Abstract
The protein microarray technology provides a versatile platform for characterization of hundreds of thousands of proteins in a highly parallel and high-throughput manner. It is viewed as a new tool that overcomes the limitation of DNA microarrays. On the basis of its application, protein microarrays fall into two major classes: analytical and functional protein microarrays. In addition, tissue or cell lysates can also be directly spotted on a slide to form the so-called “reverse-phase” protein microarray. In the last decade, applications of functional protein microarrays in particular have flourished in studying protein function and construction of networks and pathways. In this chapter, we will review the recent advancements in the protein microarray technology, followed by presenting a series of examples to illustrate the power and versatility of protein microarrays in both basic and clinical research. As a powerful technology platform, it would not be surprising if protein microarrays will become one of the leading technologies in proteomic and diagnostic fields in the next decade.
Keywords: Protein microarray, antibody microarray, reverse-phase protein microarray, protein posttranslational modification, networks, biomarkers, serum profiling
I. Introduction
The concept of microarray technology was first put forward by Ekins (1989)over 20 years ago. An ambient analyte theory was proposed that a tiny spot of a purified antibody or protein provides substantially better sensitivity than when used in conventional immunoassay formats as miniaturized features can dramatically enhance detection sensitivity. Though not exactly the same, DNA microarray technology became the first application of this theory and has been tremendously successful in gene expression profiling and other derivatized applications, such as ChIP-chip (DeRisi et al., 1997, Morley et al., 2004, Pease et al., 1994, Schadt et al., 2003, Schena et al., 1995). However, RNA expression levels do not always correlate with protein expression levels, and biological functions are carried out primarily by proteins rather than nucleic acids (Gygi et al., 1999, Lueking et al., 2005b). Therefore, it was the next logical step to develop a miniaturized protein-centered device, namely protein microarrays, for studies of protein functionalities in a high-throughput and highly flexible fashion.
A protein microarray, also known as a protein chip, is formed by immobilization of thousands of different proteins (e.g., antigens, antibodies, enzymes, substrates, etc.) in discrete spatial locations at a high-density solid surface (typically glass) (Smith et al., 2005, Tao et al., 2007). On the basis of their applications, protein microarrays can be classified into two types: analytical and functional protein microarrays (Fig. 4.1 ). Analytical protein microarrays are usually composed of well-characterized biomolecules with specific binding activities, such as antibodies, to analyze the components of complex biological samples (e.g., serum and cell lysates) or to determine whether a sample contains a specific protein of interest. They have been used for protein expression profiling, biomarker identification, cell surface marker/glycosylation profiling, clinical diagnosis, and environmental/food safety analysis (Kumble, 2003). On the other hand, functional protein microarrays are constructed by printing a large number of individually purified proteins and are mainly used to comprehensively query biochemistry properties and activities of those immobilized proteins. In principle, it is feasible to print arrays composed of virtually all annotated proteins of a given organism, effectively comprising a whole-proteome microarray. Functional protein microarrays have been successfully applied to identify protein–protein, protein–lipid, protein–antibody, protein–small molecules, protein–DNA, protein–RNA, lectin–glycan, and lectin–cell interactions, and to identify substrates or enzymes in phosphorylation, ubiquitylation, acetylation, and nitrosylation, as well as to profile immune response. In this chapter, we will mainly focus on the fabrication and application of functional protein microarrays in basic and clinical research.
Figure 4.1.
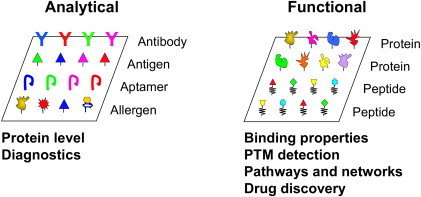
Classification of protein microarrays.
Protein microarrays are of two types: analytical and functional protein microarrays. Left: Analytical protein microarrays are constructed using biomolecule with specific binding property, such as antibodies, antigens, and aptamers. Right: Functional protein microarrays are formed by immobilization of individually purified proteins or synthetic peptides. The major applications of both types are listed below. For color version of this figure, the reader is referred to the online version of this book.
II. Fabrication of Functional Protein Microarrays
Because the biochemical properties of DNA molecules are essentially the same, the same chemistries can be applied to either immobilize to or synthesize in situ DNA strands on a solid surface (DeRisi et al., 1997, Pease et al., 1994). Therefore, the design and construction of oligonucleotide DNA microarrays are relatively straightforward. However, the protein world is manifested by much more complicated biochemistries, which are reflected by vast differences in protein size, shape/conformation, charge, stability, and hydrophobicity, to name a few. Furthermore, many proteins are known to require proper partners to be able to execute their biochemical activities. This implies that the fabrication and analysis of protein microarrays is substantially more challenging than that of DNA microarrays. Unlike DNA or RNA molecules, full-length proteins cannot be directly synthesized in vitro at high efficiency. Although in vitro synthesis of peptides has been feasible for decades, it still suffers from low yield, high cost, and effective limitation to short sequences. Moreover, the vast majority of proteins must be correctly folded and modified to be functional during and after translation, which may require a complex molecular machinery of chaperones and other accessory molecules that cannot be fully recapitulated in vitro. Therefore, the development of a high-throughput method that allows for purifying proteins under native conditions is the key to fabricate functional protein microarrays of high content.
A. High-Throughput Protein Production
Although many methods have been developed to purify proteins from both eukaryotic and prokaryotic systems, the main hurdle has been the difficulty in producing a large number of different proteins needed for construction of a truly high-content, functional protein microarray. Obviously, a readily useable high-throughput protocol for parallel production of thousands of different proteins is the key to this challenge.
An early attempt led by the Lehrach group was to express human proteins in Escherichia coli using a library consisting of random cDNAs (Bussow et al., 1998). Individual cDNA clones of this library were robotically arrayed onto polyvinylidene difluoride (PVDF) membrane laid on top of agar media and allowed to grow to full size. These cells were then lysed in situ to extract proteins. The usefulness of such an array was first demonstrated by incubation with a labeled test protein to identify interacting partners (Bussow et al., 1998). Strictly speaking, besides the fact that only one-sixth of the cDNAs are in the proper reading frames, those correctly expressed human proteins bound to the nitrocellulose membranes were not purified—the majority of the proteins in every spot were bacterial proteins. Furthermore, the proteins were neither unique nor in their native conformation, given the redundancy of the library and denaturing conditions used to break the bacteria open. Though powerful as a screening technique in early days, this particular experimental strategy had limited general applications (Holt et al., 2000, Lueking et al., 1999).
To overcome these hurdles, Zhu et al. (2001) in the Snyder group created a high-throughput protein purification protocol in the budding yeast (Fig. 4.2 ). Using a homologous recombination-based strategy, more than 5800 full-length yeast open reading frames (ORFs) were cloned into a yeast expression vector that, upon galactose induction, produces glutathione S-transferase (GST)-tagged N-terminal fusion proteins. The purification protocol took advantage of both a 96-well format and immobilized affinity chromatography. This strategy allowed parallel purification of unprecedented numbers of proteins—up to 1152 per day. The success of this approach is built upon several unique aspects. First, it utilizes a eukaryotic expression system that both generates high levels of recombinant proteins and tends to produce a high fraction of soluble proteins. Compared with bacterial expression systems, in which a large fraction of recombinant proteins end up in inclusion bodies, this is a huge advantage when a large number of eukaryotic proteins are being generated. Second, the expression of recombinant proteins is only induced over about two total cell cycles, which greatly reduces toxicity and cell death. Third, a foreign eukaryotic protein purified from yeast is more likely to be active because posttranslational modifications necessary for function are more likely to occur correctly than in either bacteria or a cell-free system. Forth, the use of an N-terminal GST tag helps protein fold correctly and therefore improve its stability and solubility. Other commonly used tags include the so-called TAP tag, MPB, and Hisx6, to name a few. In fact, the same group later went on to build a TAP-tagged yeast ORF collection and purified >5,000 yeast proteins (Gelperin et al., 2005).
Figure 4.2.
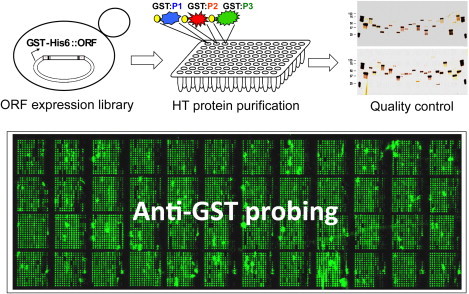
Fabrication of high-content functional protein microarrays.
Four major steps are involved to construct a functional protein microarray of high content. First, a high-quality ORF expression library is constructed to allow inducible overexpression of GST-His6 fused recombinant proteins in yeast. Second, a high-throughput protein purification protocol is applied to individually purify thousands of proteins from yeast. The purified proteins are stored in 384-well format. Third, silver stain and immunoblot analysis are employed to evaluate the quality and quantity of the purified proteins. Finally, when purified proteins pass the quality control, they are spotted in duplicate to glass slides using a robot microarrayer. The quality of printing is then tested by anti-GST probing. An image of a human protein microarray probed with anti-GST is shown in the lower panel. For color version of this figure, the reader is referred to the online version of this book.
