Abstract
Retrotransposons are ubiquitous mobile genetic elements constituting a major part of eukaryotic genomes. Yet, monitoring retrotransposition and subsequent copy number increases in multicellular eukaryotes is intrinsically difficult. By following the transgenerational accumulation of a newly activated retrotransposon EVADE (EVD) in Arabidopsis, we noticed fast expansion of activated elements transmitted through the paternal germ line but suppression when EVD-active copies are maternally inherited. This parent-of-origin effect on EVD proliferation was still observed when gametophytes carried mutations for key epigenetic regulators previously shown to restrict EVD mobility. Therefore, the main mechanism preventing active EVD proliferation seems to act through epigenetic control in sporophytic tissues in the mother plant. In consequence, once activated, this retrotransposon proliferates in plant populations owing to suppressed epigenetic control during paternal transmission. This parental gateway might contribute to the occasional bursts of retrotransposon mobilization deduced from the genome sequences of many plant species.
Keywords: epigenetics, transposable elements, retrotransposition, parent-of-origin effect
Introduction
A significant proportion of chromosomal DNA can be assigned to various families of transposable elements (TEs), with retrotransposons being the most abundant [1, 2]. Uncontrolled retrotransposon activation might threaten the genome owing to the accumulation of mutations or to negative regulatory effects on host genes residing in the vicinity of their new insertions [3]. Homologous recombination between ectopic genomic regions through related transposon sequences might also lead to further genome rearrangements [4]. Thus, mechanisms limiting increased copies of self-replicating DNA elements in host genomes are advantageous. However, it was hypothesized that some TEs could also be associated with certain benefits, for example by creating novel, favourable regulatory networks or beneficial genome rearrangements [5, 6], thus providing a selective advantage at the population level [3, 7, 8, 9]. Just how the cost/benefit balance of TEs is regulated remains largely unclear.
Recently, much emphasis has been placed on elucidating epigenetic mechanisms restricting TE activities by transcriptional and post-transcriptional gene silencing [3, 8, 10, 11, 12]. These mechanisms generally involve small RNAs [13]. For example, Piwi-interacting RNA prevent transposon proliferation in Drosophila melanogaster and mammalian germ lines [14, 15]. In plants, 21- and 24-nucleotide small interfering RNAs (siRNAs) influence transposons through post-transcriptional and transcriptional silencing mechanisms, respectively [3, 16]. While such studies address important aspects of host genome regulation, they have not defined host responses to actively mobilized retrotransposons, how active retrotransposons are transmitted or how their copy numbers increase over generations. One underlying problem has been the paucity of mobile retrotransposons to study in ‘real time’ [9]. Moreover, precise analyses of genome-wide TE copy numbers and the parental transmission of specific elements is difficult for mobile TEs belonging to families composed of hundreds of copies [17, 18].
Recently, we and others reported transpositional activation of the Arabidopsis ATCOPIA93 retrotransposon EVADE (EVD) [19, 20]. In wild-type (WT) plants, EVD transcription is suppressed by DNA methylation determined by the maintenance DNA METHYLTRANSFERASE 1 (MET1) in cooperation with the chromatin remodelling protein DECREASED DNA METHYLATION 1 (DDM1). Full-length EVD transcripts are immediately produced in met1 and ddm1 mutant plants but EVD transposition was observed only after prolonged inbreeding. In contrast, drastic EVD transposition was observed when the met1 mutation was combined with mutations impairing the biogenesis of siRNAs or the di-methylation of histone H3 in lysine 9 [19]. Increased EVD copy number was also reported in plants defective in both DDM1 function and siRNA production [21]. Importantly, although dynamic increases in EVD copies were observed after inbreeding in these strains, potential differences in parental contributions to this accumulation have not been determined.
Here, we report on parent-of-origin control of EVD proliferation. Using reciprocal crosses, we have shown that EVD proliferation occurs predominantly by pollen transmission, whereas maternal transmission largely depresses the rate of EVD accumulation. Moreover, our genetic analyses demonstrated that this maternal suppression is maintained in maternal gametes deficient in the production of siRNAs (mutation in RNA polymerases IV/V), deficient in histone H3 in lysine 9 (mutation of histone methyltransferase KYP), or lacking Agronaute 5 (AGO5). Thus, our results suggest that maternal suppression of active EVD copies might not occur in the gametes but that epigenetic mechanisms involving siRNAs and/or methylation of histone H3 on lysine 9 might act in maternal sporophytic tissues of the flower to prevent transgenerational proliferation of active EVDs.
