Abstract
The cytosolic pathogen sensor RIG-I is activated by RNAs with exposed 5′-triphosphate (5′-ppp) and terminal double-stranded structures, such as those that are generated during viral infection. RIG-I has been shown to translocate on dsRNA in an ATP-dependent manner. However, the precise role of the ATPase activity in RIG-I activation remains unclear. Using in vitro-transcribed Sendai virus defective interfering RNA as a model ligand, we show that RIG-I oligomerizes on 5′-ppp dsRNA in an ATP hydrolysis-dependent and dsRNA length-dependent manner, which correlates with the strength of type-I interferon (IFN-I) activation. These results establish a clear role for the ligand-induced ATPase activity of RIG-I in the stimulation of the IFN response.
Keywords: ATP, defective interfering RNA, interferon, oligomerization, RIG-I
Introduction
Viral infection in a healthy host results in virus-specific adaptive immune responses and immunological memory preceded by a general innate immune response. The first step is the detection of viral infection in cells, which is accomplished by a diverse array of pattern-recognition receptors that sense non-self molecules [1]. RIG-I belongs to the RIG-I-like helicase group of SF2 family of helicases that also include MDA5 and LGP2, which function as viral RNA receptors to modulate antiviral type-I interferon (IFN-I) responses during RNA virus infections [2]. RIG-I consists of two amino-terminal caspase activation and recruitment domains (CARD) in tandem, a helicase and a carboxy-terminal regulatory domain (RD). RIG-I is activated by binding to a 5′-ppp double-stranded RNA (dsRNA) ligand [3–5], resulting in the release of CARDs from an inhibitory association with the helicase domain [6], allowing not only polyubiquitination of CARD2 by an E3 ligase TRIM25 [7], but also binding of unanchored K63-linked ubiquitin chains [8], forming an activated state by promoting RIG-I tetramer formation [9]. Activated RIG-I is able to interact with its adaptor mitochondrial antiviral signal (MAVS) on mitochondria [10–13], promoting aggregation of MAVS [14] and redistribution of mitochondria [15]. Active MAVS–RIG-I signalling culminates in the activation of the antiviral IFN-I pathway [16–19].
ATP hydrolysis by RIG-I after binding to RNA is critical for downstream activation [3]. The exact function of the ATPase activity of RIG-I was not clear until it was reported that after binding to 5′-ppp on RNA, RIG-I is capable of translocating on dsRNA in an ATP hydrolysis-dependent manner [20]. However, a role for this ATP-dependent translocation on dsRNA is not clear. In addition to genomic RNAs [21], defective interfering (DI) RNAs produced during infection with RNA viruses form effective ligands for RIG-I [22]. A Sendai virus (SeV) copy-back DI RNA preferentially associates with RIG-I in SeV-infected cells and potently activates the IFN-I response [23, 24]. We investigated the process of activation of RIG-I by this ligand. Specifically, we have uncovered a previously unrecognized role of ATP hydrolysis in enhancing RIG-I oligomerization on RNA. RIG-I oligomerization was dependent on ATP concentration, ATP hydrolysis and length of the dsRNA stem. ATP-driven RIG-I oligomer formation correlated with the magnitude of IFN-I activation. The data presented here indicate that RIG-I first forms a small binding unit upon recognition of 5′-ppp dsRNA, which is independent of ATP binding. ATP hydrolysis then drives the formation of a dsRNA length-dependent oligomer of RIG-I.
Results and discussion
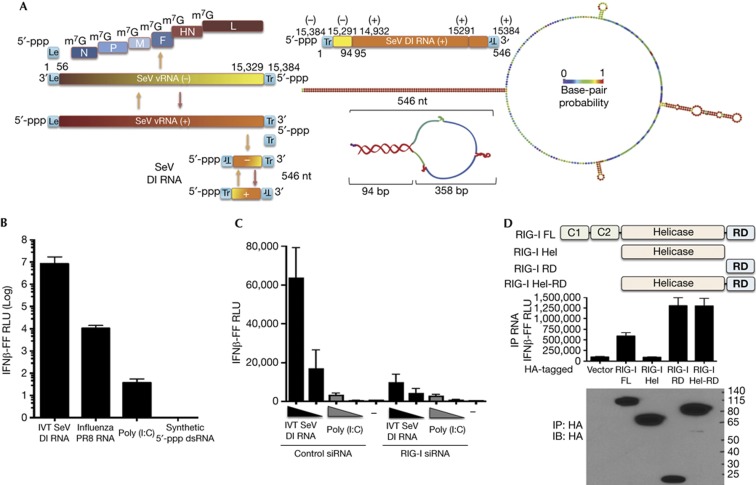
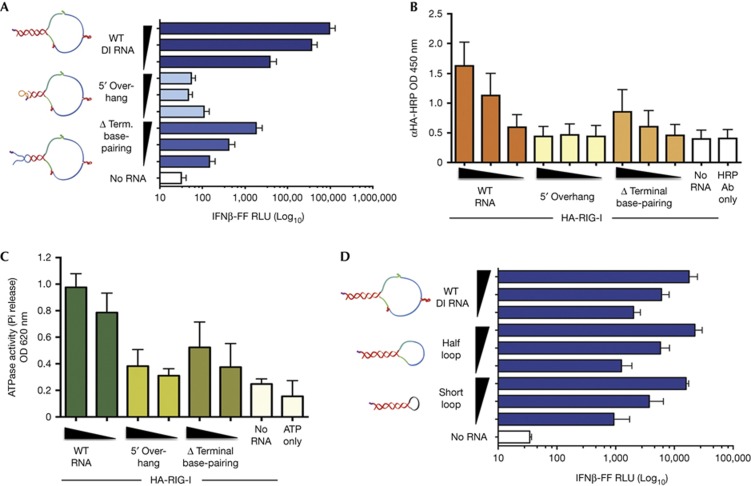
