Abstract
The bacterial stringent response links nutrient starvation with the transcriptional control of genes. This process is initiated by the stringent factor RelA, which senses the presence of deacylated tRNA in the ribosome as a symptom of amino-acid starvation to synthesize the alarmone (p)ppGpp. Here we report a cryo-EM study of RelA bound to ribosomes bearing cognate, deacylated tRNA in the A-site. The data show that RelA on the ribosome stabilizes an unusual distorted form of the tRNA, with the acceptor arm making contact with RelA and far from its normal location in the peptidyl transferase centre.
Keywords: ribosome, RelA, cryo-EM, A/T-tRNA, starvation signalling
Introduction
In order to survive, living organisms need to adapt rapidly to environmental changes. One such adaptation, the bacterial stringent response, is triggered during nutritional stress and uses the alarmone (p)ppGpp (a hyper-phosphorylated version of GDP or GTP) to promote physiological changes via transcriptional regulation [1–3]. Recently, (p)ppGpp activity has also been implicated in processes such as bacterial virulence [4] or pathogenic biofilm formation [5]. The enzymes belonging to the RelA/SpoT homologue family constitute the group of proteins that regulate the synthesis and/or degradation of (p)ppGpp [6]. Different members of this family have been found in bacteria, plant and even metazoan organisms [7, 8].
In Escherichia coli and other proteobacteria (β- and γ-proteobacteria), the factor responsible for the synthesis of (p)ppGpp in response to starvation is the ribosome-activated enzyme RelA [9]. During starvation, the resulting scarcity of amino acids results in an increase in the concentration of uncharged (that is, non-aminoacylated) transfer RNAs (tRNAs). These uncharged tRNAs could bind to a vacant ribosomal A-site if the corresponding aminoacylated species is not available and thereby stall translation (Figs 1A,B). This is the molecular event sensed by RelA, which recognizes arrested ribosomes with cognate, uncharged tRNAs in the A-site, to catalyse the transfer of the β and γ phosphates from ATP to GTP or GDP [10]. The interaction with the 3′ end is crucial for the catalytic activity of RelA in synthesizing (p)ppGpp [10–12].
Figure 1.
Biochemical characterization. (A) The normal elongation cycle of translation: a cognate, aminoacylated tRNA (purple) in complex with EF-Tu (orange) and GTP is delivered to the A-site of the ribosome (left). Following peptidyl transfer and the formation of hybrid tRNA states (middle), EF-G catalyses the translocation of tRNA and mRNA in the 30S subunit (right). (B) During starvation, an uncharged tRNA (purple) binds to the A-site of the ribosome in the absence of EF-Tu, leading to stalling. This stalled state is recognized by RelA (red). (C) Diagram representing the domain organization of the relA gene. The two constructs (NT-454-RelA and full length) used for this work are represented. (D) 4–12% SDS–PAGE loaded with the pull-downs of the full-length RelA (left) or with the truncated NT-454-RelA construct (right) with different programmed states of the 70 S. mRNA, messenger RNA; tRNA, transfer RNAs.
There is, at present, no understanding of the molecular basis for how RelA senses a ribosome stalled with an uncharged tRNA in the A-site. External factors such as RelA would normally be unable to sense the aminoacylation state of the tRNA in the A-site, as its 3′ end is buried deep in the peptidyl transferase centre. A difficulty with the structural studies on RelA is that it is prone to aggregation at working concentrations [13]. This probably reflects its role in vivo, where it is produced in very low amounts [14] and is regulated through self-oligomerization [15, 16]. Here, we establish a purification protocol and in vitro interaction assay to generate a complex suitable for structural determination using cryo-EM.