Another commonly used expression system is E. coli. The procedures for automatic high-throughput protein expression/purification using the 6xHis tag have been developed by the Zhu group (Chen et al., 2008). Once the bacterial culture is prepared, all 4000 proteins can be purified within a single day. The subsequent protein purification takes advantage of immobilized Ni-NTA affinity chromatography (Hochuli et al., 1987). The 6xHis tag usually does not alter the properties of the fusion proteins, and the increment of molecular weight is less than 1 kDa. Furthermore, it is selective and stable even under severe denaturing conditions (Joshi et al., 2000, Mukhija et al., 1995).
Despite the fact that high-throughput protein production in both prokaryotes and eukaryotes is now increasingly feasible, these protocols are labor intensive and costly. Aside from the cost of protein production, fabrication of a proteome microarray requires construction of an expressible collection of full-length ORFs, which can be both challenging and expensive when dealing with higher eukaryotes with a large number of genes, such as humans. To explore alternative approaches, several groups have attempted to test the in vitro transcription/translation systems, such as the E. coli, wheat germ, and rabbit reticulocyte systems. In these systems, proteins can be expressed directly from cDNA templates (Allen and Miller, 1999), which can be obtained through polymerase chain reaction amplification without the lengthy and costly process of subcloning. For example, the E. coli cell-free protein expression system has been used to synthesize proteins in a 96-well format (Murthy et al., 2004), and the improved wheat germ cell-free protein synthesis system has been applied to the in vitro expression of 13,364 human proteins (Goshima et al., 2008). More recently, the Felgner group has published a series of articles describing fabrication of protein microarrays in a variety of bacteria by directly spotting in vitro translated protein mixtures to glass (Crompton et al., 2010, Liang et al., 2011). However, although these systems can significantly decrease the reaction volume required for generation of recombinant proteins (Angenendt et al., 2004), the impurity of the translated proteins limits their applications.
Such systems can also be applied to directly synthesize proteins on glass slides to fabricate so-called “in situ protein microarrays.” In the Protein In Situ Array (PISA) method, proteins are expressed directly from DNA in vitro and become attached to the array surfaces through recognition of a sequence that serves as an affinity tag (He and Taussig, 2001). Similarly, in the Nucleic Acid Programmable Protein Array (NAPPA) technology, biotinylated cDNA plasmids encoding proteins as GST fusions are printed onto avidin-coated slides, together with anti-GST antibodies as the capture molecules (Ramachandran et al., 2004). The cDNA array is then incubated with rabbit reticulocyte lysate to express the proteins, which become trapped by the antibodies adjacent to each DNA spot. Recently, NAPPA has been successfully expanded to high-density arrays of 1000 different proteins (Ramachandran et al., 2008). In addition, Tao and Zhu (2006) developed a different method in which ribosomes are installed at the end of an RNA template to allow for the capture of the nascent polypeptides by a puromycin moiety that is grafted at one end of an oligonucleotide immobilized on a solid surface.
Another similar method is called DNA Array to Protein Array (DAPA), in which proteins are synthesized between two glass slides: one of which is arrayed with DNA, whereas the other carries a specific affinity reagent to capture the proteins (He et al., 2008). In this approach, tagged proteins are synthesized in parallel from the DNA array, spread across the gap between the two slides, and then bound to the tag-capturing reagents on the other slide to form a protein array. Unlike the NAPPA method in which proteins are present together with DNA and the DNA array can only be used once, DAPA generates multiple copies of “pure” protein arrays on a separate surface from the same DNA template, with at least 20 copies capable of being produced from a single template.
Because proteins must fold correctly in order to be active and because proteins are prone to inactivation due to loss of their native conformations (e.g., exposure to denaturing conditions during purification), it is better to express proteins of interest in cells and purify them under native conditions.
B. Surface Chemistry
Choosing a proper surface for protein immobilization is crucial to the success of any assay performed using protein microarrays. An ideal surface should be able to retain protein functionality with relatively high signal-to-noise ratios and possess both high protein-binding capacity and long shelf life (Smith et al., 2005, Tao et al., 2007). Glass slides covered with PVDF, nitrocellulose membrane, or polystyrene were popular for protein microarray fabrication in the early days of the technology (Bussow et al., 1998, Holt et al., 2000, Lueking et al., 1999). However, PVDF and polystyrene are relatively soft, allowing lateral spread of printed proteins, and hence limited density of proteins to be printed. Nitrocellulose membranes, in addition, tend to generate high background and low signal-to-noise ratio for most applications.
To bypass these shortcomings, researchers developed three-dimensional matrix arrays, in which glass slides are coated with polyacrylamide or agarose to form a porous hydrophilic matrix in which proteins or antibodies are trapped within the pores and lateral diffusion is restricted, reducing the size of printed protein spots and thus increasing the maximal complexity of the array (Afanassiev et al., 2000, Guschin et al., 1997). Protein activity is generally well preserved in such matrix arrays, and their protein binding capacity is relatively high. For instance, Zhu et al. (2000) utilized soft lithography to generate nanowells on a polydimethylsiloxane sheet placed on top of microscope slides. These nanowell chips were used to immobilize substrate proteins to profile phosphorylation specificity of 119 kinases encoded by budding yeast. The open structure of nanowells provides physical barriers and allows for sequential adding of different buffers, which is critical for multistep experiments. The main disadvantage of this method is the requirement of specialized equipment needed to load nanowells at high density.
Other researchers printed proteins, antigens, or antibodies directly onto plain glass slides, which are usually coated with a bifunctional cross-linker with two functional groups, one reacting with the glass surface and the other with the desired proteins. For example, Schweitzer et al. (2000) demonstrated in their study that protein microarrays fabricated on glass surface possess high sensitivities, wide dynamic range, and decent spot-to-spot reproducibility. MacBeath and Schreiber (2000) demonstrated with three proteins that thousands of protein spots could be immobilized to aldehyde-activated plain glass surfaces to form a high-density protein microarray that was suitable for a range of different classes of assays.
C. Protein Immobilization
The physical and chemical properties of different proteins vary greatly, and protein activities are closely related to their structures. Therefore, the development of a stable universal immobilization method that does not change protein structures is one of the difficulties of protein microarray fabrication. To this end, several different methods have been used for protein immobilization on solid carrier surfaces, such as noncovalent adsorption, covalent binding, and affinity capture.
Noncovalent adsorption provides both high protein capacities and low impact on protein structures but cannot control the amount and orientation of immobilized proteins. Thus, the reaction efficiency, accuracy, and reproducibility of arrays produced in this manner are variable. Covalent binding, on the other hand, results in chemically cross-linked proteins via reactive residues (e.g., lysine and cystine) to surface-grafted ligands, such as aldehyde, epoxy, reactive ester, etc. (MacBeath and Schreiber, 2000, Templin et al., 2002, Ziauddin and Sabatini, 2001). Lee et al. (2003) developed novel calix crown derivatives as a ProLinker that permits efficient immobilization of captured proteins on solid matrixes, and the immobilized proteins showed both consistent directionality and functionality. Covalent binding is suitable for immobilization of a wide range of proteins with strong conjunctions to the carrier surfaces. However, the modification of chemical groups can sometimes both alter the activities of target proteins and their binding to specific ligands.
Affinity capture is an attractive way to immobilize proteins that avoid many of the shortcomings of the previously detailed approaches. For example, biotinylated proteins have been used for protein immobilization to streptavidin-coated slides. The use of genetically encoded affinity tags, which can be fused to target proteins and bind to a specific slide surface, is an analogous approach. For example, 6xHis-tags have been utilized to immobilize proteins on nickel-NTA-coated glass slides (Zhu et al., 2001). Presumably, affinity-based protein immobilization should result in immobilization of proteins in relatively uniform orientation with minimum interruption of protein structure and thus, may be the best approach to for preserving the structure and function of printed proteins. One important caveat to bear in mind, however, is that the incorporation of affinity tags may alter the protein structures.
One way to deal with this challenge was demonstrated by Zhang et al. (2005), who developed a flexible polypeptide scaffold consisting of a surface immobilization domain and a protein capture domain, which allows much greater flexibility in the immobilization of proteins on a microarray. Wacker et al. (2004) compared the DNA-directed immobilization (DDI) method with both direct spotting and with biotin–streptavidin affinity immobilization for antibodies. DDI is based on the self-assembly of semisynthetic DNA–streptavidin conjugates that convert a DNA oligomer array into an antibody array (Niemeyer et al., 1994). DDI and direct spotting showed the highest fluorescence intensities. DDI also performed the best in spot homogeneity and intra- and interexperimental reproducibility. Moreover, DDI required the lowest amount of antibodies, at least 100-fold less than direct spotting. The drawback of DDI is that proteins have to be linked to DNA prior to immobilization, which increases the workload involved in generating microarrays.
The orientation of immobilized proteins may influence both their activity and their affinity for specific ligands. Peluso et al. (2003) compared randomly versus specifically oriented capture agents based on both full-sized antibodies and Fab’ fragments. The specific orientation of capture agents consistently increased the analyte-binding capacity of the surfaces up to 10-fold relative to surfaces with randomly oriented capture agents. When specifically oriented, Fab’ fragments formed a dense monolayer and 90% of them were active, whereas randomly attached Fab’s both packed at lower density and had lower specific activity.
III. Signal Detection
In addition to optimized surface modification and optimized reaction condition, the detection sensitivity of samples bound on microarrays is another key parameter in the design of protein microarray assays. There are two basic detection methods: label-dependent and label-free detections.
