Background: Although its expression is altered in several human diseases, the function of miR-433 is unknown.
Results: miR-433 inhibits HCC cell migration by repressing CREB1 3′-UTR activity and protein expression.
Conclusion: CREB1 is the first identified target of miR-433 in liver cancer.
Significance: This finding has broad implications given the role of CREB1 in various cancers, hepatic glucose metabolism, and liver circadian clock regulation.
Keywords: Cell Migration, Liver Cancer, MicroRNA, Post-translational Modification, Translation Regulation, HCC
Abstract
We show for the first time that potent microRNA-433 (miR-433) inhibition of expression of the cAMP response element-binding protein CREB1 represses hepatocellular carcinoma (HCC) cell migration. We identified a miR-433 seed match region in human and mouse CREB1 3′-UTRs. Overexpression of miR-433 markedly decreased human CREB1 3′-UTR reporter activity, and the inhibitory effect of miR-433 was alleviated upon mutation of its binding site. Ectopic expression of miR-433 reduced CREB1 protein levels in a variety of human and mouse cancer cells, including HeLa, Hepa1, Huh7, and HepG2. Human CREB1 protein levels in highly invasive MHCC97H cells were diminished by expression of miR-433 but were induced by miR-433 antagomir (anti-miR-433). The expression of mouse CREB1 protein negatively correlated with miR-433 levels in nuclear receptor Shp−/− liver tissues and liver tumors compared with wild-type mice. miR-433 exhibited a significant repression of MHCC97H cell migration, which was reversed by anti-miR-433. Overexpressing miR-433 inhibited focus formation dramatically, demonstrating that miR-433 may exert a tumor suppressor function. Knockdown of CREB1 by siRNAs impeded MHCC97H cell migration and invasion and antagonized the effect of anti-miR-433. Interestingly, CREB1 siRNA decreased MHCC97H cell proliferation, which was not influenced by anti-miR-433. Overexpressing CREB1 decreased the inhibitory activity of miR-433. The CpG islands surrounding miR-433 were hypermethylated, and the DNA methylation agent 5′-aza-2′-deoxycytidine, but not the histone deacetylase inhibitor trichostatin A, drastically stimulated the expression of miR-433 and miR-127 in HCC cells. The latter is clustered with miR-433. The results reveal a critical role of miR-433 in mediating HCC cell migration via CREB1.
Introduction
Several recent studies conducted in human specimens point to a physiological function of microRNA-433 (miR-433)4 in human diseases. For instance, miR-433 destabilization of HDAC6 (histone deacetylase 6) expression by targeting the HDAC6 3′-UTR has been implicated in a new form of dominant X-linked chondrodysplasia (1). The levels of miR-433 are decreased in human gastric carcinoma (2), which is associated with unfavorable outcome in overall survival (3). The expression of miR-433 is also down-regulated in visceral adipose tissue of patients with non-alcoholic steatohepatitis (4) and in hepatitis B virus-associated hepatocellular carcinoma (HCC) (5). miR-433 is aberrantly expressed in myeloproliferative neoplasms and negatively regulates CD34+ proliferation and differentiation by targeting GBP2, a member of a family of interferon-inducible GTPases (6). Although these studies provide evidence for the importance of miR-433 in various human diseases, its specific function in the liver remains elusive.
In mice, miR-433 is derived from an independent primary transcript that overlaps with primary miR-127 encoded by the miR-433-127 locus (7). Up-regulation of miR-433 is observed in the livers of nuclear receptor SHP (small heterodimer partner; Nr0b2) knock-out mice (8). At the transcriptional level, the promoter and the expression of miR-433 are repressed by SHP and activated by estrogen-related receptor γ. In particular, the miR-433 gene structure and transcriptional regulation are conserved across mammalian species (9), suggesting that miR-433 may play a similar regulatory role in humans and mice.
The transcription factor CREB1 (cAMP response element-binding protein 1) belongs to the basic leucine zipper domain (bZIP) class of proteins. It binds to the cAMP response element of its target genes to regulate gene transcription (10). Transactivation of CREB1 requires a phosphorylation event (Ser-133) in response to a wide variety of signals, in concert with dephosphorylation of cAMP-regulated transcriptional coactivators (11). Interestingly, tissue-specific coactivators can stimulate the transcriptional activities of CREB1 in a phosphorylation-independent manner (12).
CREB1 has been shown to enhance HCC progression by supporting angiogenesis and rendering HCC cells resistant to apoptosis (13). An increase in total CREB1 and phosphorylated CREB1 proteins was observed in HCC versus normal liver (14). Phosphorylated CREB1 was indirectly inhibited by miR-372 via PRKACB (15). Although CREB1 was inhibited by microRNAs (miRNAs) in other types of cancers, such as in acute myeloid leukemia by miR-34b (16), miRNAs that directly repress CREB1 in HCC remain to be identified.
