Abstract
Among prognostic factors for chronic lymphocytic leukemia (CLL), immunoglobulin heavy chain variable region (IGHV) mutation status and DNA analysis appear to be the most important. However, there is limited clinical outcome information for patients with the favorable-risk del(13q) and poor-risk unmutated IGHV. We retrospectively screened all patients with CLL at our institution between 2004 and June 2010 for del(13q) who also had an IGHV analysis. Unmutated IGHV was found in 38/79 patients; age, Rai stage, prior therapy, and time to evaluation were similar to those for patients with mutated IGHV. Unmutated patients were nearly four times more likely to harbor additional chromosomal aberrations compared to mutated patients (p < 0.001). During a median follow-up of 4.5 years, unmutated patients were more likely to demonstrate Rai stage progression (69% vs. 31%, log-rank p < 0.001) and to receive treatment (5-year cumulative probability of treatment: 65% vs. 32%, p < 0.001). Patients with unmutated CLL also had a shorter overall survival (5-year survival probability: 72% vs. 100%, p < 0.001). When limiting analysis to the 47 patients with del(13q) as a sole chromosomal abnormality, the 13 (28%) unmutated patients were more likely to demonstrate Rai progression (p < 0.001), to receive treatment (P = 0.02), and to have a shorter overall survival (P = 0.13) than the 34 mutated patients. These data suggest that del(13q) conveys an indolent course only in patients with IGHV-mutated CLL.
Keywords: CLL, del(13q), IGHV, clinical outcomes, overall survival
Introduction
Patients with chronic lymphocytic leukemia (CLL), the most common adult hematologic cancer, exhibit heterogeneous survival rates [1]. Moreover, CLL clinical grading systems poorly predict overall survival and disease aggressiveness, especially in patients with early-stage disease [2–4]. As the catalysts that lead to disease progression remain unidentified [5,6], molecular and genetic evaluations are used to predict clinical outcomes [7,8].
Based on results from the array of currently available laboratory studies of CLL prognostic factors [9–13], immunoglobulin heavy chain variable region (IGHV) mutation status [14,15] and interphase fluorescence in situ hybridization (FISH) DNA analysis appear to be the most important independent predictors of outcome [4,6,16]. In their landmark analysis, Dohner et al. reported a median survival of 2–3 years for patients with del(17p) CLL, while patients whose CLL carried a del(13q) as sole chromosomal abnormality had a median survival of 133 months, with nearly one-third of patients not requiring therapy [5]. However, there are limited prognostic data regarding patients who harbor both favorable and poor prognostic risk features [17,18]. In Krober et al.’s subset analysis of 167 patients with del(13q) CLL carried out in 2002, approximately 50% had an unmutated IGVH status. However, event-free and overall survival in this subset was not performed [17]. Accordingly, we found that many of our patients with del(13q) CLL requiring intervention exhibited a low IGHV mutation percentage. We hypothesized that a low IGHV mutation percentage, despite the presence of a del(13q), would predictably confer an aggressive CLL course. Therefore, we retrospectively analyzed the influence of IGHV mutational status on clinical outcome in patients with CLL harboring a del(13q).
Methods
Subjects
After Institutional Review Board approval (NA_00028739), we retrospectively screened all patients with CLL evaluated at our institution between 2004 and June 2010 for either homozygous del(13q) or heterozygous del(13q). At our center, standard laboratory practice for patients with CLL is to perform interphase FISH DNA analysis and IGHV mutational status analysis; however, all clinical evaluations are at the discretion of the attending physician. This report analyzes all patients identified with del(13q) on their initial FISH evaluation, who also had an IGHV sequence homology analysis. Many of these patients were already being followed before 2004, the date that IGHV mutation status became available at Johns Hopkins, and are included in the study.
FISH and IGHV analyses
All FISH was performed at a single reference laboratory by interphase analysis using probes for 6 centromere (cen)/6q23.3 (CMYB), 11cen/ 11q22.3(ATM), 12cen/12q15 (MDM2), 13q14 (D13S319)/13q34(LAMP1), 17cen/17p13.1 (p53), and 11q13 (CCND1-XT)/14q32 (IGH-XT). The 95% cut-off for homozygous del(13q) and duel del(13q) was 7.0% and 1.5%, respectively. All IGHV mutation analyses were performed at a single reference laboratory using cycle sequencing analysis. An IGHV non-homologous sequence of <2% was defined as unmutated (low mutational status), and an IGHV non-homologous sequence of ≥2% was defined as mutated (high mutational status) [14,15,19,20,21].