Results
Parent-of-origin effects on EVD accumulation
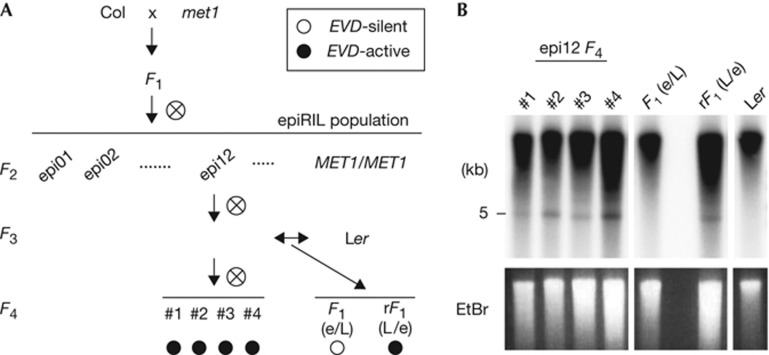
We demonstrated previously that EVD was mobilized and increased in copies during inbreeding in ∼50% of WT epigenetic recombinant inbred lines [19], here referred to as epiRILs [22], derived from a cross between the WT Columbia-0 accession (Col) and the met1-3 mutant with a complete loss of MET1 activity in the Col accession [23]. This increase was first observed in the F4 generation of epiRILs, notably in the epi12 line, and was associated with the presence of EVD extrachromosomal DNA (ecDNA) that made up 10% of the total copies of EVDs (supplementary Fig S1 online) in all studied lines [19].
We aimed to examine possible EVD activation in reciprocal F1 hybrids, as reflected by the appearance of its ecDNA. Hence, a randomly selected single F3 epi12 plant [19] with a propensity to transmit actively replicating EVD (hereafter named EVD-active) was crossed reciprocally to the Landsberg erecta (Ler) accession (EVD-silent) (Fig 1A). By Southern blot, we observed a clear parent-of-origin effect on the presence of EVD ecDNA in F1 progeny of the reciprocal crosses (Fig 1B). We detected ecDNA in the bulked progeny when epi12 pollen was used to fertilize Ler flowers, indicating completion of the replication cycle through reverse transcription. Conversely, ecDNA was absent when Ler pollen fertilized epi12 flowers. This intriguing observation prompted further investigation of a possible parent-of-origin effect on transgenerational propagation of this retrotransposon.
Figure 1.
Parent-of-origin effects on the transmission of the active EVD within inter-accession crosses. (A) Crossing scheme to obtain epi12 and its F3 cross to the Ler accession. One plant from line 12 (epi12) of the epiRIL population at the F3 generation was crossed reciprocally (double arrow) with Ler to generate F1 and reciprocal F1 (rF1) hybrids. The use of Ler (L) or the epiRIL (e) parent is indicated (maternal/paternal) for each hybrid. The inferred EVD activity levels (EVD-silent or EVD-active) are indicated by white and black circles, respectively. (B) Southern blot detection of EVD ecDNA reveals pollen transmission of EVD. Undigested genomic DNA was probed for EVD. The presence of the ∼5-kb EVD ecDNA is indicated (black arrow). Individual epiRIL siblings were investigated, while the two F1 samples and the Ler parental control each comprised a bulked lot of eight individual plants. ecDNA, extrachromosomal DNA; Ler, Landsberg erecta.