In this study, the potential role of miR-433 in regulating HCC has been investigated. We demonstrate that miR-433 functions as a potent inhibitor of HCC cell migration. CREB1 is identified as a novel direct target of miR-433 that contributes to the underlying mechanism by which miR-433 inhibits HCC cell migration. The results provide a critical foundation for further study of the in vivo function of miR-433 in HCC invasion and metastasis.
EXPERIMENTAL PROCEDURES
Plasmids and Reagents
The human miR-433 target 3′-UTRs were cloned into pMIR-REPORT (Invitrogen) using the following primers: PAK4-3′-UTR, 5′-GGGGTACCcagaaccgcaccagatga-3′ (forward) and 5′-CCGCTCGAGactaactcgaggcagggg-3′ (reverse); WTAP-3′-UTR, 5′-GGGGTACCGGAGAGGATACTGTCCAG-3′ (forward) and 5′-CCGCTCGAGTGGTCAGTACAGGTAAGAT-3′ (reverse); TLR10-3′-UTR, 5′-GACTAGTCCACAGTCCTTGGGAAGTT-3′ (forward) and 5′-CCCAAGCTTTCAAATGCTCCCTGTAATCC-3′ (reverse); CREB1-3′-UTR, 5′-GACTAGTCCTTTACTGCCACAAATCAGA-3′ (forward) and 5′-CCCAAGCTTTTTCCTTGTGCACATCTAACAC-3′ (reverse); IGFBP1-3′-UTR, 5′-CGAGCTCCaactgccagatatattttaatg-3′ (forward) and 5′-CCCAAGCTTGGCAACATCACCACAGGTA-3′ (reverse); Notch1-3′-UTR, 5′-GACTAGTCCGGAGGCCTTCAAGTAA-3′ (forward) and 5′-CCCAAGCTTCATCTTGGGACGCATCTG-3′ (reverse); E2F3-3′-UTR, 5′-CGAGCTCCATCTGTCATGCAGTGTTGTC-3′ (forward) and 5′-CCCAAGCTTCCAATGCTTCATCTAGGACC-3′ (reverse); K-ras-3′-UTR, 5′-CGAGCTCTGAAGTGCCTGTTTGGGA-3′ (forward) and 5′-CCCAAGCTTGTCACTGTAACTATTTTTATTACA-3′ (reverse); and GCLC-3′-UTR, 5′-GACTAGTAATGCCCAGAGTTACTTGGA-3′ (forward) and 5′-CCCAAGCTTGCTAGAAACCCAAGATTACCC-3′). Using a QuikChange II site-directed mutagenesis kit (Agilent Technologies), mutations in pMIR-REPORT-CREB1-3′-UTR were generated with primer CREB1-3′-UTR mut 114 (forward, 5′-AGGAGAAAGCTTGGTTCTGTGAACAACGATCAGTAAGTTATCTTTGAATATGTA-3′; and reverse, 5′-TACATATTCAAAGATAACTTACTGATCGTTGTTCACAGAACCAAGCTTTCTCCTG-3′). All constructs were confirmed by sequencing. The miR-433 expression plasmid pTarget-miR-433 was generated as described (8). siRNAs for CREB1 and miR-433 antagomir (anti-miR-433) were purchased from Qiagen. Trichostatin A and 5′-aza-2′-deoxycytidine were used as described previously (17). The pCF-CREB1 plasmid was purchased from Addgene.
Wound Healing Assay
The method used was described previously (18). Briefly, MHCC97H cells (1 × 106) were seeded overnight on 6-cm plates and transfected with plasmids or siRNAs as described in the figure legends using Lipofectamine 2000 (Invitrogen). After cells reached confluence, wounding was done by dragging a 200-μl pipette tip through the monolayer. Cells were washed and allowed to migrate for 16 h. Wound closure or cell migration images were photographed when the scrape wound was introduced (0 h) and at a designated time (8 and 16 h) after wounding using an a Leica DMI6000 inverted microscope. The relative surface traveled by the leading edge was assessed using LAS AF 6000 version 1.8.0 software. Each experiment was performed in triplicate.
Cell Viability Assay
The cell viability assay was performed following the manufacturer's instructions (Vybrant® MTT cell proliferation assay kit, Invitrogen). MHCC97H cells were transfected overnight with the indicated plasmids or anti-miRNAs. Cells (2 × 104) were seeded onto 96-well plates and allowed to grow for 48 h. 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (10 μl of a 12 mm solution) was added to each well. After incubation at 37 °C for 4 h, 100 μl of SDS/HCl solution was introduced into each well and mixed thoroughly using the pipette. The plates were returned to the incubator for another 4 h, after which absorbance at 570 nm was measured. Each group included triplicate determinations, and the experiments were repeated three times.
