Abstract
Retinoic acid-inducible gene-I (RIG-I) is an intracellular RNA sensor that activates the innate immune machinery in response to infection by RNA viruses. Here, we report the crystal structure of distinct conformations of a RIG-I:dsRNA complex, which shows that HEL2i-mediated scanning allows RIG-I to sense the length of RNA targets. To understand the implications of HEL2i scanning for catalytic activity and signalling by RIG-I, we examined its ATPase activity when stimulated by duplex RNAs of varying lengths and 5′ composition. We identified a minimal RNA duplex that binds one RIG-I molecule, stimulates robust ATPase activity, and elicits a RIG-I-mediated interferon response in cells. Our results reveal that the minimal functional unit of the RIG-I:RNA complex is a monomer that binds at the terminus of a duplex RNA substrate. This behaviour is markedly different from the RIG-I paralog melanoma differentiation-associated gene 5 (MDA5), which forms cooperative filaments.
Keywords: ATP hydrolysis, innate immunity, RNA helicase, X-ray crystallography
INTRODUCTION
RIG-I (retinoic acid-inducible gene-I), melanoma differentiation-associated gene 5 (MDA5) and laboratory of genetics and physiology 2 (LGP2), comprise the RIG-I-like receptor class of intracellular pattern recognition receptors that recognize foreign RNAs in the cytoplasm and elicit an innate immune response through the production of pro-inflammatory cytokines and type I interferons (IFNs) [1–3]. RIG-I recognizes both self and non-self RNA, including positive- and negative-stranded RNA viruses [3], synthetic poly I:C [4], and RNA aptamers lacking a 5′triphosphate [5]. The pathogen-associated molecular pattern (PAMP) of RIG-I is generally defined as duplex RNA containing a 5′triphosphate moiety [6–8], although only duplex RNA is absolutely required for RIG-I recognition [9]. The shortest RNA molecule commonly reported to activate the RIG-I signalling pathway is 5′triphosphorylated, blunt-ended 19-mer duplex RNA [10, 11]. However, the consensus, minimal functional RNA PAMP for RIG-I has not been clearly defined.
RIG-I-like receptors are part of a larger group of duplex RNA-activated ATPases (DRAs) that also includes Dicer and Dicer-related helicases [12]. All DRAs share a common superfamily 2 helicase core that consists of two RecA-like domains, HEL1 and HEL2, and a conserved insertion domain, HEL2i. Except for Dicer, DRAs contain a conserved carboxy-terminal domain (CTD) that is responsible for modulating the function of each protein and imparting substrate specificity. In RIG-I, the CTD provides the specificity for recognizing 5′triphosphates [13, 14]. RIG-I and MDA5 contain tandem caspase activation and recruitment domains (CARDs) at their amino-termini that undergo ubiquitination upon RNA binding and subsequently initiate downstream signalling by interacting with the CARD domain of the mitochondrial adaptor protein, mitochondrial antiviral-signalling protein (MAVS) [15, 16]. In the apo state, RIG-I adopts an auto-repressed conformation with the tandem CARDs partially occluded by an interaction with the HEL2i domain [17].
Structural studies of mouse, human and duck RIG-I constructs have enhanced our understanding of how RIG-I recognizes RNA and utilizes ATP [17–20], however the precise role of RNA and ATP binding in displacing the CARDs is still unclear. Intriguingly, in all of the RIG-I:RNA complex structures obtained so far, the RIG-I CTD caps the 5′ end of the RNA, regardless of the length of the bound duplex or the presence of a 5′triphosphate. Furthermore, the RIG-I helicase domain exhibits weak, micromolar affinity for blunt-ended duplex RNA that is terminated either by a 5′triphosphate or 5′hydroxyl [19, 21], suggesting that internal duplex stem binding has a lesser role in RIG-I stimulation.
Multimerization of RIG-I on RNA has been observed using a variety of in vitro techniques [22–24]. However, the fact that several proteins can bind one RNA molecule does not mean that they are all functional, it does not mean that the proteins interact with each other and it does not imply that they bind cooperatively to RNA. Interestingly, while MDA5 forms filaments through cooperative, protein–protein interactions on long duplex RNA, RIG-I prefers considerably shorter duplex RNA, suggesting that filament formation might not have an essential role in RIG-I activation [4, 25, 26]. While polyubiquitin mediated oligomerization of the CARD domains is required for interferon production [15, 27], the oligomerization state and the functional stoichiometry of the RIG-I:RNA complex is not well understood [28].
Here we conduct a multidisciplinary study of RIG-I RNA recognition to define the minimal RNA ligand that is required for RIG-I function. We provide biophysical and cell culture evidence that RIG-I surveys the cell for the 5′ terminus of dsRNA and binds the RNA molecule in a 1:1 monomeric form. We show that RIG-I ATPase activity and RIG-I-mediated IFN production are stimulated via interactions with the 10 base pairs at the 5′ ends of duplex RNA, and that a HEL2i scanning mechanism has a role in sensing and defining the correct length of duplex RNA.