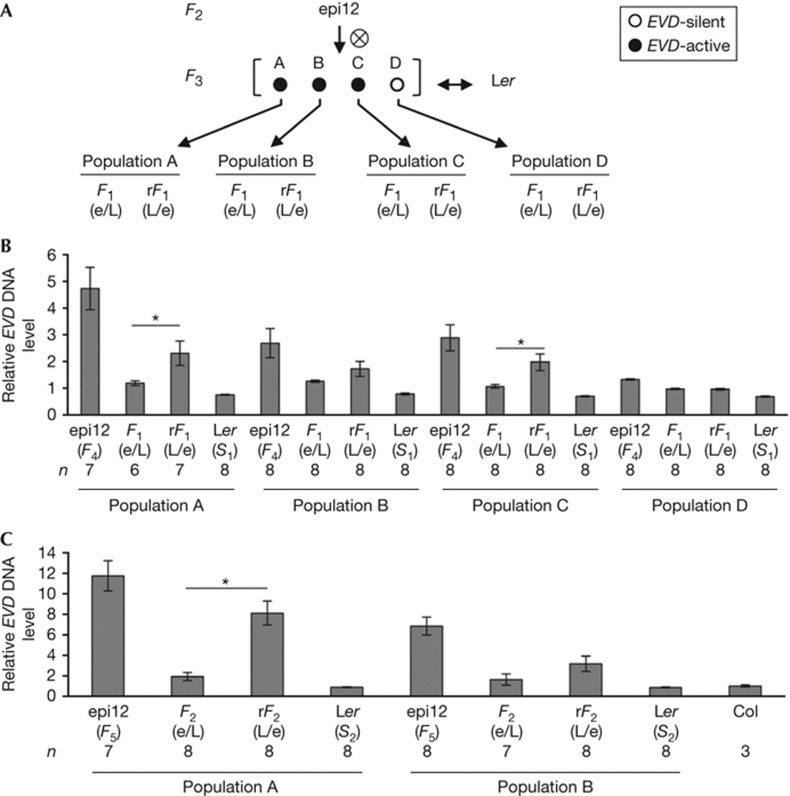
In the F3, individual epi12 plants had various levels of EVD activity. To detect EVD-active plants, we developed a long terminal repeat polymerase chain reaction (LTR-PCR) assay on the basis of the semi-quantitative detection of circular ecDNA forms (supplementary Fig S2A online). We then screened further F3 epi12 siblings and selected three EVD-active epi12 plants differing in amounts of ecDNA (A–C) for further crosses, including one EVD-silent epi12 sibling (D) as a control (supplementary Fig S2B online). These plants were crossed reciprocally to Ler to create four F1 populations A–D (Fig 2A). The relative abundance of EVD DNA in the F1 progeny was assayed using quantitative polymerase chain reaction (supplementary Fig S2C online), that reports on linear ecDNA, circular ecDNA and chromosomal EVD insertions, thus providing a general readout for progressive EVD accumulation. The EVD DNA was also determined in progeny from the self-fertilized parents (epi12 and Ler) to provide controls for relative changes in EVD copies. The highest increases in EVD copies were observed on self-fertilization of EVD-active epi12 siblings (F4, Fig 2B). There were no significant increases in EVD copies when the EVD-active epiRIL lines were fertilized with EVD-silent Ler pollen (maternal suppression phenotype). Conversely, when EVD-silent Ler plants were fertilized with EVD-active pollen increases in EVD copies were observed in all three EVD-active F1 populations derived from epi12 siblings carrying active EVD (populations A–C, Fig 2B), with two out of three (populations A and C) being significantly different at P<0.05 (Student’s t-test).
Figure 2.
Acceleration of EVD activity via pollen transmission within inter-accession crosses. (A) Crossing scheme between four epi12 F3 individuals (marked as A, B, C, D) crossed reciprocally (double arrow) to Ler parents to create populations A–D (see supplementary Fig S2B online for details). (B) Relative EVD DNA content observed in populations A–D at the F1 with the corresponding self-fertilized parental controls. The maternal and paternal contributions (as described in Fig 1) of each cross and sample size (n) are listed along the X axis below the graphs. The average relative EVD content levels (±s.e.m.) for each cross relative to the mid-parent level (set to 1 as midpoint of Col and Ler) are shown along the Y axis. The significance (P<0.05) of Student t-test comparisons of reciprocal crosses (F1 and rF1) for each population (horizontal bars) is indicated by asterisks. (C) Analyses of relative EVD DNA content in the epi12 x Ler F2 progenies of populations A and B. The maternal and paternal contribution and relative EVD DNA content for each population is shown as described in A. Significant differences between reciprocal crosses are shown as described in B. Ler, Landsberg erecta.
We next examined how the relative abundance of EVD DNA changed in the F2 progeny of self-fertilized reciprocal F1 hybrids of EVD-active populations A and B. After self-fertilization, relative abundance of EVD DNA increased to nearly twofold levels above the WT (Fig 2C) in the F2 progeny derived from F1 plants exhibiting near parental levels (see Fig 2B). In contrast, EVD DNA levels in reciprocal F1 siblings from populations A and B, differing in the initial degree of the paternal transmission (see Fig 2B), increased up to eight- and threefold levels above the WT, respectively (Fig 2C). Thus, the relative difference in EVD accumulation originating from the initial parent-of-origin effect was perpetuated in the subsequent generation, with a significant difference between the F2 and reciprocal F2 entries of population A (Student’s t-test, P<0.05; Fig 2C).
