These results indicate that the effects of Fut10 overexpression are independent of Fut9. Conversely, when we used Fut10-miRNA-expressing retroviruses to knockdown Fut10 expression in Fut9+/− and Fut9−/− primary neurospheres, the number of secondary neurospheres obtained after passage decreased when compared with Fut9+/− and Fut9−/− primary neurospheres that had been infected with control retroviruses (Fig. 7C). These results suggest that Fut10, but not Fut9, is required for the maintenance of ES cells and NSCs.
FIGURE 7.

Expression of Fut10 regulates neural precursor cell differentiation. A–C, neurospheres derived from the E15.5 medial GE were infected with retroviruses expressing Fut10, Fut10-miRNA, or Fut9. A, retrovirus-infected GFP+ cells were collected by cell sorting from secondary neurospheres 7 days after the infection. Quantitative RT-PCR was performed for Fut4, Fut7, Fut9, and Fut10 and the expression levels of those genes relative to that of β-actin are shown. B, retrovirus-infected neurospheres were passaged and the number of secondary neurospheres that were obtained from 1000 plated cells was counted. Overexpression of Fut10, but not Fut9, in neurospheres derived from the medial GE of E15.5 CD1 wild-type embryos enhanced secondary neurosphere formation. E, overexpression of Fut10 increased, and knockdown of Fut10 decreased, the number of secondary neurospheres obtained from Fut9+/− and Fut9−/− primary neurospheres. Error bars indicate S.E. from three or more independent experiments. *, p < 0.05 by one-way analysis of variance followed by post hoc Tukey's comparison.
Fut10 Is Crucial for the Radial Migration of Neural Precursor Cells
We assessed the role of Fut10 in neural development by using in utero electroporation to introduce Fut10-miRNAs into the embryonic mouse cortex. GFP+ neural precursor cells that had incorporated the control miRNA expression plasmid migrated radially from the VZ/SVZ to the cortical plate during a 3-day period (E14.5-E17.5; Fig. 8A). In contrast, radial migration was impaired in GFP+ neural precursor cells that had been electroporated with Fut10-miRNA1 (Fig. 8B). Similar results were obtained in brains that received Fut10-miRNA2 or Fut10-miRNA3 (Fig. 8C). Quantification of migration by bin counting revealed a statistically significant increase in the percentage of GFP+ cells in the basal regions of the cortex of control brains compared with that of brains that were electroporated with either Fut10-miRNA1 or Fut10-miRNA3. Additionally, the percentage of GFP+ cells in the apical regions of the cortex of control brains was significantly decreased compared with that of brains that were electroporated with either Fut10-miRNA1 or Fut10-miRNA3 (F(2,6) = 227.4, p < 0.001 for bin 6 and F(2,6) = 322.3, p < 0.001 for bin 1; Fig. 8E). We performed rescue experiments with a Fut10 expression vector and Fut10-miRNA3, which targets a 3′ UTR sequence that is absent in the Fut10 expression vector (Fig. 5A). Co-electroporation of the Fut10 expression vector with Fut10-miRNA3 restored radial migration in cortical neural precursor cells (Fig. 8D): the percentage of GFP+ cells in the basal regions of the cortex of brains that were electroporated with both vectors was increased compared with that of brains that were electroporated with Fut10-miRNA3 only. Conversely, the percentage of GFP+ cells in the apical regions of the cortex of brains that were electroporated with both Fut10-miRNA3 and Fut10 vectors was decreased compared with that of brains that were electroporated with Fut10-miRNA3 only (t(4) = 11.67, p < 0.001 for bin 6 and t(4) = 7.658, p = 0.002 for bin 1; Fig. 8E). The percentage of GFP+ cells in the basal and apical cortical regions of brains that were electroporated with both the Fut10 expression vector and Fut10-miRNA3 were similar to those of control brains (Fig. 8E).
FIGURE 8.