Results
The Rel gene and the products of its duplication, RelA and SpoT, are well conserved among bacteria [6]. The RelA gene encodes a single polypeptide of ∼750 amino acids (Fig 1C): the amino-terminal domain (residues 1-455), which is responsible for the catalytic activity; the carboxy-terminal domain (455–744), on the other hand, is involved in ribosome binding [17] and oligomerization [15, 16]. In order to establish the minimal requirements for a stable ribosome–RelA interaction, we pelleted various RelA–ribosome complexes through a sucrose cushion and analysed the pellet on SDS–polyacrylamide gel electrophoresis for the presence of RelA. We used full-length RelA protein without any purification tag (see Methods) and a C-terminal deletion mutant, termed NT-454-RelA, that encodes the catalytic N-terminal domain (Figs 1C,D). Ribosomes programmed with full occupancy of non-acylated tRNAs at the A-site are able to pull down the largest amount of full-length RelA, in line with previous data [10]. Other studies have suggested that the presence of messenger RNA alone is enough for the ribosome to activate RelA [13]. In contrast, our data suggest that the presence of an uncharged A-site tRNA promotes the efficient binding of RelA to the ribosome (Fig 1D). This binding requires the presence of the C-terminal domain of RelA as no binding was observed in its absence even in the presence of uncharged A-site tRNA. We did not see any effect of nucleotides on the binding of RelA to ribosomes; hence, all the complexes were generated in their absence.
We next proceeded with single-particle cryo-EM reconstructions of the RelA–ribosome complexes. A reconstruction using the entire data set of ∼205,000 particles revealed the heterogeneity present in the sample (Figs 2A,B). No apparent density was present for RelA, and only fragmented density was seen for the deacylated tRNA residing in the ribosomal A-site. Accordingly, we separated the heterogeneous data set into structurally more homogeneous classes, using ML3D [18], a maximum likelihood (ML)-based classification approach (see Methods), and obtained multiple classes (supplementary Figs S1, S2 online). Whereas the majority of the classes represented vacant or weakly occupied subpopulations, one sizeable class (consisting of ∼28,000 particles) showed substantial improvement in occupancy (compare Figs 2A,C) and homogeneity (compare 3D variance analyses shown in Figs 2B,D) relative to the total reconstruction. In this cryo-EM map (denoted as class 8, at ∼10.8 Å resolution, supplementary Fig S1 online), clear density could be observed for the E-, P- and A-site tRNAs (Fig 3). Density that could be identified as RelA was clearly seen, partially overlapping with the binding sites for EF-Tu, EF-G and other essential ribosomal GTPases. The rendered volume for RelA is estimated to enclose ∼80% of the mass of RelA. The remaining mass of the factor might correspond to dynamic/flexible regions, not fully stabilized.
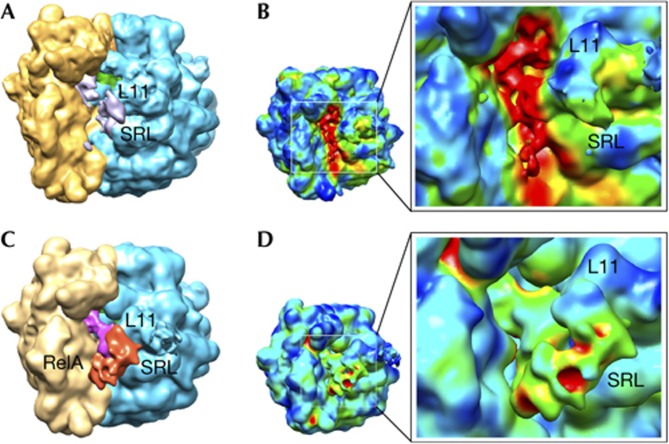
Figure 2.
Cryo-EM reconstructions using the entire data set and after classification. (A) and (B) display renderings for the cryo-EM map calculated from the entire data set collected for the 70S–RelA complex. (A) Regions identified as the 30S subunit (yellow), 50S subunit (blue) and the P-site tRNA (green) are shown. Scattered signal attributable to the deacylated A-site tRNA and RelA is shown in grey. (B) 3D variance analysis showed that the regions of higher variability (shown in red) are located in the inter-subunit space. The reconstruction exhibits considerable heterogeneity, mostly located in the region of the A-site tRNA. Panels (C) and (D) show representations for the cryo-EM map with higher occupancy for RelA after classification of the original data set. (C) Orientation and labels as in panel (A) with additional densities attributable to the deacylated A-site tRNA (magenta) and protein RelA (red). (D) Conformational variability by 3D variance analysis suggested a clear improvement in the occupancy by RelA after the sorting. Labels indicate: L11 (region that contains ribosomal protein L11 and a segment of 23S rRNA); SRL (sarcin–ricin loop); and RelA, density attributed to the stringent factor RelA. rRNA, ribosomal RNA; SRL, sarcin–ricin loop; tRNA, transfer RNA.