Statistical methods
Characteristics of patients by mutation status were compared using Wilcoxon rank-sum tests for continuous measurements and Fisher’s exact test for categorical measurements. Odds ratios from logistic regression models that adjusted for time from diagnosis to FISH analysis were used to determine whether the proportion of patients with additional chromosomal changes was different between mutation groups. Overall survival was calculated as the time from diagnosis to death or last known follow-up. Treatment-free survival was calculated as the time from diagnosis to treatment or last known follow-up at which the patient had not received treatment. Rai 3, 4, or Richter progression was calculated as the time from diagnosis to date of first progression for patients who had been Rai 1 or 2 at diagnosis. Patients currently alive or treatment-free were censored on 15 June 2010. Event-time distributions were estimated by the Kaplan–Meier method and compared by log-rank test. All analyses were completed using statistical software R version 2.11.1.
Results
Between 2004 and June 2010, FISH studies were performed on 196 patients with CLL, and 94 (48%) had a del(13q). Of these 94 patients, 79 (84%) also had an IGHV mutational percentage calculated and were selected for analysis. The median time from diagnosis to last follow-up for these patients was 4.5 (range 0–22) years. Of the 79 patients with CLL with both a del(13q) and IGHV analysis, 38 (48%) were unmutated and 41 (52%) were mutated. The median age at diagnosis of the unmutated patients was 58 (range 27–81) years, compared to a median age of 56 (range 31–71) years in mutated patients. Both groups had a similar stage distribution at diagnosis, with a documented Rai stage ≤2 in 33/38 (92%) and 34/41 (83%) unmutated and mutated patients, respectively (p = 0.225). The median time to first evaluation at Johns Hopkins for CLL was similar in both groups (p = 0.94), with over 75% in each group being treatment-naive at the time of this consultation (p = 0.79). The median time between diagnosis and first FISH analysis was 1.4 (range 0–10.2) years in the unmutated group and 2.2 (range 0–21.4) years in the mutated group (p = 0.41) (Table I).
Table I.
Characteristics of patients with del(13q) CLL, overall and by IGHV mutation percentage.
| All patients (n = 79) |
IGHV <2% (n = 38) |
IGHV ≥2% (n = 41) |
p-Value* | |
|---|---|---|---|---|
| Age at diagnosis, median (range) | 57 (27, 81) | 58 (27, 81) | 56 (31, 71) | 0.58 |
| Stage at diagnosis, n (%) | ||||
| Rai 0 | 30 (42) | 14 (39) | 16 (46) | 0.23 |
| Rai 1 | 26 (37) | 15 (42) | 11 (31) | |
| Rai 2 | 11 (15) | 4 (11) | 7 (20) | |
| Rai 3 | 3 (4) | 3 (8) | 0 (0) | |
| Rai 4 | 0 (0) | 0 (0) | 0 (0) | |
| Richter | 1 (1) | 0 (0) | 1 (3) | |
| Unknown | 8 | 2 | 6 | |
| Time from diagnosis to initial JHU consultation (years), median (range) |
0.4 (0, 16.1) | 0.6 (0, 7.6) | 0.3 (0, 16.1) | 0.94 |
| Stage at initial JHU consultation, n (%) | ||||
| Rai 0 | 28 (36) | 12 (32) | 16 (39) | 0.18 |
| Rai 1 | 25 (32) | 13 (35) | 12 (29) | |
| Rai 2 | 11 (14) | 3 (8) | 8 (20) | |
| Rai 3 | 7 (9) | 6 (16) | 1 (2) | |
| Rai 4 | 6 (8) | 3 (8) | 3 (8) | |
| Richter | 0 (0) | 0 (0) | 1 (2) | |
| Unknown | 1 | 1 | 0 | |
| Received treatment prior to JHU consultation | ||||
| No | 62 (78) | 29 (76) | 33 (80) | 0.79 |
| Yes | 17 (22) | 9 (24) | 8 (20) | |
| Time from diagnosis to FISH (years), median (range) |
1.6 (0, 21.4) | 1.4 (0, 10.2) | 2.2 (0, 21.4) | 0.41 |
| Sole del(13q), n (%) | ||||
| No | 32 (41) | 25 (66) | 7 (17) | < 0.001 |
| Yes | 47 (59) | 13 (34) | 34 (83) | |
| Trisomy 12, n (%) | ||||
| No | 77 (97) | 37 (97) | 40 (98) | >0.99 |
| Yes | 2 (3) | 1 (3) | 1 (2) | |
| del(6q), n (%) | ||||
| No | 74 (94) | 34 (89) | 41 (100) | >0.99 |
| Yes | 4 (5) | 4 (11) | 0 (0) | |
| del(11q), n (%) | ||||
| No | 61 (77) | 23 (61) | 37 (90) | < 0.001 |
| Yes | 18 (23) | 15 (39) | 4 (10) | |
| del(17p), n (%) | ||||
| No | 70 (89) | 30 (79) | 40 (98) | 0.01 |
| Yes | 9 (11) | 8 (21) | 1 (2) | |
| del(11q) or del(17p), n (%) | ||||
| No | 52 (66) | 15 (39) | 36 (88) | < 0.001 |
| Yes | 27 (34) | 23 (61) | 5 (12) |
p-Value: Fisher’s exact test or Wilcoxon rank-sum test for differences in variables between mutation groups.