RESULTS AND DISCUSSION
HEL2i movements contribute to dsRNA recognition
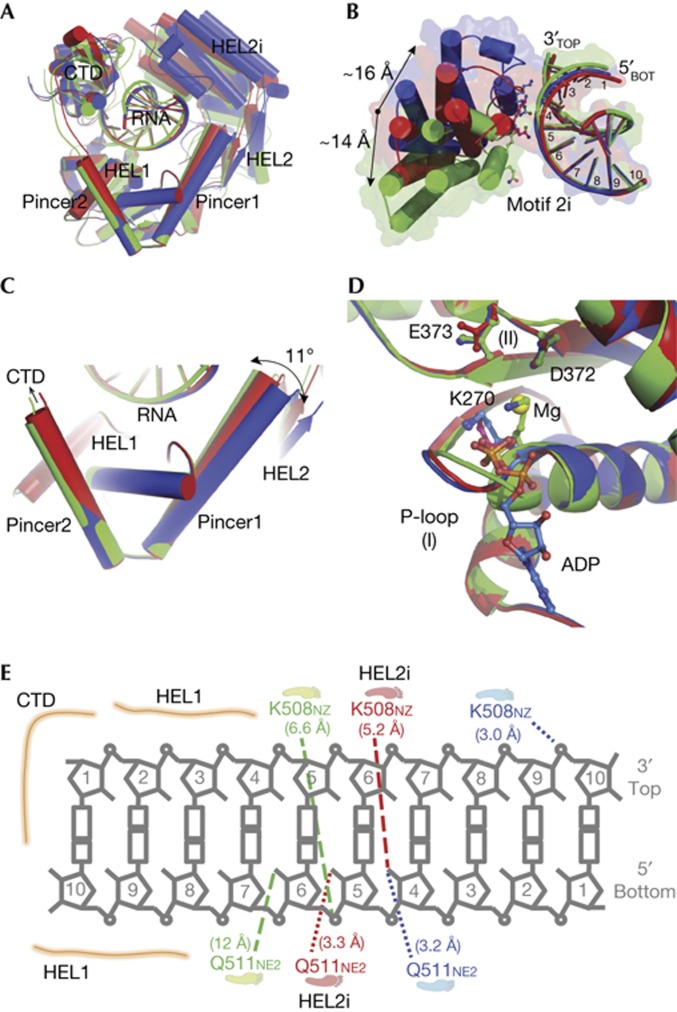
To understand the conformational changes that RIG-I undergoes during RNA recognition and surveillance, we visualized the conformations of RIG-I (ΔCARDs: 1–229) in complex with 5′OH-GC10 (Fig 1A; supplementary Table S1 online), which show well-ordered scanning movements of the HEL2i domain along the duplex RNA backbone. Conformation 1 is the binary complex of RIG-I (ΔCARDs:1–229): 5′OH-GC10, in which the ATP-binding pocket is empty and HEL2i stays in the most compact state (Fig 1A, green, pdb:3zd6). Conformation 2 is the crystal structure that we reported previously and is the ternary complex of RIG-I (ΔCARDs:1–229): 5′OH-GC10:SO42−, in which the sulphate ion occupies the ATP-binding pocket and HEL2i adopts an intermediate state (Fig 1A, red, pdb:2ykg) [20]. Conformation 3 is also a ternary complex of RIG-I (ΔCARDs:1–229): 5′OH-GC10: ADP-Mg2+, in which ADP-Mg2+ occupies the ATP-binding pocket and HEL2i adopts the most extended state (Fig 1A, blue, pdb:3zd7).
Figure 1.
The HEL2i domain scans along the duplex RNA backbone. Three distinct conformations of RIG-I (ΔCARDs: 1–229):GC10 with an empty ATP-binding pocket (green, pdb:3zd6), and in complex with a SO42− (red, pdb:2ykg) and ADP-Mg2+ (blue, pdb:3zd7). GC10 is a palindromic RNA duplex of repeating ‘GC’ with a 5′hydroxyl. (A) Alignment of the three conformations. (B) The interface between the HEL2i and the duplex RNA. Key residues in the HEL2i domain, Q511 and K508, are involved in RNA binding and are shown as sticks. Using residue E530 as the reference, there is a 14 Å movement of the HEL2i domain between conformations 1 and 2, and a 16 Å movement between conformations 2 and 3. (C) Close-up view of the pincer domain, highlighting the motions of pincer1 (the first α-helix). The change in the angle between pincer1 and pincer2 (the second α-helix) is 11°. (D) The ATP-binding pocket of the superimposed structures. Ligands (SO42− and ADP-Mg2+) and the key residues (K270 from motif I; D372 and E373 from motif II) are shown as sticks. (E) Diagram of the RIG-I:RNA duplex interface. The closest distances between the RNA and the residues K508 and Q511 from the HEL2i domain are highlighted and shown as dashed lines. CARDs, caspase activation and recruitment domains; RIG-I, retinoic acid-inducible gene-I.
An alignment of the three RIG-I:RNA structures reveals that, while the HEL1–RNA–CTD forms a rigid sandwich-like fold, the HEL2i domain of RIG-I is flexible and makes sequential contacts with several base pairs along the RNA duplex. Specifically, the HEL2i domain scans along the duplex backbone between bases four through six of the 3′ bottom strand (that is, the ‘tracking strand’ for SF2 helicase proteins) when transitioning between conformations, and then makes contact with the top strand in the extended, ADP-bound conformation (Fig 1B; supplementary Movie 1 online). Two residues of the HEL2i domain, K508 and Q511, engage the RNA duplex: Q511 does not form contacts with the RNA backbone in conformation 1; in conformation 2, Q511 interacts with the 2′-OH group of the fifth base of the bottom strand; in conformation 3, Q511 reaches the 2′OH group of the fourth base as the HEL2i domain slides along one face of the RNA duplex. K508 comes into close contact with the RNA only in the extended conformation 3, forming a salt bridge with the phosphate at position 9 on the 5′ top strand (Fig 1E).
During scanning, the pincer domain facilitates coordinated motion of HEL2–HEL2i relative to HEL1 by engaging in a swinging motion along the more N-terminal α-helix whereas the C-terminal arm of the pincer serves as an anchor by remaining rigidly stacked against HEL1 (Fig 1C). We also observe subtle changes in the ATP-binding pocket among the conformations, including movements of the phosphate-binding loop and K270 of motif I, suggesting that the pincer and HEL2i motions might be linked to ATP-binding and hydrolysis (Fig 1D) [29]. Collectively, these conformations show dynamic opening and closing motions of the HEL2i domain along a 10 base pair stretch of RNA. This led us to further investigate two questions: (1) How important are more RNA pairings that extend beyond this central core of 10 base pairs at the helical terminus? That is, do more base pairs contribute to duplex RNA binding, stimulation of in vitro ATPase activity, or RIG-I-mediated IFN production? (2) How many RIG-I molecules are necessary per RNA molecule to activate both ATPase activity and an IFN response?