Cell Migration and Invasion Assays
The cell migration assays were described previously (19). Briefly, after transfection, MHCC97H cells were serum-starved for 24 h, and 5 × 104 cells were seeded on Transwell inserts (8-μm pore size, Cultrex 96-well cell migration assay, catalog no. 3465-096-K, Trevigen). After 16 h, the non-migrated cells were removed from the insert with a cotton swab. The migrated cells were fixed for 10 min in 3.7% (v/v) formaldehyde in PBS before staining with 0.1% crystal violet for 15 min, followed by washing with PBS. Images were taken with an Olympus MicroFire/Qcam CCD 1 × 81 microscope. The crystal violet-stained cells were counted. Cell invasion assays were performed as described for the migration assays, except that cells were seeded on precoated inserts with basement membrane extract solution (Cultrex 96-well BME cell invasion assay, catalog no. 3455-096-K, Trevigen). Invading cells were fixed and stained. Images were taken, and cells were counted.
Focus Formation
HepG2 cells were transfected with the pTarget or pTarget-miR-433 plasmid (2 μg) for 24 h, and 2.5 × 104 cells were seeded in 0.4% top agar onto 6-well plates coated with 0.7% bottom agar. Ten days later, six areas per plate were chosen randomly, and the number of visible colonies was counted and used for statistical analysis.
Analysis of miRNA Expression
The method used was as described previously (20). Briefly, MHCC97H cells were grown to 60–70% confluence and transfected with the indicated siRNAs or anti-miRNAs, or MHCC97H, HepG2, Huh7, and CCL13 cells were treated with the indicated doses of 5′-aza-2′-deoxycytidine or trichostatin A for 48 h. Total RNAs including miRNAs were isolated using a miRNeasy mini kit (Qiagen) and reverse-transcribed using a miRNA reverse transcription kit (Qiagen). Mature miR-433 expression was quantified by quantitative PCR (qPCR) using a miRNA primer assay kit (Qiagen). All kits were used according to the manufacturer's instructions. U6 transcript was used as an internal control to normalize RNA input.
Transient Transfection and Luciferase Assays
Transient transfection and luciferase reporter assays were performed following standard protocols as described previously (21, 22). In brief, HeLa or MHCC97H cells were transfected with the indicated plasmids using Lipofectamine 2000. Luciferase activities were measured and normalized against β-gal activities (Promega). Three independent experiments were done, each involving triplicate transfections.
Western Blotting
Western blotting was done as described previously (23, 24). In brief, transfection was performed with Lipofectamine LTX (Invitrogen) according to the manufacturer's instructions. Forty-eight hours post-transfection, proteins were recovered and subjected to SDS-PAGE. The separated proteins were transferred to a PVDF membrane and incubated in blocking buffer. The membrane was then incubated with anti-CREB1 (Cell Signaling Technology) or anti-β-actin antibodies overnight at 4 °C, followed by incubation with horseradish peroxidase-conjugated secondary antibody at room temperature for 1 h. Protein bands were visualized with SuperSignal West Pico chemiluminescent substrate (Pierce).
Bisulfite Genomic Sequencing Analysis
Briefly, genomic DNA was isolated using a DNeasy blood & tissue kit (Qiagen) from cultured MHCC97H, HepG2, Huh7, or CCL13 cells. Bisulfite-modified DNA from an EZ DNA Methylation-DirectTM kit (Zymo Research Corp.) was used for PCR using primers specific for the modified DNA. Four sets of primers were designed: F1 (5′-TTTAGGATTTGGGGATTTATTAGGT-3′) and R1 (5′-AAAAACCACCTCCCTACCCTAC-3′); F2 (5′-GGTATTTGGGTGAGTAGTTGGG-3′) and R2 (5′-CTAAAAAAACACCCTAACCCTAACA-3′); F3 (5′-TAGTTTGGAGTTAGGGGTAGGGTAT-3′) and R3 (5′-CTCAATAAATCAAAAAAAACACCTC-3′); and F4 (5′-TTTGGTGGTTTTGGTGGATATATAT-3′) and R4 (5′-CCCACCTCTTCCTACTACTCATAATAA-3′). PCR products were subcloned into the pGEM-T Easy vector (Promega). Six clones from each DNA template were sequenced at the University of Utah HSC DNA Sequencing Core Research Facility to determine the methylation status.
Statistical Analysis
All experiments were repeated at least three times. Statistical analyses were carried out using Student's unpaired t test; p < 0.01 was considered statistically significant.