Sporophytic parental control of EVD
We repeated the reciprocal crosses using the isogenic Col background to test whether the Ler genome itself influences EVD mobility (Fig 3A). Moreover, we examined whether the maternal suppression phenotype is affected by mutations in particular epigenetic regulators by using mutants previously shown to promote EVD transposition [19]. Specifically, mutants lacking KYP or the common subunit of polymerase IV/V (NRPD2A) when combined with met1-3 permitted massive EVD transposition [19]. Given EVD pollen transmission and the reported activity of TE-derived small RNA produced in the vegetative nucleus in reinforcing silence in male gametes [24], we also sought to determine whether perturbing a pollen-specific function would result in higher relative increases in EVD DNA during male or female transgenerational transmission. For this purpose, we selected a mutant allele of AGO5, an Argonaute family member known to be highly expressed in sperm cell nuclei [25].
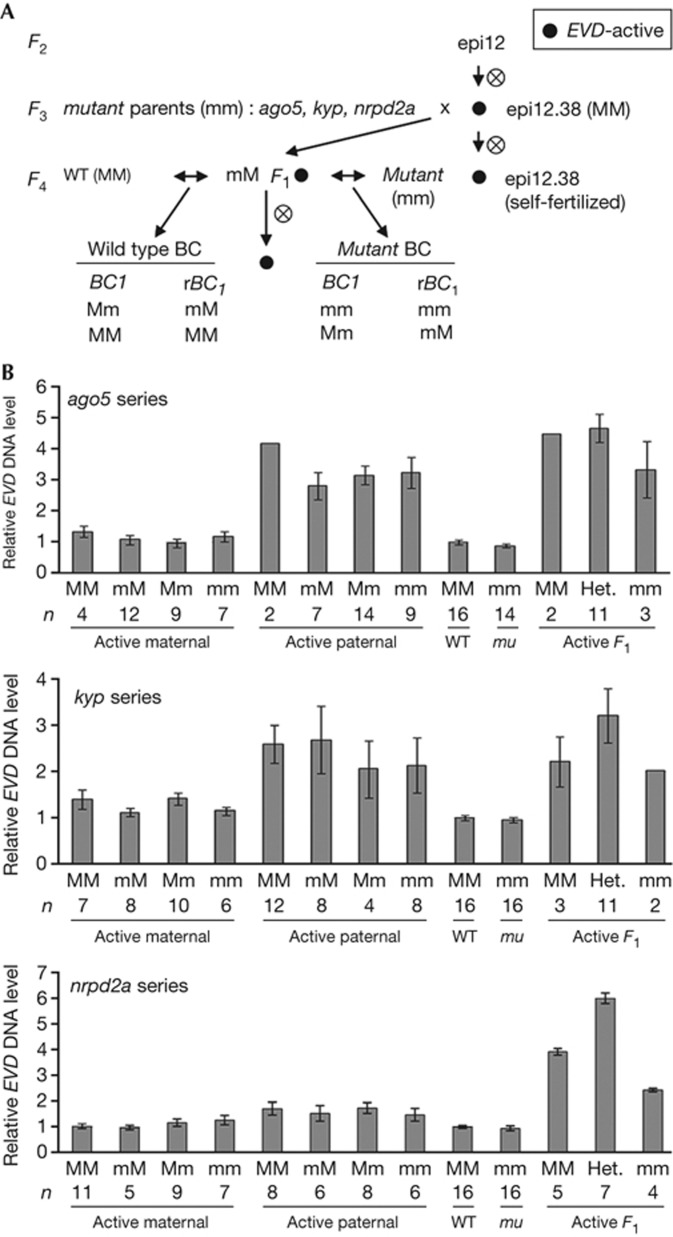
Figure 3.
EVD transmission rates in EVD-active first-generation reciprocal backcross (BC1) populations. (A) Crossing scheme between one F3 generation epi12 individual (epi12.38, paternal parent) and ago5, kyp and nrpd2a mutants (as maternal parents) to create EVD-active backcross populations. Resultant F1 individuals selected as either EVD-active or EVD-silent (see supplementary Fig S3 online) was backcrossed reciprocally to the WT Col parent and the selected mutant parents (ago5, kyp and nrpd2a) to create the respective backcross populations. Shown here are the EVD-active populations; see supplementary Fig S4 online for EVD-silent population results. (B) Relative EVD DNA content levels (Y axis) shown for each entry (X axis) for the mutant populations: ago5 (upper panel), kyp (middle panel), and nrpd2a (lower panel). The parent-of-origin effect (active maternal, active paternal or active F1), sample size (n), and allelic origin at each respective epigenetic regulator for WT (M) and mutant (m) alleles are indicated with respect to parental contributions (maternal/paternal). Heterozygous F2 generation progeny (Het.) comprise both mM and Mm progeny. WT, wild type.