RIG-I binds duplex RNA termini as a monomer
To study RIG-I binding to the internal duplex RNA regions, we synthesized a family of structurally well-defined RNA hairpins in which the duplex length was varied, but one terminus was blocked by the presence of a structured, RNA tetraloop. We then employed a hydrodynamic method, sedimentation velocity (SV), to monitor populations of RIG-I and RIG-I:RNA complexes that form in the solution using hairpin duplexes of 10, 20 and 30 base pairs in length, each bearing a single 5′triphosphorylated end (5′ppp10L, 5′ppp20L, and 5′ppp30L, supplementary Table S2 online). In addition, we examined RIG-I binding to a 22mer duplex RNA that contains two 5′triphosphorylated ends (5′pppGC22). We observed that, at micromolar concentrations of protein and RNA, RIG-I formed 1:1 complexes with each hairpin tested, regardless of duplex length (Fig 2). Specifically, we determined peak s20,w (standardized to 20 °C and water) values of 6.0 for RIG-I alone, and 6.2, 6.4 and 6.9 for excess RIG-I with hairpins of lengths 10, 20 and 30, respectively. By contrast, the complex of RIG-I with 5′pppGC22 had a s20,w of 9.3, indicating a 2:1 protein:RNA stoichiometry.
Figure 2.
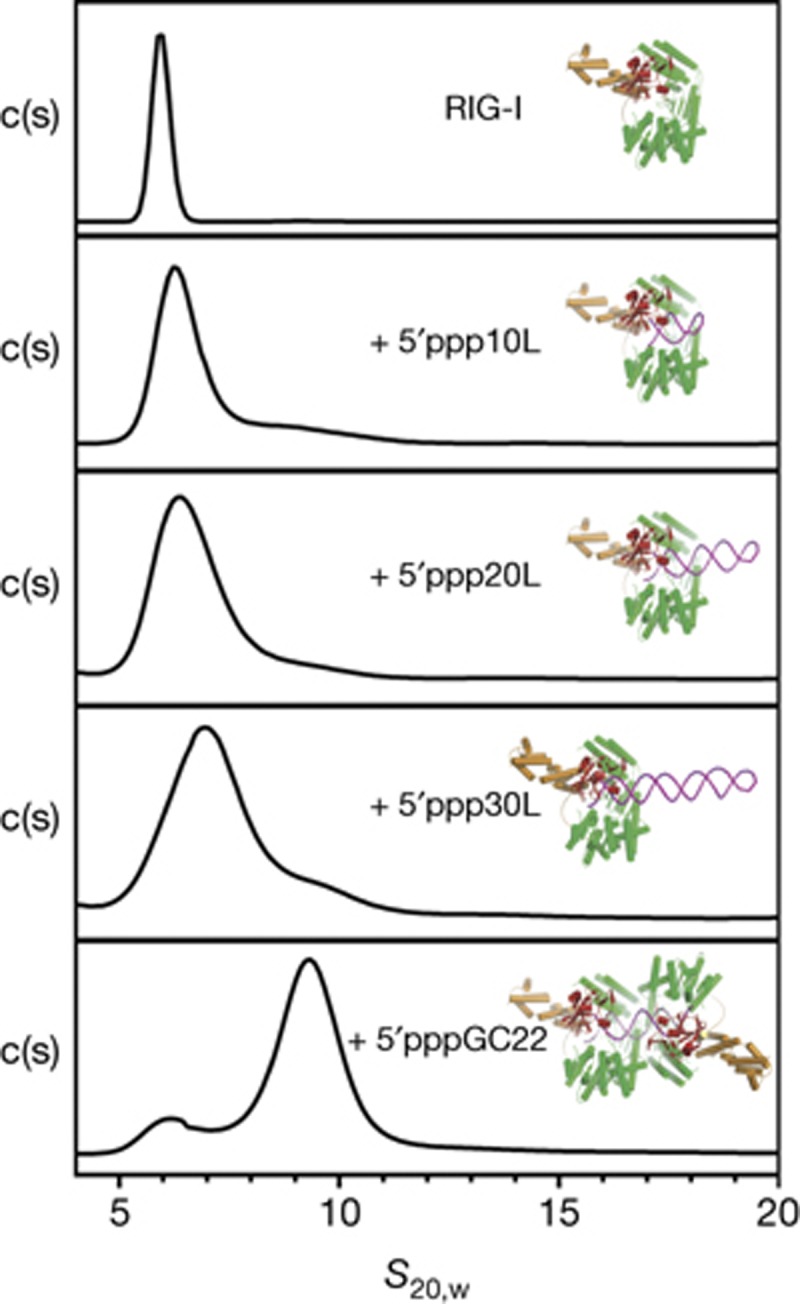
RIG-I binds hairpins with one triphosphate with a 1:1 stoichiometry. Hydrodynamic analysis of RIG-I in complex with 5′ppp10L, 5′ppp20L, 5′ppp30L and 5′pppGC22 RNA. c(s) distributions for each SV experiment were plotted against the sedimentation coefficient (s20,w). Peak s20,w values for each distribution are 6.0 for RIG-I alone and 6.2, 6.4, 6.9 and 9.3 for RIG-I: 5′ppp10L, 5′ppp20L, 5′ppp30L and 5′pppGC22 complexes, respectively. Estimated molecular weights from Sedfit are 106 kDa (f/f0=1.31), 113 kDa (f/f0=1.31), 121 kDa (f/f0=1.53), 133 kDa (f/f0=1.57) and 228 kDa (f/f0=1.45), respectively. Models of RIG-I bound to each RNA construct are shown next to each c(s) distribution. RIG-I, retinoic acid-inducible gene-I; SV, sedimentation velocity.
Kowalinksi et al [17] also demonstrated that RIG-I binds with a 2:1 stoichiometry to a longer dsRNA that has two blunt termini (61mer). This is consistent with our SV analysis, and taken together, these results show that RIG-I specifically recognizes the base-paired terminus of duplex RNA, and that RIG-I does not form protein–protein-mediated oligomers even in the presence of RNA (and ADP/ATP analogs, as shown in Luo et al [29]). Internal binding within the duplex is neither strongly favourable nor required for strong monomeric binding at the 5′ end.