CLL, chronic lymphocytic leukemia; IGHV, immunoglobulin heavy chain variable region; JHU, Johns Hopkins University; FISH, fluorescence in situ hybridization.
Unmutated patients are more likely to have additional adverse cytogenetic abnormalities
Thirteen of 38 (34%) unmutated patients had del(13q) as a sole chromosomal aberration on their initial FISH examination, compared to 34/41 (83%) of mutated patients. Twenty-five of 38 (66%) unmutated patients had additional abnormalities, including 15/38 (39%) with an 11q deletion and 8/38 (21%) with a 17p deletion, compared to only 7/41 (17%) mutated patients with additional chromosomal changes, including 3/41 (7%) with 11q deletion and 1/41 (2%) with a 17p deletion. Mutated patients were significantly less likely than unmutated patients to have additional adverse changes at diagnosis, regardless of the length of time between diagnosis and FISH analysis, including 11q (odds ratio = 0.09, 95% confidence interval [CI] 0.02–0.4, p < 0.001), 17p (odds ratio = 0.10,95% CI 0.01–0.88,p = 0.01), and either 11q or 17p (odds ratio = 0.07, 95% CI 0.02–0.24, p < 0.001).
Serial FISH analyses were not routinely performed during follow-up. Only 12 (32%) unmutated patients had repeat testing. Interestingly, two of these patients acquired a del(17p) and one patient acquired a del(11q). In contrast, none of the seven (17%) mutated patients who underwent repeat FISH analyses developed additional chromosomal abnormalities during follow-up.
Unmutated patients are more likely to show Rai stage migration
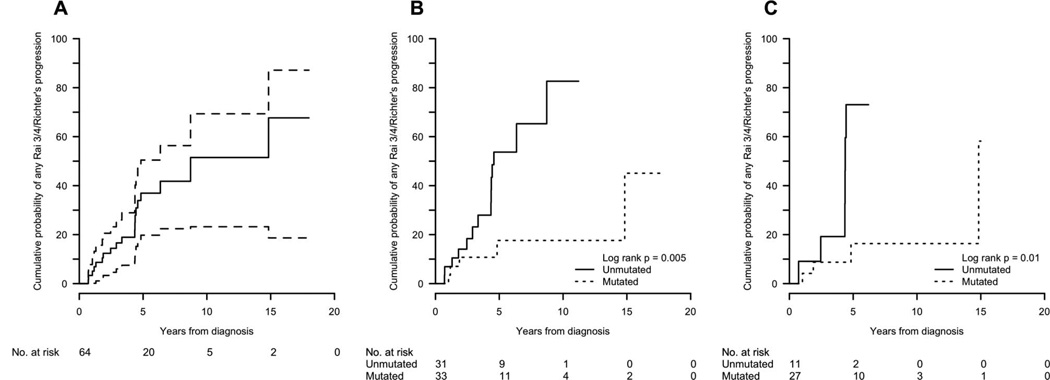
Rai stage progression was more common in the unmutated patients than in the mutated patients (69% vs. 31%, log-rank p < 0.001), as was progression to either Rai stage 3/4 disease or Richter transformation (53% vs. 22%, log-rank p = 0.005), with one of the mutated patients undergoing a Richter transformation 21 years after diagnosis; 32 (78%) mutated patients remained Rai stage 2 or less (Table II, Figure 1).
Table II.
Overall clinical events of patients with del(13q) CLL from time of diagnosis to last follow-up, overall and by IGHV mutation percentage.
| All patients (n = 79) |
IGHV <2% (n = 38) |
IGHV ≥2% (n = 41) |
p-Value | |
|---|---|---|---|---|
| Time from diagnosis to last follow-up (years), median (range) | 4.5 (0–22) | 4.5 (0–13) | 4.3 (0.4–22) | 0.14* |
| Maximum Rai stage, n (%) | ||||
| Rai 0 | 15 (19) | 3 (8) | 12 (30) | NA† |
| Rai 1 | 16 (21) | 7 (18) | 9 (22) | |
| Rai 2 | 18 (23) | 8 (21) | 10 (25) | |
| Rai 3 | 8 (10) | 8 (21) | 0 (0) | |
| Rai 4 | 17 (22) | 10 (26) | 7 (18) | |
| Richter | 4 (5) | 2 (5) | 2 (5) | |
| Unknown | 1 | 0 | 1 | |
| Rai progression, n (%) | ||||
| No | 35 (49) | 11 (31) | 24 (69) | <0.001‡ |
| Yes | 36 (51) | 25 (69) | 11 (31) | |
| Unknown | 8 | 2 | 6 | |
| Rai 3/4 or Richter at any time, n (%) | ||||
| No | 48 (62) | 18 (47) | 32 (78) | 0.005‡ |
| Yes | 30 (38) | 20 (53) | 9 (22) | |
| Unknown | 1 | 0 | 0 | |
| Ever received CLL treatment, n (%) | ||||
| No | 39 (49) | 13 (34) | 26 (63) | <0.001‡ |
| Yes | 40 (51) | 25 (66) | 15 (37) |
p-Value: Fisher’s exact test or Wilcoxon rank-sum test for differences in variables between mutation groups.