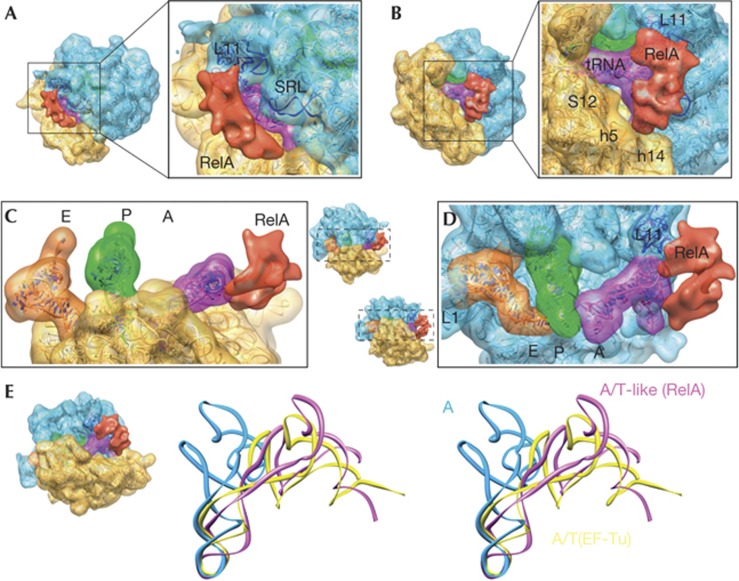
Figure 3.
Three-dimensional cryo-EM structure of the 70S–RelA complex. (A–D) Cryo-EM reconstruction showing the regions corresponding to the 30S subunit (yellow), the 50S subunit (blue) and tRNAs in the A (magenta), P (green) and E (orange) sites. RelA is shown in red. The molecular structures fitted to the density are shown as ribbon diagrams. The isolated densities for the subunits, tRNAs and RelA are rendered using the same density threshold value. (E) Stereo views of the A-site tRNA in the current structure (magenta, denoted as A/T-like), the A/T state of aminoacyl–tRNA in complex with EF-Tu in the ribosome (yellow) [21] and the ‘classical’ A-site tRNA (blue) [38]. In each panel, the orientations of the subunits are shown as thumbnails. The following features are also labelled: L1 (stalk that contains ribosomal protein L1); SRL (sarcin–ricin loop); L11 (region that contains ribosomal protein L11 and a segment of 23S rRNA); S12 (ribosomal protein S12); h5 and h14 (helices from 16S rRNA). rRNA, ribosomal RNA; SRL, sarcin–ricin loop; tRNA, transfer RNAs
In the map, the density attributable to RelA can roughly be divided into two separate domains connected by a linker region: one domain contacts the 50S via the L11 lobe, which consists of the N-terminal domain of L11 and nucleotides 1051–1108 of 23S ribosomal RNA (rRNA) (Figs 3A,B); the other domain settles in a cavity formed by helices h5 (region comprising nucleotides 358–359 and 55–56) and h14 (nucleotides 340–342) of 16S rRNA (Fig 3B). The sarcin–ricin loop, implicated in ribosome-associated GTPase activation for several translation factors [19] is not seen in interaction distance with RelA (Fig 3A). The density for RelA is also seen in close contact with the acceptor stem of the A-site tRNA, which adopts a highly distorted conformation to simultaneously bind RelA through its acceptor arm and the decoding centre in the 30S subunit through its anticodon end (Figs 3C,D). We note that the tRNA CCA end region is seen close to RelA; however, at the density threshold used in rendering Fig 3, both regions are not in direct contact. The contact between the CCA end and the density for RelA is only visible at lower thresholds (below 1σ). The possible sub-stoichiometric nature of the complex and the limited resolution of the map do not allow an unambiguous location of the 3′-CCA end, single-stranded RNA portion of the deacylated tRNA, during the flexible fitting (see below). However, the distorted structure of the tRNA renders the interaction between the acceptor end and RelA possible—that is, the sensing of an uncharged tRNA on the ribosome, which is the result of amino-acid starvation.
