Background: Cdc13 in Candida species is small and missing conserved domains.
Results: A second Cdc13 homologue (Cdc13B) was shown to form heterodimers with Cdc13 and to mediate overlapping functions in telomere protection.
Conclusion: Cdc13 has undergone duplication and functional specialization in a branch of Saccharomycotina yeast.
Significance: Understanding the assembly and mechanisms of fungal CST complex provides insights on the evolution and function of this crucial telomere complex.
Keywords: Chromosomes, Chromosomes/Non-histone Chromosomal Proteins, DNA-Protein Interaction, Telomerase, Telomeres, CST, Cdc13, OB Fold, Dimerization
Abstract
The budding yeast G-tail binding complex CST (Cdc13-Stn1-Ten1) is crucial for both telomere protection and replication. Previous studies revealed a family of Cdc13 orthologues (Cdc13A) in Candida species that are unusually small but are nevertheless responsible for G-tail binding and the regulation of telomere lengths and structures. Here we report the identification and characterization of a second family of Cdc13-like proteins in the Candida clade, named Cdc13B. Phylogenetic analysis and sequence alignment indicate that Cdc13B probably arose through gene duplication prior to Candida speciation. Like Cdc13A, Cdc13B appears to be essential. Deleting one copy each of the CDC13A and CDC13B genes caused a synergistic effect on aberrant telomere elongation and t-circle accumulation, suggesting that the two paralogues mediate overlapping and nonredundant functions in telomere regulation. Interestingly, Cdc13B utilizes its C-terminal OB-fold domain (OB4) to mediate self-association and binding to Cdc13A. Moreover, the stability of the heterodimer is evidently greater than that of either homodimer. Both the Cdc13 A/A homodimer and A/B heterodimer, but not the B/B homodimer, recognized the telomere G-tail repeat with high affinity and sequence specificity. Our results reveal novel evolutionary elaborations of the G-tail-binding protein in Saccharomycotina yeast, suggesting a drastic remodeling of CDC13 that entails gene duplication, fusion, and functional specialization. The repeated and independent duplication of G-tail-binding proteins such as Cdc13 and Pot1 hints at the evolutionary advantage of having multiple G-tail-binding proteins.
Introduction
Telomeres, the protective nucleoprotein structures located at the ends of linear eukaryotic chromosomes, are critical for chromosome stability. Dysfunctional telomeres often suffer DNA loss, end-to-end fusion, excessive recombination, and other abnormal transactions that ultimately result in genomic instability and rearrangements (1–3). In most organisms, telomere DNAs consist of copies of a short asymmetric sequence that is G-rich for the 3′-end-bearing strand (G-strand). Because the G-strand is typically longer than the complementary C-strand, most chromosomes terminate in 3′-overhangs commonly referred to as G-tails. Proteins and complexes that interact with G-tails are known to play key roles in telomere protection and maintenance.
The G-tails of budding yeast are capped by a three-subunit complex comprising Cdc13, Stn1, and Ten1 (CST)2 (4). Initially discovered in Saccharomyces cerevisiae, the CST subunits from this budding yeast have been subjected to extensive characterization. ScCdc13 is a multifunctional protein that probably comprises four OB-fold domains as well as a recruitment domain (Fig. 1A). The most N-terminal OB-fold domain (OB1) mediates ScCdc13 dimerization as well as its binding to Pol1 (the catalytic subunit of Pol α) (5). In addition, OB1 has been reported to possess a low affinity DNA binding activity (6). The second OB fold (OB2) also forms dimers and appears to modulate interaction with Stn1 (7). The two C-terminal OB folds (named DBD and OB4) are responsible for high affinity DNA binding and interaction with Stn1, respectively (8). In addition, the recruitment domain of Cdc13, located in between OB1 and OB2, interacts with the telomerase regulatory subunit Est1 to promote the recruitment of telomerase to chromosome ends and the activation of telomerase (9, 10). The Stn1 and Ten1 subunits of CST resemble the Rpa2 and Rpa3 subunits of RPA (the major ssDNA-binding complex in eukaryotes) with respect to their three-dimensional structures and low affinity DNA binding activities (3, 11–13). Stn1 also interacts with the Pol12 subunit of Pol α to promote telomere capping through a mechanism that remains incompletely understood (14, 15).
FIGURE 1.
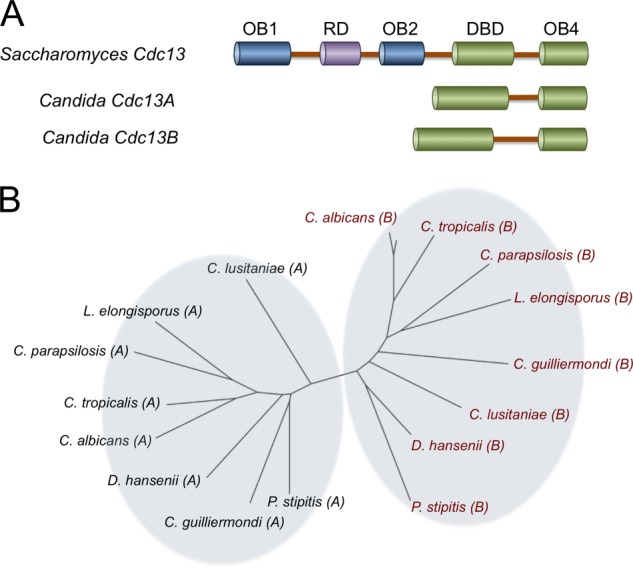
The domain organization and phylogenetic relationship of Cdc13 homologues in Saccharomycotina. A, the domain organizations of Cdc13, Cdc13A, and Cdc13B proteins from Saccharomyces and Candida species are illustrated. B, the relationship between the Candida Cdc13A and Cdc13B family members was analyzed using the neighbor-joining method, and the results were plotted using FigTree. The Cdc13A and Cdc13B family members are designated by A and B in parentheses following the species names. The accession codes for the Cdc13A members are as follows: Debaryomyces hansenii, XP_461188; C. albicans, XP_719034; Candida parapsilosis, CPAG_03609; Lodderomyces elongisporus, XP_001526643; C. guilliermondii, XP_001486879, PGUG_00256; C. lusitaniae, CLUG_03319; C. tropicalis, CTRG_04305; and P. stipitis, XP_001384972. The accession codes for the Cdc13B members are as follows: D. hansenii, DEHA0F12859g; C. albicans, CAWG_05171; C. parapsilosis, CPAG_02733; L. elongisporus, LELG_04784.1; C. guilliermondii, PGUG_00873; C. lusitaniae, CLUG_05491.1; C. tropicalis, CTRG_02892; and P. stipitis, XP_001384748.