A. Label-Dependent Detection Methods
Radioisotopes and fluorescent dyes are the two most common labeling methods for signal detection in protein microarray assays. Fluorescent dyes, such as Cy-3/5 and their equivalent, have been used as a popular labeling method. Because most good dyes have relatively narrow excitation and emission spectra, multicolor scheme can be readily implemented for simultaneous detection and direct comparison of different samples, both reducing cost and avoiding chip-to-chip variation. Semiconductor quantum dot labeling, which is brighter and more stable than organic dyes, has also been applied to protein microarrays (Shingyoji et al., 2005, Zajac et al., 2007).
In addition to fluorescent labeling, Huang (2001) detected multiple cytokines on an antibody array with enhanced chemiluminescence, providing an alternative detection method. Enzymatic signal amplification is also a valuable labeling method. Rolling circle amplification (RCA) has been developed for protein microarray assays. For low abundance protein samples, the sensitivity of traditional fluorescence or chemiluminescence detection is relatively low, whereas RCA can detect captured proteins at femtomole level and is promising to improve the sensitivity of fluorescent detection (Lizardi et al., 1998, Schweitzer et al., 2000, Schweitzer et al., 2002, Shao et al., 2003, Zhou et al., 2004). Tyramide signal amplification is another way to amplify signals with enzymes, which utilizes the horseradish peroxidase conjugated on secondary antibodies to convert the labeled substrates (tyramide) into short-lived extremely reactive intermediates, which then very rapidly react with and covalently bind to adjacent proteins (Varnum et al., 2004).
For certain types of biochemical assays, especially enzymatic reactions, use of radioisotopes is the only detection method available (see below for more details). They still offer the most sensitive and reliable detection of posttranslation modification events when there is a lack of high-quality and high-affinity detection reagents, such as antibodies. We and others have successfully applied 32P-, 33P-, and 14C-labeled substrates to detect protein phosphorylation and acetylation events (Lin et al., 2009, Lu et al., 2011, Ptacek et al., 2005, Zhu et al., 2009).
B. Label-Free Detection Methods
One obvious disadvantage of label-dependent detection is the requirement of either manipulating structure of a probe or a specific antibody. It is not amenable to real-time label-free detection, which can provide important information when analyzing reaction dynamics. Therefore, label-free detection methods have also been investigated for protein microarrays. Optical techniques of various types are emerging as an important tool for mentoring the dynamics of biomolecule interactions on a solid surface. For instance, Imaging Surface Plasmon Resonance Spectroscopy (SPR) (Nelson et al., 1999, Thiel et al., 1997), Imaging Optical Ellipsometry (OE) (Wang and Jin, 2003), and Reflectometric Interference Spectroscopy (Piehler et al., 1997) are three label-free optical techniques that in essence measure the same optical dielectric response of a thin film and therefore detect changes of physical or chemical properties of the thin film, such as thickness and mass density during biochemical reactions.
As compared with the above three methods, the oblique-incidence reflectivity difference (OIRD) technique is a more sensitive form of ellipsometry that measures the difference in reflectivity between s- and p-polarized light (Chen et al., 2001, Landry et al., 2004). Recently, the OIRD technique has been applied to detect DNA hybridization and protein–protein interactions in a microarray format in a real-time fashion, and these studies demonstrated its potential as a high-throughput detection method that can obtain association and dissociation rates of biomolecule interactions (Lu et al., 2010, Wang et al., 2010). This is an extremely sensitive detection method: it has a time resolution of 20 μs, a space resolution (i.e., thickness) of 0.4 nm, and a detection limit of 14 fg of protein per spot. In addition, it also shares other advantages of the SPR and OE methods, such as noncontacting damage- and label-free detection (Fei et al., 2008, Lu et al., 2010).
The principle of OIRD-based detection is illustrated in Fig. 4.3 . First, a p-polarized He-Ne laser beam (λ = 632.8 nM) passes through a photoelastic modulator to induce oscillation between p- and s-polarization at a frequency of 50 kHz. Second, after passing through a phase shifter, the resultant beam is incident on the microarray surface at an oblique angle theta (θ inc). Finally, the first I(Ω) and second harmonics I(2Ω) of the reflected beam intensity are simultaneously monitored by two digital lock-in amplifiers (Lu et al., 2010). The difference caused by changes in reflectivity between the s- and p-polarized light, namely the OIRD signal, is Δp − Δs, composed of both real and imaginary components. Because the imaginary component, which is proportional to the first I(Ω), is more sensitive, the OIRD signal is determined as “Im{Δp − Δs}” (Formula I), which is dependent on the incident angle (θ inc) and the dielectric constants of the ambient, protein, and substrate of the microarray (i.e., glass) (Wen et al., 2010).
Figure 4.3.
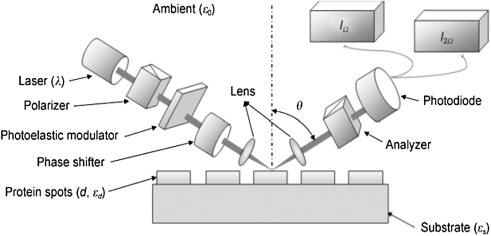
Principle of the OIRD method.
First, a p-polarized He-Ne laser beam (λ = 632.8 nM) passes through a photoelastic modulator to induce oscillation between p- and s-polarization at a frequency of 50 kHz. Second, after passing through a phase shifter, the resultant beam is incident on the microarray surface at an oblique angle theta (θinc). Finally, the first I(Ω) and second harmonics I(2Ω) of the reflected beam intensity are simultaneously monitored by two digital lock-in amplifiers.
Formula I
| (1) |
The OIRD system is attractive in several ways. First, it was first designed in the format of microarray and, therefore, readily to be applied to protein microarray assays. Second, it is promising to be developed as an extremely high-throughput method as it has already been able to detect approximately 10,000 protein spots at once (Lv et al., personal communication). Third, unlike the SPR system, it is not restricted to a particular surface type, which makes it much more flexible for various types of biochemical assays.
Finally, mass spectrometry has also been used for detecting ligands bound to individual proteins printed on protein microarrays, with such approaches as MALDI-MS, SELDI-TOF-MS, and MALDI-TOF-MS used for this purpose (Diamond et al., 2003, Evans-Nguyen et al., 2008, Gavin et al., 2005). The analysis is rapid and simple, requires small sample amount, and can be used for direct detection of analytes bound from complex samples, such as urine, serum, plasma, and cell lysates. Atomic force microscopy (AFM) uses surface topological changes to identify the analytes bound on the array (Lee et al., 2002, Yan et al., 2003). More specifically, AFM detects the increase in height of the proteins/antibodies on the array and thus is able to measure binding interactions.
IV. Applications of Functional Protein Microarrays
A. Development of new Assays
Unlike the DNA/oligo microarray or analytical protein microarrays, functional protein microarrays provide a flexible platform that allows development and detection of a wide range of protein biochemical properties. To date, well-developed assays include detection of various types of protein–ligand interactions, such as protein–protein, protein–DNA, protein–RNA, protein–lipid, protein–drug, and protein–glycan interactions (Chen et al., 2008, Hall et al., 2004, Ho et al., 2006, Hu et al., 2009, Huang et al., 2004, Kung et al., 2009, MacBeath and Schreiber, 2000, Popescu et al., 2007, Zhu et al., 2001, Zhu et al., 2007), and identification of substrates of various classes of enzymes, such as protein kinase, ubiquitin E3 ligase, and acetyltransferase, to name a few(Lin et al., 2009, Lu et al., 2008, Ptacek et al., 2005, Schnack et al., 2008, Zhu et al., 2000).
During the development of various assay types, it became obvious that surface chemistry plays an important role in the success of a new assay (Table 4.1 ). For example, protein–DNA interactions were first performed on the yeast proteome microarrays on a nitrocellulose surface (i.e., FAST slide) with randomly shared yeast genomic DNA fragments that were labeled with Cy5 (Hall et al., 2004). Later, Hu et al. (2009) also found that the FAST slide, among other tested surfaces, produced the best signal-to-noise ratio for DNA-binding assays. In another example, when our group was developing protein acetylation reactions using 14C-labeled Ac-CoA as a donor, we first tested the NuA4 acetylation reaction using histone H3 and H4 as substrates on FAST slides, as well as aldehyde- and Ni-NTA-coated slides (Lin et al., 2009, Lu et al., 2011). The results were clear that both FAST and nickel surfaces worked, but FAST surface produced better signal-to-noise ratios (Fig. 4.4 ). However, FAST surface was not suitable for phosphorylation reactions because the background noises were too high (Ptacek et al., 2005, Zhu et al., 2009). Another interest case is to develop an assay for profiling cell surface glycans on a lectin microarray. We and others found that so far the only proper surface for this type of binding assays is a commercial Schott slide, although the exact surface chemistry is not revealed (Hsu et al., 2006, Pilobello et al., 2007, Tao et al., 2008). Several reasons may be accounted for the importance of surface chemistry. First, for low-affinity binding assays (e.g., protein–DNA interactions), a porous surface (e.g., FAST) is likely to retain more proteins and hence improving sensitivity. Second, when radioisotope-labeled small molecules are used, it is important to completely remove unincorporated radioisotopes from the surface to reduce background noise. This might explain why phosphorylation does not work well on FAST surface. Third, in the case of using live cells to probe a lectin microarray, the grafted chemical ligands must not be too repulsive to cells. Other factors, such as protein conformation and stability, can also be affected by surface chemistry. Therefore, whenever a novel assay is to be developed, a variety of surfaces should be tested first in a pilot study.