RIG-I ATPase activity is dependent on poly I:C ends
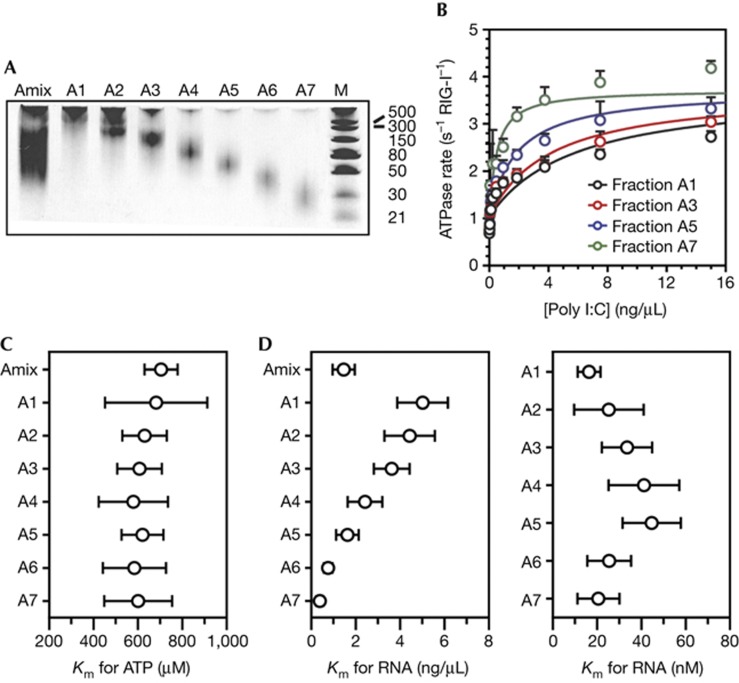
To calibrate our findings with those in the literature, we examined RIG-I:RNA interactions using a polymer that is more typically used in studies of RIG-I. Specifically, we analysed RNA-stimulated ATPase activity using poly I:C, which is a synthetic analogue of double-stranded RNA that is commonly used for experimental stimulation of an IFN response. The ATPase activity of RIG-I is strictly dependent on the concentration of RNA, therefore the enzymatic activity of RIG-I can be used as a metric for productive binding to poly I:C, or any other RNA polymer. Poly I:C is a mixture of lengths and RNA conformational states, and we therefore hypothesized that RIG-I ATPase activity will be more efficiently stimulated by shorter poly I:C fragments because they have more accessible ends per base pair. To test this hypothesis, we conducted analysis of RIG-I ATPase stimulation by low-molecular weight (LMW) poly I:C (supplementary Fig S1 online), which is a mixture of ∼25–500 base pair fragments. To reduce heterogeneity of the poly I:C sample, we fractionated the poly I:C on an analytical Superdex 200 column to create seven fractions of decreasing size (Fig 3A). We estimated the mean length of each fraction, making the assumption that each fraction was a discrete size, and thereby converted between ng/μl and nanomolar amounts of poly I:C strands (supplementary Table S3 online). Individual fractions were tested for the ability to stimulate RIG-I ATPase activity by varying the poly I:C fraction concentration at 5 mM ATP (Km,RNA, Fig 3B) or by varying the ATP concentration at 15 ng/μl poly I:C fraction (Km,ATP, Fig 3C). Remarkably, we found a clear trend demonstrating that shorter poly I:C fragments stimulated RIG-I ATPase activity more effectively. We then plotted the Km,RNA for every fraction in terms of both ng/μl and nM poly I:C strands (Fig 3D). Whereas the Km,RNA spanned a 10-fold range when expressed in ng/μl, the Km,RNA varied approximately two-fold or less when expressed in molarity of poly I:C strands. In fact, we observed an identical Km,RNA value of 20 nM for both fractions A1 and A7, which are at two extremes in terms of length, and the Km,RNA values for the other fractions were similar to this, within error. Furthermore, Km,ATP values for each fraction of poly I:C (at saturating number of ends) were between ∼600 and 700 μM ATP (Fig 3C). These data demonstrate that RIG-I ATPase activity is dependent on the number of duplex ends that are available in each poly I:C fraction, and they corroborate the view that internal duplex regions are not critical for the enzymatic function of RIG-I.
Figure 3.
RIG-I is stimulated by the ends of poly I:C. (A) LMW poly I:C was fractionated on an analytical Superdex 200 size exclusion chromatography column and separated into seven fractions on a 15% polyacrylamide, 4M urea semi-denaturing gel stained with ethidium bromide (marker is in base pairs). (B) ATPase activity of RIG-I stimulated by 0–15 ng/μl of the poly I:C fractions A1, A3, A5 and A7 at 5 mM ATP. The data were fit to the quadratic form of the Briggs–Haldane equation with the assumption that the kcat values are the same for all the fractions. (C) The Km,ATP for RIG-I stimulated by 15 ng/μl of the poly I:C fractions A1–A7 while varying the ATP concentration from 0–5 mM ATP. (D) The calculated Km,RNA for RIG-I stimulated by 0–15 ng/μl of the poly I:C fractions A1–A7 at 5 mM ATP. The Km values in the left panel are in ng/μl and values in the right panel are in nM for all seven fractions on the basis of the estimated sizes of each fraction. Error bars for the poly I:C data report the standard deviation across six experiments. LMW, low molecular weight; RIG-I, retinoic acid-inducible gene-I.
The minimal RNA for stimulating RIG-I ATPase activity
To more precisely define the minimal duplex length for enzymatic activation of RIG-I, we measured the steady-state kinetic parameters for RIG-I activation by RNA hairpins and double-stranded duplexes ranging in size from 8 to 30 base pairs, with and without a 5′triphosphate (RNAs listed in supplementary Table S2 online). In order to evaluate the respective roles of ATP and RNA in activation of the RIG-I:RNA complex, we measured the Michaelis constant for ATP (Km,ATP) at saturating RNA (500 nM) and the Michaelis constant for RNA (Km,RNA) at saturating ATP (5 mM) for each RNA construct (kcat, Km,ATP and Km,RNA summary in supplementary Table S4 online).