Interestingly, recent analysis of Cdc13 homologues reveals a high degree of evolutionary divergence. In particular, we and others have identified and characterized a family of Cdc13 homologues in Candida species that are noticeably smaller than ScCdc13. These homologues lack the N-terminal half of the prototypical Cdc13 and contain just two OB-fold domains: DBD and OB4 (Fig. 1A) (11, 16, 17). Despite this difference, we showed that the Candida albicans Cdc13 (CaCdc13) indeed associates with telomere DNA in vivo and is required for normal telomere length regulation (18). We further demonstrated high affinity telomere DNA binding by the Candida tropicalis Cdc13 (CtCdc13) in vitro and found that this binding activity requires dimerization of CtCdc13 through its OB4 domain (18). We therefore concluded that the Candida Cdc13 proteins are indeed orthologous to the C-terminal half of ScCdc13 (i.e. they share a common ancestor and perform similar functions). Nevertheless, the lack of the N-terminal half of Cdc13 poses a conundrum, indicating that the functions mediated by the missing domains such as Pol1 binding and telomerase recruitment must either be absent or taken over by other proteins in Candida species. In the present report, we have identified and characterized a second family of Cdc13-like proteins in the Candida species named Cdc13B. We show here that Cdc13B is required for telomere regulation but is functionally distinct from the initially characterized Candida Cdc13 (henceforth referred to as Cdc13A). Like Cdc13A, Cdc13B utilizes the OB4 domain for binding to itself and Stn1. However, unlike Cdc13A, Cdc13B binds telomere G-strand with low affinity and moderate sequence specificity. Surprisingly, Cdc13A and Cdc13B can form heterodimers, and each protein exhibits a preference for heterodimerization over homodimerization. The Cdc13A/Cdc13B dimer (A/B dimer) binds telomere G-tails with high affinity and sequence specificity in vitro, suggesting that it could constitute the predominant form of Cdc13 in vivo. We propose an evolutionary model to account for the presence of two small Cdc13 paralogues in the Candida clade and that of just one large Cdc13 in Saccharomyces. Our findings provide insights on the assembly and DNA binding mechanisms of CST as well as its unusual elaboration in a fungal branch.
EXPERIMENTAL PROCEDURES
Construction and Growth of Candida Strains
The C.albicans strain BWP17 (ura3Δ::λimm434/ura3Δ::λimm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG) was used as the parental strain (19). The heterozygous strain CDC13B+/− was generated by subjecting BWP17 to one round of transformation and 5-FOA selection using a CDC13B::hisG-URA3-hisG cassette (containing ∼650 bp of CDC13B upstream and ∼630 bp of the downstream sequence) (20, 21). The upstream and downstream fragments were generated using the CDC13B-CUB1/CDC13B-CUB2 and CDC13B-CUB3/CDC13B-CUB4 primer pairs, respectively (Table 1). To make the strains with C-terminally tagged Cdc13B, we first constructed a tagging plasmid by replacing the GFP fragment in pGFP-URA with a TAP (tandem affinity purification) fragment derived by PCR from pBS1479 (22, 23). This plasmid, named pTAP-URA3, contains the TAP tag followed by the C. albicans ADH2 terminator and the URA3 selectable marker. A cassette consisting of the following elements was then obtained by PCR using the appropriate primers (CDC13B-(3601–3683)-TAP-F and CDC13B-(3691–3775)-R1-R; see Table 1) and pTAP-URA3 as the template: ∼100 bp of the C terminus of CaCDC13B, the TAP tag, the ADH2 terminator, the URA3 marker, and ∼100 bp of the DNA downstream of the CaCDC13B termination codon. This tagging cassette was used to transform the CDC13B+/− heterozygous strains. Correct transformants were selected on SD-Ura plates and identified by PCR. The transformants were passaged in YPD (1% yeast extract, 2% peptone, 2% glucose) prior to telomere analysis.
TABLE 1.
Proteins and protein domains tested for interactions in this study
| Proteins and protein domains | Amino acids |
|---|---|
| CaCdc13A | 1–447 |
| CaCdc13A-OB4 | 294–447 |
| CaCdc13B | 1–561 |
| CaCDC13B-OB4 | 395–561 |
| CaStn1 | 1–585 |
Sequence Analysis
Cdc13, Cdc13A, and Cdc13B homologues from Saccharomycotina yeast were identified in the NCBI and Broad Institute databases by BLAST or psi-BLAST searches. The multiple sequence alignment was generated using the PROMALS server and displayed using BoxShade.
Telomere Analyses
The telomere length analysis and the two-dimensional gel analysis of circular and linear telomeric DNA were performed as described previously (24).
Chromatin Immunoprecipitation
The assays were performed as described previously (18) except that the enrichment of telomeric DNA was measured by PCR amplification (95 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s for 25 cycles) of a 300-bp subtelomeric fragment on chromosome 3. The forward and reverse primers were Chr3-F and Chr3-R, respectively (Table 2). Pilot titration experiments indicated that for the range of DNA concentrations analyzed in the assays, the PCR signals were roughly proportional to the amount of starting DNAs.
TABLE 2.
Oligonucleotides used in this study
| CDC13B-CUB1 | AATGGTACCCGGTACTTGGCCCAACTGTTG |
| CDC13B-CUB2 | TTACTCGAGGGTAAAAAGGCGACGAACACG |
| CDC13B-CUB3 | AATCCGCGGCAAGAGCATATATGCAGCTCG |
| CDC13B-CUB4 | TTAGAGCTCGCTTACATCAGATTAACCTCTG |
| Chr3-F | TCTAATACACAAACTATTCACTTCTTC |
| Chr3-R | AGTCAGAGAGATGTCAATGGC |
| CDC13B-up-Sac1 | ATTTGAGCTCATGACAAGCAACAAGAAGTTGTTG |
| CDC13B-dn-st-Xho | AATCTCGAGCTATATAATTTGGTTAATTGGAGAGTC |
| CDC13B-(3601–3683)-TAP-F | ATATCAAAAGGAAAATCAGGACACTTCCCAACAACTTTAAGTATACTTATTGGACCATTGACTCTCCAATTAACCAAATTATAATGGAAAAGAGAAGATGGAAAAAGA |
| CDC13B-(3691–3775)-R1-R | GACTAATGAAATAAATTTAACTAACAATCGAAGTCTTTCTTATAATTGTTATTTAACATAGATAGTTCATATCATGTACTATAAATCTAGAAGGACCACCTTTGATTG |
| Non-Tel 1 | AATCTCGAGTTACTTGTCATCGTCATCCTTGTAATCATTATTGCATGTTTGAGTTGATTG |
| Non-Tel 2 | AAACTCGAGCTAATCCACAGCAAAGAAATGATTATC |
Gel Electrophoretic Mobility Shift Analysis
To analyze the DNA binding activity of Cdc13B, the entire ORF (amino acids 1–561) was cloned in between the SacI and XhoI sites of the pSMT3 vector to enable the expression of the His6-SUMO-Cdc13B fusion protein. Because of the atypical translation of the CUG codon in Candida species, the CTG triplets encoding amino acids 175 and 383 of CaCdc13B were mutated to TCG to enable the expression of wild type proteins in Escherichia coli (25). Following induction, extracts were prepared and the fusion proteins purified with Ni-NTA chromatography as described previously (24). In some assays, the fusion protein was first cleaved by the ULP1 protease and then subjected to DNA binding analysis. To analyze the activity of the A/A, A/B, and B/B complexes, we co-expressed appropriate His6-SUMO and GST-FLAG fusion proteins in E. coli and then purified the respective complexes by Ni-NTA and M2 affinity chromatography. Binding reactions contained 50 mm Tris-HCl, pH 7.5, 2 mm MgCl2, 50 mm NaCl, 0.1 mm EDTA, 1 mm DTT, 40 ng/μl poly(dI-dC), 10% glycerol, and specified concentrations of probe and proteins. Following incubation at 25 °C for 20 min, the reaction mixtures were subjected to gel electrophoresis through a 5% nondenaturing polyacrylamide gel (acrylamide:bis = 44:1) to resolve the free probe from the DNA-protein complex.