SeV Cantell strain DI RNA is a 546-nt-long copy-back, non-coding viral RNA produced during infection of cells and consists of a predicted stem-loop structure with a 94-bp stem and 358-b loop (Fig 1A). Compared to other ligands of RIG-I, influenza RNA, short poly (I:C) and a synthetic 19-mer 5′-ppp dsRNA, the in vitro transcribed (IVT) SeV DI RNA robustly activates IFN-I at the amount tested (50 ng) (Fig 1B). The remarkably higher stimulation of IFN-I by this RNA compared to other RIG-I ligands prompted us to investigate the specific requirements of this RNA to be a potent IFN-I inducer. Using short interfering RNA-mediated knockdown of RIG-I, we show that IVT DI RNA activates IFN-I in a RIG-I-dependent manner, whereas a mixture of short and long poly (I:C) does not depend on RIG-I to activate IFN-I (Fig 1C). Using an RNA pull-down assay (supplementary Fig S1A online), we show that this RNA interacts primarily with the RD domain of RIG-I (Fig 1D), requiring the basic cleft residues K858 and K861 (supplementary Fig S1B online), which have been implicated in binding to the 5′-ppp on RNA ligands of RIG-I [25, 26]. To uncover a mechanism for the high potency of this RNA in RIG-I-mediated IFN-I stimulation, we first generated structure mutants that either shield the 5′-ppp (5′-overhang) or lack terminal base pairing, as 5′-ppp and dsRNA have been suggested to be criticial features for interaction with RIG-I [4, 5]. We found that these mutant RNAs do not activate IFN-I as well as wild-type (WT) RNA (Fig 2A). They also do not bind to RIG-I as well as WT RNA (Fig 2B; supplementary Fig S2A online) and further do not optimally activate the ATPase of RIG-I, which might explain the defect in IFN-I activation (Fig 2C). Reducing the size of the loop to half or to a short loop did not have any impact on IFN-I activation (Fig 2D). These results indicate that the terminal 5′-ppp and dsRNA moieties but not the loop are critical features of this RNA for interaction with RIG-I and activation of IFN-I.
Figure 1.
SeV DI RNA is a potent RIG-I-dependent inducer of IFN-I. (A) Graphic illustrates different RNAs produced during infection of SeV. SeV DI produced from the anti-genomic positive sense (+) RNA consists of both negative and positive sense sequences of the genomic and anti-genomic RNAs resulting in a copy-back structure. The right panel shows the 546-nt SeV DI RNA mapped to the genomic/anti-genomic sequence and a predicted structure of the RNA (RNAfold). Colours on the DI RNA representations indicate base-pair probability (as indicated, red=1; purple=0). (B) 293T-IFNβ-FF-Luc cells were transfected with 50 ng of indicated RNAs and a luciferase assay was performed 24 h later to measure IFNβ promoter-driven luciferase activity. (C) 293T-IFNβ-FF-Luc cells were transfected with control or RIG-I short interfering RNA and 24 h later transfected with 5 and 1 ng of IVT DI RNA and 200 and 40 ng poly (I:C), followed by luciferase assay 24 h later to measure IFNβ promoter-driven luciferase activity. (D) Lysates from 293T cells expressing FL RIG-I or truncated RIG-I Hel, RIG-I C-terminal RD domain or RIG-I Hel-RD were incubated with 0.25 μg of RNA, RIG-I–RNA complexes were immunoprecipitated, and RNA and protein fractions were isolated. RNA was transfected into 293T-IFNβ-FF-Luc cells and 24 h later IFNβ promoter-driven luciferase activity was measured by luciferase assay. Protein fractions were subjected to immunoblotting using HA antibody to assess pull-down efficiency. Data are representative of at least three independent experiments and error bars indicate mean±s.d. DI, defective interfering; FL, full length; Hel, Helicase domain; Hel-RD, Helicase regulatory domain; IFN-I, type-I interferon; IVT, in vitro transcribed; RD, regulatory domain; SeV, Sendai virus.
Figure 2.