Knockdown of Fut10 in the cortex impairs radial migration. A–E, Fut10-miRNAs (miRNA1, miRNA2, and miRNA3) or control miRNA vectors were electroporated into the dorsal cortex of E14.5 embryos in utero and the brains were analyzed at E17.5. A, GFP+ neural precursor cells that received control miRNA migrated radially to the outer cortical plate (CP) during the 3-day period after electroporation. B and C, because the three Fut10-miRNAs showed similar phenotypes, only the results of electroporation with Fut10-miRNA1 (B) and Fut10-miRNA3 (C) are shown. D, co-electroporation of Fut10-miRNA3 and a Fut10 expression plasmid restored radial migration. Scale bar = 250 μm. IMZ, intermediate zone. E, the entire cortex of E17.5 brains that had been electroporated at E14.5 was divided into 6 bins and the GFP+ cells in each bin were counted. Error bars indicate S.D. from three independent experiments. F–L, Fut10-miRNA1 or control miRNA vectors were electroporated into the dorsal cortex of E14.5 embryos in utero, and then the embryos were fixed and the brains were analyzed 24 h later. Double label immunofluorescence of coronal cryosections was used to detect expression of GFP and BrdU (F and G), Tbr2 (H and I), or Pax6 (J and K). Arrowheads indicate double-positive cells. L, quantification of BrdU/GFP (upper graph), Tbr2/GFP (middle graph), or Pax6/GFP (bottom graph) double-positive cells. Scale bar = 50 μm. Error bars indicate S.E. and n values are shown within the columns. *, p < 0.05 by one-way analysis of variance followed by post hoc Dunnett's comparison (E) or by Student's t test (L).
As paired box 6 (Pax6)+ neural precursor cells in the VZ differentiate, they exit the cell cycle, migrate into the SVZ, and start to express eomesodermin homolog (Xenopus laevis) (Tbr2). We first examined the incorporation of bromodeoxyuridine (BrdU), which had been administered 2 h prior to perfusion, in the cortex of embryos that had previously (24 h) been electroporated with Fut10-miRNA1 (Fig. 8, F and G). After Fut10 knockdown, GFP+ neural precursor cells incorporated less BrdU (Fig. 8L). We also determined the proportion of GFP+/Tbr2+ (Fig. 8, H and I) and GFP+/Pax6+ (Fig. 8, J and K) cells in the cortex of Fut10-miRNA1- or control miRNA-electroporated brains. We found more GFP+, Tbr2+ cells and less GFP+, Pax6+ cells in the Fut10-miRNA1-electroporated cortex than in controls (Fig. 8L). These results suggest that the expression of Fut10 is crucial for preventing neural precursor cells from differentiating and migrating radially to the cortical plate.
DISCUSSION
In this study, we provide evidence that Fut10 is involved in an α1,3-fucosyltransferase activity with stringent substrate specificity, and that this activity synthesizes LeX structures on bisecting N-glycans of glycoproteins. First, the LeX-harboring bisecting N-glycan, A2G2Fo2FB, was detected in the VZ/SVZ of the GE of E15.5 mouse brains, where Fut10, but not Fut9, is expressed. In contrast, in the dorsal cortex, where Fut9 mRNA is abundant, LeX is found predominantly on biantennary N-glycans, not bisecting N-glycans. Second, overexpression of Fut10 in neurospheres increased the amount of an N-glycan that harbors 2 or 3 outer fucoses. Third, an in vitro α1,3-fucosyltransferase assay revealed that although Fut10 could increase the amount of A2G1Fo(6)FB and A2G1Fo(6)B, which are both LeX-containing bisecting N-glycans, by fucosylating the N-glycans of glycoproteins, it could not fucosylate the oligosaccharide lacto-N-tetraose or PA-tagged bisecting N-glycans such as A2G2FB. These findings suggest that the Fut10-related α1,3-fucosyltransferase activity requires the peptide portion of the glycoprotein in addition to the bisecting N-glycan.