In these crosses (Fig 3A), we used a single epi12 F3 individual, epi12.38, with moderate EVD activity (supplementary Fig S3A online) as the pollen donor to fertilize each selected mutant (supplementary Figs S3B,C online). We selected F1 individuals with EVD-silent or EVD-active copies according to quantitative polymerase chain reaction results in the ago5, kyp and nrpd2a populations (supplementary Fig S3C online). These selected F1 progeny were backcrossed reciprocally to both the WT and respective mutant parent (Fig 3A and supplementary Fig S4A online) to examine the effect of these epigenetic regulators on EVD inheritance. In agreement with the previous results, self-fertilization of EVD-active F1 individuals produced two- to over fourfold increases in EVD copies relative to the WT level, while copies of EVD-silent classes did not differ from the WT level (Fig 3B and supplementary Fig S4 online). By genotyping the BC1 at the AGO5, KYP or NRPD2A loci pertaining to each population, we partitioned seedlings into WT or mutant classes. Given that reciprocal crosses were performed, this also permitted us to address parent-of-origin effects in all genotyped and partitioned populations. Abundance of EVD DNA in the EVD-silent class stayed at the WT level, regardless of crossing direction or gametophytic genotype (supplementary Fig S4B online). The BC1 progeny of EVD-active populations exhibited parent-of-origin effects that reflected maternal suppression or paternal transmission (Fig 3B). This effect was less pronounced in the nrpd2a population, illustrating that rates for which EVD transmission occurs can vary (see also Fig 2), as reported for DNA transposons [26]. The presence of EVD neo-insertions was confirmed by transposon display analysis (supplementary Fig S5 online) in a subset of lines, but we acknowledge our method will only display neo-insertions near DraI recognition motifs. As mutant or WT gametes did not differ in EVD levels within BC1 derived populations, we conclude that the loss of AGO5, KYP or NRPD2A activity alone in the male and female gametophytes is not sufficient to explain the parent-of-origin effects on transgenerational propagation of EVD.
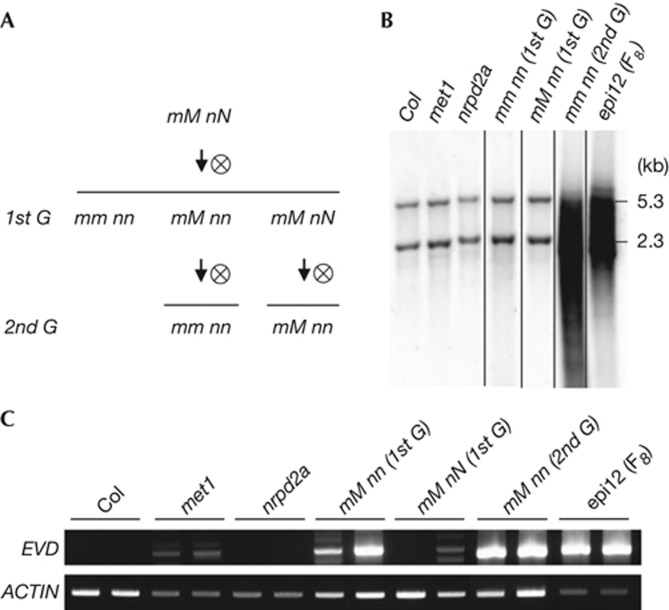
To further examine if parent-of-origin effects depend on parental sporophytic tissues rather than Pol IV/V activities within the gametophytes, we examined EVD DNA levels across several generations of plants segregating for MET1 and/or Pol IV/V mutations (Fig 4A). Southern blot analyses showed WT EVD patterns in both met1 and nrpd2a parents (Fig 4B). In the first generation when the two mutations were combined, there was no increase in EVD copy number (Fig 4B), which is consistent with the above results showing no gametophytic role for Pol IV/V activities (Fig 3). In contrast, a met1 nrpd2a double mutant selected in the second generation (from a met1 heterozygous nrpd2a homozygous parent, thus devoid of Pol IV/V activities during sporophytic growth) showed a massive increase in EVD copy number (Fig 4B), consistent with our previous report [19]. This observation together with the parent-of-origin effects described above points towards a suppressive effect of Pol IV/V activities within the maternal sporophytic tissues in transgenerational EVD proliferation. To further examine EVD activation events in the maternal sporophyte, we compared EVD activities in heterozygous met1 plants that acquired homozygosity of the nrpd2a mutation at different generations in respect to the F1 hybrid. For this, we used the long terminal repeat polymerase chain reaction assay to reveal EVD activity through detection of its ecDNA copies, which cannot be clearly revealed by Southern blot (Fig 4B). Consistent with our previous observation, we detected initiation of EVD life cycle only in plants homozygous for the nrpd2a mutation (Fig 4C). As we repeatedly observed that progenies of such EVD-active plants will accumulate new chromosomal insertions ([19] and supplementary Fig S5 online), we conclude that sporophytic epigenetic control contributes to the early dynamics of EVD inheritance.