Exposed 5′-ppp and terminal dsRNA, but not loop structure, are important features of SeV DI RNA for RIG-I-mediated IFN-I activation. (A) 25 fmol and five-fold dilutions of WT, 5′ overhang and Δ terminal base-pairing RNAs were transfected or not into 293T-IFNβ-FF-Luc cells and 24 h later IFNβ promoter-driven luciferase activity was measured by a luciferase assay. (B) 500 fmol and three-fold dilutions of Biotin-UTP-labelled WT, 5′ overhang and Δ terminal base-pairing RNAs were immobilized onto NeutrAvidin-coated wells and incubated with lysates from HA-RIG-I-expressing 293T cells. The levels of bound RIG-I were determined by measuring the absorbance/HRP activity of HA-HRP antibody. (C) 0.5 μg of purified His-HA-RIG-I was incubated with 100 or 50 fmol of WT, 5′ overhang, Δ terminal base-pairing or no RNA in the presence of 0.5 mM ATP and 2.5 mM Mg2+ at 37 °C for 25 min. Released phosphates were measured using Malachite Green-based reagent at an absorbance of 620 nm. (D) 25 fmol and five-fold dilutions of WT, half-loop and short loop RNAs were transfected or not into 293T-IFNβ-FF-Luc cells and 24 h later IFNβ promoter-driven luciferase activity was measured by a luciferase assay. Data are representative of at least three independent experiments and error bars indicate mean±s.d. Log10 in Figs 2A and 2D refers to the scale on the x-axis. 5′-ppp, 5′-triphosphate; DI, defective interfering; dsRNA, double-stranded RNA; HRP, horseradish peroxidase; IFNβ, interferon β; SeV, Sendai virus; WT, wild type.
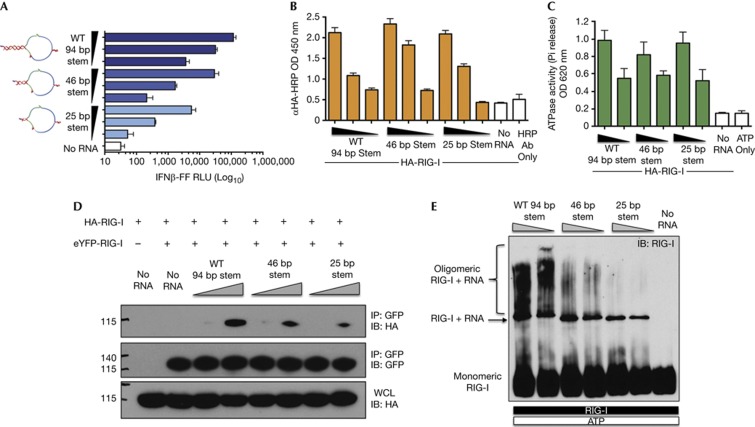
Although any RNA sequence with terminal 5′-ppp and panhandle structures should, in theory, induce the same response as IVT DI RNA, we sought to determine the importance of the length of the dsRNA stem in the panhandle. Truncating the 94-bp dsRNA stem from the non-terminal side to 46 and 25 bp drastically reduced IFN-I activation in an RNA length-dependent manner (Fig 3A). Compared to WT RNA, the 25- and 46-bp RNAs showed ∼100 and ∼10-fold reduction in IFN-I activation, respectively. Thus, the length of the dsRNA stem is critical for its potent activity. Surprisingly, we found that 46- and 25-bp RNAs bind to RIG-I and activate its ATPase to the same level as WT RNA (Figs 3B,C). Thus, to test if smaller stem RNAs have a defect in association of multiple RIG-I molecules on RNA, we performed a co-immunoprecipitation experiment of HA and eYFP-tagged RIG-I expressing cell lysates in the presence of RNA (Fig 3D). We found that higher levels of HA-RIG-I co-immunoprecipitated with eYFP-RIG-I in the presence of WT RNA compared to 46- and 25-bp stem RNAs, suggesting that the long stem of WT RNA allows greater association of multiple RIG-I molecules. To strengthen our findings, we analysed RIG-I–RNA complexes by nativePAGE after incubating RIG-I and RNAs in vitro in the presence of Mg-ATP. Upon immunoblotting for RIG-I, we discovered that RIG-I is able to form high-molecular-weight complexes in the presence of WT RNA (Fig 3E). These oligomers do not appear when no RNA is added, ruling out non-specific aggregation of the protein. We found a dramatic reduction in high-molecular-weight oligomers in the presence of 46- and 25-bp RNAs, which was also RNA-length dependent. Interestingly, the levels of the minimal binding unit of RIG-I remained essentially unchanged on the different RNAs, which confirms the binding data (Fig 3B) and explains the similar overall ATPase stimulation of RIG-I (Fig 3C). The defective formation of high-molecular-weight oligomers of RIG-I by shorter stem RNAs correlates with the levels of IFN-I activation, thereby suggesting dsRNA length-dependent oligomer formation as a mechanism for the high immunostimulatory activity of SeV DI RNA.