In previous studies, an anti-CD15 LeX antibody did not detect any LeX antigens on Fut10-overexpressing COS cells (14). A possible explanation for this result is that COS cells express very low levels of bisecting N-glycans. Alternatively, immunohistochemical analysis might be less sensitive than our HPLC system for detecting LeX-containing bisecting N-glycans. It is also possible that the CD15 antibody binds less preferentially to the LeX residues of bisecting N-glycans, because the GlcNAc that is attached to the inner mannose with an α1,4-linkage may make it more difficult for the antibody to access the LeX antigen. Recently, Mollicone et al. (16) reported that the enzymatic activity of Fut10 transfers a fucose onto the innermost GlcNAc of the core chitobiose. This type of activity is clearly distinct from the LeX-synthesizing α1,3-fucosyltransferase activity detected in our study. Their enzymatic assay conditions may not have been optimized to detect α1,3-fucosyltransferase activities that synthesize LeX at the peripheral GlcNAc residue, because they did not use bisecting N-glycans. It remains to be determined whether or not Fut10 itself is an α1,3-fucosyltransferase because neither Mollicone et al. (16) nor we used purified preparations of Fut10 to detect the enzymatic activity. Fut10 may function as a chaperone to another α1,3-fucosyltransferase as does core 1 synthase-specific molecular chaperone (Cosmc) to core 1 β3-galactosyltransferase, both of which show 26% homology (26, 27).
The expression of LeX in various types of stem cells and its subsequent down-regulation in cells that are differentiating suggests that LeX plays an important role in maintaining stem cell in the undifferentiated state. However, analyses of mice deficient for other α1,3-fucosyltransferase genes argue against this possibility: Fut4 and Fut7 double knock-out mice show only subtle immunological defects (28), and mice that lack Fut9, which is responsible for the synthesis of most of the LeX antigens on glycoproteins and glycolipids, develop normally until adulthood (11) and the only neurological phenotypes these mice exhibit are a few behavioral changes (12). The expression of Fut9 and the LeX antigens it synthesizes in the outer cortical plate and the mantle zone of the ventral forebrain of E15.5 embryos suggests a role for Fut9 in migrating neuroblasts rather than in the self-renewing NSCs in the VZ/SVZ. In addition, our findings that the number of NSCs and their self-renewal capability are increased in the cortical VZ/SVZ of Fut9−/− brains suggests that the expression of LeX synthesized by Fut9 facilitates the differentiation and migration of neural precursor cells. This notion appears inconsistent with the fact that neurosphere-forming NSCs are enriched in LeX-positive cells from the adult mouse subependyma (8). However, this inconsistency disappears if one considers that neurosphere-forming NSCs express LeX epitopes, which are synthesized by Fut10-related α1,3-fucosyltransferase activity. Indeed, we demonstrated that immuno-positivity by mouse monoclonal anti-LeX antibody remains in the VZ/SVZ of Fut9−/− brains. We propose that different LeX-containing epitopes synthesized by Fut9 and Fut10 could be required for distinct cellular functions such as promoting their differentiation and maintaining NSCs in an undifferentiated state.