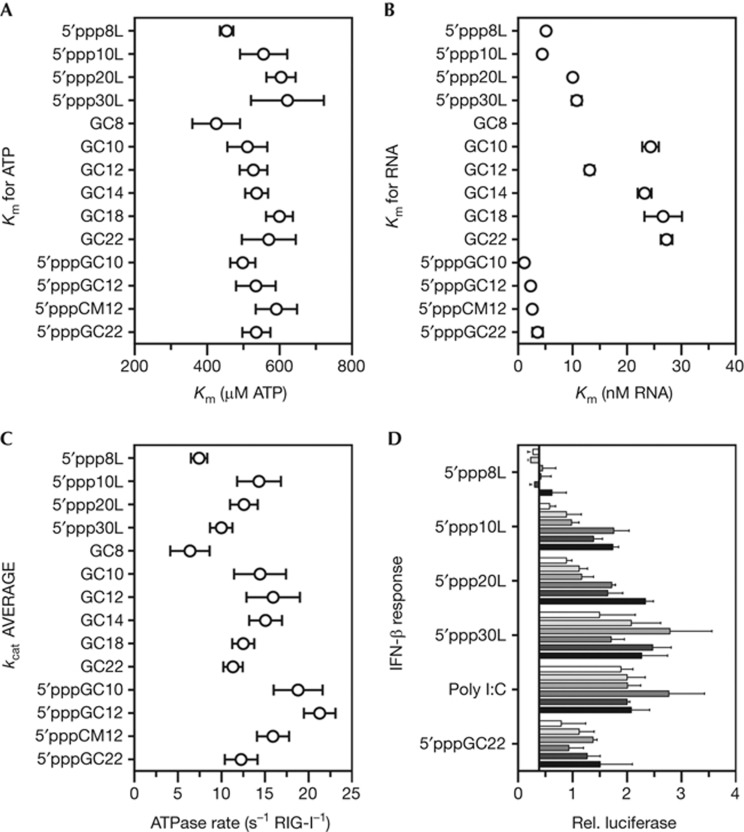
Four 5′triphosphorylated RNA hairpins with duplex lengths of 8, 10, 20 and 30 base pairs were tested for stimulation of RIG-I ATPase activity (Fig 4; supplementary Fig S2 online). The 5′ppp8L and 5′ppp10L hairpins displayed the largest disparity in kcat, doubling from 7.45 s−1 to 14.32 s−1 (ATP molecules hydrolysed per second per molecule of RIG-I) on the addition of only two base pairs. The two larger constructs, 5′ppp20L and 5′ppp30L, were slightly less effective at stimulation than 5′ppp10L, with kcat values of 12.59 s−1 and 9.98 s−1, respectively. It is interesting that, although 5′ppp8L was crystallized with RIG-I (ΔCARDs:1–229) [29], the 5′ppp8L hairpin is sub-optimal for stimulation of RIG-I ATPase activity. This result is intriguing in the context of our structural data, as it provides further support for the hypothesis that two extra base pairs beyond the footprint of RIG-I, as in the 5′ppp10L hairpin, likely provide the HEL2i domain with the required room for full flexibility and the coordinated internal motions that lead to efficient ATP hydrolysis.
Figure 4.
In vitro and cell culture activities of RIG-I on short duplex RNAs. (A) The Km,ATP of RIG-I stimulated by a library of duplex RNA constructs at ATP concentrations varying between 0 and 5 mM. (B) The Km,RNA of RIG-I stimulated by a library of duplex RNA constructs at RNA concentrations varying between 0 and 500 nM. The Km,RNA for the GC8 duplex is ∼100 nM and did not fit on the scale. (C) The kcat summary averaged from the Km,ATP and Km,RNA experiments. (D) RIG-I stimulated IFN-β production was measured in 293T cells. RIG-I was stimulated by 5′triphosphorylated hairpins (20–650 nM) and the positive controls, poly I:C (15–500 ng/well) and 5′pppGC22 (20–650 nM). The increase in RNA concentration is indicated by a darkening color gradient. The relative luciferase is the firefly luciferase (IFN-β reporter) divided by the constitutively expressed Renilla luciferase. Error bars for the ATPase data report the standard deviation from at least three measurements. Error bars for the cell culture data report the standard error of the mean from three measurements. IFN-β, interferon-β; RIG-I, retinoic acid-inducible gene-I.
In order to more comprehensively evaluate the RNA length dependence for RIG-I ligands, six ‘GC’ palindromic, blunt-ended, 5′hydroxyl RNA duplexes with lengths of 8, 10, 12, 14, 18 and 22 were tested for stimulation of RIG-I ATPase activity (Fig 4; supplementary Fig S2 online). Remarkably, the kcat values at saturating ATP and RNA concentrations followed the same trends as the 5′triphosphorylated hairpins. The peak ATPase activity occurred with stimulation from GC12, albeit with negligible differences in comparison to stimulation from GC10 or GC14. Taken together, the length dependence of the kcat for the palindromic, 5′hydroxyl duplexes were qualitatively similar to the 5′triphosphorylated hairpins. In each case, robust ATPase activity for RIG-I stimulation was observed with RNA of at least 10 base pairs in length, regardless of the presence of a 5′-triphosphate moiety, and activity declined slightly with increasing duplex length.
5′ppp enhances RNA binding, but not ATP hydrolysis
The trends in kcat values for the 5′triphosphorylated hairpins and the 5′hydroxyl duplexes were similar despite tighter RNA binding (reflected by smaller Km,RNA values) by the 5′triphosphorylated hairpins. This finding reveals that the 5′triphosphate might function primarily at the step of binding and that it does not have a major impact on ATP hydrolysis. To further investigate the function of the triphosphate, three ‘GC’ palindromic blunt-ended RNA duplexes with 5′triphosphates of lengths 10, 12 and 22 were tested for stimulation of RIG-I ATPase activity, as well as a 5′triphosphorylated 12-mer, 5′pppCM12, containing a palindromic but non-uniform sequence including all four nucleotides (Fig 4; supplementary Fig S2 online). Although the kcat for 5′pppGC10 and 5′pppGC12 were marginally higher than GC10 and GC12, the measured kcat for 5′pppCM12 was identical to GC12, and the kcat for 5′pppGC22 was within the experimental error of the kcat for GC22. These data further demonstrate that the triphosphate has a minimal effect on the kcat values for RIG-I ATP hydrolysis when all ligands are saturating.