Figure 3.
dsRNA length-dependent oligomerization is a mechanism for the high immunostimulatory activity of SeV DI RNA. (A) 5 ng and five-fold dilutions of WT, 46- and 25-bp stem RNAs were transfected or not into 293T-IFNβ-FF-Luc cells and 24 h later IFNβ promoter-driven luciferase activity was measured by a luciferase assay. (B) 500 fmol and three-fold dilutions of Biotin-UTP-labelled WT, 46- and 25-bp stem RNAs were immobilized onto NeutrAvidin-coated wells and incubated with lysates from HA-RIG-I expressing 293T cells. The levels of bound RIG-I were determined by measuring the absorbance/HRP activity of HA-HRP antibody. (C) 100 and 50 fmol of RNAs was incubated with 0.5 μg of RIG-I in the presence of 0.5 mM ATP and 2.5 mM Mg2+ at 37 °C for 25 min. Released phosphates were measured by a colorimetric ATPase assay at absorbance 620 nm. (D) Lysates from cells expressing eYFP-RIG-I or HA-RIG-I were mixed and incubated with 1.25 and 5 pmol of WT, 46- and 25-bp stem RNA or no RNA, eYFP-RIG-I was immunoprecipitated with GFP antibody and eYFP-RIG-I and HA-RIG-I levels were assessed by SDS–PAGE and immunoblotting with GFP or HA antibodies. Levels of input HA-RIG-I in WCL were determined by SDS–PAGE and immunoblotting with HA antibody. (E) 1 μg of RIG-I was incubated with 1.25 or 0.625 pmol of WT, 46- and 25-bp stem RNAs or no RNA in the presence of 0.5 mM ATP and 2.5 mM Mg2+ for 25 min at 37 °C and RIG-I complexes were analysed by NativePAGE and immunoblotting. Data are representative of at least three independent experiments and error bars indicate mean±s.d. Log10 in Fig 3A refers to the scale on the x-axis. DI, defective interfering; dsRNA, double-stranded RNA; GFP, green fluorescent protein; HRP, horseradish peroxidase; SeV, Sendai virus; WCL, whole cell lysates; WT, wild type.
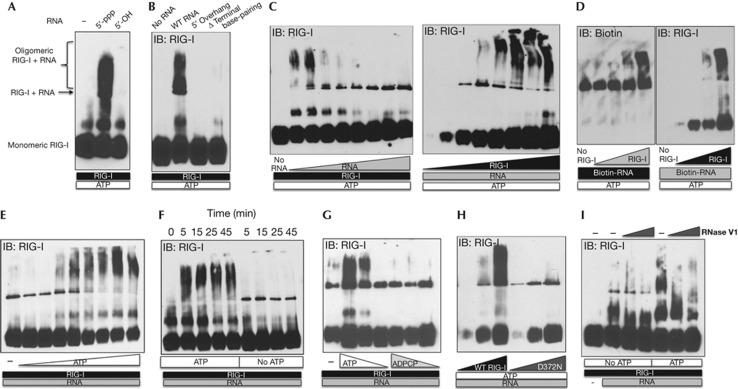
There is limited evidence in the literature for ligand and co-factor requirements for RIG-I multimerization. Binder et al [27] showed some evidence of RIG-I oligomerization on dsRNA by scanning force microscopy and size-exclusion chromatography. However, the role of 5′-ppp on RNA was not tested and the experiments were performed in the absence of ATP, which is an essential co-factor for activation. First, we asked whether the terminal 5′-ppp binding is required for RIG-I oligomerization on RNA. To this end, we tested RIG-I oligomerization on RNA treated with a phosphatase. We confirmed that the 5′-OH RNA was not able to bind to RIG-I (supplementary Fig S2B online) or activate IFN-I (supplementary Fig S2C online). NativePAGE analyses revealed that the 5′-OH RNA is not able to induce oligomerization of RIG-I like 5′-ppp RNA, suggesting that binding to 5′-ppp is critical for formation of RIG-I oligomers (Fig 4A). In accordance with this data, 5′-overhang RNA also did not allow oligomer formation and neither did the Δ terminal base-pairing RNA, confirming that both 5′-ppp and dsRNA motifs are important for oligomerization of RIG-I (Fig 4B). Furthermore, we found that oligomerization is enhanced at high RIG-I to RNA ratios (Fig 4C). Utilizing Biotin-labelled RNA in oligomerization reactions and immunoblotting for Biotin in nativePAGE separated RIG-I–RNA complexes, we confirmed the presence of RNA in the oligomers (Fig 4D). We ensured that Biotin labelling of RNA had no impact on its immunostimulatory potential (supplementary Fig S2D online).
Figure 4.