Table 4.1.
Effects of surface chemistry to protein microarray assays
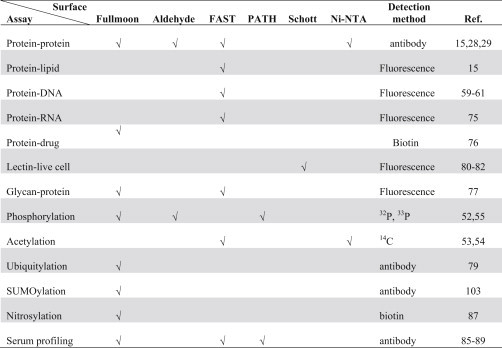 |
Figure 4.4.
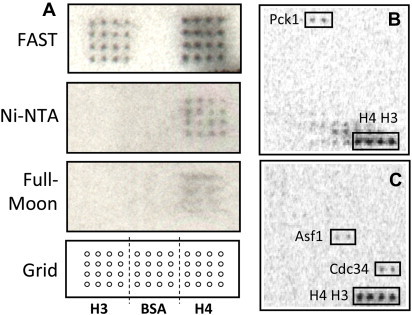
Effects of surface chemistry on assay development.
(A) A pilot experiment to optimize the reaction conditions for protein acetylation in a microarray format. In each reaction, the yeast NuA4 acetyltransferase complex was added to an acetylation reaction mixture containing 14C-Ac-CoA and incubated with histone H3 and H4 spotted on three different surfaces. Bovine serum albumin (BSA) was also included as a negative control. The acetylation signals were detected by long exposure to X-ray film. (B, C) Examples of newly identified non-histone substrates. For color version of this figure, the reader is referred to the online version of this book.
Application of these assays has had a profound impact on a wide range of research areas. This is especially true when they are used in large-scale high-throughput projects, exemplified in both network construction and biomarker identification (see below and Table 4.2 ).
Table 4.2.
Application of functional protein microarrays in large-scale projects
| Type of assay | Type of array | Type of probe | No. of probe | Application | Reference |
|---|---|---|---|---|---|
| Protein–peptide interaction | Human SH2 and PTB domain array | Peptide | 61 | Protein interaction network | (Jones et al., 2006) |
| Protein–DNA interaction | Yeast TF array | DNA motif | 75 | Protein–DNA interaction Network | (Ho et al., 2006) |
| Human TF array | DNA motif | 460 | Protein–DNA interaction Network | (Hu et al., 2009) | |
| Kinase assay | Yeast proteome array | Protein kinase | 87 | Phosphorylation network | (Ptacek et al., 2005) |
| Antigen–antibody interaction | Coronavirus array | SARS patient sera | 602 | Biomarker identification | (Hu et al., 2007) |
| E. coli proteome array | Inflammatory bowel disease (IBD) patient sera | 134 | Biomarker identification | (Xie et al., 2010) | |
| Human protein array | Ovarian cancer patient sera | 60 | Biomarker identification | (Jones et al., 2006) | |
| Human protein array | Alopecia areata patient sera | 44 | Biomarker identification | (Foster et al., 2009) | |
| Human protein array | Autoimmune hepatitis (AIH) patient sera | 278 | Biomarker identification | (Robinson et al., 2002) |
B. Detection of Protein-Binding Properties
1. Protein–protein interaction
Among the first applications of protein microarrays was in the analysis of protein–protein and protein–lipid interactions, where test ligands were directly or indirectly labeled with fluorescent dyes. For example, Zhu et al. (2001) developed the first proteome microarray composed of approximately 5,800 recombinant yeast proteins (>85% of the yeast proteome) and identified binding partners of calmodulin and phosphatidylinositides (PIPs). They first incubated the microarrays with biotinylated bovine calmodulin and discovered 39 new calmodulin binding partners. In addition, using liposomes as a carrier for various PIPs, they identified more than 150 binding proteins, more than 50% of which were known membrane-associated proteins. Popescu et al. (2007) developed a protein microarray containing 1,133 Arabidopsis thaliana proteins and also used it to globally identify proteins bind to calmodulins or calmodulin-like proteins in Arabidopsis. A large number of previously known and novel targets were identified, including transcription factors (TFs), receptor and intracellular protein kinases, F-box proteins, RNA-binding proteins, and proteins of unknown function. Alternative approaches to identifying protein–protein interactions, such as the yeast two-hybrid system and protein complex purification coupled with mass spectrometry analysis, are well-established, however, and are used as standard high-throughput methods to detect protein–protein interactions in higher eukaryotes (Krogan et al., 2006, Vidal et al., 1996). Thus, while protein microarray-based approaches provide a rapid approach to characterizing protein–protein interactions, they have much competition in this arena.
2. Protein–peptide interaction
MacBeath et al. fabricated protein domain microarrays to investigate protein–peptide interactions in a semiquantitative fashion that might play an important role in signaling (Jones et al., 2006). They constructed an array by printing 159 human Src homology 2 (SH2) and phosphotyrosine binding (PTB) domains on the aldehyde-modified glass substrates and incubated the arrays with 61 peptides representing tyrosine phosphorylation sites on the four ErbB receptors. Eight concentrations of each peptide (10–5 mM) were tested in the assay, allowing quantitative measurement of the binding affinity of each peptide to its protein ligand.
3. Protein–DNA interaction
Protein microarrays have also been applied extensively and productively to characterize protein–DNA interactions (PDIs). In an earlier study, Snyder et al. screened for novel DNA-binding proteins by probing the yeast proteome microarrays with fluorescent labeled yeast genomic DNA (Hall et al., 2004). Of the approximately 200 positive proteins, half were not previously known to bind to DNA. By focusing on a single yeast gene, ARG5,6, encoding two enzymes involved in arginine biosynthesis, they discovered that it bound to a specific DNA motif and associated with specific nuclear and mitochondrial loci in vivo.
In a later report, the Snyder and Johnston groups constructed a protein microarray with 282 known and predicted yeast TFs to identify their interactions with 75 evolutionarily conserved DNA motifs (Ho et al., 2006). Over 200 specific PDIs were identified, and more than 60% of them are previously unknown. The binding site of a previously uncharacterized DNA-binding protein, Yjl103p, was defined, and a number of its target genes were identified, many of which are involved in stress response and oxidative phosphorylation.
Our team developed a bacterial proteome microarray composed of 4,256 proteins encoded by the E. coli K12 strain (approximately 99% coverage of the proteome) using a bacterial high-throughput protein purification protocol (Chen et al., 2008). To demonstrate the usefulness, end-labeled double-stranded DNA probes carrying a basic or mismatched base pairs were used to identify proteins involved in DNA damage recognition. A small number of proteins were specifically recognized by each type of the probes with high affinity. Two of them, YbaZ and YbcN, were further characterized to encode base-flipping activity using biochemical assays.
Recently, our group also undertook a large-scale analysis of human PDIs using a protein microarray composed of 4,191 unique human proteins in full length, including approximately 90% of the annotated TFs and a wide range of other protein categories, such as RNA-binding proteins, chromatin-associated proteins, nucleotide-binding proteins, transcription coregulators, mitochondrial proteins, and protein kinases (Hu et al., 2009). The protein microarrays were probed with 400 predicted and 60 known DNA motifs, and a total of 17,718 PDIs were identified. Many known PDIs and a large number of new PDIs for both well-characterized and predicted TFs were recovered, and new consensus sites for over 200 TFs were determined, which doubled the number of previously reported consensus sites for human TFs (Hu et al., 2009, Xie et al., 2010). Surprisingly, over 300 proteins that were previously unknown to specifically interact with DNA showed sequence-specific PDIs, suggesting that many human proteins may bind specific DNA sequences as a moonlighting function. To further investigate whether the DNA-binding activities of these unconventional DNA binding proteins (uDBPs) were physiologically relevant, we carried out in-depth analysis on a well-studied protein kinase, Erk2, to determine the potential mechanism behind its DNA-binding activity. Using a series of in vitro and in vivo approaches, such as electrophoretic mobility shift assay (EMSA), luciferase assay, mutagenesis, and chromatin immunoprecipitation (ChIP), we demonstrated that the DNA-binding activity of Erk2 is independent of its protein kinase activity and it acts as a transcription repressor of transcripts induced by interferon gamma signaling (Hu et al., 2009). Other than Erk2, many other uDBPs show sequence-specific DNA-binding activity, and more intriguingly, many of their consensus sequences are highly similar to those recognized by annotated TFs (Fig. 4.5 ). This observation suggests that these uDBPs may synergistically work with the TFs to achieve highly accurate transcription regulation.
Figure 4.5.
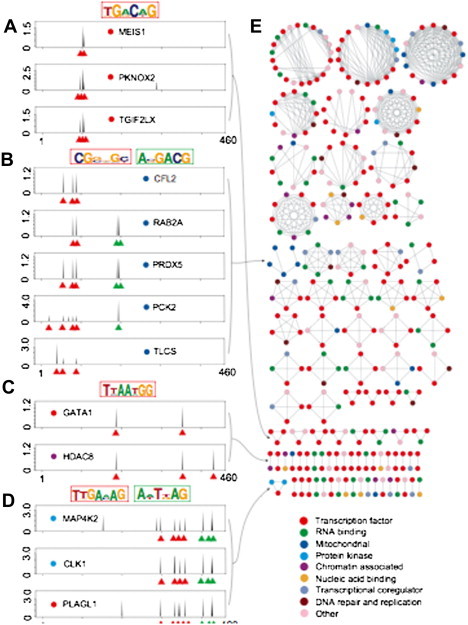
Similar consensus sites are recognized by both TFs and uDBPs.