RESULTS AND DISCUSSION
CREB1 Is a Direct Target Gene of miR-433
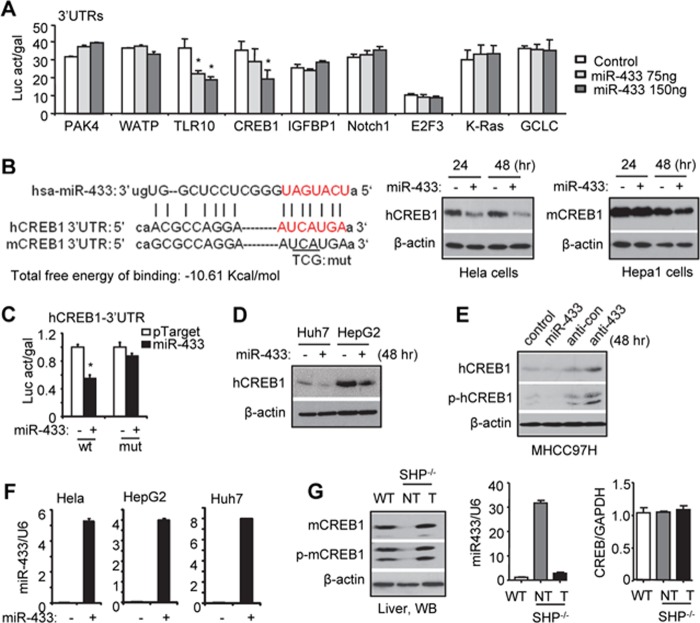
To determine the potential function of miR-433 in HCC and the underlying mechanism, the identification of the miR-433 downstream target genes is essential. Using TargetScan and miRanda, we predicted multiple putative targets of miR-433 based on the conserved seed region between miR-433 and the 3′-UTR of each gene (data not shown). We cloned the 3′-UTRs of nine genes into the respective luciferase reporters. Transient transfections showed that ectopic expression of miR-433 repressed the 3′-UTR reporter activities of TLR10 (Toll-like receptor 10) and CREB1 in a dose-dependent manner (Fig. 1A), but exerted no effects on other genes. To the best of our knowledge, no functional association of TLR10 with HCC has been reported thus far. Therefore, we focused our studies on characterizing miR-433 inhibition of CREB1, the role of which in HCC had been suggested previously (13, 14).
FIGURE 1.
CREB1 is a direct target gene of miR-433. A, transient transfection assays. HeLa cells were cotransfected with pTarget (Control), pTarget-miR-433 (miR-433), and 3′-UTRs of the indicated genes. Luciferase activity (Luc act) was normalized to β-gal activity (gal). *, p < 0.01. B, left, sequence alignment of miR-433 and the 3′-UTR of CREB1 in humans and mice. The seed match region is indicated in red. mut, mutant. Middle and right, Western blotting was performed to detect endogenous hCREB1 (middle) and mCREB1 (right) protein expression in human HeLa and mouse Hepa1 cells that were transfected with pTarget-miR-433 (2 μg) or control pTarget (2 μg). C, transient transfection assays. HeLa cells were cotransfected with the wild-type (wt; 25 ng) or mutant (mut; 25 ng) hCREB1 3′-UTR, pTarget-miR-433 (150 ng), or control pTarget (150 ng). The luciferase activity assay was performed, and the activity was normalized to β-gal activity. D, Western blotting was performed to determine endogenous hCREB1 proteins in Huh7 and HepG2 cells that were transfected with pTarget-miR-433 (2 μg) or control pTarget (2 μg). E, Western blotting was performed to determine endogenous hCREB1 and phosphorylated hCREB1 (p-hCREB1) proteins in MHCC97H cells that were transfected with pTarget, pTarget-miR-433, anti-miR control (anti-con), or anti-miR-433. β-Actin served as an internal control. F, qPCR analysis of miR-433 expression. G, left, Western blotting (WB) was performed to determine endogenous mCREB1 and phosphorylated mCREB1 (p-mCREB1) proteins in the livers of wild-type (WT) and Shp−/− (non-tumor (NT)) mice and in Shp−/− liver tumors (T). Right, qPCR analysis of miR-433 expression and Creb1 mRNA. Error bars represent S.E.
A conserved miR-433 seed match region was observed in human (hCREB1) and mouse (mCREB1) CREB1 3′-UTRs (Fig. 1B, left), and the free energy of binding (−10.61 kcal/mol) has a probability of 80% for binding, indicating that miR-433 may inhibit hCREB1 and mCREB1 proteins. Indeed, overexpression of miR-433 reduced the levels of hCREB1 and mCREB1 proteins in human HeLa and mouse Hepa1 cells, respectively, although stronger inhibition was observed in HeLa cells (Fig. 1B, right). To determine whether the predicted miR-433-binding site is responsible for its inhibitory effect, we mutated several nucleotides in the miR-433-binding sequence within the hCREB1 3′-UTR (Fig. 1B, left). As expected, the inhibition by miR-433 was abolished by the mutant hCREB1 3′-UTR reporter (Fig. 1C).
We extended this finding to other human HCC cells. hCREB1 protein was expressed more abundantly in HepG2 than in Huh7 cells, and overexpressing miR-433 reduced hCREB1 protein in both cells (Fig. 1D). In addition, both hCREB1 and phosphorylated hCREB1 proteins were repressed by miR-433 and induced by anti-miR-433 in MHCC97H cells (Fig. 1E). The intracellular levels of miR-433 after transfection are shown in Fig. 1F. Overall, the results provide convincing evidence that CREB1 is a direct target of miR-433 regardless of HCC cell origins.