Co-expression and Pulldown Assays
The DNAs encoding full-length CaCdc13A, CaCdc13B, and CaStn1, as well as various domains (see Table 1 for the amino acids included in each expression construct) were amplified by PCR and cloned into the pSMT3 vector (26) and the pGEX4T-2 vector (GE Healthcare) to enable their expression as His6-SUMO and GST-FLAG fusion proteins, respectively. Each His6-SUMO fusion protein was expressed alone or co-expressed with a GST-FLAG fusion protein in E. coli BL21(DE3). The growth and induction protocols as well as the extract preparation procedures were as described previously (18). For anti-FLAG pulldown assays, ∼500 μl of 10 mg/ml extract was incubated with 20 μl of M2-agarose beads (Sigma) in FLAG(250) buffer (50 mm Tris-HCl, pH 7.5, 250 mm NaCl, 10% glycerol, 0.1% Nonidet P-40, 2.5 mm MgCl2, and 1 mm DTT). Following incubation with constant mixing on a rotator at 4 °C for 2 h, the beads were washed five times with 0.5 ml of FLAG(150) buffer (same as FLAG(250) except that it contained 150 mm NaCl), and then the M2-bound proteins were eluted with 60 μl of FLAG(150) containing 0.2 mg/ml 3× FLAG peptide. The eluates were analyzed by SDS-polyacrylamide gel electrophoresis followed by staining with Coomassie Brilliant Blue R-250 or Western blotting. The GST pulldown assays were carried out using glutathione-Sepharose (GE Healthcare). The binding and washing buffers were identical to those of the M2 pulldown assays, and the elution buffer consisted of FLAG(150) supplemented with 15 mm reduced glutathione.
Glycerol Gradient
Approximately 200 μl of the indicated protein complex (∼0.2–2 μm) purified by affinity chromatography was applied to a 5-ml glycerol gradient (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 2 mm DTT, 0.1% Triton X-100, and 15–30% glycerol). The gradient was subjected to centrifugation in an AH-650 rotor (Sorvall) at 42,000 rpm for 20 h, and 20–25 fractions were collected and analyzed by SDS-PAGE and Western blotting.
RESULTS
Candida Species Contain a Second Family of Cdc13-like Proteins
Iterative psi-BLAST searches at the NCBI database uncovered a second family of Cdc13-like proteins in Candida species, which we named Cdc13B. In sequence analysis, most Cdc13B align well with the DBD and OB4 domains of Candida Cdc13A as well as Saccharomyces Cdc13 (Fig. 1, supplemental Fig. 1, and data not shown). The only exception is Candida guilliermondi Cdc13B, which evidently lacks the OB4 domain (supplemental Fig. 1). Overall, the α-helices and β-strands of the two OB folds constitute the most conserved regions in the sequence alignments, implying that these domains in all three protein families are structurally similar (supplemental Fig. 1 and data not shown). Conversely, the region in between the two OB folds is poorly conserved and variable in length (e.g. ∼80 amino acids long in Candida lusitaniae Cdc13B and ∼460 amino acids long in Pichia stipitis Cdc13B) and may thus function partly as a flexible linker. Notably, a CDC13B ORF is present in each of the fully sequenced Candida genomes. Moreover, within this clade, the CDC13A and CDC13B genes form well defined monophyletic groups, consistent with the duplication of an ancestral gene prior to Candida speciation. By contrast, we were unable to detect any CDC13B-like genes in the Saccharomyces and Kluyveromyces clades, suggesting that either the duplication did not take place or that the duplicated gene was lost in these lineages.
Cdc13B and Cdc13A Mediate Overlapping but Nonredundant Functions in Telomere Length Regulation
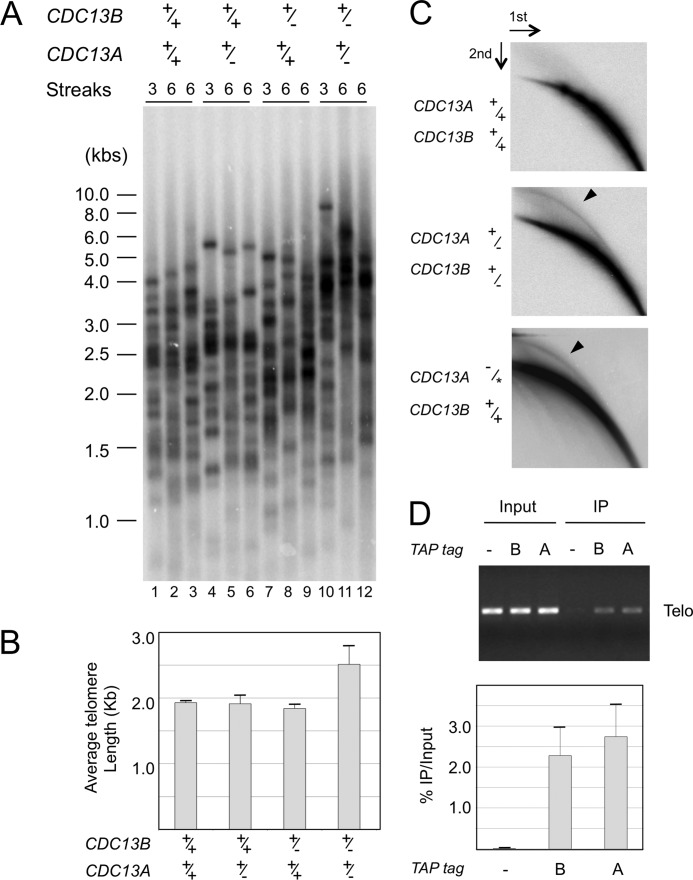
To address the function of Cdc13B, we attempted to generate a homozygous null strain by performing two rounds of URA-Blaster-mediated gene replacement. Although we were able to obtain several heterozygous clones, none of the more than 30 transformants screened after the second round of gene replacement was a homozygous null mutant, suggesting that CDC13B is essential for cell viability, just like CDC13A (18). However, we were able to construct and characterize a CDC13A+/− CDC13B+/− combination mutant. We subjected the individual and compound heterozygous deletion strains to both growth and telomere length analyses. Upon repeated passages in YPD medium, none of the mutants exhibited an obvious growth defects or senescence (data not shown). However, although neither the CDC13A+/− nor the CDC13B+/− strain exhibited changes in telomere lengths, the combination mutant was found to contain longer and more heterogeneous telomeres (Fig. 2, A and B). The synergistic effect of deleting one copy each of CDC13A and CDC13B suggests that these genes mediate overlapping functions in telomere length regulation.
FIGURE 2.
The role of Cdc13B in telomere regulation. A, the indicated strains were passaged in YPD and their telomeres subjected to Southern analysis. B, the average telomere lengths ± S.D. in the indicated strains were determined from three independent transformants after six streaks and plotted. C, the levels of t-circles in the wild type, a compound heterozygous, and a CDC13A-TAP strain (18) were assayed by two-dimensional gel electrophoresis and Southern analysis. The TAP-tagged allele of Cdc13A is designated by an asterisk. The percentages of total telomere hybridization signals in the circular DNA arcs (marked by arrowheads in two blots) are 0.4, 2.0, and 2.3%, respectively. D, the wild type (−) and two TAP-tagged strains (one carrying just one copy of CDC13A-TAP (lane A) and the other carrying one copy of CDC13B-TAP (lane B)) were subjected to chromatin immunoprecipitation analysis. The levels of telomeric DNA in 2% of the Input and 20% of the immunoprecipitation (IP) samples were estimated by PCR amplification of a subtelomeric fragment on chromosome 3. A representative assay is shown at the top, and the calculated percentages of telomeric DNA trapped in the immunoprecipitation samples are plotted at the bottom. Data (average ± S.D.) are from three independent experiments.