miRNA-mediated knockdown of Fut10 expression in ES cells as well as NSCs attenuated the self-renewal capacity of both stem cell populations. Although we could not exclude the possibility that Fut10 enhances stemness through a nonenzymatic action, it is likely that the unique LeX-containing bisecting N-glycans synthesized by Fut10 modify the function of the glycoproteins they are attached to, including those expressed on stem cells. Several glycoproteins that have been identified in neurospheres contain LeX antigens (29, 30). Moreover, blocking the N-glycans on the EGF receptor was reported to modify EGF signaling and affect the proliferation and differentiation of EGF receptor-positive cells. The erythroagglutinating phytohemagglutinin lectin from Phaseolus vulgaris (E-PHA) binds specifically to the bisecting N-glycans on the EGF receptor in U373 MG cells. In doing so, E-PHA prevents EGF from binding to its receptor and suppresses EGF-induced autophosphorylation of the receptor (31). It would be intriguing to determine whether the glycoproteins present on the EGF receptor express LeX and contain bisecting N-glycans. Knockdown of Fut10 reduced the number of GFP+ cells that migrated radially into basal regions of the cortex. This defect could occur if the differentiation of neural precursor cells was suppressed or, alternatively, if the migratory ability of the cells was impaired independent of their state of differentiation. We think the latter possibility is more likely for the following reasons. First, knockdown of Fut10 in neurospheres attenuated their capacity to self-renew. Second, compared with electroporation of control miRNAs, electroporating Fut10-miRNAs into the embryonic cortex induced less neural precursor cells of the VZ/SVZ to express Pax6 as opposed to Tbr2, which is a marker for differentiating cells in the SVZ.
Acknowledgments
We thank J. Miyazaki for plasmids, N. Suzuki for breeding the Fut9 mice, and I. Ito, K. Inaba, R. Taguchi, and M. Tomoeda for technical assistance.
This work was supported by Grants-in-Aid for Exploratory Research (21650092) (to S. H.) and Scientific Research on Priority Area (20019032) (to K. I.) from the Ministry of Education, Culture, Sports, Science and Technology of Japan and grants from the Sankyo Foundation of Life Science and the Astellas Foundation for Research on Metabolic Disorders (to S. H.).
- LeX
- Lewis X
- ES
- embryonic stem
- NSC
- neural stem cells
- Fut
- fucosyltransferase
- VZ
- ventricular zone
- GE
- ganglionic eminence
- DIG
- digoxygenin
- PA
- fluorophore 2-aminopyridine
- SVZ
- subventricular zone
- LTA
- L. tetragonolobus agglutinin
- Pax6
- paired box 6
- Tbr2
- eomesodermin homolog (X. laevis)
- E-PHA
- erythroagglutinating phytohemagglutinin lectin from Phaseolus vulgaris
- miRNA
- microRNA
- E
- embryonic day.
REFERENCES
- 1. Solter D., Knowles B. B. (1978) Monoclonal antibody defining a stage-specific mouse embryonic antigen (SSEA-1). Proc. Natl. Acad. Sci. U.S.A. 75, 5565–5569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bird J. M., Kimber S. J. (1984) Oligosaccharides containing fucose linked α(1–3) and α(1–4) to N-acetylglucosamine cause decompaction of mouse morulae. Dev. Biol. 104, 449–460 [DOI] [PubMed] [Google Scholar]
- 3. Muramatsu T. (1988) Alterations of cell-surface carbohydrates during differentiation and development. Biochimie 70, 1587–1596 [DOI] [PubMed] [Google Scholar]