(A–D) Examples of proteins sharing similar DNA binding profiles. Each peak represents normalized signal intensity of a specific DNA motif probe, with individual motifs organized along the X-axis by sequence similarity. Binding peaks used to generate the major logos (outlined in red) are indicated by red triangles. For proteins that recognize more than one logo (outlined in blue), binding peaks for the second logo are indicated in blue. (E) Correlation network for proteins with highly similar DNA binding profiles (see Supplementary Methods for construction of the network). Protein class is indicated by colored dots. For interpretation of the references to color in this figure legend, the reader is referred to the online version of this book.
4. Protein–Small molecule interaction
Discovering new drug molecules and drug targets is another field in which protein microarrays have shown its potential. For example, Huang et al. (2004) incubated biotinylated small-molecule inhibitors of rapamycin (SMIRs) on the yeast proteome microarrays and obtained the binding profiles of the SMIRs across the entire yeast proteome. They identified candidate target proteins of the SMIRs, including Tep1p, a homologue of the mammalian PTEN tumor suppressor, and Ybr077cp (Nir1p), a protein of previously unknown function, both of which are validated to associate with PI(3,4)P2, suggesting a novel mechanism by which phosphatidylinositides might modulate the target of rapamycin (TOR) pathway.
5. Protein–RNA interaction
The yeast proteome microarray has been used to identify specific RNA-binding proteins for antiviral activities (Zhu et al., 2007). In these experiments, arrays were incubated with a fluorescently tagged small RNA hairpin containing a clamped adenine motif, which is required for the replication of Brome Mosaic Virus (BMV), a plant-infecting RNA virus that can also replicate in the budding yeast. Two of the candidate proteins, Pseudouridine Synthase 4 (Pus4) and the Actin Patch Protein 1 (App1), were further characterized in Nicotiana benthamiana. Both of them modestly reduced BMV genomic plus-strand RNA accumulation and dramatically inhibited the spread of BMV in plants.
6. Protein–Glycan interaction
Protein glycosylation, a general posttranslational modification of proteins involved in cell membrane formation, is crucial to dictate proper conformation of many membrane proteins, retain stability on some secreted glycoproteins, and play a role in cell–cell adhesion. To further understand the roles of protein glycosylation in yeast, the Zhu and Snyder groups profiled yeast protein glycosylation on a yeast proteome microarray using fluorescently labeled lectins, such as Concanavalin A (ConA) and Wheat-Germ Agglutinin (WGA) (Kung et al., 2009). This experiment was based on the assumption that yeast proteins purified from their original host should maintain most of their PTMs. A total of 534 proteins were identified, 406 of which were previously not known to be glycosylated. Many proteins in the secretory pathway were identified, as well as other functional classes of proteins, including TFs and mitochondrial proteins. Upon treatment with tunicamycin, an inhibitor of N-linked protein glycosylation, two of the four mitochondrial proteins identified showed partial distribution to the cytosol and reduced localization to the mitochondria, suggesting a new role of protein glycosylation in mitochondrial protein function and localization.
C. Protein Posttranslational Modifications
Protein posttranslational modifications (PTMs) are one of the most important mechanisms to regulate protein activities. Among hundreds of PTMs identified so far, the reversible protein (de)phosphorylation, (de)ubiquitylation, (de)SUMOylation, and (de)acetylation, as well as glycosylation, are perhaps the most well studied. To fully understand the biological functions of these PTMs, an important step is to identify their downstream targets at the systems level. The recent advance in the “shotgun” tandem mass spectrometry (MS/MS) technique has identified many PTM sites in mammalian proteomes; however, such a bottom–up approach does not help to connect these identified PTM sites to their upstream modification enzymes. Therefore, we and others have been developing various types of enzymatic reactions on the functional protein microarrays to identify direct in vitro targets of these enzymes.
1. Protein phosphorylation
Protein phosphorylation plays a central role in almost, if not all, aspects of cellular processes. The application of protein microarray technology to protein phosphorylation was first demonstrated by Zhu et al. (2000). They immobilized 17 different substrates on a nanowell protein microarray followed by individual kinase assays with almost all of the yeast kinases (119/122). This approach allowed them to determine the substrate specificity of the yeast kinome and identify new tyrosine phosphorylation activity.
In a later report, Snyder’s group accomplished a large scale “Phosphorylome Project” using the yeast proteome microarrays (Ptacek et al., 2005). Eighty-seven purified yeast kinases or kinase complexes were individually incubated on the yeast proteome arrays in a kinase buffer in the presence of 33P-γ-ATP, and a total of 1,325 distinct protein substrates were identified, representing a total of 4,129 phosphorylation events (Fig. 4.6 ). These results provided a global network that connect kinases to their potential substrates and offered a new opportunity to identify new signaling pathways or cross talk between pathways. Several smaller scale studies of kinase–substrate interactions have been reported in higher eukaryotes. For instance, a commercially available human protein microarray composed of approximately 3,000 individual proteins was used to identify substrates of cyclin-dependent kinase 5 (Cdk5), a serine/threonine kinase that plays an important role during central nerve system development (Schnack et al., 2008).
Figure 4.6.
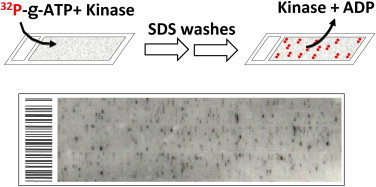
In vitro kinase assays on protein microarrays.
Recombinant kinase proteins were overexpressed and purified from yeast. Each kinase was added to a kinase reaction mixture and incubated on a pre-blocked protein microarray in the presence of radiolabeled ATP. The reaction was terminated by 0.5% SDS washes, followed by PBS washes to assure complete removal of unincorporated ATP and the added kinase. The lower panel shows a portion of an image after exposing a phosphorylated protein microarray to X-ray film. For color version of this figure, the reader is referred to the online version of this book.
2. Protein ubiquitylation
Ubiquitylation is one of the most prevalent PTMs and controls almost all types of cellular events in eukaryotes. To establish a protein microarray-based approach for identification of ubiquitin E3 ligase substrates, Lu et al. (2008) developed an assay for yeast proteome microarrays that utilizes a HECT-domain E3 ligase, Rsp5, in combination with the E1 and E2 enzymes. More than 90 new substrates were identified, eight of which were validated as in vivo substrates of Rsp5. Further in vivo characterization of two substrates, Sla1 and Rnr2, demonstrated that Rsp5-dependent ubiquitylation affects either posttranslational process of the substrate or subcellular localization.
3. Protein acetylation
Histone acetylation and deacetylation, which are catalyzed by histone acetyltransferases (HATs) and histone deacetylases (HDACs), respectively, are emerging as critical regulators of chromatin structure and transcription. However, it has been hypothesized that many HATs and HDACs might also modify nonhistone substrates. For example, the core enzyme, Esa1, of the essential nucleosome acetyltransferase of H4 (NuA4) complex, is the only essential HAT in yeast, which strongly suggested that it may target additional nonhistone proteins that are crucial for cell to survive. To identify nonhistone substrates of the NuA4 complex, Lin et al. (2009) established and performed acetylation reactions on the yeast proteome microarrays using the NuA4 complex in the presence of [14C]-Acetyl-CoA as a donor. Surprisingly, 91 proteins were found to be readily acetylated by the NuA4 complex on the array (examples are shown in Fig. 4.4). To further validate these in vitro results, 20 of them were randomly chosen and 13 of them showed Esa1-dependent acetylation in cells. One of them, phosphoenolpyruvate carboxykinase (Pck1p), was further characterized to explore the possible link between acetylation and metabolism. Mass spectrometry assay revealed Lys19 and 514 as the acetylation sites of Pck1p, and mutagenesis analyses demonstrated that acetylation on K514 is critical to enhance Pck1p’s enzyme activity and results in longer life span for yeast cells growing under starvation. This study offers a molecular link between the HDAC Sir2 and yeast longevity.
In a more recent study, Lu et al. (2011) focused on in-depth characterization of another nonhistone substrate, Sip2. Sip2 is one of three regulatory β subunits of Snf1 complex (yeast homolog of AMP-activated protein kinase), and its protein level decreases as cells age. We used mutants at four acetylation sites, K12, 16, 17, and 256, to study acetyl-Sip2 function. Sip2 acetylation, controlled by antagonizing NuA4 acetyltransferase and Rpd3 deacetylase, enhances interaction with kinase Snf1, the catalytic α subunit of Snf1 complex. Sip2–Snf1 interaction inhibits Snf1 activity, thus decreasing the phosphorylation of a downstream target, Sch9, and ultimately leading to impaired growth but extends yeast replicative life span. We also demonstrate that the antiaging effect of Sip2 acetylation is independent of nutrient availability and TORC1 activity. Therefore, intrinsic aging stress, signaled via the Sip2–Snf1 acetylation, constitutes a second TORC1-independent pathway regulating Sch9 activity that controls life span in yeast.