Longer and more heterogeneous telomeres are often associated with the accumulation of extrachromosomal telomere circles (t-circles) (24, 27). Consistent with this notion, we found elevated levels of t-circles only in the compound heterozygous strain (Fig. 2C and data not shown). The amount of t-circles in this strain is similar to that found in a strain carrying a single copy of CDC13A-TAP, which was also reported previously to contain long and heterogeneous telomeres (Fig. 2C and Ref. 18).
To determine whether Cdc13B functions directly at telomeres, we tagged the intact CDC13B locus of the CDC13B+/− strain with a TAP tag. Western analysis indicated that the tagged protein was expressed at appreciable levels (data not shown). Chromatin immunoprecipitation assays revealed significant association of Cdc13B-TAP with telomere DNA, to the same extent as the previously characterized Cdc13A-TAP, suggesting that both proteins act directly at telomeres (Fig. 2D).
Cdc13B Binds Telomere G-strand with Low Affinity and Sequence Specificity
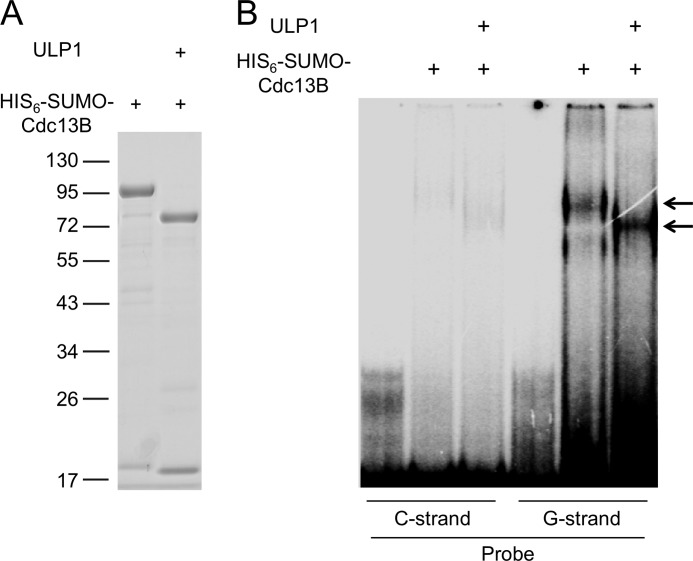
The presence of a DBD-like domain in Cdc13B (supplemental Fig. 1) suggests that this protein may be capable of binding to G-tails, just like Cdc13A (18). We expressed Cdc13B as a His6-SUMO fusion protein and subjected the purified protein to electrophoretic mobility shift assays (EMSA). Despite using a variety of assay conditions and buffers, we were able to observe only low affinity binding of the fusion protein to G-tails (Fig. 3, Kd ≫ 1 μm). As expected, removing the His6-SUMO tag from the fusion protein by ULP1 treatment resulted in a shift in the mobility of the DNA-protein complex, indicating that the complex was due to Cdc13B rather than a contaminant.
FIGURE 3.
Low affinity DNA binding by C. albicans Cdc13B. A, the purified His6-SUMO-CaCdc13B fusion protein was analyzed by SDS-PAGE and Coomassie staining. The identity of the protein was confirmed by ULP1 cleavage, which as predicted released the His6-SUMO tag. B, the intact His6-SUMO-CaCdc13B fusion protein and the ULP1-treated protein (1 μm) were subjected to EMSA using single-stranded oligonucleotide probes (7.5 nm) that consisted of two copies of the C. albicans telomere G- and C-strand repeats (CaC2, (CATCCGTACACCAAGAAGTTAGA)2; CaG2, (TCTAACTTCTTGGTGTACGGATG)2). Note that the fraction of probe in the complexes represents less than 1% of the total probe used in these assays.
Cdc13B Binds Stn1 and Can Form a Homo-oligomer as Well as a Hetero-oligomer with Cdc13A
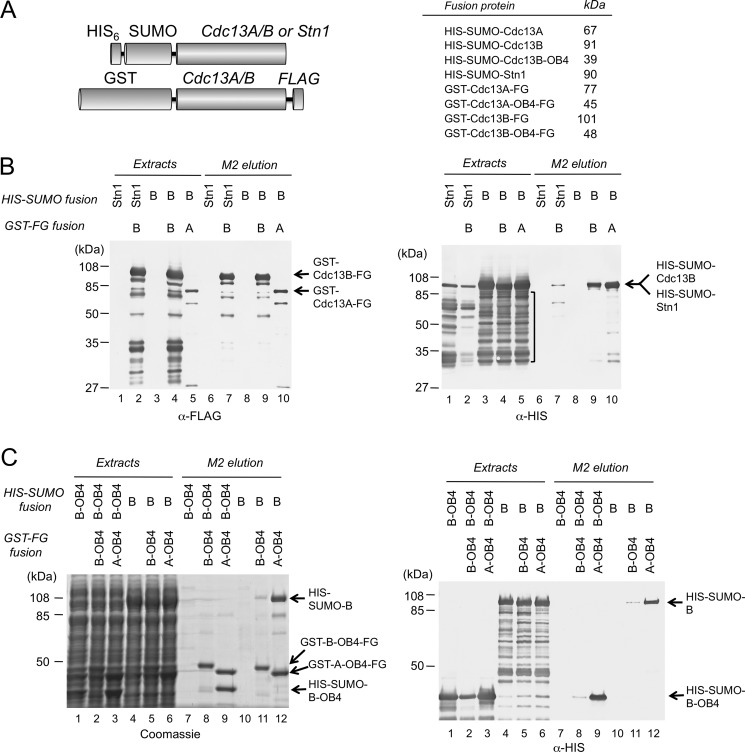
Next we analyzed the ability of Cdc13B to interact with Stn1, which is an established activity of Cdc13A (13). His6-SUMO-Stn1 was expressed alone or co-expressed with GST-Cdc13B-FLAG in E. coli (Fig. 4A), and the extracts were subjected to affinity purification using M2 (anti-FLAG)-agarose. A significant amount of Stn1 fusion was detected in the affinity eluate only when it was co-expressed with the Cdc13B fusion, indicating that the two proteins can form a complex, just like Cdc13A and Stn1 (Fig. 4B, both panels, lanes 1 and 2).
FIGURE 4.
Cdc13B-Stn1, Cdc13B-Cdc13A, and Cdc13B-Cdc13B interactions. A, left, the fusion proteins used in the co-expression and anti-FLAG affinity purification analyses are illustrated. Right, the expected sizes of the various fusion proteins are listed. B, left, the levels of the GST-FLAG fusion proteins (the bait proteins) in the indicated cell extracts and elution fractions were analyzed by Western using anti-FLAG antibodies. Right, the levels of the His6-SUMO fusion proteins (the target proteins) in the indicated cell extracts and elution fractions were analyzed by Western using anti-His6 antibodies. High levels of proteolytic fragments (indicated by a vertical bracket) were detected for both His6-SUMO-Stn1 and His6-SUMO-Cdc13B in crude extracts. C, left, the protein contents of the indicated cell extracts and anti-FLAG elution fractions were analyzed by SDS-PAGE and Coomassie staining. Right, the levels of the His6-SUMO fusion proteins (the target proteins) in the indicated cell extracts and elution fractions were analyzed by Western using anti-His6 antibodies.