We next established a direct association of miR-433 with CREB1 protein in vivo in mice. Both mCREB1 and phosphorylated mCREB1 proteins were reduced to undetectable levels in Shp−/− livers and were induced drastically in liver tumors developed in Shp−/− mice (Fig. 1G) (25). Such changes negatively correlated with a marked induction of miR-433 in Shp−/− livers (8) and its down-regulation in Shp−/− tumors.
miR-433 Functions as a Potent Inhibitor of HCC Cell Migration and Growth
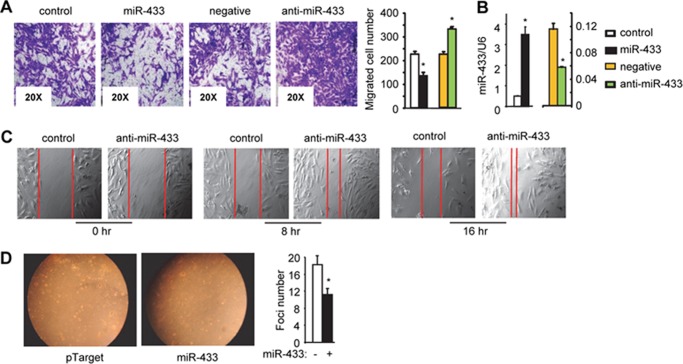
We compared the migration potential of HCC cells of different origin. HepG2 and Huh7 cells exhibited very poor ability to migrate, and MHCC97H cells appeared to be the most appropriate model for migration/invasion assays. Following our published method (19), we found that ectopic expression of miR-433 using miR-433 expression plasmids significantly attenuated and miR-433 knockdown by its antagomir (anti-miR-433) enhanced the migration potential of MHCC97H cells (Fig. 2A). The levels of miR-433 after overexpression or knockdown are presented in Fig. 2B.
FIGURE 2.
miR-433 inhibits HCC cell migration. A, Transwell cell migration assay. MHCC97H cells were transfected with pTarget (control; 2 μg), pTarget-miR-433 (miR-433; 2 μg), anti-miR negative control (negative; 20 nm), or miR-433 inhibitor (anti-miR-433; 20 nm). Migration assays were performed, and migrated cells were counted. *, p < 0.01. B, qPCR analysis of miR-433 levels. *, p < 0.05. C, Wound healing assay. MHCC97H cells were transfected with anti-miR negative control (20 nm) or miR-433 inhibitor (20 nm). Images were taken at 0, 8, and 16 h post-scratch. Red lines indicate the wound gap. D, focus formation assay. HepG2 cells were transfected with the pTarget or pTarget-miR-433 (miR-433) plasmid (2 μg) and seeded in 0.4% top agar onto 6-well plates coated with 0.7% bottom agar. Ten days later, colonies were counted from triplicate plates, and one representative plate is shown. Statistical results represent the mean ± S.D. of focus counts. *, p < 0.01, pTarget-miR-433 versus pTarget. Error bars represent S.E.
An alternative approach using a scratch wound healing assay further demonstrated the potent effect of miR-433 in inhibiting HCC cell migration. Knockdown of endogenous miR-433 markedly facilitated the closure of the wound field gap when examined after both 8 and 16 h compared with the control group (Fig. 2C). Focus formation assays showed that overexpression of miR-433 significantly inhibited the number of foci (Fig. 2D), which supports a role of miR-433 as a potential tumor suppressor.
miR-433 Inhibition of HCC Cell Migration and Invasion Is Mediated by CREB1
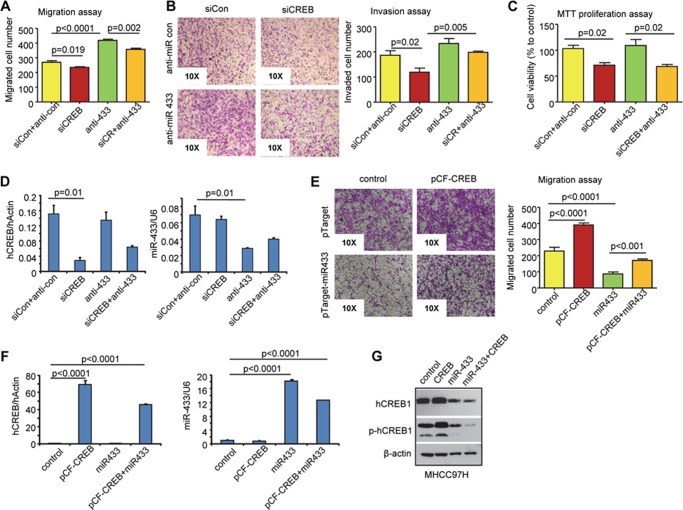
To determine a functional relationship between miR-433 and CREB1, we transfected MHCC97H cells with siRNA against CREB1 (siCREB1) alone or in combination with anti-miR-433 and examined the effects on HCC cell migration, invasion, and proliferation. siCREB1 significantly decreased the migration rate of cells (Fig. 3A). In contrast, anti-miR-433 increased HCC cell migration drastically. In the presence of cotransfected siCREB1, the effect of anti-miR-433 was markedly diminished. Similarly, cells transfected with siCREB1 exhibited decreased invasion, and the potency of anti-miR-433 to induce cell invasion was attenuated (Fig. 3B). It should be noted that CREB1 knockdown by siCREB1 reduced cell proliferation, whereas anti-miR-433 showed no effect (Fig. 3C). The knockdown efficiency of siCREB1 and anti-miR-433 is presented in Fig. 3D. anti-miR-433 did not affect CREB1 mRNA expression, suggesting that miR-433 does not regulate CREB1 gene transcription (Fig. 3D, left) and vice versa (Fig. 3D, right). In addition, overexpression of CREB1 reversed the inhibitory activity of miR-433 (Fig. 3, E and F). The CREB1 protein is presented in Fig. 3G. Overall, the results suggest that miR-433 inhibition of HCC cell migration and invasion is mediated, at least in part, by CREB1. In addition, the function of CREB1 in HCC cell proliferation is not shared by miR-433.