Maximum Rai stage is time-dependent per patient and a formal comparison between groups was not done,
p-Value: log-rank tests for differences in time-to-event outcomes between mutation groups.
CLL, chronic lymphocytic leukemia; IGHV, immunoglobulin heavy chain variable region.
Figure 1.
Cumulative probability of progression to Rai 3/4 or Richter transformation: (A) all patients; (B) all patients by IGHV mutation status; (C) del(13q)-only patients by IGHV mutation status.
Unmutated patients are more likely to receive treatment and have shorter survival
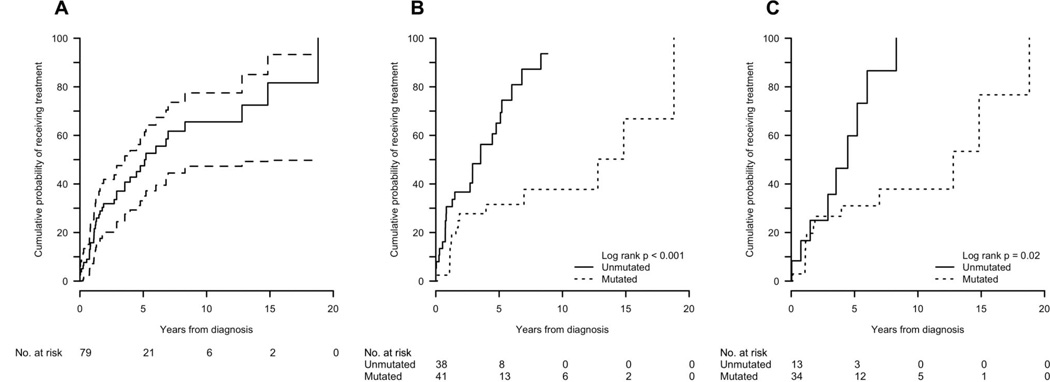
No patient died without first receiving CLL treatment. For the entire group, the median time to treatment was 2.7 (range 0–18.8) years and the cumulative probability of receiving treatment (CPT) at 5 years was 48%. The most common reasons for treatment initiation were either bulky adenopathy or progression to an advanced Rai stage. Among the unmutated patients, the median time to treatment initiation was 2.0 (range 0–8.8) years and their CPT at 5 years was 65%. Six unmutated patients underwent allogeneic transplant, of whom three have died and three remain disease-free at 9, 18, and 21 months. In contrast, among the mutated patients, the median time to treatment initiation was 3.8 (range 0.1–18.8) years and their CPT at 5 years was 32% (log-rank p < 0.001) (Table III, Figure 2).
Table III.
Cumulative probability of treatment (CPT), overall survival (OS), and cumulative probability of Rai 3, Rai 4, or Richter (CPR) at 5 years after diagnosis for the whole cohort and by mutational analysis and genotype*.
| n | Median | 5-year | HR | 95% CI | p-Value† | |
|---|---|---|---|---|---|---|
| CPT, all patients | 79 | 5.1 | 48 (33, 59) | |||
| TFS, by genotype | ||||||
| Complex genotype with del(13q) | 32 | 3.54 | 61 (35, 77) | 1.00 | — | |
| del (13q) only | 47 | 6.96 | 39 (21, 53) | 0.54 | (0.29, 1.02) | 0.06 |
| CPT, by mutational percentage | ||||||
| IGHV mutation < 2% | 38 | 3.54 | 65 (42, 79) | 1.00 | — | |
| IGHV mutation ≥2% | 41 | 12.75 | 32 (14, 46) | 0.3 | (0.15, 0.60) | < 0.001 |
| CPT, del(13q) as sole deletion | ||||||
| IGHV mutation < 2% | 13 | 4.46 | 60 (10, 82) | 1.00 | — | |
| IGHV mutation ≥2% | 34 | 12.75 | 31 (12, 46) | 0.33 | (0.13, 0.83) | 0.02 |
| OS, all patients | 79 | NR | 88 (79, 97) | |||
| OS, by genotype | ||||||
| Complex genotype with del(13q) | 32 | 11.22 | 78 (63, 98) | 1.00 | — | |
| del(13q) only | 47 | NR | 94 (85, 100) | 0.21 | (0.05, 0.84) | 0.027 |
| OS, by mutation percentage | ||||||
| IGHV mutation < 2% | 38 | 11.22 | 72 (57, 91) | 1.00 | — | |
| IGHV mutation ≥2% | 41 | NR | 100 (100, 100) | 0.06 | (0.01, 0.5) | < 0.001 |
| OS, del(13q) only | ||||||
| IGHV mutation < 2% | 13 | NR | 79 (56, 100) | 1.00 | — | |
| IGHV mutation ≥2% | 34 | NR | 100 (100, 100) | 0.17 | (0.01, 1.8) | 0.13 |
| CPR, all patients | 64 | 8.7 | 37 (20, 50) | |||
| CPR, by genotype | ||||||
| Complex genotype with del(13q) | 26 | 6.4 | 38 (10, 57) | 1.00 | — | |
| del(13q) only | 38 | 14.8 | 36 (13, 54) | 0.66 | (0.27, 1.6) | 0.37 |
| CPR, by mutation percentage | ||||||
| IGHV mutation < 2% | 31 | 4.6 | 54 (26, 71) | 1.00 | — | |
| IGHV mutation ≥2% | 33 | NR | 18 (0, 33) | 0.23 | (0.08, 0.72) | 0.005 |
| CPR, del(13q) only | ||||||
| IGHV mutation < 2% | 11 | 4.4 | 73 (13, 92) | 1.00 | — | |
| IGHV mutation ≥2% | 27 | 14.8 | 16 (0, 32) | 0.18 | (0.04, 0.72) | 0.01 |
Values for median survival are in years. Other values are probability (95% CI).