ATP hydrolysis drives oligomerization of RIG-I on 5′-ppp dsRNA. (A,B) 0.2 μg of indicated IVT RNAs was incubated with 1 μg of RIG-I in the presence of 1 mM ATP and 2.5 mM Mg2+ for 25 min at 37 °C, analysed by NativePAGE and immunoblotted for RIG-I. (C) Left panel: 0.5 μg of RIG-I was incubated with 0.05, 0.075, 0.1, 0.175, 0.25, 0.5, 1 and 2 μg of RNA or no RNA in the presence of 1 mM ATP and 2.5 mM Mg2+ for 25 min at 37 °C and analysed. Right panel: 0.5 μg of RNA was incubated with 0.05, 0.1, 0.25, 0.5, 1, 2, 3, 4 or 5 μg of RIG-I in the presence of 1 mM ATP and 2.5 mM Mg2+ for 25 min at 37 °C and analysed. (D) 0.2 μg of Biotin-RNA was incubated with 0.05, 0.25, 0.5 and 1 μg of RIG-I in the presence of 1 mM ATP and 2.5 mM Mg2+ for 25 min at 37 °C, analysed by nativePAGE and immunoblotted for Biotin-RNA and RIG-I. (E) 2 μg of RIG-I was incubated with 0.3 μg of RNA in the absence or presence of 0.05, 0.2, 0.5, 1, 2, 3, 4 and 5 mM of ATP and 2.5 mM Mg2+ for 25 min at 37 °C and analysed. (F) 2 μg of RIG-I was incubated with 0.3 μg of RNA with or without 2 mM ATP for 5, 15, 25 and 45 min at 37 °C and RIG-I oligomerization was analysed. (G) 2 μg of RIG-I was incubated with 0.3 μg of RNA in the presence of 1, 0.5 and 0.25 mM of ATP or ADPCP for 25 min at 37 °C and native complexes were analysed. (H) 0.25 μg of RNA was incubated with 0.05, 0.3 and 1 μg of either WT or D372N RIG-I with 1 mM of ATP and 2.5 mM Mg2+ for 25 min at 37 °C and analysed. (I) 0.2 μg of RNA was incubated with 1 μg of RIG-I in the presence or absence of 1 mM ATP and 2.5 mM Mg2+ for 15 min at 37 °C, followed by addition of 0, 0.01 and 0.1 units of RNase V1 at RT for 15 min and RIG-I oligomerization was analysed. Data are representative of two to three independent experiments. 5′-ppp, 5′-triphosphate; dsRNA, double-stranded RNA; IVT, in vitro transcribed.
ATP is essential for RIG-I activation. ATP hydrolysis allows translocation of RIG-I on RNA [20]. However, the outcome of this ATPase-driven translocase activity is unknown. Using electrophoretic mobility shift assay and electron microscopy, it was recently reported that RIG-I is not able to form filaments on RNA like MDA5 [28]. However, curiously, the authors performed these experiments using ADP-AlF4, which mimics the ADP-Pi state. Here, we investigated the role of ATP in the oligomerization process of RIG-I. We incubated RIG-I and RNA either in the absence or in the presence of increasing concentrations of ATP up to 4 mM. We found an ATP-dose-dependent increase in the formation of RIG-I–RNA oligomers (Fig 4E). A time-course experiment revealed that the oligomeric complexes of RIG-I–RNA are formed within 5 min in the presence of ATP (Fig 4F). Interestingly, in the absence or at lower concentrations of ATP, only the minimal RNA-binding unit of RIG-I is formed but not the high-molecular-weight oligomers (Figs 4E,F). Next, we tested if ATP binding was sufficient for oligomerization or if ATP hydrolysis was required. To this end, we utilized a non-hydrolysable analogue of ATP, β,γ-methyleneadenosine 5′-triphosphate (ADPCP), and the D372N ATPase mutant of RIG-I. We found that WT RIG-I is not able to form oligomers on RNA in the presence of ADPCP (Fig 4G) and the D372N RIG-I is not able to form oligomers on RNA in the presence of ATP (Fig 4H). Thus, formation of an initial binding unit of RIG-I on RNA does not require ATP, but ATP hydrolysis by RIG-I is crucial for formation of higher-order structures on RNA. This is in contrast to MDA5, where ATP hydrolysis drives dissociation of MDA5 oligomers on dsRNA [28, 29]. Post-treatment with dsRNA-specific RNAse V1 disrupted RIG-I oligomers on RNA, suggesting that the oligomers are dynamic and transient in nature and the dsRNA is exposed enough to be vulnerable to an endonuclease (Fig 4I). Jiang et al [9] have recently shown that RIG-I tetramer formation on RNA is driven by binding of N-terminal CARDs to polyubiquitin chains. As we used purified components in our analysis of native RIG-I/RNA complexes, oligomerization of RIG-I was detected in the absence of polyubiquitin binding or supplementation. However, it is likely that binding to polyubiquitin or other co-factors further stabilizes and promotes the formation of RIG-I multimers in vivo.