We also observed only small changes in the measured Km,ATP (the apparent binding constant for ATP) for the RNA constructs tested (Fig 4A). Specifically, the Km,ATP were between ∼500 and 600 μM, implying that the conformational changes in RIG-I that are required for catalysis are not influenced by the presence of a triphosphate moiety or duplex length. This observation is corroborated in part by structural evidence showing that RIG-I binds 5′triphosphorylated RNA identically to 5′hydroxyl RNA [29].
By contrast, the Km,RNA (the apparent binding constant for RNA) directly correlated with the presence of a 5′triphosphate on the RNA hairpin or duplex (Fig 4B). The Km,RNA for the four 5′triphosphorylated duplexes were between 1.2 and 3.6 nM, and the Km,RNA for the four 5′triphosphorylated hairpins were between 5.2 and 10.8 nM. However, the 5′hydroxyl RNA duplexes yielded Km,RNA values between 20 and 30 nM, with the exception of GC8 (91 nM) and GC12 (13 nM). These data underscore the fact that any RNA duplex of the appropriate length (>10 bp) can fully stimulate the ATPase activity of RIG-I, but a duplex containing a 5′-triphosphate binds RIG-I with higher affinity and will therefore stimulate ATPase activity at lower RNA concentrations. This finding is an important distinction that explains why only trace amounts of viral RNA might be required to activate the interferon-β (IFN-β) response in infected cells. Interestingly, the poly I:C fractions exhibited a similar range of Km,RNA values as those observed for the 5′hydroxyl duplex RNA, suggesting that RIG-I functions on poly I:C in a manner that is similar to any other RNA duplex that lacks a 5′-triphosphate.
1:1 RIG-I:RNA binding is sufficient to stimulate IFN-β
Our in vitro SV and RNA-stimulated ATPase studies provide strong evidence that RIG-I activation requires only the 5′ terminus of duplex RNA, along with an adjacent 10–12 base pairs. And while RIG-I in vitro activity is typically associated with RNA binding or ATPase activity, the direct relationship to interferon stimulation is not clear. Therefore, it was important to test the relevance of our in vitro results in cell culture. To accomplish this, we measured the ability of 5′triphosphorylated hairpins and poly I:C fractions to stimulate a RIG-I-mediated IFN-β response in 293T cells (Fig 4D).
Remarkably, we found that three of the four hairpins—5′ppp10L, 5′ppp20L and 5′ppp30L—stimulated an IFN-β response comparable to the positive controls, LMW poly I:C and 5′pppGC22 (mock control in supplementary Fig S3 online). Both LMW poly I:C and short 19 bp+ RNA duplexes have been shown to be good activators of RIG-I [4, 10]. The IFN-β stimulation from the 5′ppp10L construct is of particular interest because it strongly supports the idea that RIG-I does not survey the cell as an oligomer, that RIG-I does not need to oligomerize on a target RNA duplex strand to elicit an IFN-β response, and that RIG-I does not need to translocate on duplex RNA regions to elicit an IFN-β response. Consistent with these findings, even the shortest poly I:C fragments fully stimulated the IFN response in cells (supplementary Fig S4 online). The slightly better IFN production, especially at lower RNA concentrations, for 5′ppp20L and 5′ppp30L, can be attributed to the fact that they are more stable duplexes that likely have a longer half-life in the cell. The lack of an IFN-β response for 5′ppp8L (and the suboptimal ATPase activity) suggests that HEL2i contacts with the 9th and 10th base pair are required for interferon stimulation, and further corroborates the hypothesis that HEL2i flexibility has a functional role. It is therefore evident that the minimum requirement for RIG-I to initiate an interferon response is a single-RNA duplex terminus (preferably containing a 5′-triphosphate) at the end of at least 10 base pairs.
Model for RNA surveillance by RIG-I
Recent structural studies have shed new light on RNA surveillance by RIG-I. In all cases, RIG-I is shown to bind RNA molecules as a monomer and to interact specifically with the terminus of an RNA duplex. Indeed, it has been called an end-capper [17]. Intriguingly, RIG-I is observed to bind all blunt RNA termini in much the same way, without regard to RNA sequence or the presence of a 5′-triphosphate [19, 20]. While these crystallographic observations are useful, they do not establish the minimal length of RIG-I PAMPs in solution for binding, for ATPase activity, and ultimately for signalling in the cell. In addition, the issue of cooperative RIG-I multimerization on RNA has not been squarely addressed. Given the importance of these issues, and of RIG-I activation in general, we decided to use a combination of techniques to define the minimal RNA PAMP that is required for full activation of RIG-I in vitro and in mammalian cells.