p-Value: log-rank test for differences in time-to-event outcomes between mutation groups.
CPT, treatment-free survival; IGHV, immunoglobulin heavy chain variable region; HR, hazard ratio; CI, confidence interval.
Figure 2.
Cumulative probability of receiving treatment: (A) all patients; (B) all patients by IGHV mutation status; (C) del(13q)-only patients by IGHV mutation status.
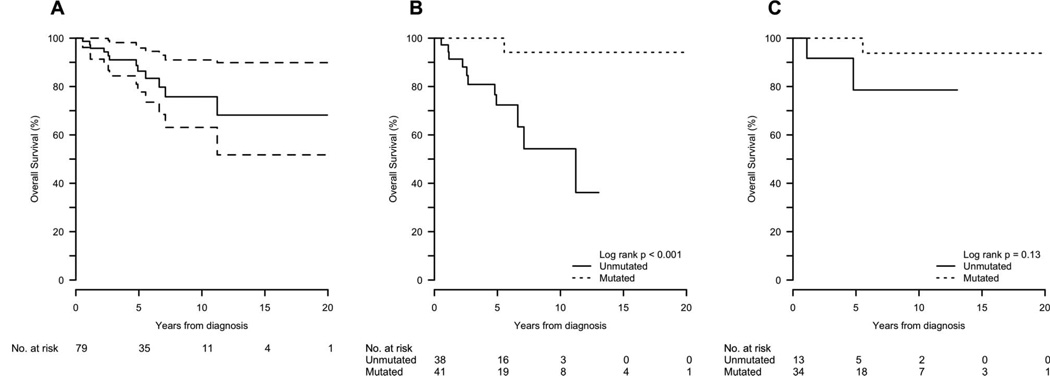
The probability of overall survival (OS) at 5 years was 86% for the entire group. The median survival for the unmutated patients was 11.2 years, with a 5-year OS probability of 75%. For mutated patients, the median survival was not reached, and the 5-year OS probability was 100% (log-rank p < 0.001). Two patients in the mutated group underwent allogeneic transplant, and both are alive and disease-free, although one is only 60 days post-transplant (Table III, Figure 3). All 12 patients died from CLL disease progression.
Figure 3.
Overall survival: (A) all patients; (B) all patients by IGHV mutation status; (C) del(13q)-only patients by IGHV mutation status.
Unmutated IGHV remains poor prognostic factor when del(13q) is sole chromosomal abnormality
When limiting analysis to patients with del(13q) as their sole chromosomal aberration, 13/47 patients had an unmutated and 34/47 patients had a mutated IGHV (Table IV). In the unmutated group, during a median of 5.0 (range 0.0–13.0) years after diagnosis, nine (75%) had stage progression, seven (54%) to Rai ≥3 disease including a Richter transformation, and in the mutated group, during a median of 5.0 (range 0.4–22) years after diagnosis, 12 (41%) had stage progression, with eight (24%) progressing to Rai ≥3 disease including a Richter transformation (Table V).
Table IV.
Characteristics of del(13q)-only patients by IGHV mutation status.