FIGURE 3.
siCREB1 antagonizes the effect of anti-miR-433 on HCC cell migration and invasion. A–C, MHCC97H cells were transfected with siCREB1 (human; siCR; 20 nm), control siRNA (siCon; 20 nm), anti-miR-433 siRNA (anti-433; 20 nm), or anti-miR-433 control siRNA (anti-con; 20 nm). Transwell cell migration assay (A), invasion assay (B), and 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) proliferation assay (C) were performed. D, qPCR analysis of endogenous CREB1 and miR-433. E, MHCC97H cells were transfected with pCF-CREB1, pTarget-miR-433, and their respective controls, and Transwell cell migration assay was conducted. F, qPCR analysis of CREB1 and miR-433 expression. G, Western blotting was performed to determine hCREB1 and phosphorylated hCREB1 (p-hCREB1) protein levels. Error bars represent S.E.
miR-433 Expression Is Regulated by DNA Methylation but Not Deacetylation in HCC Cells
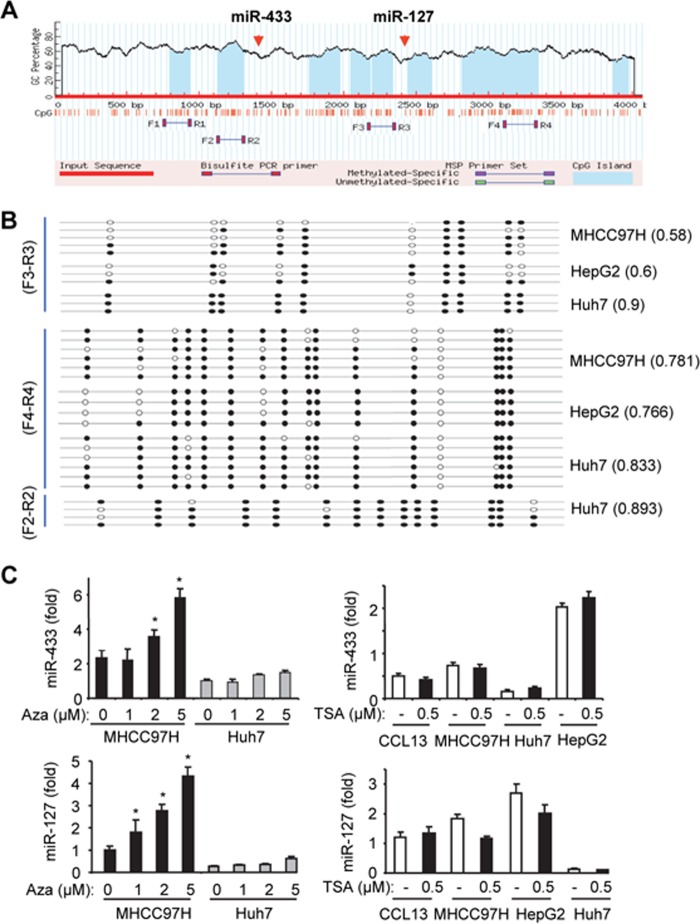
The genomic locus embedding miR-433 and its twin miR-127 is surrounded by several CpG islands (Fig. 4A). We designed several primer sets for methylation analysis in MHCC97H, HepG2, and Huh7 cells. The number of methylated cytosines using the F3/R3 primers appeared to be the highest in Huh7 cells compared with MHCC97H and HepG2 cells (Fig. 4B). Interestingly, all three HCC cell lines showed a similar CpG methylation profile using the F4/R4 primers. In addition, the methylation profile was indistinguishable as analyzed by the F2/R2 versus F4/R4 primers in Huh7 cells.
FIGURE 4.