An atomic model was built for the entire complex by flexible fitting of atomic structures (see Methods). The overall conformation of the A-site tRNA resembles that previously reported for the A/T state of the aminoacyl tRNA in complex with EF-Tu in the ribosome [20, 21]. Both structures (Fig 3E) are characterized by a bend in the anticodon stem loop and a displacement of the D-stem, suggesting that uncharged tRNA can sample the conformational space accessible to aminoacyl tRNA during accommodation [22]. However, because the interactions with RelA are different from those with EF-Tu, the tRNA distortions are slightly different from those of the A/T state [21]. Whereas the distortion in the tRNA begins at the vicinity of base pair 30–40 in a similar way, the bending of nucleotides 25–30 and 40–48 is more abrupt, with resulting differences in the conformation of the D-loop and especially the D-arm. There is also a significant change in the T-loop conformation, which is shifted towards the L11 region (Figs 3D,E). At the elbow, nucleotides around 54–60 of the T-loop make contact with the density attributed to RelA. The structure also shows that the anticodon loop is close enough to interact with ribosomal protein S12 (residues 78–80) and helix H69 (nucleotides 1913–1916), whereas portions of the elbow including nucleotide 56 contact the region around nucleotide A1067 of 23S rRNA, implicated in the GTPase activity of ribosomal factors [23]. Early reports showed an association between RelA and the thiostrepton-binding site on the ribosome, as well as the in vitro inhibition of RelA-dependent (p)ppGpp synthesis by the antibiotic [24, 25]. Thiostrepton binding involves nucleotides A1067 and A1095 of 23S rRNA and the N-term domain of L11 [26]; hence, the interference of the antibiotic with RelA function can be rationalized in structural terms.
Discussion
There are divergent views from in vitro biochemical studies [13] and in vivo labelling systems [27] on whether (p)ppGpp is produced by RelA on or off the ribosome. In both scenarios, however, the activation of the factor leads to a low-affinity conformation and its subsequent dissociation from the ribosome, which could be a reason for the sub-stoichiometric occupancy in our preparations. Thus, the conformation we observe is likely to be that of the quasi-activated state before its release from the ribosome. Despite the possible transient nature of the ribosome–RelA interaction and the resulting sub-stoichiometric occupancy, it has been possible to determine its structure by using advanced cryo-EM in combination with sorting algorithms.
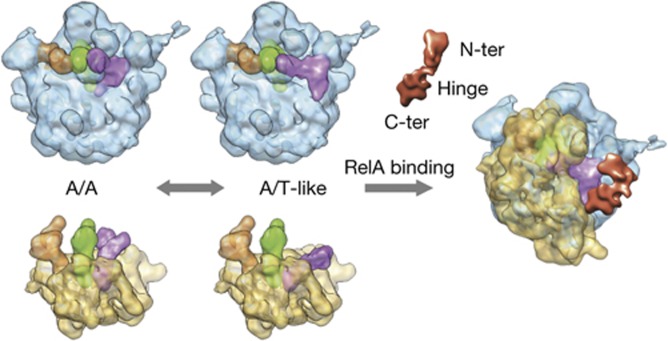
The stabilization of an unusual conformation of the uncharged A-site tRNA by RelA allows us to propose a mechanism for its activation (Fig 4). The initial binding of RelA to the stalled ribosome is likely to involve the shoulder region of the small subunit via the C-terminal domain, which represents the regulatory and ribosome-binding domain as shown previously [15, 28]. Along these lines, the binding observed in the assays even in the absence of A-site tRNA (Fig 1D) is regarded as the initial survey of the ribosome complex by RelA to sample the A-site [13, 27]. The ability to contact the CCA end of a non-acylated tRNA that has bounced backwards after not being able to be anchored on the P-site owing to the absence of peptidyl transfer allows a long-lasting interaction, which results in mutual stabilization. The conformation of the tRNA is stabilized in a pseudo A/T state that evocates the one induced by the ternary complex with acylated tRNAs during the delivery of cognate tRNAs in a standard elongation step. A key step in its activation is likely to be its interaction with the 3′ end of the uncharged A-site tRNA, which is only made possible because its interactions with RelA are sufficiently strong enough to stabilize the distorted conformation of the tRNA. Note that, in the proposed model, the deacylated tRNA moves from the A/A state to an A/T-like conformation