| del(13q)-only patients (n = 47) |
IGHV <2% (n = 13) |
IGHV ≥2% (n = 34) |
p-Value* | |
|---|---|---|---|---|
| Age at diagnosis, median (range) | 58 (33, 74) | 60 (34, 74) | 57 (33, 71) | 0.31 |
| Stage at diagnosis, n (%) | ||||
| Rai 0 | 18 (38) | 4 (33) | 14 (48) | 0.47 |
| Rai 1 | 17 (36) | 6 (50) | 10 (34) | |
| Rai 2 | 4 (9) | 1 (8) | 4 (14) | |
| Rai 3 | 1 (2) | 1 (8) | 0 (0) | |
| Rai 4 | 0 (0) | 0 (0) | 0 (0) | |
| Richter | 1 (2) | 0 (0) | 1 (3) | |
| Unknown | 6 | 1 | 5 | |
| Time from diagnosis to initial JHU consultation (years), median (range) |
0.3 (0, 16.1) | 0.2 (0, 4.4) | 0.4 (0, 16.1) | 0.3 |
| Stage at initial JHU consultation, n (%) | ||||
| Rai 0 | 18 (39) | 4 (33) | 13 (38) | 0.22 |
| Rai 1 | 17 (37) | 4 (33) | 11 (32) | |
| Rai 2 | 4 (9) | 1 (8) | 5 (15) | |
| Rai 3 | 3 (7) | 3 (25) | 1 (3) | |
| Rai 4 | 3 (7) | 0 (0) | 3 (9) | |
| Richter | 1 (2) | 0 (0) | 1 (3) | |
| Unknown | 1 | 1 | 0 | |
| Received treatment prior to JHU consultation | ||||
| No | 40 (85) | 12 (92) | 28 (82) | 0.66 |
| Yes | 7 (15) | 1 (8) | 6 (18) | |
| Time from diagnosis to FISH (years), median (range) | 2.5 (0.0, 21.4) | 1.5 (0.1, 10.2) | 3.3 (0, 21.4) | 0.26 |
p-Value: Fisher’s exact test or Wilcoxon rank-sum test for differences in variables between mutation groups.
IGHV, immunoglobulin heavy chain variable region; JHU, Johns Hopkins University; FISH, fluorescence in situ hybridization.
Table V.
Clinical events of del(13q)-only patients from time of diagnosis to last follow-up, overall and by IGHV mutation percentage.
| All patients (n = 47) |
IGHV <2% (n = 13) |
IGHV ≥2% (n = 34) |
p-Value | |
|---|---|---|---|---|
| Time from diagnosis to last follow-up (years), median (range) | 5.0 (0, 22) | 5.0 (0, 13) | 5.0 (0.4, 22) | 0.48* |
| Maximum Rai stage, n (%) | ||||
| Rai 0 | 11 (24) | 1 (8) | 10 (30) | NA† |
| Rai 1 | 12 (27) | 2 (15) | 9 (27) | |
| Rai 2 | 9 (20) | 3 (23) | 6 (18) | |
| Rai 3 | 3 (7) | 3 (23) | 0 (0) | |
| Rai 4 | 10 (21) | 3 (23) | 6 (18) | |
| Richter | 2 (4) | 1 (8) | 2 (6) | |
| Unknown | 1 | 0 | 1 | |
| Rai progression, n (%) | ||||
| No | 22 (51) | 3 (25) | 19 (61) | <0.001‡ |
| Yes | 21 (49) | 9 (75) | 12 (39) | |
| Unknown | 4 | 1 | 3 | |
| Rai 3/4 or Richter at any time, n (%) | ||||
| No | 31 (67) | 6 (46) | 25 (76) | 0.01‡ |
| Yes | 14 (33) | 7 (54) | 8 (24) | |
| Unknown | 1 | 0 | 1 | |
| Ever received CLL treatment | ||||
| No | 25 (53) | 4 (31) | 21 (62) | 0.02‡ |
| Yes | 22 (47) | 9 (69) | 13 (38) |
p-Value: Fisher exact test or Wilcoxon rank-sum test for differences in variables between mutation groups.
Maximum Rai stage is time-dependent per patient and a formal comparison between groups was not done.
p-Value: log-rank test for differences in time-to-event outcomes between mutation groups.
IGHV, immunoglobulin heavy chain variable region; CLL, chronic lymphocytic leukemia.
In the unmutated group, treatment was initiated in 9/13 (69%), including two allogeneic transplants, and two have died (one of whom was transplanted). The unmutated, del(13q)-only group had a CPT at 5 years of 60%, compared to 31% in the mutated group (log-rank p=0.02). Thirteen of the 34 (38%) mutated patients with del(13q) as sole abnormality have received treatment, including two allogeneic transplants. Only one death in the mutated-CLL group has occurred (non-transplanted patient). The OS at 5 years in the del(13q)-only group, unmutated versus mutated, was 79% vs. 100%, respectively (p = 0.13) (Table III, Figure 3). Also, although multiple FISH analyses were not routinely performed, it is intriguing that cytogenetic progression occurred twice in the sole del(13q), IGHV-unmutated cohort and not in the patients with mutated IGHV.
Discussion
Cytogenetic abnormalities [5], usually determined by FISH because of the low proliferative rate of CLL, and IGHV mutational status [6,14,19–21] appear to be the most important independent prognostic factors in this disease. IGHV mutational status and cytogenetics also commonly track with one another: 17p deletions and 11q deletions are associated with an unmutated IGHV, while del(13q) is associated with a mutated IGHV [17,20]. However, there are limited data regarding the 25–50% of patients with CLL who harbor a del(13q) together with an unmutated IGHV [17,18]. As in previous reports [17], we found that nearly 50% (38/79) of our patients with del(13q) CLL had an unmutated IGHV. The incidence decreased to 28% (13/47) when limiting del(13q) as a sole abnormality to unmutated disease, similar to other reports in such patients [17,20]. Conversely, when limiting analysis to patients with CLL with multiple cytogenetic abnormalities on first FISH examination, most (25/32 or 78%) had unmutated disease.