Figure 4.
Initiation of EVD proliferation is controlled by parental sporophyte. (A) F1 plants met1 (±) nrpd2a (±) (mM nN) originating from a cross between met1 and nrpd2a parents were self-fertilized to produce segregating 1st and 2nd generation plants (1st and 2nd G, respectively). (B) Southern blot analysis of individual plants with SspI-digested DNA hybridized to an EVD-specific probe as previously reported [19]. (C) LTR-PCR showing EVD extrachromosomal forms diagnostic for EVD life cycle initiation in individual plants. Actin was a loading control and epi12 (F8) a positive control for EVD activity. LTR-PCR, long terminal repeat polymerase chain reaction; met1, methyltransferase 1.
Discussion
Most retrotransposons are transcriptionally silenced by DNA methylation with the developmental control of epigenome reprogramming likely contributing to retrotransposon ‘immunity’. For example, active DNA demethylation in plant companion cells during gametogenesis and the movement of 21-nucleotide siRNAs from the vegetative cell to the germ cell is believed to reinforce continued transposon silencing in gametes [24, 27, 28, 29, 30]. Yet, host responses to actively replicating retrotransposons remain less clear. A notable example however is the maize DNA transposon MuDR where increased transmission rates in progeny inheriting MuDR-1 from the male parent relative to the female parent was reported [31–33].
Our analysis of reciprocal EVD transmission during sexual reproduction in A. thaliana has likewise shown the rate of increase in EDV copy levels is affected by a parent-of-origin effect. The initial results with F1 inter-accession hybrids (Fig 1) could have been due to structural variation between the Col and Ler genomes leading to limited homology-dependent recognition of active elements. This possibility is not likely given the parental effect persisted in intra-accession Col × Col crosses (Fig 3B).
Our results are consistent with a mechanism for the maternal suppression of EVD activity, possibly dependent on the RNA dependent RNA Polymerase 6 (RDR6)–RdDM pathway functioning to re-silence transcriptionally activate TEs [34] and/or diminished canonical RdDM activity in microspores and sperm cells [27]. Given our previous results demonstrating a crucial role of the common subunit of Pol IV/V (NRPD2A) and KYP in preventing EVD mobilization in self-fertilized mutant backgrounds [19], we were surprised the maternal suppression persisted in gametophytes with NRPD2A and KYP mutant genotypes. Therefore, NRPD2A and KYP most likely contribute to sporophytic maternal suppression of EVD replication before gametophyte development. If small RNAs are produced within maternal sporophytic tissue of EVD-active individuals, AGO9-mediated transposon silencing acting in the somatically derived megaspore mother cell could be activated [35]. We speculate such pre-meiotic silencing would facilitate the maternal EVD suppression observed here, independent of the subsequent genotype of the maternal gametophyte. Indeed, NRPD2A activity is involved in restricting the movement and transgenerational accumulation of the heat-activated ONSEN retrotransposon within sporophytic tissues before gametogenesis [36]. In this case, however, as retrotransposition took place before separation of male and female organs in the flower, the parental effect was not expected and thus was not analysed.
Because the observed maternal restriction of EVD level occurs only within the maternal germ line but does not affect paternally transmitted EVD activity, we conclude that maternally-produced small RNAs do not protect the embryo against EVD proliferation after fertilization. This conclusion is supported by data showing that de novo DNA methyltransferases are expressed during female gametogenesis [37] but are barely detectable in microspores and sperm cells [27], despite that RdDM activity becomes highly active during embryogenesis resulting in DNA methylation levels equivalent to the adult sporophytic phase [27, 37]. This example of retrotransposon control is in contrast to the transposon immunity provided by maternal-derived Piwi-interacting RNA that repress paternally active TEs in Drosophila [38, 39].
In summary, we report that after loss of transcriptional silencing and subsequent activation of EVD retrotransposition, transgenerational proliferation depends on the source of the parental gametes. In contrast to the suppressive effect conferred by epigenetic control within the maternal sporophytic tissue, the rapid expansion observed through paternal germ line transmission illustrates how retrotransposons can amplify and proliferate within the plant populations through pollen dispersal. This proliferation in turn supports the accumulation of de novo variation, and the cost/benefit balance of TEs continues to influence genome evolution as natural selection proceeds.