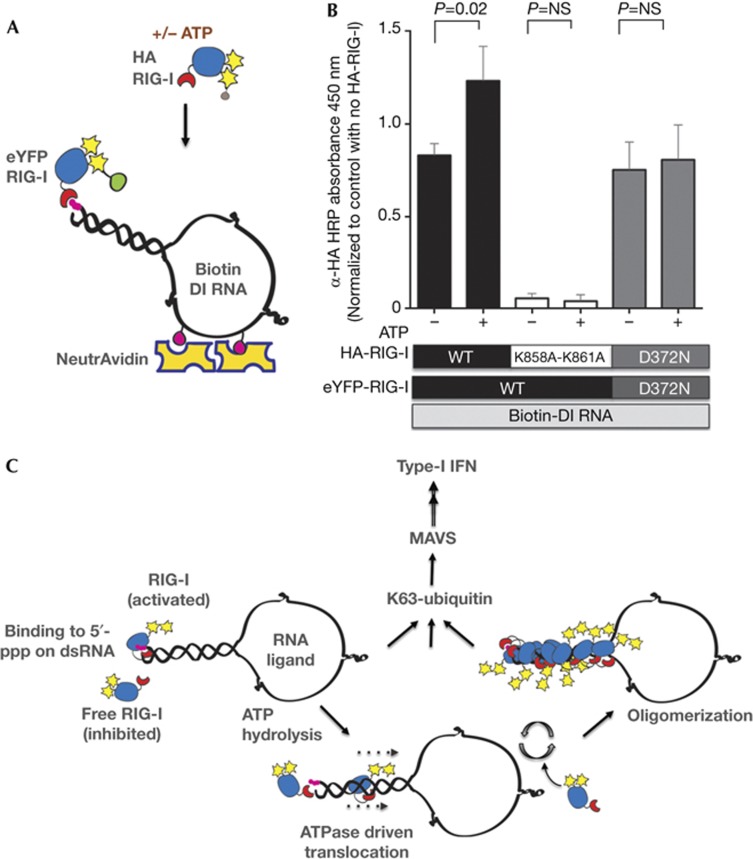
In order to understand the mechanism of oligomerization, we investigated the requirement for loading of an additional unit of RIG-I on preformed minimal RNA-binding units of RIG-I by utilizing differentially tagged RIG-I proteins in an RNA-binding experiment either in the presence or in the absence of ATP. NeutrAvidin-immobilized Biotin-RNA was first saturated with eYFP-RIG-I, followed by addition of limited amounts of HA-RIG-I either in the presence or in the absence of ATP (Fig 5A). We found that addition of ATP enhances binding of HA-RIG-I to eYFP-RIG-I–RNA complexes (Fig 5B), suggesting that ATP hydrolysis exposes the 5′-ppp to allow the additional unit of RIG-I to bind to the same RNA. Importantly, we found that a K858A-K861A 5′-ppp-binding mutant is not able to bind to eYFP–RIG-I–RNA complexes both in the presence and in the absence of ATP, confirming that 5′-ppp exposure by bound RIG-I and 5′-ppp binding by incoming RIG-I molecules are required for recruitment of additional molecules of RIG-I. Additionally, we show that substituting WT RIG-I with D372N ATPase mutant prevents loading of additional molecules of RIG-I in the presence of ATP. Based on other studies and the data presented here, we propose a model of RIG-I oligomerization where each RIG-I molecule binds to 5′-ppp and, using ATP hydrolysis, enters the RNA probably by translocation. As multiple RIG-I molecules enter the RNA in this manner, the result is formation of an oligomer. These large oligomers of RIG-I on RNA possibly induce more robust aggregation of MAVS, which was recently shown to produce prion-like fibrils in the presence of strong RIG-I ligands such as SeV RNA [14], thereby leading to a robust activation of IFN-I (Fig 5C).
Figure 5.
Mechanism for ATP-driven formation of RIG-I oligomers on RNA. (A) Schematic of the experiment highlighting immobilization of Biotin-labelled RNA onto NeutrAvidin and saturation with eYFP-RIG-I, followed by binding of HA-RIG-I in the presence or absence of ATP. (B) 80 ng of Biotin-labelled DI RNA was immobilized on NeutrAvidin-coated wells, washed and 1 μg of eYFP-RIG-I added (WT or D372N mutant). Following washes, 0.2 μg of HA-RIG-I (WT, K858A-K861A or D372N) was added in the presence or absence of 1 mM ATP as indicated. Bound HA-RIG-I was detected using α-HA-HRP antibody. Data are representative of three independent experiments and error bars indicate mean±s.d. The P-value was calculated using a Student’s unpaired t test. (C) A proposed model of RIG-I activation. Upon binding to 5′-ppp on dsRNA ligand, RIG-I is activated with a conformation change and ATP hydrolysis. Our data suggest that ATP hydrolysis by RNA-bound RIG-I allows exposure of 5′-ppp and recruitment of additional RIG-I molecules on long dsRNA forming RIG-I oligomers. As demonstrated previously, upon K63-linked polyubiquitination or polyubiquitin binding, RIG-I is further activated and binds MAVS to induce IFN-I production. 5′-ppp, 5′-triphosphate; DI, defective interfering; dsRNA, double-stranded RNA; HRP, horseradish peroxidase; IFN-I, type-1 interferon; MAVS, mitochondrial antiviral signal; NS, not significant; WT, wild type.