In this cohort of patients with del(13q) CLL, the median time to treatment initiation was significantly shorter in unmutated patients. Further, only a single death occurred in patients with mutated IGHV, while nearly a quarter of the patients with low mutational IGHV died at less than 5 years of median follow-up. Importantly, the aggressive phenotype associated with del(13q) and unmutated IGHV was not limited to those with additional poor-risk chromosomal aberrations; in fact, the median time to treatment and overall survival were similar in unmutated patients with 13q deletion as sole abnormality and in those with additional cytogenetic abnormalities. Although the IGHV mutational status does not change during disease evolution [1], the cut-off for the degree of IGHV sequence homology that separates unmutated and mutated disease is arbitrary [14,19,22]. While the most commonly used cut-off for non-homology is <2%, some authors have used 3% [17] or even 5% [23]. Using these other percentage cut-offs to distinguish between mutated and unmutated disease did not significantly alter our findings (data not shown). When limiting analysis to patients with mutated IGHV, 34/41 (83%) had del(13q)-only disease and 7/41 (17%) suffered additional chromosomal abnormalities. No difference was observed in respect to age at diagnosis, stage at presentation, maximum Rai stage, or ever receiving treatment (Supplementary table).
A retrospective analysis of patients seen at a large tertiary referral facility has the potential for substantial selection bias, including that patients with more aggressive disease may be more likely to be referred. Despite a slightly younger age at diagnosis compared to a community practice, the individuals in this analysis do not represent an advanced, chemotherapy-refractory group, and clearly the patients with mutated IGHV had excellent outcomes. Nearly 80% of the patients had not received prior therapy for CLL, including 76% (29/38) of the unmutated and 80% (33/41) of the mutated patients. Moreover, two-thirds of both the mutated and unmutated patients presented to our center with Rai stage 0 or 1 disease (Tables I and IV).
Although del(13q) in CLL is generally considered to correlate with a favorable prognosis, these findings suggest that this association may only be true for patients with a mutated IGHV: del(13q) as the sole abnormality or in combination with other chromosomal aberrations did not convey an indolent course for our patients with CLL with an unmutated IGHV. These data suggest that both IGHV mutational status and cytogenetics should be assessed in patients with CLL. The ultimate prognostic and therapeutic implications of IGHV mutational status in patients with CLL with a del(13q) require further prospective studies.
Footnotes
Potential conflict of interest: Disclosure forms provided by the authors are available with the full text of this article at www.informahealthcare.com/lal.
References
- 1.Dighiero G. Unsolved issues in CLL biology and management. Leukemia. 2003;17:2385–2391. doi: 10.1038/sj.leu.2403154. [DOI] [PubMed] [Google Scholar]
- 2.Rai KR, Sawitsky A, Cronkite EP, Chanana AD, Levy RN, Pasternack BS. Clinical staging of chronic lymphocytic leukemia. Blood. 1975;46:219–234. doi: 10.1182/blood-2016-08-737650. [DOI] [PubMed] [Google Scholar]
- 3.Montserrat E, Sanchez-Bisono J, Vinolas N, Rozman C. Lymphocyte doubling time in chronic lymphocytic leukaemia: analysis of its prognostic significance. Br J Haematol. 1986;62:567–575. doi: 10.1111/j.1365-2141.1986.tb02969.x. [DOI] [PubMed] [Google Scholar]
- 4.Codony C, Crespo M, Abrisqueta P, Montserrat E, Bosch F. Gene expression profiling in chronic lymphocytic leukaemia. Best Pract Res Clin Haematol. 2009;22:211–222. doi: 10.1016/j.beha.2009.05.006. [DOI] [PubMed] [Google Scholar]
- 5.Dohner H, Stilgenbauer S, Benner A, et al. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med. 2000;343:1910–1916. doi: 10.1056/NEJM200012283432602. [DOI] [PubMed] [Google Scholar]
- 6.Dewald GW, Brockman SR, Paternoster SF, et al. Chromosome anomalies detected by interphase fluorescence in situ hybridization: correlation with significant biological features of B-cell chronic lymphocytic leukaemia. Br J Haematol. 2003;121:287–295. doi: 10.1046/j.1365-2141.2003.04265.x. [DOI] [PubMed] [Google Scholar]