4. S-Nitrosylation
S-nitrosylation is independent of enzyme catalysis but is an important PTM that affects a wide range of proteins involved in many cellular processes. Recently, Foster et al. (2009) developed a protein microarray-based approach to detect proteins reactive to S-nitrosothiol (SNO), the donor of NO+ in S-nitrosylation, and to investigate determinants of S-nitrosylation. S-nitrosocysteine (CysNO), a highly reactive SNO, was added to the yeast proteome microarray, and the nitrosylated proteins were then detected using a modified biotin switch technique. The top 300 proteins with the highest relative signal intensity were further analyzed, and the results revealed that proteins with active-site Cys thiols residing at N-termini of alpha helices or within catalytic loops were particularly prominent. However, substantial variations of S-nitrosylation were observed even within these protein families, indicating that secondary structure or intrinsic nucleophilicity of Cys thiols was not sufficient to interpret the specificity of S-nitrosylation. Further analyses revealed that NO-donor stereochemistry and structure had significant impact on S-nitrosylation efficiency.
D. Applications in Clinical Research
1. Biomarker identification
Though the applications described above are most useful in basic research, functional protein microarrays may have enormous impacts on clinical diagnosis and prognosis. When proteins on a functional protein microarray are viewed as potential antigens that may or may not associated with a particular disease, it becomes a powerful tool in biomarker identification. The principle is straightforward: when an autoantibody presented in human sera associated with a human disease (e.g., autoimmune diseases) recognizes a human protein spotted on the array, it can be readily detected with fluorescently labeled anti-human immunoglobulin antibodies (e.g., anti-IgG) and a profile of autoantibodies associated with a disease thus created, providing a rapid approach to identifying potential disease biomarkers (Fig. 4.7 ). For example, Robinson et al. (2002) reported the first application of protein microarray technology to profile multiple human disease sera. They constructed a microarray with 196 biomolecules shown to be autoantigens in eight human autoimmune diseases, including proteins, peptides, enzyme complexes, ribonucleoprotein complexes, DNA, and posttranslationally modified antigens. The arrays were incubated with patient sera to study the specificity and pathogenesis of autoantibody responses and were used to identify and define relevant autoantigens in human autoimmune diseases, including systemic lupus erythematosus and rheumatoid arthritis.
Figure 4.7.
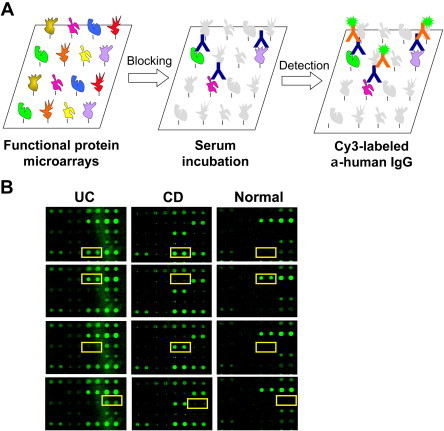
Biomarker identification using protein microarrays.
Proteins spotted on a functional microarray can be viewed as potential autoantigens that may be associated with a particular disease (e.g., autoimmune diseases). (A) To identify such autoantigens, a protein microarray is blocked, incubated with diluted serum sample, washed, and a fluorescently labeled anti-human IgG is used to detect captured autoantibodies. Following statistic analyses (e.g., SAM) can be used to identify potential autoantigens associated with the disease of interest. (B). Examples of biomarker identification in inflammatory bowel diseases (Xie et al., 2010). UC, ulcerative colitis; CD, Crohn’s disease; normal, healthy subjects. For color version of this figure, the reader is referred to the online version of this book.
Hu et al. (2007) reported a new approach for high-throughput characterization of monoclonal antibody target specificity using a protein microarray composed of 1,058 unique human liver proteins. They immunized mice with live cells from human livers, isolated 54 hybridomas with binding activities to human cells, and identified the corresponding antigens for five monoclonal antibodies via screening on the protein microarray. Expression profiles of the corresponding antigens of the five antibodies were characterized by using tissue microarrays, and one of the antigens, eIF1A, was found to be expressed in normal human liver but not in hepatocellular carcinoma. Other applications include biomarker identification for ovarian cancer (Hudson et al., 2007), inflammatory bowel disease (Chen et al., 2009), alopecia areata (Lueking et al., 2005a), and autoimmune hepatitis (Song et al., 2010).
Protein microarrays can also be used for detection of infectious diseases. Zhu et al. (2006) developed a coronavirus protein microarray for the diagnosis of severe acute respiratory syndrome (SARS), which included all the SARS-CoV proteins as well as proteins from five additional coronaviruses that can infect humans (HCoV-229E and HCoV-OC43), cows (BCV), cats (FIPV), and mice (MHVA59). These microarrays could quickly distinguish patient serum samples as SARS positive or SARS negative based on the presence of human IgG and IgM antibodies against SARS-CoV proteins, with a 94% accuracy compared with standard diagnostic methods. Patients carrying antibodies against other coronavirus proteins were also identified. The advantages of this microarray-based assay to standard ELISA-based diagnostic methods include at least 100-fold higher sensitivity and the need for substantially less sample for analysis.
2. Pathogen–host interactions
Another interesting application of the functional protein microarray is to elucidate the molecular mechanism as how a pathogen (e.g., a virus) hijacks the host pathways and machineries for its own replication. The application of high-throughput approaches has uncovered many new host factors that regulate the life cycle of infecting viruses, such as global RNAi-based screens (Brass et al., 2009, Karlas et al., 2010, Shapira et al., 2009). However, correlating this information with a fundamental underling mechanism is often challenging. Our group in collaboration with the Hayward group hypothesized that conserved proteins from related viruses would tend to target the same host pathways using similar mechanisms (Li et al., in press). Herpesviruses all encode conserved serine/threonine kinases that play an important role in virus replication and spread. We utilized a human protein microarray to identify shared host targets of the conserved kinases encoded by four human herpesviruses and discovered that the DNA damage pathway was statistically enriched for shared substrates. Using the gamma herpesvirus Epstein–Barr virus (EBV), we demonstrated that the EBV kinase activates an upstream mediator of the DNA damage response, the histone acetyltransferase TIP60. EBV also utilizes the chromatin remodeling function of TIP60 in a positive feedback loop to enhance expression of EBV genes needed for virus replication. Identification of key cellular targets of the conserved herpesvirus kinases will facilitate the development of broadly effective antiviral strategies. This work provides a new paradigm for the discovery of key virus–host interactions.
V. Outlook
Recent years have witnessed a rapid growth in using functional protein microarrays for basic research (Tao et al., 2007). Although the technology is still at a relatively early stage of development, it has become obvious that the protein microarray platform can and will act as a versatile tool suitable for the large-scale high-throughput biology, especially in the areas of profiling PTMs and in analysis of signal transduction networks and pathways (Hu et al., 2009, Ptacek et al., 2005). As another crucial proteomics technology, recent progress in mass spectrometry has allowed global profiling of PTMs using a shotgun approach. For example, the Zhao, Mann, and Guan groups recently identified numerous acetylated lysine residues in metabolic enzymes in mice and human cells without knowing the upstream HATs (Choudhary et al., 2009, Kim et al., 2006, Zhao et al., 2010). In parallel, our team also identified many yeast metabolic enzymes as substrates of the NuA4 acetylation complex without knowing the actual modified sites (Lin et al., 2009, Lu et al., 2011). Therefore, we envision that the combination of the two technologies will have enormous potential to both identify critical regulatory PTMs at the resolution of modified individual amino acids and to identify the enzymes that mediate these effects. Another emerging direction is in the forefront of understanding the molecular mechanisms of pathogen–host interactions. In the same manner in which we identified host proteins that recognized the SLD loop of the BMV virus, functional protein microarrays (e.g., a human protein microarray) can be used to discover those host proteins targeted by pathogens (e.g., HIV, HCV, and SARS-CoV). The identification of the host targets of a virus will provide alternative therapeutics that cannot be rapidly evaded via mutation of the viral genomes (Brass et al., 2009). In conclusion, the potential of functional protein microarrays is only just now starting to reveal itself. It is expected that it will become an indispensable and invaluable tool in proteomics and systems biology research.
Acknowledgements
We thank Professor H. Lu for discussion of the OIRD technology and the National Institutes of Health for funding support.