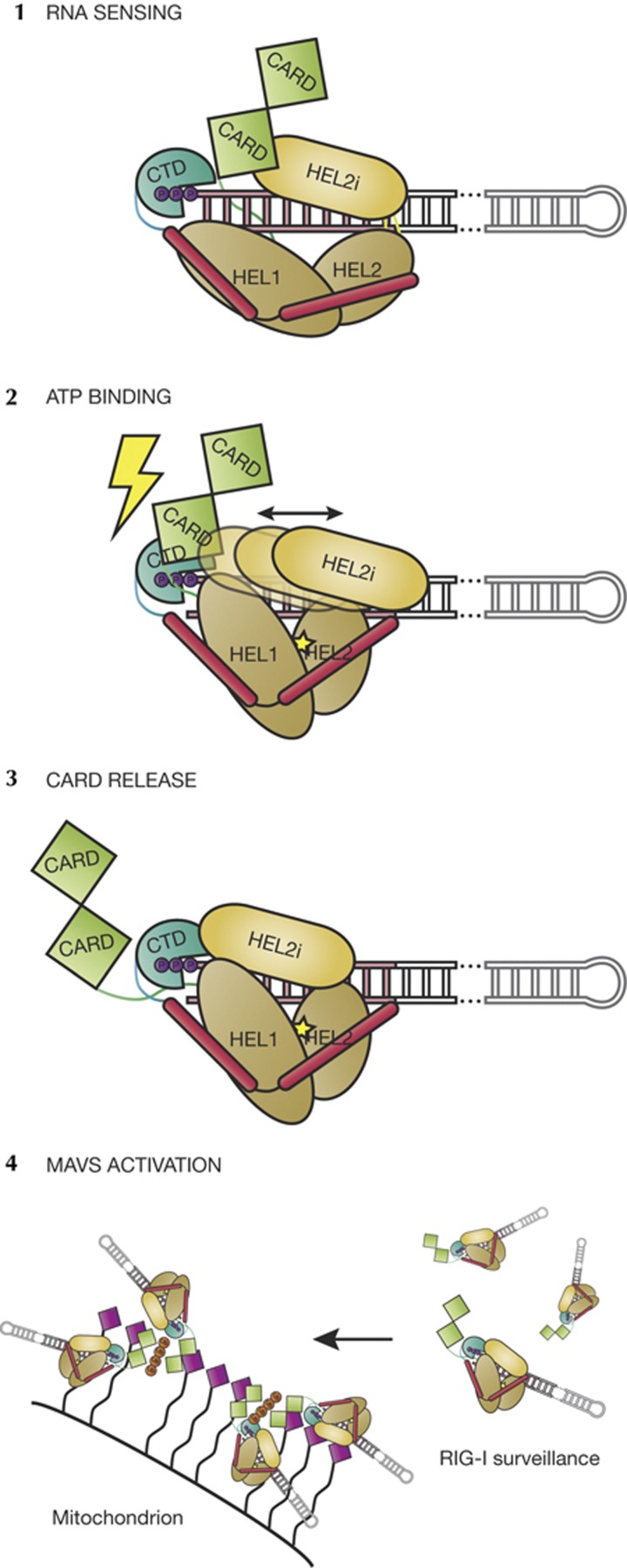
Our findings indicate that we have defined the minimal RNA PAMP that is required for activation in vitro and in cell culture, and that determinants under all conditions agree: the RIG-I monomer is activated upon binding the blunt terminus of a RNA duplex. The protein interacts with the 10 base pairs adjacent to the 5′ end with an affinity that is enhanced by the presence of a 5′triphosphate. Collectively, the available data in the literature suggest that these 1:1 RIG-I:RNA(end) complexes might then oligomerize into higher order complexes via the CARD domains, resulting in a model that is consistent with findings on downstream events that have been reported by others (Fig. 5) [15, 30].
Figure 5.
Model of RIG-I activation. (1) RNA binding is the first trigger of the RIG-I-mediated interferon response. The CTD binds firmly to the 5′ end of the duplex RNA. The CARD domains rest on the HEL2i domain [17] and likely are not displaced upon RNA binding. (2) ATP binding serves as the second trigger, whereupon HEL1 and HEL2 close and HEL2 initiates contacts with the tracking strand, creating a clash between the CTD and the CARDs [12]. HEL2i scanning might be directly linked to ATP binding and hydrolysis, or it might move stochastically. (3) Once the CARD domains are released, a 1:1:1 RIG-I:RNA:ATP ternary complex is competent for signalling and activation of MAVS. (4) Ubiquitin-mediated multimerization (tetraubiquitin shown in orange) of RIG-I through the CARD domains might be required for MAVS activation [15, 30]. CARD, caspase activation and recruitment domain; CTD, carboxy-terminal domain; MAVS, mitochondrial antiviral-signalling protein; RIG-I, retinoic acid-inducible gene-I.
Here we identify the minimal determinants for functional RNA recognition by RIG-I, and we demonstrate that RIG-I uses its functional domains collaboratively to accomplish specific antiviral surveillance in a complex intracellular environment.
METHODS
RIG-I purification. Purification of the full-length human RIG-I and the N-terminal CARDs (residues 1–229) deletion construct was described previously [20, 29].
RNA synthesis and transcription. The 5′OH ‘GC’ palindromic RNA duplexes were made by RNA synthesis (Sigma). The 5′ppp ‘GC’ palindromic RNA duplexes were made by in vitro runoff transcription using DNA templates (SIGMA) with a 2′O-methyl modification on the penultimate nucleotide of the template strand [31]. The LMW poly I:C was ordered from Invivogen.
Crystallization, data collection, structure determination and refinement. The crystallization and data collection of RIG-I (ΔCARDs:1–229) binary and ternary complexes were performed as described previously, with modifications [20]. Structures were determined by molecular replacement using pdb:2ykg as a model.
Analytical ultracentrifugation-SV experiments. Mixtures were loaded into SV chambers and equilibrated at 20 °C for 1 h before beginning the experiment. The sedimentation of the RIG-I:hairpin complexes was monitored by absorbance at 260 nm, and the protein without RNA was monitored by absorbance at 280 nm.
NADH-coupled ATPase experiments. ATPase activity of RIG-I was measured with the NADH-coupled ATPase assay adapted from previously described protocols [20].
Cell culture IFN-β response. 293T cells transfected with pUNO-RIG-I, pRL-TK and IFN-ß/firefly luciferase reporter were seeded at 15,000 cells per well, with each well containing 5 μl of Lyovec (Invivogen) and either RNA hairpin or poly I:C. For luciferase measurements, the Promega Dual Luciferase Reporter assay system was used to quantify the cellular levels of firefly and Renilla luciferase.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
We thank Brett Lindenbach for the HEK293T cells. We thank scientists from APS NECAT 24-ID for assistance. We thank members of the Pyle lab for their input on the manuscript. This research was funded by the Howard Hughes Medical Institute.
Author contributions: A.K., D.L. and A.M.P. designed the experiments. A.K., D.L., D.C.R. and S.C.D. performed protein expression and purification. D.L. performed the RNA work. D.L. performed crystallization, X-ray diffraction data collection and structural analysis. A.K. ran SV experiments with help from D.L. and A.K. and D.L. set up cell culture assays. A.K. and D.C.R. set up ATPase experiments with help from D.L. and S.C.D. A.K., D.L. and A.M.P. wrote the manuscript with input from all authors.
Footnotes
The authors declare that they have no conflict of interest.