before contacting RelA. Although never observed directly, previously published crystallographic ribosomal structures seem to support this notion, as the spontaneous deacylation of A-site tRNAs led to structures in which the anticodon stem loop was ordered and visible but the rest of the body was smeared out because of the reduction of affinity for the PTC and subsequent disordering of the acceptor arm (for example see [29]). Concomitantly, as a result of the interaction of the elbow region of the tRNA with the L11 region of the 50S subunit, the latter becomes more ordered and facilitates the interaction of L11 with the N-terminal domain of RelA involved in ppGpp synthesis [17] leading to the activation of the factor. Thus, the distorted tRNA configuration allows RelA to sense when there is an uncharged tRNA in the A-site of the ribosome. Presumably, if the tRNA were charged, the probability of it sampling a conformation required for interaction with RelA would be smaller. All in all, the data seem to favour a mechanism in which the discrimination of deacylated tRNAs is based on the A/T-like configuration that provides the scaffold to promote the activated stringent factor conformation. Such a situation can only be reached by tRNAs without amino acid, and hence not fully engaged on the A-site. Previous data demonstrated the critical importance of a free CCA end in the activation of the factor [10–12]. Indeed, binding experiments using the non-hydrolizable Phe-NH-tRNAPhe (supplementary Fig S4 online) show that in vitro the presence of deacylated tRNA promotes the maximum binding of RelA to the ribosome. In our cryo-EM density (Fig 3), the CCA end region is seen in interaction distance with RelA; however, at the threshold level used in the rendering, both regions are not in direct contact. The level of involvement of the direct recognition of the deacylation state of the 3′ end by RelA will definitively require higher-resolution studies. In addition, snapshots of different stages along RelA functioning are still needed to establish the whole picture using structural data.
Figure 4.
Cartoon-like schematic representation of RelA activation. See the text for details on the proposed model for the activation of RelA in binding to stalled ribosomes bearing deacylated tRNA.
Methods
Sample preparation and binding assay. The RelA ORF was amplified by PCR from genomic DNA extracted from the E. coli strain BL21 and cloned in the T7-derived expression vector pRAT-4 [30] modified to in frame add a octahistidine tag at the N-terminal followed by a cleavage sequence for the tobacco etch virus protease. The cloned construct was designed so that after tobacco etch virus treatment only two extra amino acids (glycine+alanine) would remain on the protein of interestRelA was overexpressed in E. coli for 4 h at 30 °C following induction by isopropyl-β-D-thiogalactoside at a concentration of 0.5 mM. Cells were collected by centrifugation at 4 °C and resuspended in cold buffer L (20 mM Hepes–KOH (pH 7.45), 1 M KCl, 1 mM Mg-acetate, 20 mM imidazol, 4 mM ß-mercaptoethanol and protease inhibitors) and disrupted by sonication. The supernatant of the lysate after centrifugation was loaded on a 5 ml HisTrapHP (GE Healthcare) column. After washing with buffer L, the His-tagged-RelA protein was eluted by a 100 ml linear gradient of 20–500 mM imidazole. A peak for RelA (confirmed by mass spectrometry) eluted with small contamination from endogenous E. coli proteins. The peak was dialysed against 2 L of buffer D (20 mM Tris–HCl pH 7.5, 500 mM KCl, 1 mM Mg-acetate and 2 mM DTT) at 4 °C for 16 h in the presence of purified tobacco etch virus protease at 1/200th the mass ratio of RelA. A second purification step on the 5 ml HisTrapHP column equilibrated in 20 mM Tris–HCl (pH 7.5), 500 mM KCl, 1 mM Mg-acetate and 2 mM DTT was performed in a gradient of 0–500 mM imidazole. The untagged RelA protein eluted as a single, 99% pure, polypeptide. A final dialysis step in storage buffer G500 (5 mM Hepes–KOH pH 