Methods
ATPase activity assay. Purified RIG-I was incubated in 50-μl reactions with RNAs in the presence of 0.5 mM ATP and 2.5 mM MgCl2 in 50 mM Tris, 150 mM NaCl and 1 mM DTT (protein buffer) at 37 °C for 20 min. Released phosphates were measured using a Malachite green-based colorimetric assay (Novus Biologicals) at 620 nm using a plate reader (Biotek), following the manufacturer’s instructions. RNA-induced ATPase activity for WT and D372N RIG-I was confirmed (supplementary Fig S3C online).
NativePAGE of RIG-I–RNA complexes. Purified RIG-I was incubated with RNA in amounts as indicated in the presence of given ATP concentrations with 2.5 mM MgCl2 in protein buffer at 37 °C for 25 min in 50-μl reactions, unless otherwise indicated. Reactions were stopped using 4 × NativePAGE sample buffer and analysed by NativePAGE on a 3–12% Bis-Tris gel (Life), transferred onto PVDF membranes (Life), fixed with 8% acetic acid in H2O, and immunoblotting was performed using anti-Biotin HRP (Abcam) or RIG-I antibodies.
RNA ELISA for RIG-I binding. RNA preparation is described in supplementary information online. Eighty nanograms of Biotin-UTP-labelled RNAs was added to NeutrAvidin-coated wells (supplementary Fig S2A online). Following RNA immobilization, wells were washed and incubated with lysates from cells expressing HA-RIG-I or 0.2 μg of purified HA-RIG-I. The washing buffer comprised 50 mM Tris, pH 7.5, 150 mM NaCl, 1 mM MgCl2 and 0.05% NP-40, and the binding buffer consisted of washing buffer supplemented with RNase inhibitor (Ambion) and 1 mM DTT. Bound RIG-I was detected using HA-HRP antibody (Cell Signaling) and TMB (Pierce) by measuring absorbance at 450 nm (Biotek) after stopping the reactions with 0.5 M HCl. In a modified protocol using differentially tagged RIG-I to understand the mechanism of formation of RIG-I oligomers on RNA, 80 ng of Biotin-RNA was first immobilized as given above, followed by addition of 1 μg of purified eYFP-RIG-I to saturate the bound RNA. Following washes, 0.20 μg of HA-RIG-I was added with or without 1 mM ATP, incubated at 37 °C for 20 min, washed and binding was measured as described above. Protein purification is described in supplementary information online. To test the RNA-binding ability of purified proteins, a monoclonal RIG-I antibody was used to determine protein bound to RNA by ELISA (supplementary Fig S3A online). The purity of proteins was checked by Coomassie Brilliant Blue (EZBlue, SIGMA) staining (supplementary Fig S3B online). Additional methods are described in supplementary information online.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
J.R.P. is supported by National Institutes of Health T32 training grant 5T32AI007647-13. We would like to thank Florian Krammer, Saad Rahmat, Shashank Tripathi, Mirco Schmolke, Mila Ortigoza, Balaji Manicassamy, Melissa Uccellini, Amzie Pavlisin, Ricardo Rajsbaum, Gijs Versteeg, Estanislao Nistal-Villan, Ana Fernandez-Sesma, Benjamin R. tenOever, Megan L. Shaw, Viviana A. Simon and Peter Palese for helpful discussions; Juan Ayllon for the stable IFNβ-FF-Luc 293T cell line and the Center for Therapeutic Antibody Development for the RIG-I antibody; and Richard Cadagan and Osman Lizardo for their excellent technical assistance. These studies have been partly supported by the National Institute of Allergy and Infectious Diseases grant U19 AI083025 to A.G.-S. and T.H.
Author contributions: J.R.P. designed and performed the experiments and wrote the manuscript; J.R.P., A.J., Y.-Y.C., A.B., T.H. and A.G.-S. analysed the data; A.G.-S. supervised all aspects of the study.
Footnotes
The authors declare that they have no conflict of interest.