Another established property of both the Cdc13 proteins in Saccharomyces and the small Cdc13A proteins in Candida species is dimerization (5–7, 18). To assess the oligomerization property of Cdc13B, we co-expressed various combinations of His6-SUMO and GST-FLAG fusion proteins containing either Cdc13A or Cdc13B in E. coli and subjected the extracts to M2 (anti-FLAG) affinity purification and Western analysis. As shown in Fig. 4B, both GST-Cdc13A-FLAG and GST-Cdc13B-FLAG associated with substantial amounts of His6-SUMO-Cdc13B, indicating that Cdc13B can bind to itself as well as the Cdc13A paralogue (lanes 9 and 10). Moreover, C-terminal truncation of Cdc13B appears to abolish these interactions, as the majority of His6-SUMO-Cdc13B proteolytic fragments (marked by a bracket) were not recovered by M2 affinity purification (Fig. 4B, lanes 9 and 10). This observation implicates the C-terminal OB4 domain of Cdc13B in Cdc13A/Cdc13B (A/B) and Cdc13B/Cdc13B (B/B) complex formation.
To address the role of the OB4 domain more directly, we performed two additional sets of co-expression/purification analysis. In one set, full-length Cdc13B (His6-SUMO-Cdc13B) was synthesized alone or together with the OB4 domain of Cdc13A or Cdc13B (GST-Cdc13AOB4-FLAG or GST-Cdc13BOB4-FLAG) (Fig. 4C, lanes 4–6 and 10–12, and Table 1). In another set, just the OB4 domain of Cdc13B (His6-SUMO-Cdc13BOB4) was co-expressed with the same domain of Cdc13A or Cdc13B (GST-Cdc13AOB4-FLAG or GST-Cdc13BOB4-FLAG) (Fig. 4C, lanes 1–3 and 7–9). As predicted, both the A/A and A/B complexes can be detected in the two sets of assays, implying that the OB4 domains are sufficient for homo- and hetero-oligomer formation (Fig. 4C, lanes 8–9 and 11–12). More surprisingly, the levels of the A/B complex were reproducibly and substantially higher than those of the B/B complex in the M2 elution, suggesting that Cdc13B is more efficient at forming hetero-oligomers than homo-oligomers.
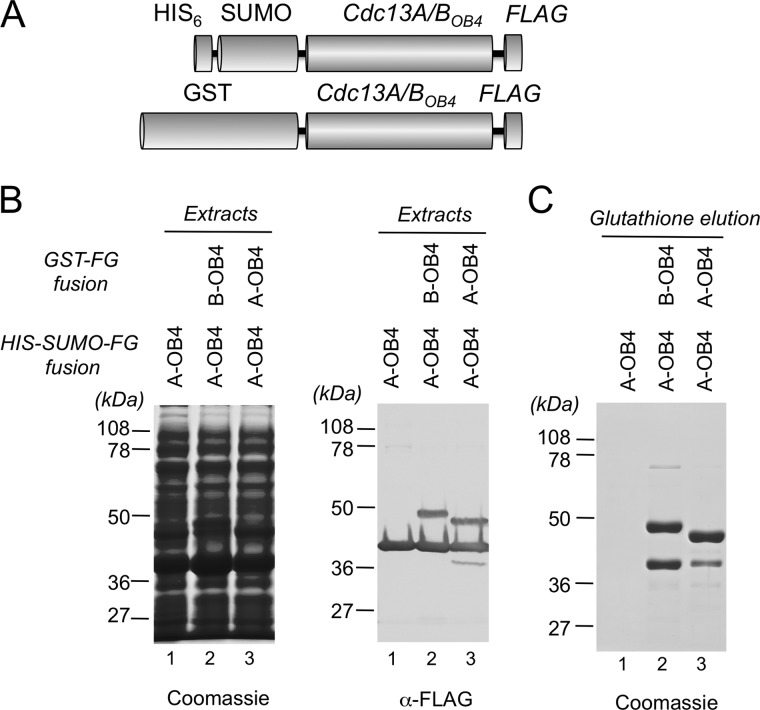
We then carried out a “reciprocal” experiment in which GST-tagged Cdc13AOB4 or Cdc13BOB4 was used to pull down His6-SUMO-tagged Cdc13AOB4 (Fig. 5A). Both the bait and target proteins also contained the FLAG tag, allowing us to determine simultaneously their expression levels, which were found to be comparable (Fig. 5B). Consistent with the previous experiment, the efficiency of the A/B complex formation was again higher than that of the A/A complex (Fig. 5C, lanes 2 and 3). Taken together, our results indicate that C. albicans Cdc13A and Cdc13B have a greater propensity to form hetero-oligomers than homo-oligomers.
FIGURE 5.
The interactions between the OB4 domains of Cdc13A and Cdc13B. A, the fusion proteins used in the co-expression and GST affinity purification analyses are illustrated. B, the total proteins and the levels of bait and target proteins in the cell extracts were analyzed by SDS-PAGE followed by Coomassie staining and anti-FLAG Western, respectively. C, the levels of the bait and target proteins in the GST affinity elution fractions were analyzed by SDS-PAGE and Coomassie staining.
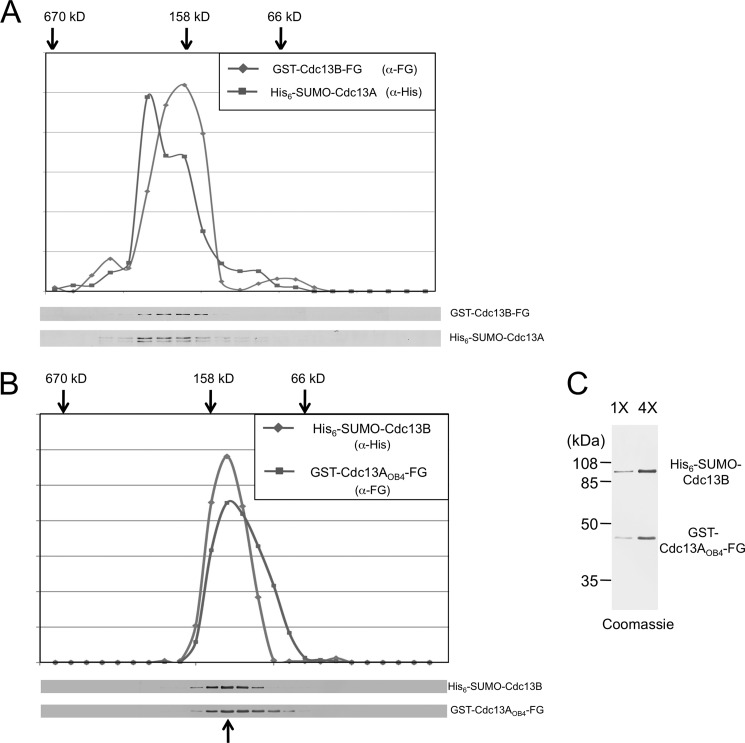
To assess the stoichiometry of the complex, we performed glycerol gradient sedimentation analyses. Because the A/B complex appears to be more stable, we first isolated the complex comprising full-length His6-SUMO-Cdc13A and GST-Cdc13B-FLAG by sequential Ni-NTA, M2, and glutathione affinity chromatography and then characterized the sedimentation behavior of the complex. Each protein exhibited a peak signal at ∼160 kDa, consistent with the predicted size for a mixed dimer (158 kDa, Fig. 6A). The analysis of the “full-length” A/B complex was performed three times using materials obtained by different combinations of affinity steps, and the ∼160-kDa peaks for both subunits were reproducibly observed (data not shown). We then tested a number of A/B complexes bearing truncated Cdc13A or Cdc13B and found that the complex comprising GST-Cdc13AOB4-FLAG and His6-SUMO-Cdc13B behaved homogeneously as a ∼140-kDa complex (Fig. 6B). This estimated size was again close to that predicted for a heterodimer (125 kDa). Coomassie staining of the two proteins in the putative dimer peak fraction was also consistent with the two proteins being at a ratio of 1:1 (Fig. 6C). Taken together, our observations suggest that the simplest form of the A/B complex is likely to be a heterodimer.