7.45, 500 mM KCl, 10 mM NH4Cl, 10 mM Mg-acetate, 5 mM β-mercaptoethanol) yielded a protein with better solubility properties than previously reported for a C-terminal His-tagged version [13]. The NT-454-RelA mutant was cloned and purified following a similar procedure. The NT-454-RelA mutant was more soluble and could be concentrated up to 100 μM in buffer G (5 mM Hepes–KOH pH 7.45, 50 mM KCl, 10 mM NH4Cl, 10 mM Mg-acetate and 5 mM β-mercaptoethanol) without aggregationRibosomes from Thermus thermophilus HB8, tRNAfMet and tRNAPhe were produced as previously reported [21]. messenger RNAs with sequences 5′-GGCAAGGAGGUAAAAAUGUUCAAA-3′ for the full A-site version and 5′-GGCAAGGAGGUAAAAAUGA-3′ for the version without a codon on the A-site (the codons for fMet and Phe are underlined) were chemically synthesized (Dharmacon)RelA was incubated with ribosomes and the final concentrations of the different components were as follows: 4 μM 70 S, 8 μM messenger RNA, 12 μM tRNAfMet, 12 μM tRNAPhe and 4 μM of RelA (or NT-454-RelA) in buffer G. The 50 μl samples were loaded on top of a 1.5 ml cushion consisting of 1.1 M sucrose and centrifuged for 14 h at 45,000 r.p.m. and 4 °C. The supernatants were quickly removed and the pellets were resuspended in 50 μl of fresh buffer G. A last centrifugation step was performed on a benchtop centrifuge at 12,000 r.p.m. for 5 min. For analysis, 0.25 OD260 nm per reaction was loaded on an SDS–polyacrylamide gel electrophoresis 4–12% gradient gel.
Cryo-EM and image processing. Carbon-coated holey grids were prepared following standard procedures. Low-dose images were taken on Kodak SO-163 films using a JEM-2200FS electron microscope operating at 200 kV and a calibrated magnification of 40,000. Micrographs were scanned on a Z/I Photoscan scanner with a step size of 7 μm, resulting in a final pixel size of 1.75 Å on the object scale. A total of 272 micrographs were selected and assigned to one of 36 defocus groups ranging from 1–4 μm underfocus. In all, 205,502 individual ribosome particles were finally selected by combining automated particle picking [31], multivariate data analysis [32] and visual inspectionTo separate the heterogeneous data set into structurally homogeneous classes, ML3D, a maximum likelihood (ML)-based classification approach, was used [18]. Preliminary runs made use of decimated particles (70 × 70 pixels, 5.25 Å/pixel). An angular sampling of 8° was used during the optimization process. Individual angular refinements of the subsets and of the total number of images were carried out using undecimated, original-sized particles in SPIDER [33]The bootstrapping method was used to compute the 3D variance map [34] with 15,000 bootstrap reconstructions for each subset. As introduced previously [35], the number of bootstrap reconstructions was chosen such that the cross-correlation coefficient between two half-set variance maps was at least 0.97Atomic models were obtained using MDFF, a molecular dynamics-based flexible fitting method [36]. The starting structure was the crystal structure of the ribosome bound to EF-Tu and A/T-tRNA [21]. Structural restraints were imposed during the simulations. In the visualization of cryo-EM densities and atomic models, the programme Chimera was used [37]. The RelA-bound ribosome structure has been deposited in the 3D-EM database under the accession code EMD-2373.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
We thank Hstau L. Liao for help during 3D variance analysis. X.A. is a recipient of a ‘Ramon y Cajal’ fellowship from the Spanish Government (RYC-2009-04885). This study was supported by the following grants: BFU2012-34873 from the Spanish Ministry of Economy and Competitiveness (to M.V.); PI2011-23 from the Department of Education, Universities and Research of the Basque Country (to X.A.); a Ramon Areces Foundation postdoctoral Fellowship (to I.S.F.); and grants from the UK Medical Research Council (U105184332), the Wellcome Trust, the Agouron Institute and the Louis-Jeantet Foundation (to V.R.).