References
- Ranjan P, Bowzard JB, Schwerzmann JW, Jeisy-Scott V, Fujita T, Sambhara S (2009) Cytoplasmic nucleic acid sensors in antiviral immunity. Trends Mol Med 15: 359–368 [DOI] [PubMed] [Google Scholar]
- Zou J, Chang M, Nie P, Secombes CJ (2009) Origin and evolution of the RIG-I like RNA helicase gene family. BMC Evol Biol 9: 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T (2004) The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol 5: 730–737 [DOI] [PubMed] [Google Scholar]
- Schlee M et al. (2009) Recognition of 5' triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity 31: 25–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A et al. (2009) 5'-triphosphate RNA requires base-paired structures to activate antiviral signaling via RIG-I. Proc Natl Acad Sci USA 106: 12067–12072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalinski E, Lunardi T, Mccarthy AA, Louber J, Brunel J, Grigorov B, Gerlier D, Cusack S (2011) Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell 147: 423–435 [DOI] [PubMed] [Google Scholar]
- Gack MU et al. (2007) TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature 446: 916–920 [DOI] [PubMed] [Google Scholar]
- Zeng W, Sun L, Jiang X, Chen X, Hou F, Adhikari A, Xu M, Chen ZJ (2010) Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell 141: 315–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Kinch LN, Brautigam Ca, Chen X, Du F, Grishin NV, Chen ZJ (2012) Ubiquitin-induced oligomerization of the RNA sensors RIG-I and MDA5 activates antiviral innate immune response. Immunity 36: 959–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth RB, Sun L, Ea C-K, Chen ZJ (2005) Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell 122: 669–682 [DOI] [PubMed] [Google Scholar]
- Xu L-G, Wang Y-Y, Han K-J, Li L-Y, Zhai Z, Shu H-B (2005) VISA is an adapter protein required for virus-triggered IFN-β signaling. Mol Cell 19: 727–740 [DOI] [PubMed] [Google Scholar]
- Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp J (2005) Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437: 1167–1172 [DOI] [PubMed] [Google Scholar]
- Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, Akira S (2005) IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol 6: 981–988 [DOI] [PubMed] [Google Scholar]
- Hou F, Sun L, Zheng H, Skaug B, Jiang Q-X, Chen Zhijian J (2011) MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell 146: 448–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onoguchi K, Onomoto K, Takamatsu S, Jogi M, Takemura A, Morimoto S, Julkunen I, Namiki H, Yoneyama M, Fujita T (2010) Virus-infection or 5'ppp-RNA activates antiviral signal through redistribution of IPS-1 mediated by MFN1. PLoS Pathog 6: e1001012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servant MJ, Grandvaux N, Hiscott J (2002) Multiple signaling pathways leading to the activation of interferon regulatory factor 3. Biochem Pharmacol 64: 985–992 [DOI] [PubMed] [Google Scholar]
- Sharma S, tenOever BR, Grandvaux N, Zhou G-P, Lin R, Hiscott J (2003) Triggering the interferon antiviral response through an IKK-related pathway. Science 300: 1148–1151 [DOI] [PubMed] [Google Scholar]
- Panne D (2008) The enhanceosome. Curr Opin Struct Biol 18: 236–242 [DOI] [PubMed] [Google Scholar]
- Dixit E et al. (2010) Peroxisomes are signaling platforms for antiviral innate immunity. Cell 141: 668–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myong S, Cui S, Cornish PV, Kirchhofer A, Gack MU, Jung JU, Hopfner K-P, Ha T (2009) Cytosolic viral sensor RIG-I is a 5'-triphosphate-dependent translocase on double-stranded RNA. Science 323: 1070–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehwinkel J et al. (2010) RIG-I detects viral genomic RNA during negative-strand RNA virus infection. Cell 140: 397–408 [DOI] [PubMed] [Google Scholar]
- Baum A, García-Sastre A (2011) Differential recognition of viral RNA by RIG-I. Virulence 2: 166–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum A, Sachidanandam R, García-Sastre A (2010) Preference of RIG-I for short viral RNA molecules in infected cells revealed by next-generation sequencing. Proc Natl Acad Sci USA 107: 16303–16308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahle L, Garcin D, Kolakofsky D (2006) Sendai virus defective-interfering genomes and the activation of interferon-beta. Virology 351: 101–111 [DOI] [PubMed] [Google Scholar]
- Lu C, Xu H, Ranjith-Kumar CT, Brooks MT, Hou TY, Hu F, Herr AB, Strong RK, Kao CC, Li P (2010) The structural basis of 5' triphosphate double-stranded RNA recognition by RIG-I C-terminal domain. Structure 18: 1032–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui S, Eisenächer K, Kirchhofer A, Brzózka K, Lammens A, Lammens K, Fujita T, Conzelmann KK, Krug A, Hopfner KP (2008) The C-terminal regulatory domain is the RNA 5'-triphosphate sensor of RIG-I. Mol Cell 29: 169–179 [DOI] [PubMed] [Google Scholar]
- Binder M, Eberle F, Seitz S, Mücke N, Hüber CM, Kiani N, Kaderali L, Lohmann V, Dalpke A, Bartenschlager R (2011) Molecular mechanism of signal perception and integration by the innate immune sensor retinoic acid-inducible gene-I (RIG-I). J Biol Chem 286: 27278–27287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peisley A, Lin C, Wu B, Orme-Johnson M, Liu M, Walz T, Hur S (2011) Cooperative assembly and dynamic disassembly of MDA5 filaments for viral dsRNA recognition. Proc Natl Acad Sci USA 108: 21010–21015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berke IC, Modis Y (2012) MDA5 cooperatively forms dimers and ATP sensitive filaments upon binding to dsRNA. EMBO J 31: 1714–1726 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.