Methods
Plant Material. The epiRILs [22] and mutant lines (ago5-1, kyp-7 nrpd2a-2) are all in the Columbia background. See supplementary information online for EVD-active and EVD-silent lines selection.
EVD DNA Content Analyses. For Southern blot analysis, DNA was extracted from leaf tissue with the DNeasy MaxiPrep kit (Qiagen) and treated as previously described [19] using 5 μg of DNA. For DNA content analysis DNA was extracted from individual mature leaf tissue as previously described [22] and quantified by quantitative polymerase chain reaction. Assays are described in supplementary information online.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Acknowledgments
We thank L. Broger for technical assistance and M Lieberman-Lazarovich, V. Walbot, and P. King for useful comments. This work was supported by grants from the Swiss National Science Foundation (3100A0-102107), the EU (TAGIP; FP6 No.018785) and RECBREED (FP7 No.227190) and the Agropolis Foundation (RETROCROP 1202-041).
Author contributions: J.R., M.M. and J.P. conceived the experiments; J.R., M.M., and J.N. performed the experiments; J.R., M.M., J.N. and J.P. analysed the data; J.R., M.M., and J.P wrote the manuscript.
Footnotes
The authors declare that they have no conflict of interest.
References
- Kumar A, Bennetzen JL (1999) Plant retrotransposons. Ann Review Genetics 33: 479–532 [DOI] [PubMed] [Google Scholar]
- Kidwell MG, Lisch D (1997) Transposable elements as sources of variation in animals and plants. Proc Nat Acad Sci 94: 7704–7711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin RK, Martienssen R (2007) Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet 8: 272–285 [DOI] [PubMed] [Google Scholar]
- Kidwell MG, Lisch DR (2000) Transposable elements and host genome evolution. Trends Ecol Evol 15: 95–99 [DOI] [PubMed] [Google Scholar]
- Feschotte C (2008) Transposable elements and the evolution of regulatory networks. Nat Rev Genet 9: 397–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisch D (2013) How important are transposons for plant evolution? Nat Rev Genet 14: 49–61 [DOI] [PubMed] [Google Scholar]
- Fedoroff NV (2012) Presidential address. Transposable elements, epigenetics and genome evolution. Science 338: 758–767 [DOI] [PubMed] [Google Scholar]
- Lisch D, Bennetzen JL (2011) Transposable element origins of epigenetic gene regulation. Curr Opin Plant Biol 14: 156–161 [DOI] [PubMed] [Google Scholar]
- Tenaillon MI, Hollister JD, Gaut BS (2010) A triptych of the evolution of plant transposable elements. Trends Plant Sci 15: 471–478 [DOI] [PubMed] [Google Scholar]
- Lisch D (2012) Regulation of transposable elements in maize. Curr Opin Plant Biol 15: 511–516 [DOI] [PubMed] [Google Scholar]
- Lisch D, Slotkin RK (2011) Strategies for silencing and escape: the ancient struggle between transposable elements and their hosts. Int Rev Cell Mol Biol 292: 119–152 [DOI] [PubMed] [Google Scholar]
- McCue AD, Slotkin RK (2012) Transposable element small RNAs as regulators of gene expression. Trends Genet 28: 616–623 [DOI] [PubMed] [Google Scholar]
- Czech B, Hannon GJ (2011) Small RNA sorting: matchmaking for argonautes. Nat Rev Genet 12: 19–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senti KA, Brennecke J (2010) The piRNA pathway: a fly's perspective on the guardian of the genome. Trends Genet 26: 499–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi MC, Sato K, Pezic D, Aravin AA (2011) PIWI-interacting small RNAs: the vanguard of genome defence. Nat Rev Mol Cell Biol 12: 246–258 [DOI] [PubMed] [Google Scholar]
- Law JA, Jacobsen SE (2010) Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet 11: 204–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang W, Schulman AH (2008) BARE retrotransposons produce multiple goups of rarely polyadenylated transcripts from two differentially regulated promoters. Plant J 56: 40–50 [DOI] [PubMed] [Google Scholar]
- Le QH, Melayah D, Bonnivard E, Petit M, Grandbastien MA (2007) Distribution dynamics of the Tnt1 retrotransposon in tobacco. Mol Genet Genomics 278: 639–651 [DOI] [PubMed] [Google Scholar]
- Mirouze M et al. (2009) Selective epigenetic control of retrotransposition in Arabidopsis. Nature 461: 427–430 [DOI] [PubMed] [Google Scholar]