DNA methylation regulates miR-433 expression in HCC cells. A, miR-433 resides in the CpG-rich region in the genomic locus. The genomic sequence containing miR-433 and miR-127 was uploaded and analyzed using MethPrimer. The blue areas indicate predicted CpG islands. Primers were designed to cover the CpG islands surrounding miR-433 and miR-127 for methylation analysis. B, methylation profile in MHCC97H, HepG2, and Huh7 cells using primer sets F3/R3, F4/R4, and F2/R2. Black and white circles denote methylated and unmethylated cytosines in specific CpGs, respectively. Three to six clones from each cell line were analyzed. The numbers following each cell line indicate methylated CpGs out of the total CpGs that were sequenced. C, qPCR analysis of miR-433 (upper) and miR-127 (lower) levels in HCC cells treated with the demethylating agent 5′-aza-2′-deoxycytidine (Aza; 24 h; left) or the histone deacetylase inhibitor trichostatin A (TSA; 24 h; right). Data are expressed as means ± S.D. of triplicate assays. *, p < 0.01, significant difference versus the untreated group. Error bars represent S.E.
Treatment with the DNA methylation inhibitor 5′-aza-2′-deoxycytidine resulted in a dose-dependent induction of miR-433 in MHCC97H cells but not in Huh7 cells (Fig. 4C, upper left), suggesting that miR-433 is silenced by DNA methylation in MHCC97H cells. In contrast, treatment of HCC cells with the histone deacetylase inhibitor trichostatin A did not induce miR-433 expression (Fig. 4C, upper right), suggesting that histone deacetylation may not be involved in regulating miR-433 expression. Similar results were observed with miR-127 (Fig. 4C, lower), the paired gene that is clustered with miR-433 (7, 8).
Conclusion
This is the first study showing that miR-433 is a potent inhibitor of HCC cell migration through its action on CREB1, a well known oncogene in a variety of cancers. The activation of CREB1 has a major impact on cell growth, proliferation, and survival (26). Given that CREB1 is also a critical component of the liver circadian clock (27) and regulator of hepatic glucose metabolism (28), miR-433 may be significant in a variety of conditions beyond HCC migration. The results provide new insights into the function of miR-433 via its regulation of CREB1 and suggest potential avenues for therapeutic intervention. Understanding the in vivo role of miR-433 by using appropriate miR-433 gene mutant mouse models warrants future studies.
Acknowledgment
We acknowledge the use of core facilities (Cell Imaging Core) supported by National Institutes of Health Grant P30 CA042014 to the Huntsman Cancer Institute.
This work was supported, in whole or in part, by National Institutes of Health DK080440 (to L. W.).
- miR-433
- microRNA-433
- HCC
- hepatocellular carcinoma
- miRNA
- microRNA
- anti-miR-433
- miR-433 antagomir
- qPCR
- quantitative PCR
- hCREB1
- human CREB1
- mCREB1
- mouse CREB1
- siCREB1
- CREB1 siRNA.
REFERENCES
- 1. Simon D., Laloo B., Barillot M., Barnetche T., Blanchard C., Rooryck C., Marche M., Burgelin I., Coupry I., Chassaing N., Gilbert-Dussardier B., Lacombe D., Grosset C., Arveiler B. (2010) A mutation in the 3′-UTR of the HDAC6 gene abolishing the post-transcriptional regulation mediated by hsa-miR-433 is linked to a new form of dominant X-linked chondrodysplasia. Hum. Mol. Genet. 19, 2015–2027 [DOI] [PubMed] [Google Scholar]
- 2. Luo H., Zhang H., Zhang Z., Zhang X., Ning B., Guo J., Nie N., Liu B., Wu X. (2009) Down-regulated miR-9 and miR-433 in human gastric carcinoma. J. Exp. Clin. Cancer Res. 28, 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ueda T., Volinia S., Okumura H., Shimizu M., Taccioli C., Rossi S., Alder H., Liu C. G., Oue N., Yasui W., Yoshida K., Sasaki H., Nomura S., Seto Y., Kaminishi M., Calin G. A., Croce C. M. (2010) Relation between microRNA expression and progression and prognosis of gastric cancer: a microRNA expression analysis. Lancet Oncol. 11, 136–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Estep M., Armistead D., Hossain N., Elarainy H., Goodman Z., Baranova A., Chandhoke V., Younossi Z. M. (2010) Differential expression of miRNAs in the visceral adipose tissue of patients with non-alcoholic fatty liver disease. Aliment. Pharmacol. Ther. 32, 487–497 [DOI] [PubMed] [Google Scholar]
- 5. Wang W., Zhao L. J., Tan Y. X., Ren H., Qi Z. T. (2012) Identification of deregulated miRNAs and their targets in hepatitis B virus-associated hepatocellular carcinoma. World J. Gastroenterol. 18, 5442–5453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lin X., Rice K. L., Buzzai M., Hexner E., Costa F. F., Kilpivaara O., Mullally A., Soares M. B., Ebert B. L., Levine R., Licht J. D. (2013) miR-433 is aberrantly expressed in myeloproliferative neoplasms and suppresses hematopoietic cell growth and differentiation. Leukemia 27, 344–352 [DOI] [PubMed] [Google Scholar]