- 4. Toumadje A., Kusumoto K., Parton A., Mericko P., Dowell L., Ma G., Chen L., Barnes D. W., Sato J. D. (2003) Pluripotent differentiation in vitro of murine ES-D3 embryonic stem cells. In Vitro Cell Dev. Biol. Anim. 39, 449–453 [DOI] [PubMed] [Google Scholar]
- 5. Yamamoto M., Boyer A. M., Schwarting G. A. (1985) Fucose-containing glycolipids are stage- and region-specific antigens in developing embryonic brain of rodents. Proc. Natl. Acad. Sci. U.S.A. 82, 3045–3049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Allendoerfer K. L., Magnani J. L., Patterson P. H. (1995) FORSE-1, an antibody that labels regionally restricted subpopulations of progenitor cells in the embryonic central nervous system, recognizes the LeX carbohydrate on a proteoglycan and two glycolipid antigens. Mol. Cell Neurosci. 6, 381–395 [DOI] [PubMed] [Google Scholar]
- 7. Capela A., Temple S. (2006) LeX is expressed by the principle progenitor cells in the embryonic nervous system, is secreted into their environment and binds Wnt-1. Dev. Biol. 291, 300–313 [DOI] [PubMed] [Google Scholar]
- 8. Capela A., Temple S. (2002) LeX/SSEA-1 is expressed by adult mouse CNS stem cells, identifying them as nonependymal. Neuron 35, 865–875 [DOI] [PubMed] [Google Scholar]
- 9. Gooi H. C., Feizi T., Kapadia A., Knowles B. B., Solter D., Evans M. J. (1981) Stage-specific embryonic antigen involves α1–3 fucosylated type 2 blood group chains. Nature 292, 156–158 [DOI] [PubMed] [Google Scholar]
- 10. Nishihara S., Iwasaki H., Nakajima K., Togayachi A., Ikehara Y., Kudo T., Kushi Y., Furuya A., Shitara K., Narimatsu H. (2003) α1,3-Fucosyltransferase IX (Fut9) determines Lewis X expression in brain. Glycobiology 13, 445–455 [DOI] [PubMed] [Google Scholar]
- 11. Kudo T., Kaneko M., Iwasaki H., Togayachi A., Nishihara S., Abe K., Narimatsu H. (2004) Normal embryonic and germ cell development in mice lacking α1,3-fucosyltransferase IX (Fut9) which show disappearance of stage specific embryonic antigen 1. Mol. Cell Biol. 24, 4221–4228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kudo T., Fujii T., Ikegami S., Inokuchi K., Takayama Y., Ikehara Y., Nishihara S., Togayachi A., Takahashi S., Tachibana K., Yuasa S., Narimatsu H. (2007) Mice lacking α1,3-fucosyltransferase IX demonstrate disappearance of Lewis x structure in brain and increased anxiety-like behaviors. Glycobiology 17, 1–9 [DOI] [PubMed] [Google Scholar]
- 13. Roos C., Kolmer M., Mattila P., Renkonen R. (2002) Composition of Drosophila melanogaster proteome involved in fucosylated glycan metabolism. J. Biol. Chem. 277, 3168–3175 [DOI] [PubMed] [Google Scholar]
- 14. Baboval T., Smith F. I. (2002) Comparison of human and mouse Fuc-TX and Fuc-TXI genes, and expression studies in the mouse. Mamm. Genome 13, 538–541 [DOI] [PubMed] [Google Scholar]
- 15. Patnaik S. K. (2007) Characterization of Fut10 and Fut11, putative α1–3/4 fucosyltransferase genes important for vertebrate development. Nature Precedings http://precedings.nature.com/documents/141/version/1
- 16. Mollicone R., Moore S. E., Bovin N., Garcia-Rosasco M., Candelier J. J., Martinez-Duncker I., Oriol R. (2009) Activity, splice variants, conserved peptide motifs, and phylogeny of two new α1,3-fucosyltransferase families (FUT10 and FUT11). J. Biol. Chem. 284, 4723–4738 [DOI] [PubMed] [Google Scholar]
- 17. Reynolds B. A., Weiss S. (1992) Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science 255, 1707–1710 [DOI] [PubMed] [Google Scholar]
- 18. Hitoshi S., Kippin T., van der Kooy D. (2011) in Neurogenesis in the Adult Brain II: Clinical Implications (Seki T., Sawamoto K., Parent J. M., Alvarez-Buylla A., eds) pp. 189–207, Springer, New York [Google Scholar]