- Tsukahara S, Kobayashi A, Kawabe A, Mathieu O, Miura A, Kakutani T (2009) Bursts of retrotransposition reproduced in Arabidopsis. Nature 461: 423–426 [DOI] [PubMed] [Google Scholar]
- Zemach A, Kim MY, Hsieh PH, Coleman-Derr D, Eshed-Williams L, Thao K, Harmer SL, Zilberman D (2013) The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell 153: 193–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinders J, Wulff BB, Mirouze M, Mari-Ordonez A, Dapp M, Rozhon W, Bucher E, Theiler G, Paszkowski J (2009) Compromised stability of DNA methylation and transposon immobilization in mosaic Arabidopsis epigenomes. Genes Dev 23: 939–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saze H, Mittelsten Scheid O, Paszkowski J (2003) Maintenance of CpG methylation is essential for epigenetic inheritance during plant gametogenesis. Nat Genet 34: 65–69 [DOI] [PubMed] [Google Scholar]
- Slotkin RK, Vaughn M, Borges F, Tanurdzic M, Becker JD, Feijo JA, Martienssen RA (2009) Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell 136: 461–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges F, Gomes G, Gardner R, Moreno N, McCormick S, Feijo JA, Becker JD (2008) Comparative transcriptomics of Arabidopsis sperm cells. Plant Physiol 148: 1168–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Miura A, Bender J, Jacobsen SE, Kakutani T (2003) Role of CG and non-CG methylation in immobilization of transposons in Arabidopsis. Curr Biol 13: 421–426 [DOI] [PubMed] [Google Scholar]
- Calarco JP et al. (2012) Reprogramming of DNA methylation in pollen guides epigenetic inheritance via small RNA. Cell 151: 194–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh TF, Ibarra CA, Silva P, Zemach A, Eshed-Williams L, Fischer RL, Zilberman D (2009) Genome-wide demethylation of Arabidopsis endosperm. Science 324: 1451–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarra CA et al. (2012) Active DNA demethylation in plant companion cells reinforces transposon methylation in gametes. Science 337: 1360–1364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosher RA, Melnyk CW, Kelly KA, Dunn RM, Studholme DJ, Baulcombe DC (2009) Uniparental expression of PolIV-dependent siRNAs in developing endosperm of Arabidopsis. Nature 460: 283–286 [DOI] [PubMed] [Google Scholar]
- Lisch D, Chomet P, Freeling M (1995) Genetic characterization of the Mutator system in maize: behavior and regulation of Mu transposons in a minimal line. Genetics 139: 1777–1796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinard PS, Robertson DS, Schnable PS (1993) Genetic isolation, cloning, and analysis of a mutator-induced, dominant antimorph of the maize amylose extender1 locus. Plant Cell 5: 1555–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walbot V (1986) Inheritance of mutator activity in Zea mays as assayed by somatic instability of the bz2-mu1 allele. Genetics 114: 1293–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuthikattu S, McCue AD, Panda K, Fultz D, Defraia C, Thomas EN, Slotkin RK (2013) The initiation of epigenetic silencing of active transposable elements is triggered by RDR6 and 21-22 nucleotide small interfering RNAs. Plant Physiol 162: 116–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmedo-Monfil V, Duran-Figueroa N, Arteaga-Vazquez M, Demesa-Arevalo E, Autran D, Grimanelli D, Slotkin RK, Martienssen RA, Vielle-Calzada JP (2010) Control of female gamete formation by a small RNA pathway in Arabidopsis. Nature 464: 628–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H, Gaubert H, Bucher E, Mirouze M, Vaillant I, Paszkowski J (2011) An siRNA pathway prevents transgenerational retrotransposition in plants subjected to stress. Nature 472: 115–119 [DOI] [PubMed] [Google Scholar]
- Jullien PE, Susaki D, Yelagandula R, Higashiyama T, Berger F (2012) DNA methylation dynamics during sexual reproduction in Arabidopsis thaliana. Curr Biol 22: 1825–1830 [DOI] [PubMed] [Google Scholar]
- Blumenstiel JP, Hartl DL (2005) Evidence for maternally transmitted small interfering RNA in the repression of transposition in Drosophila virilis. Proc Natl Acad Sci USA 102: 15965–15970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Malone CD, Aravin AA, Sachidanandam R, Stark A, Hannon GJ (2008) An epigenetic role for maternally inherited piRNAs in transposon silencing. Science 322: 1387–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.