- 7.Shanafelt TD, Witzig TE, Fink SR, et al. Prospective evaluation of clonal evolution during long-term follow-up of patients with untreated early-stage chronic lymphocytic leukemia. J Clin Oncol. 2006;24:4634–4641. doi: 10.1200/JCO.2006.06.9492. [DOI] [PubMed] [Google Scholar]
- 8.Shanafelt TD, Jenkins G, Call TG, et al. Validation of a new prognostic index for patients with chronic lymphocytic leukemia. Cancer. 2009;115:363–372. doi: 10.1002/cncr.24004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crespo M, Bosch F, Villamor N, et al. ZAP-70 expression as a surrogate for immunoglobulin-variable-region mutations in chronic lymphocytic leukemia. N Engl J Med. 2003;348:1764–1775. doi: 10.1056/NEJMoa023143. [DOI] [PubMed] [Google Scholar]
- 10.Marjanovic G, Stanojevic I, Jankovic-Velickovic L, Hubl W, Marjanovic V. Detection of ZAP-70 in patients with chronic lymphocytic leukemia. J BUON. 2008;13:543–546. [PubMed] [Google Scholar]
- 11.Erikci AA, Karagoz B, Ozyurt M, Ozturk A, Kilic S, Bilgi O. HLA-G expression in B chronic lymphocytic leukemia: a new prognostic marker? Hematology. 2009;14:101–105. doi: 10.1179/102453309X385197. [DOI] [PubMed] [Google Scholar]
- 12.Gattei V, Bulian P, DelPrincipe MI, et al. Relevance of CD49d protein expression as overall survival and progressive disease prognosticator in chronic lymphocytic leukemia. Blood. 2008;111:865–873. doi: 10.1182/blood-2007-05-092486. [DOI] [PubMed] [Google Scholar]
- 13.Carlucci F, Marinello E, Tommassini V, Pisano B, Rosi F, Tabucchi A. A 57-gene expression signature in B-cell chronic lymphocytic leukemia. Biomed Pharmacother. 2009;63:663–671. doi: 10.1016/j.biopha.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 14.Hamblin TJ, Davis Z, Gardiner A, Oscier DG, Stevenson FK. Unmutated Ig V(H) genes are associated with a more aggressive form of chronic lymphocytic leukemia. Blood. 1999;94:1848–1854. [PubMed] [Google Scholar]
- 15.Hamblin TJ, Orchard JA, Ibbotson RE, et al. CD38 expression and immunoglobulin variable region mutations are independent prognostic variables in chronic lymphocytic leukemia, but CD38 expression may vary during the course of the disease. Blood. 2002;99:1023–1029. doi: 10.1182/blood.v99.3.1023. [DOI] [PubMed] [Google Scholar]
- 16.Nelson BP, Gupta R, Dewald GW, Paternoster SF, Rosen ST, Peterson LC. Chronic lymphocytic leukemia FISH panel: impact on diagnosis. Am J Clin Pathol. 2007;128:323–332. doi: 10.1309/21TN2RUWKR827UW2. [DOI] [PubMed] [Google Scholar]
- 17.Krober A, Seiler T, Benner A, et al. V(H) mutation status, CD38 expression level, genomic aberrations, and survival in chronic lymphocytic leukemia. Blood. 2002;100:1410–1416. [PubMed] [Google Scholar]
- 18.Van Dyke DL, Shanafelt TD, Call TG, et al. A comprehensive evaluation of the prognostic significance of 13q deletions in patients with B-chronic lymphocytic leukaemia. Br J Haematol. 2010;148:544–550. doi: 10.1111/j.1365-2141.2009.07982.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Damle RN, Wasil T, Fais F, et al. Ig V gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia. Blood. 1999;94:1840–1847. [PubMed] [Google Scholar]
- 20.Kharfan-Dabaja MA, Chavez JC, Khorfan KA, Pinilla-Ibarz J. Clinical and therapeutic implications of the mutational status of IgVH in patients with chronic lymphocytic leukemia. Cancer. 2008;113:897–906. doi: 10.1002/cncr.23671. [DOI] [PubMed] [Google Scholar]
- 21.Trojani A, Montillo M, Nichelatti M, et al. ZAP-70, IgVh, and cytogenetics for assessing prognosis in chronic lymphocytic leukemia. Cancer Biomark. 2010;6:1–9. doi: 10.3233/CBM-2009-0114. [DOI] [PubMed] [Google Scholar]
- 22.Hamblin TJ, Davis ZA, Oscier DG. Determination of how many immunoglobulin variable region heavy chain mutations are allowable in unmutated chronic lymphocytic leukaemia - long-term follow up of patients with different percentages of mutations. Br J Haematol. 2008;140:320–323. doi: 10.1111/j.1365-2141.2007.06928.x. [DOI] [PubMed] [Google Scholar]
- 23.Lin K, Sherrington PD, Dennis M, Matrai Z, Cawley JC, Pettitt AR. Relationship between p53 dysfunction, CD38 expression, and IgV(H) mutation in chronic lymphocytic leukemia. Blood. 2002;100:1404–1409. doi: 10.1182/blood-2001-11-0066. [DOI] [PubMed] [Google Scholar]