FIGURE 6.
Glycerol gradient sedimentation analysis of the Cdc13A/Cdc13B complexes. A, the complex comprising His6-SUMO-Cdc13A and GST-Cdc13B-FLAG was purified by sequential Ni-NTA, M2-agarose, and glutathione-Sepharose affinity chromatography and then subjected to glycerol gradient analysis. The distribution of each protein across the gradient fractions was determined by Western analysis (bottom) and plotted (top). The peak positions for the thyroglubulin (670 kDa), aldolase (158 kDa), and BSA (66 kDa) standards as determined in a parallel gradient are marked by arrows. B, the same as A except that the complex comprising GST-Cdc13AOB4-FLAG and His6-SUMO-Cdc13B was purified by Ni-NTA and M2-agarose and analyzed. Note that the standards in this experiment migrate to slightly different positions from those in A. C, the peak fraction from the gradient shown in B (as marked by an arrow at bottom) was subjected to SDS-PAGE and Coomassie staining. Two different amounts of the fraction were analyzed. Assuming that the staining was proportional to the size of the protein, the ratio of the two proteins is ∼1:1.
The Cdc13A/B Dimer Binds the Cognate Telomere G-strand with High Affinity and Sequence Specificity
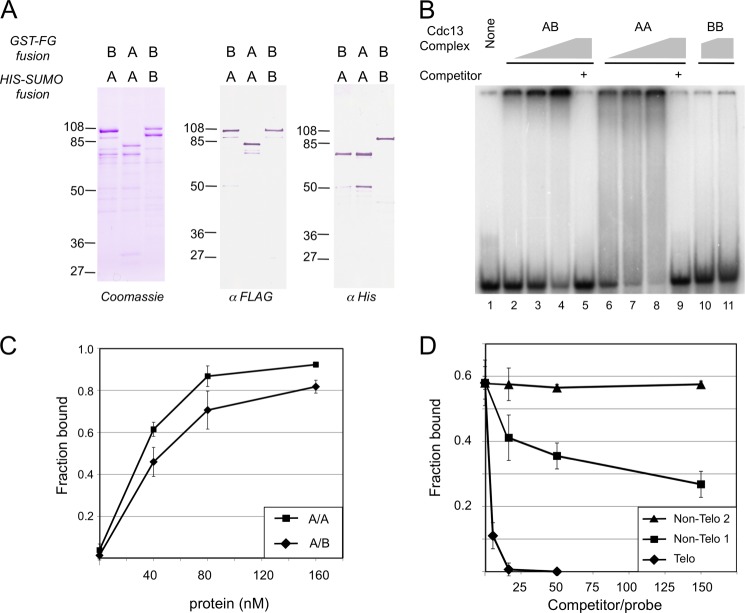
The propensity of C. albicans Cdc13A and Cdc13B to form heterodimers prompted us to compare the DNA binding properties of the homodimers with that of the heterodimer. For this analysis, we co-expressed appropriate combinations of His6-SUMO and GST-FLAG fusion proteins and isolated the A/A, A/B, and B/B complexes by sequential Ni-NTA and M2 affinity chromatography. As expected, each preparation contained an approximately equal molar amount of the two fusion proteins that reacted with the relevant antibodies in Western analysis (Fig. 7A). In gel mobility shift assays, both the A/A and A/B complex substantially reduced the levels of free probes (46-nt oligonucleotides with two copies of the C. albicans telomere G-strand repeat) with an ∼Kd of 40 and 55 nm, respectively (Fig. 7B, lanes 1–9, and 7C). Both the A/A and A/B complex appeared to experience aggregation; the majority of shifted probes for both the A/A and A/B complexes migrated minimally into the gel. In contrast, the B/B complex interacted weakly with the same probe (Fig. 7B, lanes 10 and 11). Inspection of the pattern of shifted probes revealed an interesting difference between the A/A and A/B samples; although a significant proportion of the A/A-DNA complex migrated as a heterogeneous smear throughout the lane, little smear was observed in the A/B-DNA samples. Although the reason for the discrepancy is not clear, one possible explanation is that the A/A-DNA complex may be unstable and dissociate during electrophoresis.
FIGURE 7.
DNA binding activities of the Cdc13 A/A, A/B, and B/B dimers. A, the purified A/B, A/A, and B/B dimers were analyzed by SDS-PAGE followed by Coomassie staining and Western analyses using the indicated antibodies. B, the purified dimers were tested for binding to CaG2 ((TCTAACTTCTTGGTGTACGGATG)2, 7.5 nm) in gel mobility shift assays. The concentrations of proteins used were as followed: A/B, 40 nm, 80 nm, 160 nm, 160 nm; A/A, 40 nm, 80 nm, 160 nm, 160 nm; B/B, 80 nm, 160 nm. Samples 5 and 9 also contained a 150-fold molar excess of cold CaG2 as competitors. C, the fractions of probes bound by increasing concentrations of the A/A and A/B complexes were calculated and plotted. Data (averages ± S.D.) were derived from three independent sets of assays. D, gel mobility shift assays were performed using the CaG2 probe (7.5 nm) and the A/B complex (70 nm). Increasing concentrations of unlabeled CaG2 and two non-telomeric oligos were added to the assays as competitors. The fractions of probes bound in the assays were determined and plotted against the competitor/probe ratios. Data (averages ± S.D.) were derived from three independent sets of assays.
We than analyzed the DNA binding specificity of the A/B complex by competition assays. As shown Fig. 7D, the complex exhibited substantial preference for the cognate telomere repeat in comparison with two non-telomeric oligonucleotides (Non-Telo 1 and Non-Telo 2). More specifically, the concentrations of the non-telomeric oligos needed to inhibit 50% complex formation are at least 50-fold higher than that of the telomere oligo (Fig. 7D). Taken together with previous results on the C. tropicalis Cdc13A/A complex, these observations indicate that both the A/A and A/B complex probably have the requisite affinity and sequence specificity to interact with the telomere G-strand in vivo.
DISCUSSION
We have uncovered a second Cdc13 homologue in Candida species and shown that the two paralogues perform overlapping and nonredundant functions in telomere regulation. Unexpectedly, the two paralogues manifest a propensity to form heterodimers, and the heterodimers interact with the cognate telomere repeat with high affinity and sequence specificity. Candida species may thus carry a variant CST complex with distinct composition and properties. The evolutionary and mechanistic implications of these findings are discussed below.