References
- Afanassiev V., Hanemann V., Wolfl S. Preparation of DNA and protein micro arrays on glass slides coated with an agarose film. Nucl. Acids Res. 2000;28:E66. doi: 10.1093/nar/28.12.e66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen S.V., Miller E.S. RNA-binding properties of in vitro expressed histidine-tagged RB69 RegA translational repressor protein. Anal. Biochem. 1999;269:32–37. doi: 10.1006/abio.1999.4025. [DOI] [PubMed] [Google Scholar]
- Angenendt P. Cell-free protein expression and functional assay in nanowell chip format. Anal. Chem. 2004;76:1844–1849. doi: 10.1021/ac035114i. [DOI] [PubMed] [Google Scholar]
- Brass A.L. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell. 2009;139:1243–1254. doi: 10.1016/j.cell.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussow K. A method for global protein expression and antibody screening on high-density filters of an arrayed cDNA library. Nucl. Acids Res. 1998;26:5007–5008. doi: 10.1093/nar/26.21.5007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F., Lu H., Chen Z., Zhao T., Yang G. Optical real-time monitoring of the laser molecular-beam epitaxial growth of perovskite oxide thin films by an oblique-incidence reflectance-difference technique. JOSA. B. 2001;18(7):1031–1035. [Google Scholar]
- Chen C.S. A proteome chip approach reveals new DNA damage recognition activities in Escherichia coli. Nat. Methods. 2008;5:69–74. doi: 10.1038/NMETH1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C.S. Identification of novel serological biomarkers for inflammatory bowel disease using Escherichia coli proteome chip. Mol. Cell. Proteomics. 2009;8:1765–1776. doi: 10.1074/mcp.M800593-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary C. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- Crompton P.D. A prospective analysis of the Ab response to Plasmodium falciparum before and after a malaria season by protein microarray. Proc. Natl. Acad. Sci. U. S. A. 2010;107(15):6958–6963. doi: 10.1073/pnas.1001323107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRisi J.L., Iyer V.R., Brown P.O. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- Diamond D.L. Use of ProteinChip array surface enhanced laser desorption/ionization time-of-flight mass spectrometry (SELDI-TOF MS) to identify thymosin beta-4, a differentially secreted protein from lymphoblastoid cell lines. J. Am. Soc. Mass Spectrom. 2003;14:760–765. doi: 10.1016/s1044-0305(03)00265-4. [DOI] [PubMed] [Google Scholar]
- Ekins R.P. Multi-analyte immunoassay. J. Pharm. Biomed. Anal. 1989;7:155–168. doi: 10.1016/0731-7085(89)80079-2. [DOI] [PubMed] [Google Scholar]
- Evans-Nguyen K.M., Tao S.C., Zhu H., Cotter R.J. Protein arrays on patterned porous gold substrates interrogated with mass spectrometry: detection of peptides in plasma. Anal. Chem. 2008;80:1448–1458. doi: 10.1021/ac701800h. [DOI] [PubMed] [Google Scholar]
- Fei Y.Y. A novel high-throughput scanning microscope for label-free detection of protein and small-molecule chemical microarrays. Rev. Sci. Instrum. 2008;79(1):013708. doi: 10.1063/1.2830286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster M.W., Forrester M.T., Stamler J.S. A protein microarray-based analysis of S-nitrosylation. Proc. Natl. Acad. Sci. U. S. A. 2009;106:18948–18953. doi: 10.1073/pnas.0900729106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin I.M., Kukhtin A., Glesne D., Schabacker D., Chandler D.P. Analysis of protein interaction and function with a 3-dimensional MALDI-MS protein array. Biotechniques. 2005;39:99–107. doi: 10.2144/05391RR02. [DOI] [PubMed] [Google Scholar]
- Gelperin D.M. Biochemical and genetic analysis of the yeast proteome with a movable ORF collection. Genes Dev. 2005;19:2816–2826. doi: 10.1101/gad.1362105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima N. Human protein factory for converting the transcriptome into an in vitro-expressed proteome. Nat. Methods. 2008;5:1011–1017. doi: 10.1038/nmeth.1273. [DOI] [PubMed] [Google Scholar]
- Guschin D. Manual manufacturing of oligonucleotide, DNA, and protein microchips. Anal. Biochem. 1997;250:203–211. doi: 10.1006/abio.1997.2209. [DOI] [PubMed] [Google Scholar]
- Gygi S.P., Rochon Y., Franza B.R., Aebersold R. Correlation between protein and mRNA abundance in yeast. Mol. Cell Biol. 1999;19:1720–1730. doi: 10.1128/mcb.19.3.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall D.A. Regulation of gene expression by a metabolic enzyme. Science. 2004;306:482–484. doi: 10.1126/science.1096773. [DOI] [PubMed] [Google Scholar]
- He M., Taussig M.J. Single step generation of protein arrays from DNA by cell-free expression and in situ immobilisation (PISA method) Nucl. Acids Res. 2001;29 doi: 10.1093/nar/29.15.e73. E73-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M. Printing protein arrays from DNA arrays. Nat. Methods. 2008;5:175–177. doi: 10.1038/nmeth.1178. [DOI] [PubMed] [Google Scholar]
- Ho S.W., Jona G., Chen C.T., Johnston M., Snyder M. Linking DNA-binding proteins to their recognition sequences by using protein microarrays. Proc. Natl. Acad. Sci. U. S. A. 2006;103:9940–9945. doi: 10.1073/pnas.0509185103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochuli E., Dobeli H., Schacher A. New metal chelate adsorbent selective for proteins and peptides containing neighbouring histidine residues. J. Chromatogr. 1987;411:177–184. doi: 10.1016/s0021-9673(00)93969-4. [DOI] [PubMed] [Google Scholar]
- Holt L.J., Bussow K., Walter G., Tomlinson I.M. By-passing selection: direct screening for antibody-antigen interactions using protein arrays. Nucl. Acids Res. 2000;28:E72. doi: 10.1093/nar/28.15.e72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu K.L., Pilobello K.T., Mahal L.K. Analyzing the dynamic bacterial glycome with a lectin microarray approach. Nat. Chem. Biol. 2006;2(3):153–157. doi: 10.1038/nchembio767. [DOI] [PubMed] [Google Scholar]
- Hu S. A protein chip approach for high-throughput antigen identification and characterization. Proteomics. 2007;7:2151–2161. doi: 10.1002/pmic.200600923. [DOI] [PubMed] [Google Scholar]
- Hu S. Profiling the human protein-DNA interactome reveals ERK2 as a transcriptional repressor of interferon signaling. Cell. 2009;139:610–622. doi: 10.1016/j.cell.2009.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J. Finding new components of the target of rapamycin (TOR) signaling network through chemical genetics and proteome chips. Proc. Natl. Acad. Sci. U. S. A. 2004;101:16594–16599. doi: 10.1073/pnas.0407117101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang R.P. Detection of multiple proteins in an antibody-based protein microarray system. J. Immunol. Methods. 2001;255:1–13. doi: 10.1016/s0022-1759(01)00394-5. [DOI] [PubMed] [Google Scholar]
- Hudson M.E., Pozdnyakova I., Haines K., Mor G., Snyder M. Identification of differentially expressed proteins in ovarian cancer using high-density protein microarrays. Proc. Natl. Acad. Sci. U. S. A. 2007;104:17494–17499. doi: 10.1073/pnas.0708572104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones R.B., Gordus A., Krall J.A., MacBeath G. A quantitative protein interaction network for the ErbB receptors using protein microarrays. Nature. 2006;439:168–174. doi: 10.1038/nature04177. [DOI] [PubMed] [Google Scholar]
- Joshi B., Janda L., Stoytcheva Z., Tichy P. PkwA, a WD-repeat protein, is expressed in spore-derived mycelium of Thermomonospora curvata and phosphorylation of its WD domain could act as a molecular switch. Microbiology. 2000;146(Pt 12):3259–3267. doi: 10.1099/00221287-146-12-3259. [DOI] [PubMed] [Google Scholar]
- Karlas A. Genome-wide RNAi screen identifies human host factors crucial for influenza virus replication. Nature. 2010;463:818–822. doi: 10.1038/nature08760. [DOI] [PubMed] [Google Scholar]
- Kim S.C. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol. Cell. 2006;23:607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- Krogan N.J. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- Kumble K.D. Protein microarrays: new tools for pharmaceutical development. Anal. Bioanal. Chem. 2003;377:812–819. doi: 10.1007/s00216-003-2088-6. [DOI] [PubMed] [Google Scholar]
- Kung L.A. Global analysis of the glycoproteome in Saccharomyces cerevisiae reveals new roles for protein glycosylation in eukaryotes. Mol. Syst. Biol. 2009;5:308. doi: 10.1038/msb.2009.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry J.P., Zhu X.D., Gregg J.P. Label-free detection of microarrays of biomolecules by oblique-incidence reflectivity difference microscopy. Opt. Lett. 2004;29(6):581–583. doi: 10.1364/ol.29.000581. [DOI] [PubMed] [Google Scholar]
- Lee K.B., Park S.J., Mirkin C.A., Smith J.C., Mrksich M. Protein nanoarrays generated by dip-pen nanolithography. Science. 2002;295:1702–1705. doi: 10.1126/science.1067172. [DOI] [PubMed] [Google Scholar]
- Lee Y. ProteoChip: a highly sensitive protein microarray prepared by a novel method of protein immobilization for application of protein-protein interaction studies. Proteomics. 2003;3:2289–2304. doi: 10.1002/pmic.200300541. [DOI] [PubMed] [Google Scholar]