- 19. Nagy A., Rossant J., Nagy R., Abramow-Newerly W., Roder J. C. (1993) Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc. Natl. Acad. Sci. U.S.A. 90, 8424–8428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hitoshi S., Alexson T., Tropepe V., Donoviel D., Elia A. J., Nye J. S., Conlon R. A., Mak T. W., Bernstein A., van der Kooy D. (2002) Notch pathway molecules are essential for the maintenance, but not the generation, of mammalian neural stem cells. Genes Dev. 16, 846–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Naruse M., Nakahira E., Miyata T., Hitoshi S., Ikenaka K., Bansal R. (2006) Induction of oligodendrocyte progenitors in dorsal forebrain by intraventricular microinjection of FGF-2. Dev. Biol. 297, 262–273 [DOI] [PubMed] [Google Scholar]
- 22. Tanabe K., Ikenaka K. (2006) In-column removal of hydrazine and N-acetylation of oligosaccharides released by hydrazinolysis. Anal. Biochem. 348, 324–326 [DOI] [PubMed] [Google Scholar]
- 23. Yoshimura T., Yamada G., Narumi M., Koike T., Ishii A., Sela I., Mitrani-Rosenbaum S., Ikenaka K. (2012) Detection of N-glycans on small amounts of glycoproteins in tissue samples and sodium dodecyl sulfate-polyacrylamide gels. Anal. Biochem. 423, 253–260 [DOI] [PubMed] [Google Scholar]
- 24. Yan L., Wilkins P. P., Alvarez-Manilla G., Do S. I., Smith D. F., Cummings R. D. (1997) Immobilized Lotus tetragonolobus agglutinin binds oligosaccharides containing the Le(x) determinant. Glycoconj. J. 14, 45–55 [DOI] [PubMed] [Google Scholar]
- 25. Nishihara S., Iwasaki H., Kaneko M., Tawada A., Ito M., Narimatsu H. (1999) α1,3-Fucosyltransferase 9 (FUT9; Fuc-TIX) preferentially fucosylates the distal GlcNAc residue of polylactosamine chain while the other four α1,3FUT members preferentially fucosylate the inner GlcNAc residue. FEBS Lett. 462, 289–294 [DOI] [PubMed] [Google Scholar]
- 26. Kudo T., Iwai T., Kubota T., Iwasaki H., Takayma Y., Hiruma T., Inaba N., Zhang Y., Gotoh M., Togayachi A., Narimatsu H. (2002) Molecular cloning and characterization of a novel UDP-Gal:GalNAcα peptide β1,3-galactosyltransferase (C1Gal-T2), an enzyme synthesizing a core 1 structure of O-glycan. J. Biol. Chem. 277, 47724–47731 [DOI] [PubMed] [Google Scholar]
- 27. Ju T., Cummings R. D. (2002) A unique molecular chaperone Cosmc required for activity of the mammalian core 1 β3-galactosyltransferase. Proc. Natl. Acad. Sci. U.S.A. 99, 16613–16618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Weninger W., Ulfman L. H., Cheng G., Souchkova N., Quackenbush E. J., Lowe J. B., von Andrian U. H. (2000) Specialized contributions by α(1,3)-fucosyltransferase-IV and FucT-VII during leukocyte rolling in dermal microvessels. Immunity 12, 665–676 [DOI] [PubMed] [Google Scholar]
- 29. Yanagisawa M., Taga T., Nakamura K., Ariga T., Yu R. K. (2005) Characterization of glycoconjugate antigens in mouse embryonic neural precursor cells. J. Neurochem. 95, 1311–1320 [DOI] [PubMed] [Google Scholar]
- 30. Yagi H., Saito T., Yanagisawa M., Yu R. K., Kato K. (2012) Lewis X-carrying N-glycans regulate the proliferation of mouse embryonic neural stem cells via the Notch signaling pathway. J. Biol. Chem. 287, 24356–24364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rebbaa A., Yamamoto H., Moskal J. R., Bremer E. G. (1996) Binding of erythroagglutinating phytohemagglutinin lectin from Phaseolus vulgaris to the epidermal growth factor receptor inhibits receptor function in the human glioma cell line, U373 MG. J. Neurochem. 67, 2265–2272 [DOI] [PubMed] [Google Scholar]