- 7. Song G., Wang L. (2008) miR-433 and miR-127 arise from independent overlapping primary transcripts encoded by the miR-433–127 locus. PLoS ONE 3, e3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Song G., Wang L. (2008) Transcriptional mechanism for the paired miR-433 and miR-127 genes by nuclear receptors SHP and ERRγ. Nucleic Acids Res. 36, 5727–5735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Song G., Wang L. (2009) A conserved gene structure and expression regulation of miR-433 and miR-127 in mammals. PLoS ONE 4, e7829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. De Cesare D., Fimia G. M., Sassone-Corsi P. (1999) Signaling routes to CREM and CREB: plasticity in transcriptional activation. Trends Biochem. Sci. 24, 281–285 [DOI] [PubMed] [Google Scholar]
- 11. Altarejos J. Y., Montminy M. (2011) CREB and the CRTC co-activators: sensors for hormonal and metabolic signals. Nat. Rev. Mol. Cell Biol. 12, 141–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fimia G. M., De Cesare D., Sassone-Corsi P. (1999) CBP-independent activation of CREM and CREB by the LIM-only protein ACT. Nature 398, 165–169 [DOI] [PubMed] [Google Scholar]
- 13. Abramovitch R., Tavor E., Jacob-Hirsch J., Zeira E., Amariglio N., Pappo O., Rechavi G., Galun E., Honigman A. (2004) A pivotal role of cyclic AMP-responsive element binding protein in tumor progression. Cancer Res. 64, 1338–1346 [DOI] [PubMed] [Google Scholar]
- 14. Kovach S. J., Price J. A., Shaw C. M., Theodorakis N. G., McKillop I. H. (2006) Role of cyclic-AMP responsive element binding (CREB) proteins in cell proliferation in a rat model of hepatocellular carcinoma. J. Cell. Physiol. 206, 411–419 [DOI] [PubMed] [Google Scholar]
- 15. Wang J., Liu X., Wu H., Ni P., Gu Z., Qiao Y., Chen N., Sun F., Fan Q. (2010) CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res. 38, 5366–5383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pigazzi M., Manara E., Baron E., Basso G. (2009) miR-34b targets cyclic AMP-responsive element binding protein in acute myeloid leukemia. Cancer Res. 69, 2471–2478 [DOI] [PubMed] [Google Scholar]
- 17. He N., Park K., Zhang Y., Huang J., Lu S., Wang L. (2008) Epigenetic inhibition of nuclear receptor small heterodimer partner is associated with and regulates hepatocellular carcinoma growth. Gastroenterology 134, 793–802 [DOI] [PubMed] [Google Scholar]
- 18. Song G., Zhang Y., Wang L. (2009) MicroRNA-206 targets notch3, activates apoptosis, and inhibits tumor cell migration and focus formation. J. Biol. Chem. 284, 31921–31927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang Y., Yang Z., Whitby R., Wang L. (2011) Regulation of miR-200c by nuclear receptors PPARα, LRH-1 and SHP. Biochem. Biophys. Res. Commun. 416, 135–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Song G., Wang L. (2009) Nuclear receptor SHP activates miR-206 expression via a cascade dual inhibitory mechanism. PLoS ONE 4, e6880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yang Z., Zhang Y., Wang L. (2012) Mdm2 is a novel activator of ApoCIII promoter which is antagonized by p53 and SHP inhibition. Biochem. Biophys. Res. Commun. 417, 744–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang Y., Bonzo J. A., Gonzalez F. J., Wang L. (2011) Diurnal regulation of the early growth response 1 (Egr-1) protein expression by hepatocyte nuclear factor 4α (HNF4α) and small heterodimer partner (SHP) cross-talk in liver fibrosis. J. Biol. Chem. 286, 29635–29643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang Y., Wang L. (2011) Nuclear receptor SHP inhibition of Dnmt1 expression via ERRγ. FEBS Lett. 585, 1269–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhou T., Zhang Y., Macchiarulo A., Yang Z., Cellanetti M., Coto E., Xu P., Pellicciari R., Wang L. (2010) Novel polymorphisms of nuclear receptor SHP associated with functional and structural changes. J. Biol. Chem. 285, 24871–24881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhang Y., Xu P., Park K., Choi Y., Moore D. D., Wang L. (2008) Orphan receptor small heterodimer partner suppresses tumorigenesis by modulating cyclin D1 expression and cellular proliferation. Hepatology 48, 289–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Siu Y. T., Jin D. Y. (2007) CREB–a real culprit in oncogenesis. FEBS J. 274, 3224–3232 [DOI] [PubMed] [Google Scholar]
- 27. Asher G., Schibler U. (2011) Crosstalk between components of circadian and metabolic cycles in mammals. Cell Metab. 13, 125–137 [DOI] [PubMed] [Google Scholar]
- 28. Jitrapakdee S. (2012) Transcription factors and coactivators controlling nutrient and hormonal regulation of hepatic gluconeogenesis. Int. J. Biochem. Cell Biol. 44, 33–45 [DOI] [PubMed] [Google Scholar]