Potential Functions of Cdc13B
The discovery of Cdc13B proteins in Candida spp. provides a possible resolution to the conundrum posed by the small size of Cdc13A proteins in this group of fungi. That is, the functions served by the N-terminal half of Cdc13 in Saccharomyces and Kluyveromyces yeast could conceivably be performed by Cdc13B. These functions include, among others, dimerization, binding to Pol1, and binding to Est1 (5, 9, 28). Disruption of these interactions in Saccharomyces causes complex and disparate effects on telomere lengths, making it difficult to gauge potential conservation of functions from the existing telomere length data on our Cdc13B mutants. Thus, whether Cdc13B can indeed execute some or all of the functions ascribed to the N-terminal half of large Cdc13 is a question that can only be resolved by detailed investigation of more discrete mutations.
Homo-dimerization and Hetero-dimerization of Cdc13A and Cdc13B through the OB4 Domain
We have shown earlier that the OB4 domain of Candida glabrata Cdc13 and those of C. albicans and C. tropicalis Cdc13A can self-associate to form dimers or other higher order structures (18). In keeping with this observation, we now demonstrate that Cdc13B can also self-associate through its OB4 domain. Perhaps more surprisingly, both Cdc13A and Cdc13B exhibited a greater propensity to form heterodimers than homodimers, suggesting that the heterodimer constitutes the predominant form of Candida Cdc13 in vivo. Because the A/B dimer binds telomere G-strand with high affinity and sequence specificity, it has the requisite biochemical property to localize to telomeres. By contrast, the low DNA binding affinity of the B/B complex suggests that it may not represent a physiologically relevant form of Cdc13. Likewise, even though the A/A complex can certainly bind telomere DNA, several observations suggest that it may be unstable and less capable of functioning at telomeres than the A/B complex. In particular, the A/A-DNA complex appears to disassemble more quickly in gel mobility shift assays, resulting in substantial heterogeneity in the migration of the probe. This can be rationalized by the dissociation of the A/A dimer during electrophoresis and by the inability of the monomers to bind DNA stably (18). Clearly, more studies are necessary to ascertain the physiologically relevant form of Cdc13 in vivo. Moreover, the Cdc13 dimer is presumably just a part of a larger CST complex, and how the unusual A/B dimer in Candida interacts with Stn1 and Ten1 is clearly an interesting question for future investigation.
Evolution of Cdc13 Homologues in Saccharomycotina Yeast
The discovery and characterization of Cdc13B in the current report suggests a plausible model for Cdc13 evolution in the Saccharomycotina subphylum of budding yeast (Fig. 8). In this model, a single, 2-OB-fold CDC13 gene in the ancestral budding yeast is presumed to undergo gene duplication followed by fixation to yield two paralogues. In the Candida clade, subfunctionalization of the two paralogues resulted in a pair of telomere-capping proteins that share overlapping but nonredundant activities. By contrast, in the common ancestor of Saccharomyces and Kluyveromyces yeast, the two paralogues underwent gene fusion to yield a 4-OB-fold Cdc13, which in turn evolved a specialized function for its individual domains. This scenario is motivated by several key findings in the current study as well as previous observations. First, the existence of two Cdc13-like proteins with common biochemical properties (e.g. binding to each other and Stn1) in the Candida clade argues strongly for duplication of an ancestral gene. Second, the large Cdc13 proteins in Saccharomyces appear to result from the fusion of a pair of duplicated genes; the high resolution structure of ScCdc13 OB1 bears unmistakable resemblances to its DBD, and the OB2 structure is most similar to OB4 of C. glabrata Cdc13 (5, 7). The notion of gene fusion could also account for the absence of Cdc13B in Saccharomyces, assuming that the aforementioned gene duplication transpired in the common precursor of the Saccharomyces and Candida lineages. Although it is possible to imagine alternative evolutionary pathways, our model is relatively parsimonious and invokes just two major events to account for the disparate collection of present day Cdc13 in Saccharomycotina yeast. More importantly, the model makes testable predictions about the structure of Cdc13 orthologues in other related Saccharomycotina yeast. Specifically, if the contention that the ancestral yeast harbors a 2-OB-fold Cdc13 is true, then other early branching species in the subphylum, such as Yarrowia lipolytica, are also more likely to harbor such an orthologue rather than the large 4-OB-fold Cdc13 found in Saccharomyces and Kluyveromyces. Further studies will be necessary to test the validity of this and related hypotheses.
FIGURE 8.
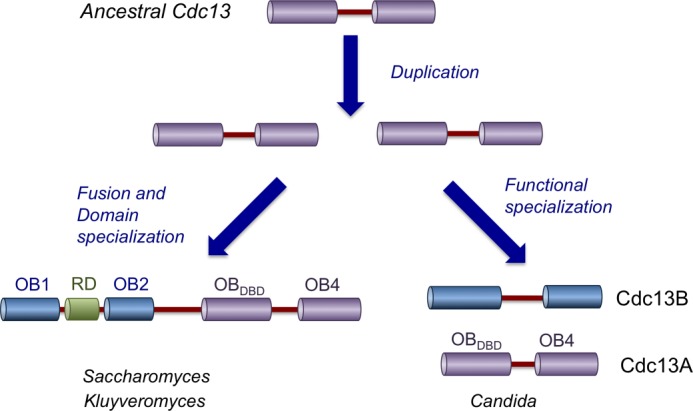
A model for Cdc13 evolution in Saccharomycotina yeast. In the proposed model, a 2-OB-fold Cdc13 was duplicated, and the resulting paralogues were both retained in the descendants. In the Candida clade, the two genes underwent functional specialization (neofunctionalization or subfunctionalization) and became the present day CDC13A and CDC13B. In the Saccharomyces and Kluyveromyces lineages, the duplicated CDC13 became fused to each other, resulting in gene products that are twice the size of the ancestral protein. Again, neofunctionalization or subfunctionalization of individual OB-fold domains is presumed to be responsible for the disparate functions of these domains in the present day CDC13. See “Discussion” for more details.
Recurrent Duplication of G-tail-binding Proteins
Fixation of duplicated G-tail-binding proteins (GTBP) appears to be a remarkably common evolutionary event. Previous studies have uncovered recurrent duplication of the POT1 gene in diverse organisms including plants, mammals, and worms (29–31). Our discovery of CDC13 gene duplication reinforces the impression that the existence of multiple GTBP can confer evolutionary advantages to an organism. What could be the potential benefits? All of the known G-tail-binding proteins are modular polypeptides that interact with a multiplicity of factors to confer telomere protection and regulate both telomere G- and C-strand synthesis. If one surface of the polypeptide is responsible for binding several target proteins, it may be difficult to fine-tune the affinity of interactions to achieve optimal functions. The duplication of a GTBP followed by subfunctionalization may enable each GTBP to achieve optimal binding to its specific interaction partner, thus enhancing organismal fitness. In this regard, it will be interesting to determine whether Candida Cdc13A and Cdc13B use similar surfaces to bind distinct target proteins.
This work was supported, in whole or in part, by National Institutes of Health Grant GM062631. This work was also supported by National Science Foundation Grant MCB-1157305.

This article contains supplemental Fig. 1.