References
- Abdullah Z et al. (2012) RIG-I detects infection with live Listeria by sensing secreted bacterial nucleic acids. EMBO J 31: 4153–4164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Takahasi K, Fujita T (2011) RIG-I-like receptors: cytoplasmic sensors for non-self RNA. Immunol Rev 243: 91–98 [DOI] [PubMed] [Google Scholar]
- Ramos HJ, Gale M Jr (2011) RIG-I like receptors and their signaling crosstalk in the regulation of antiviral immunity. Curr Opin Virol 1: 167–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H, Takeuchi O, Mikamo-Satoh E, Hirai R, Kawai T, Matsushita K, Hiiragi A, Dermody TS, Fujita T, Akira S (2008) Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med 205: 1601–1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang SY, Sun HY, Lee KH, Oh BH, Cha YJ, Kim BH, Yoo JY (2012) 5'-Triphosphate-RNA-independent activation of RIG-I via RNA aptamer with enhanced antiviral activity. Nucleic Acids Res 40: 2724–2733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornung V et al. (2006) 5'-Triphosphate RNA is the ligand for RIG-I. Science 314: 994–997 [DOI] [PubMed] [Google Scholar]
- Pichlmair A, Schulz O, Tan CP, Naslund TI, Liljestrom P, Weber F, Reis e Sousa C (2006) RIG-I-mediated antiviral responses to single-stranded RNA bearing 5'-phosphates. Science 314: 997–1001 [DOI] [PubMed] [Google Scholar]
- Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T (2004) The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol 5: 730–737 [DOI] [PubMed] [Google Scholar]
- Lu C, Ranjith-Kumar CT, Hao L, Kao CC, Li P (2011) Crystal structure of RIG-I C-terminal domain bound to blunt-ended double-strand RNA without 5' triphosphate. Nucleic Acids Res 39: 1565–1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlee M et al. (2009) Recognition of 5' triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity 31: 25–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A et al. (2009) 5'-triphosphate RNA requires base-paired structures to activate antiviral signaling via RIG-I. Proc Natl Acad Sci USA 106: 12067–12072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo D, Kohlway A, Pyle AM (2012a) Duplex RNA activated ATPases (DRAs): platforms for RNA sensing, signaling and processing. RNA Biol 10: 111–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Xu H, Ranjith-Kumar CT, Brooks MT, Hou TY, Hu F, Herr AB, Strong RK, Kao CC, Li P (2010) The structural basis of 5' triphosphate double-stranded RNA recognition by RIG-I C-terminal domain. Structure 18: 1032–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y et al. (2010) Structural and functional insights into 5'-ppp RNA pattern recognition by the innate immune receptor RIG-I. Nat Struct Mol Biol 17: 781–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Kinch LN, Brautigam CA, Chen X, Du F, Grishin NV, Chen ZJ (2012) Ubiquitin-induced oligomerization of the RNA sensors RIG-I and MDA5 activates antiviral innate immune response. Immunity 36: 959–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth RB, Sun L, Ea CK, Chen ZJ (2005) Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell 122: 669–682 [DOI] [PubMed] [Google Scholar]
- Kowalinski E, Lunardi T, McCarthy AA, Louber J, Brunel J, Grigorov B, Gerlier D, Cusack S (2011) Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell 147: 423–435 [DOI] [PubMed] [Google Scholar]
- Civril F, Bennett M, Moldt M, Deimling T, Witte G, Schiesser S, Carell T, Hopfner KP (2011) The RIG-I ATPase domain structure reveals insights into ATP-dependent antiviral signalling. EMBO Rep 12: 1127–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F, Ramanathan A, Miller MT, Tang GQ, Gale M Jr, Patel SS, Marcotrigiano J (2011) Structural basis of RNA recognition and activation by innate immune receptor RIG-I. Nature 479: 423–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo D, Ding SC, Vela A, Kohlway A, Lindenbach BD, Pyle AM (2011) Structural insights into RNA recognition by RIG-I. Cell 147: 409–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vela A, Fedorova O, Ding SC, Pyle AM (2012) The thermodynamic basis for viral RNA detection by the RIG-I innate immune sensor. J Biol Chem 287: 42564–42573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckham SA, Brouwer J, Roth A, Wang D, Sadler AJ, John M, Jahn-Hofmann K, Williams BR, Wilce JA, Wilce MC (2013) Conformational rearrangements of RIG-I receptor on formation of a multiprotein:dsRNA assembly. Nucleic Acids Res 41: 3436–3445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder M, Eberle F, Seitz S, Mucke N, Huber CM, Kiani N, Kaderali L, Lohmann V, Dalpke A, Bartenschlager R (2011) Molecular mechanism of signal perception and integration by the innate immune sensor retinoic acid-inducible gene-I (RIG-I). J Biol Chem 286: 27278–27287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng M, Ding Z, Xu L, Kong L, Wang W, Jiao S, Shi Z, Greene MI, Cong Y, Zhou Z (2012) Structural and biochemical studies of RIG-I antiviral signaling. Protein cell 4: 142–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berke IC, Modis Y (2012) MDA5 cooperatively forms dimers and ATP-sensitive filaments upon binding double-stranded RNA. EMBO J 31: 1714–1726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berke IC, Yu X, Modis Y, Egelman EH (2012) MDA5 assembles into a polar helical filament on dsRNA. Proc Natl Acad Sci USA 109: 18437–18441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H (2013) Higher-order assemblies in a new paradigm of signal transduction. Cell 153: 287–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolakofsky D, Kowalinski E, Cusack S (2012) A structure-based model of RIG-I activation. RNA 18: 2118–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo D, Kohlway A, Vela A, Pyle AM (2012b) Visualizing the determinants of viral RNA recognition by innate immune sensor RIG-I. Structure 20: 1983–1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gack MU et al. (2007) TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature 446: 916–920 [DOI] [PubMed] [Google Scholar]
- Kao C, Zheng M, Rudisser S (1999) A simple and efficient method to reduce nontemplated nucleotide addition at the 3 terminus of RNAs transcribed by T7 RNA polymerase. RNA 5: 1268–1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.