Author contributions: I.S.F. and A.C.K. carried out the purification and preparation of the ribosomal complexes; X.A. and D.G.C. conducted the cryo-EM experiments; X.A. determined the structures. All authors discussed the experimental results and wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Stent GS, Brenner S (1961) A genetic locus for the regulation of ribonucleic acid synthesis. Proc Natl Acad Sci USA 47: 2005–2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potrykus K, Cashel M (2008) (p)ppGpp: still magical? Annu Rev Microbiol 62: 35–51 [DOI] [PubMed] [Google Scholar]
- Srivatsan A, Wang JD (2008) Control of bacterial transcription, translation and replication by (p)ppGpp. Curr Opin Microbiol 11: 100–105 [DOI] [PubMed] [Google Scholar]
- Klinkenberg LG, Lee JH, Bishai WR, Karakousis PC (2010) The stringent response is required for full virulence of Mycobacterium tuberculosis in guinea pigs. J Infect Dis 202: 1397–1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He H, Cooper JN, Mishra A, Raskin DM (2012) Stringent response regulation of biofilm formation in Vibrio cholerae. J Bacteriol 194: 2962–2972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson GC, Tenson T, Hauryliuk V (2011) The RelA/SpoT homolog (RSH) superfamily: distribution and functional evolution of ppGpp synthetases and hydrolases across the tree of life. PLoS One 6: e23479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braeken K, Moris M, Daniels R, Vanderleyden J, Michiels J (2006) New horizons for (p)ppGpp in bacterial and plant physiology. Trends Microbiol 14: 45–54 [DOI] [PubMed] [Google Scholar]
- Sun D et al. (2010) A metazoan ortholog of SpoT hydrolyzes ppGpp and functions in starvation responses. Nat Struct Mol Biol 17: 1188–1194 [DOI] [PubMed] [Google Scholar]
- Haseltine WA, Block R, Gilbert W, Weber K (1972) MSI and MSII made on ribosome in idling step of protein synthesis. Nature 238: 381–384 [DOI] [PubMed] [Google Scholar]
- Haseltine WA, Block R (1973) Synthesis of guanosine tetra- and pentaphosphate requires the presence of a codon-specific, uncharged transfer ribonucleic acid in the acceptor site of ribosomes. Proc Natl Acad Sci USA 70: 1564–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinali G, Liou R, Ofengand J (1978) Role of the aminoacyl end of transfer RNA in the allosteric control of guanosine pentaphosphate synthesis by the stringent factor-ribosome complex of Escherichia coli. Biochemistry 17: 2761–2768 [DOI] [PubMed] [Google Scholar]
- Sprinzl M, Richter D (1976) Free 3'-OH group of the terminal adenosine of the tRNA molecule is essential for the synthesis in vitro of guanosine tetraphosphate and pentaphosphate in a ribosomal system from Escherichia coli. Eur J Biochem 71: 171–176 [DOI] [PubMed] [Google Scholar]
- Wendrich TM, Blaha G, Wilson DN, Marahiel MA, Nierhaus KH (2002) Dissection of the mechanism for the stringent factor RelA. Mol Cell 10: 779–788 [DOI] [PubMed] [Google Scholar]
- Pedersen FS, Kjeldgaard NO (1977) Analysis of the relA gene product of Escherichia coli. Eur J Biochem 76: 91–97 [DOI] [PubMed] [Google Scholar]
- Gropp M, Strausz Y, Gross M, Glaser G (2001) Regulation of Escherichia coli RelA requires oligomerization of the C-terminal domain. J Bacteriol 183: 570–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Ishiguro EE (2001) Dimerization of the RelA protein of Escherichia coli. Biochem Cell Biol 79: 729–736 [DOI] [PubMed] [Google Scholar]
- Schreiber G, Metzger S, Aizenman E, Roza S, Cashel M, Glaser G (1991) Overexpression of the relA gene in Escherichia coli. J Biol Chem 266: 3760–3767 [PubMed] [Google Scholar]
- Scheres SH, Gao H, Valle M, Herman GT, Eggermont PP, Frank J, Carazo JM (2007) Disentangling conformational states of macromolecules in 3D-EM through likelihood optimization. Nat Methods 4: 27–29 [DOI] [PubMed] [Google Scholar]
- Voorhees RM, Schmeing TM, Kelley AC, Ramakrishnan V (2010) The mechanism for activation of GTP hydrolysis on the ribosome. Science 330: 835–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle M et al. (2003) Incorporation of aminoacyl-tRNA into the ribosome as seen by cryo-electron microscopy. Nat Struct Biol 10: 899–906 [DOI] [PubMed] [Google Scholar]