- CST
- Cdc13-Stn1-Ten1
- OB
- oligonucleotide/oligosaccharide
- DBD
- DNA-binding domain
- Ni-NTA
- nickel-nitrilotriacetic acid
- TAP
- tandem affinity purification
- t-circle
- telomere circle
- A/B
- Cdc13A/Cdc13B
- A/A
- Cdc13A/Cdc13A
- B/B
- Cdc13B/Cdc13B
- GTBP
- G-tail-binding protein
- 5-FOA
- 5-fluoroorotic acid.
REFERENCES
- 1. de Lange T. (2009) How telomeres solve the end-protection problem. Science 326, 948–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. O'Sullivan R. J., Karlseder J. (2010) Telomeres: protecting chromosomes against genome instability. Nat. Rev. Mol. Cell Biol. 11, 171–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Giraud-Panis M. J., Teixeira M. T., Géli V., Gilson E. (2010) CST meets shelterin to keep telomeres in check. Mol. Cell 39, 665–676 [DOI] [PubMed] [Google Scholar]
- 4. Lundblad V. (2006) Budding yeast telomeres, in Telomeres and Telomerase (de Lange T., Lundblad V., Blackburn E., eds) 2nd Ed., pp. 345–386, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 5. Sun J., Yang Y., Wan K., Mao N., Yu T. Y., Lin Y. C., DeZwaan D. C., Freeman B. C., Lin J. J., Lue N. F., Lei M. (2011) Structural bases of dimerization of yeast telomere protein Cdc13 and its interaction with the catalytic subunit of DNA polymerase α. Cell Res 21, 258–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mitchell M. T., Smith J. S., Mason M., Harper S., Speicher D. W., Johnson F. B., Skordalakes E. (2010) Cdc13 N-terminal dimerization, DNA binding, and telomere length regulation. Mol. Cell. Biol. 30, 5325–5334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mason M., Wanat J. J., Harper S., Schultz D. C., Speicher D. W., Johnson F. B., Skordalakes E. (2013) Cdc13 OB2 dimerization required for productive Stn1 binding and efficient telomere maintenance. Structure 21, 109–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mitton-Fry R. M., Anderson E. M., Theobald D. L., Glustrom L. W., Wuttke D. S. (2004) Structural basis for telomeric single-stranded DNA recognition by yeast Cdc13. J. Mol. Biol. 338, 241–255 [DOI] [PubMed] [Google Scholar]
- 9. Pennock E., Buckley K., Lundblad V. (2001) Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell 104, 387–396 [DOI] [PubMed] [Google Scholar]
- 10. Chan A., Boulé J. B., Zakian V. A. (2008) Two pathways recruit telomerase to Saccharomyces cerevisiae telomeres. PLoS Genet. 4, e1000236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sun J., Yu E. Y., Yang Y., Confer L. A., Sun S. H., Wan K., Lue N. F., Lei M. (2009) Stn1-Ten1 is an Rpa2-Rpa3-like complex at telomeres. Genes Dev. 23, 2900–2914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gao H., Cervantes R. B., Mandell E. K., Otero J. H., Lundblad V. (2007) RPA-like proteins mediate yeast telomere function. Nat. Struct. Mol. Biol. 14, 208–214 [DOI] [PubMed] [Google Scholar]
- 13. Lue N. F., Zhou R., Chico L., Mao N., Steinberg-Neifach O., Ha T. (2013) The telomere capping complex CST has an unusual stoichiometry, makes multipartite interaction with G-tails, and unfolds higher-order g-tail structures. PLoS Genet. 9, e1003145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Puglisi A., Bianchi A., Lemmens L., Damay P., Shore D. (2008) Distinct roles for yeast Stn1 in telomere capping and telomerase inhibition. EMBO J. 27, 2328–2339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Petreaca R. C., Chiu H. C., Eckelhoefer H. A., Chuang C., Xu L., Nugent C. I. (2006) Chromosome end protection plasticity revealed by Stn1p and Ten1p bypass of Cdc13p. Nat. Cell Biol. 8, 748–755 [DOI] [PubMed] [Google Scholar]
- 16. Lue N. F. (2010) Plasticity of telomere maintenance mechanisms in yeast. Trends Biochem. Sci. 35, 8–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mandell E. K., Gelinas A. D., Wuttke D. S., Lundblad V. (2011) Sequence-specific binding to telomeric DNA is not a conserved property of the Cdc13 DNA binding domain. Biochemistry 50, 6289–6291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yu E. Y., Sun J., Lei M., Lue N. F. (2012) Analyses of Candida Cdc13 orthologues revealed a novel OB fold dimer arrangement, dimerization-assisted DNA binding, and substantial structural differences between Cdc13 and RPA70. Mol. Cell. Biol. 32, 186–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wilson R. B., Davis D., Mitchell A. P. (1999) Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J. Bacteriol. 181, 1868–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fonzi W. A., Irwin M. Y. (1993) Isogenic strain construction and gene mapping in Candida albicans. Genetics 134, 717–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Enloe B., Diamond A., Mitchell A. (2000) A single-transformation gene function test in diploid Candida albicans. J. Bacteriol. 182, 5730–5736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rigaut G., Shevchenko A., Rutz B., Wilm M., Mann M., Séraphin B. (1999) A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17, 1030–1032 [DOI] [PubMed] [Google Scholar]
- 23. Gerami-Nejad M., Berman J., Gale C. A. (2001) Cassettes for PCR-mediated construction of green, yellow, and cyan fluorescent protein fusions in Candida albicans. Yeast 18, 859–864 [DOI] [PubMed] [Google Scholar]
- 24. Yu E. Y., Yen W. F., Steinberg-Neifach O., Lue N. F. (2010) Rap1 in Candida albicans: an unusual structural organization and a critical function in suppressing telomere recombination. Mol. Cell. Biol. 30, 1254–1268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Santos M. A., Keith G., Tuite M. F. (1993) Non-standard translational events in Candida albicans mediated by an unusual seryl-tRNA with a 5′-CAG-3′ (leucine) anticodon. EMBO J. 12, 607–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mossessova E., Lima C. D. (2000) Ulp1-SUMO crystal structure and genetic analysis reveal conserved interactions and a regulatory element essential for cell growth in yeast. Mol. Cell 5, 865–876 [DOI] [PubMed] [Google Scholar]
- 27. Pickett H. A., Cesare A. J., Johnston R. L., Neumann A. A., Reddel R. R. (2009) Control of telomere length by a trimming mechanism that involves generation of t-circles. EMBO J. 28, 799–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wu Y., Zakian V. A. (2011) The telomeric Cdc13 protein interacts directly with the telomerase subunit Est1 to bring it to telomeric DNA ends in vitro. Proc. Natl. Acad. Sci. U.S.A. 108, 20362–20369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hockemeyer D., Daniels J. P., Takai H., de Lange T. (2006) Recent expansion of the telomeric complex in rodents: two distinct POT1 proteins protect mouse telomeres. Cell 126, 63–77 [DOI] [PubMed] [Google Scholar]
- 30. Shakirov E. V., Surovtseva Y. V., Osbun N., Shippen D. E. (2005) The Arabidopsis Pot1 and Pot2 proteins function in telomere length homeostasis and chromosome end protection. Mol. Cell. Biol. 25, 7725–7733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Raices M., Verdun R. E., Compton S. A., Haggblom C. I., Griffith J. D., Dillin A., Karlseder J. (2008) C. elegans telomeres contain G-strand and C-strand overhangs that are bound by distinct proteins. Cell 132, 745–757 [DOI] [PubMed] [Google Scholar]