- Schmeing TM, Voorhees RM, Kelley AC, Gao YG, Murphy FVt, Weir JR, Ramakrishnan V (2009) The crystal structure of the ribosome bound to EF-Tu and aminoacyl-tRNA. Science 326: 688–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitford PC, Geggier P, Altman RB, Blanchard SC, Onuchic JN, Sanbonmatsu KY (2010) Accommodation of aminoacyl-tRNA into the ribosome involves reversible excursions along multiple pathways. RNA 16: 1196–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saarma U, Remme J, Ehrenberg M, Bilgin N (1997) An A to U transversion at position 1067 of 23S rRNA from Escherichia coli impairs EF-Tu and EF-G function. J Mol Biol 272: 327–335 [DOI] [PubMed] [Google Scholar]
- Sy J (1974) Reversibility of the pyrophosphoryl transfer from ATP to GTP by Escherichia coli stringent factor. Proc Natl Acad Sci USA 71: 3470–3473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith I, Paress P, Pestka S (1978) Thiostrepton-resistant mutants exhibit relaxed synthesis of RNA. Proc Natl Acad Sci USA 75: 5993–5997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosendahl G, Douthwaite S (1994) The antibiotics micrococcin and thiostrepton interact directly with 23S rRNA nucleotides 1067A and 1095A. Nucleic Acids Res 22: 357–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- English BP, Hauryliuk V, Sanamrad A, Tankov S, Dekker NH, Elf J (2011) Single-molecule investigations of the stringent response machinery in living bacterial cells. Proc Natl Acad Sci USA 108: E365–E373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Ishiguro EE (2001) Involvement of the N terminus of ribosomal protein L11 in regulation of the RelA protein of Escherichia coli. J Bacteriol 183: 6532–6537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selmer M, Dunham CM, Murphy FVt, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V (2006) Structure of the 70S ribosome complexed with mRNA and tRNA. Science 313: 1935–1942 [DOI] [PubMed] [Google Scholar]
- Peranen J, Rikkonen M, Hyvonen M, Kaariainen L (1996) T7 vectors with modified T7lac promoter for expression of proteins in Escherichia coli. Anal Biochem 236: 371–373 [DOI] [PubMed] [Google Scholar]
- Rath BK, Frank J (2004) Fast automatic particle picking from cryo-electron micrographs using a locally normalized cross-correlation function: a case study. J Struct Biol 145: 84–90 [DOI] [PubMed] [Google Scholar]
- Shaikh TR, Trujillo R, LeBarron JS, Baxter WT, Frank J (2008) Particle-verification for single-particle, reference-based reconstruction using multivariate data analysis and classification. J Struct Biol 164: 41–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaikh TR, Gao H, Baxter WT, Asturias FJ, Boisset N, Leith A, Frank J (2008) SPIDER image processing for single-particle reconstruction of biological macromolecules from electron micrographs. Nat Protoc 3: 1941–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penczek PA, Yang C, Frank J, Spahn CM (2006) Estimation of variance in single-particle reconstruction using the bootstrap technique. J Struct Biol 154: 168–183 [DOI] [PubMed] [Google Scholar]
- Agirrezabala X, Schreiner E, Trabuco LG, Lei J, Ortiz-Meoz RF, Schulten K, Green R, Frank J (2011) Structural insights into cognate versus near-cognate discrimination during decoding. EMBO J 30: 1497–1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trabuco LG, Villa E, Mitra K, Frank J, Schulten K (2008) Flexible fitting of atomic structures into electron microscopy maps using molecular dynamics. Structure 16: 673–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25: 1605–1612 [DOI] [PubMed] [Google Scholar]
- Voorhees RM, Weixlbaumer A, Loakes D, Kelley AC, Ramakrishnan V (2009) Insights into substrate stabilization from snapshots of the peptidyl transferase center of the intact 70S ribosome. Nat Struct Mol Biol 16: 528–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.