- Li R. Conserved herpesvirus kinases regulate TIP60 to promote virus replication. Cell Host Microbe. 2011;10:390–400. doi: 10.1016/j.chom.2011.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang L. Identification of potential serodiagnostic and subunit vaccine antigens by antibody profiling of toxoplasmosis cases in Turkey. Mol. Cell. Proteomics. 2011;10 doi: 10.1074/mcp.M110.006916. M110.006916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y.Y. Protein acetylation microarray reveals that NuA4 controls key metabolic target regulating gluconeogenesis. Cell. 2009;136:1073–1084. doi: 10.1016/j.cell.2009.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lizardi P.M. Mutation detection and single-molecule counting using isothermal rolling-circle amplification. Nat. Genet. 1998;19:225–232. doi: 10.1038/898. [DOI] [PubMed] [Google Scholar]
- Lu J.Y. Functional dissection of a HECT ubiquitin E3 ligase. Mol. Cell. Proteomics. 2008;7:35–45. doi: 10.1074/mcp.M700353-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H. Detection of the specific binding on protein microarrays by oblique-incidence reflectivity difference method. J. Opt. 2010;12:095301. (pp. 5) [Google Scholar]
- Lu J.-Y. Acetylation of AMPK controls intrinsic aging independently of caloric restriction. Cell. 2011;146:1–11. doi: 10.1016/j.cell.2011.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lueking A. Protein microarrays for gene expression and antibody screening. Anal. Biochem. 1999;270:103–111. doi: 10.1006/abio.1999.4063. [DOI] [PubMed] [Google Scholar]
- Lueking A. Profiling of alopecia areata autoantigens based on protein microarray technology. Mol. Cell. Proteomics. 2005;4:1382–1390. [Google Scholar]
- Lueking A., Cahill D.J., Mullner S. Protein biochips: a new and versatile platform technology for molecular medicine. Drug Discov. Today. 2005;10:789–794. doi: 10.1016/S1359-6446(05)03449-5. [DOI] [PubMed] [Google Scholar]
- MacBeath G., Schreiber S.L. Printing proteins as microarrays for high-throughput function determination. Science. 2000;289:1760–1763. doi: 10.1126/science.289.5485.1760. [DOI] [PubMed] [Google Scholar]
- Morley M. Genetic analysis of genome-wide variation in human gene expression. Nature. 2004;430:743–747. doi: 10.1038/nature02797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhija R., Rupa P., Pillai D., Garg L.C. High-level production and one-step purification of biologically active human growth hormone in Escherichia coli. Gene. 1995;165:303–306. doi: 10.1016/0378-1119(95)00525-b. [DOI] [PubMed] [Google Scholar]
- Murthy T.V. Bacterial cell-free system for high-throughput protein expression and a comparative analysis of Escherichia coli cell-free and whole cell expression systems. Protein Expr. Purif. 2004;36:217–225. doi: 10.1016/j.pep.2004.04.002. [DOI] [PubMed] [Google Scholar]
- Nelson B.P., Frutos A.G., Brockman J.M., Corn R.M. Near-Infrared Surface Plasmon Resonance Measurements of Ultrathin Films. 1. Angle Shift and SPR Imaging Experiments. Anal. Chem. 1999;71(18):3928–3934. [Google Scholar]
- Niemeyer C.M., Sano T., Smith C.L., Cantor C.R. Oligonucleotide-directed self-assembly of proteins: semisynthetic DNA–streptavidin hybrid molecules as connectors for the generation of macroscopic arrays and the construction of supramolecular bioconjugates. Nucl. Acids Res. 1994;22:5530–5539. doi: 10.1093/nar/22.25.5530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh Y.H. Chip-based analysis of SUMO (small ubiquitin-like modifier) conjugation to a target protein. Biosens. Bioelectron. 2007;22(7):1260–1267. doi: 10.1016/j.bios.2006.05.023. [DOI] [PubMed] [Google Scholar]
- Pease A.C. Light-generated oligonucleotide arrays for rapid DNA sequence analysis. Proc. Natl. Acad. Sci. U. S. A. 1994;91:5022–5026. doi: 10.1073/pnas.91.11.5022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peluso P. Optimizing antibody immobilization strategies for the construction of protein microarrays. Anal. Biochem. 2003;312:113–124. doi: 10.1016/s0003-2697(02)00442-6. [DOI] [PubMed] [Google Scholar]
- Piehler J. Label-free monitoring of DNA-ligand interactions. Anal. Biochem. 1997;249(1):94–102. doi: 10.1006/abio.1997.2160. [DOI] [PubMed] [Google Scholar]
- Pilobello K.T., Slawek D.E., Mahal L.K. A ratiometric lectin microarray approach to analysis of the dynamic mammalian glycome. Proc. Natl. Acad. Sci. U. S. A. 2007;104(28):11534–11539. doi: 10.1073/pnas.0704954104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popescu S.C. Differential binding of calmodulin-related proteins to their targets revealed through high-density Arabidopsis protein microarrays. Proc. Natl. Acad. Sci. U. S. A. 2007;104:4730–4735. doi: 10.1073/pnas.0611615104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptacek J. Global analysis of protein phosphorylation in yeast. Nature. 2005;438:679–684. doi: 10.1038/nature04187. [DOI] [PubMed] [Google Scholar]
- Ramachandran N. Self-assembling protein microarrays. Science. 2004;305:86–90. doi: 10.1126/science.1097639. [DOI] [PubMed] [Google Scholar]
- Ramachandran N. Next-generation high-density self-assembling functional protein arrays. Nat. Methods. 2008;5:535–538. doi: 10.1038/nmeth.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson W.H. Autoantigen microarrays for multiplex characterization of autoantibody responses. Nat. Med. 2002;8:295–301. doi: 10.1038/nm0302-295. [DOI] [PubMed] [Google Scholar]
- Schadt E.E. Genetics of gene expression surveyed in maize, mouse and man. Nature. 2003;422:297–302. doi: 10.1038/nature01434. [DOI] [PubMed] [Google Scholar]
- Schena M., Shalon D., Davis R.W., Brown P.O. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- Schnack C., Hengerer B., Gillardon F. Identification of novel substrates for Cdk5 and new targets for Cdk5 inhibitors using high-density protein microarrays. Proteomics. 2008;8:1980–1986. doi: 10.1002/pmic.200701063. [DOI] [PubMed] [Google Scholar]
- Schweitzer B. Inaugural article: immunoassays with rolling circle DNA amplification: a versatile platform for ultrasensitive antigen detection. Proc. Natl. Acad. Sci. U. S. A. 2000;97:10113–10119. doi: 10.1073/pnas.170237197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweitzer B. Multiplexed protein profiling on microarrays by rolling-circle amplification. Nat. Biotechnol. 2002;20:359–365. doi: 10.1038/nbt0402-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao W. Optimization of Rolling-Circle Amplified Protein Microarrays for Multiplexed Protein Profiling. J. Biomed. Biotechnol. 2003;2003:299–307. doi: 10.1155/S1110724303209268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapira S.D. A physical and regulatory map of host-influenza interactions reveals pathways in H1N1 infection. Cell. 2009;139:1255–1267. doi: 10.1016/j.cell.2009.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shingyoji M., Gerion D., Pinkel D., Gray J.W., Chen F. Quantum dots-based reverse phase protein microarray. Talanta. 2005;67:472–478. doi: 10.1016/j.talanta.2005.06.064. [DOI] [PubMed] [Google Scholar]
- Smith M.G., Jona G., Ptacek J., Devgan G., Zhu H., Zhu X., Snyder M. Global analysis of protein function using protein microarrays. Mech. Ageing Dev. 2005;126:171–175. doi: 10.1016/j.mad.2004.09.019. [DOI] [PubMed] [Google Scholar]
- Song Q. Novel autoimmune hepatitis-specific autoantigens identified using protein microarray technology. J. Proteome Res. 2010;9:30–39. doi: 10.1021/pr900131e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao S.C., Zhu H. Protein chip fabrication by capture of nascent polypeptides. Nat. Biotechnol. 2006;24:1253–1254. doi: 10.1038/nbt1249. [DOI] [PubMed] [Google Scholar]
- Tao S.C., Chen C.S., Zhu H. Applications of protein microarray technology. Comb. Chem. High Throughput Screen. 2007;10:706–718. doi: 10.2174/138620707782507386. [DOI] [PubMed] [Google Scholar]
- Tao S.C. Lectin microarrays identify cell-specific and functionally significant cell surface glycan markers. Glycobiology. 2008;18:761–769. doi: 10.1093/glycob/cwn063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templin M.F. Protein microarray technology. Trends Biotechnol. 2002;20:160–166. doi: 10.1016/s0167-7799(01)01910-2. [DOI] [PubMed] [Google Scholar]
- Thiel A.J., Frutos A.G., Jordan C.E., Corn R.M., Smith L.M. In situ surface plasmon resonance imaging detection of DNA hybridization to oligonucleotide arrays on gold surfaces. Anal. Chem. 1997;69(24):4948–4956. [Google Scholar]
- Varnum S.M., Woodbury R.L., Zangar R.C. A protein microarray ELISA for screening biological fluids. Methods Mol. Biol. 2004;264:161–172. doi: 10.1385/1-59259-759-9:161. [DOI] [PubMed] [Google Scholar]
- Vidal M., Brachmann R.K., Fattaey A., Harlow E., Boeke J.D. Reverse two-hybrid and one-hybrid systems to detect dissociation of protein-protein and DNA-protein interactions. Proc. Natl. Acad. Sci. U. S. A. 1996;93:10315–10320. doi: 10.1073/pnas.93.19.10315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacker R., Schroder H., Niemeyer C.M. Performance of antibody microarrays fabricated by either DNA-directed immobilization, direct spotting, or streptavidin-biotin attachment: a comparative study. Anal. Biochem. 2004;330:281–287. doi: 10.1016/j.ab.2004.03.017. [DOI] [PubMed] [Google Scholar]
- Wang Z.H., Jin G. A label-free multisensing immunosensor based on imaging ellipsometry. Anal. Chem. 2003;75(22):6119–6123. doi: 10.1021/ac0347258. [DOI] [PubMed] [Google Scholar]
- Wang X. Label-free and high-throughput detection of protein microarrays by oblique-incidence reflectivity difference method. Chin. Phys. Lett. 2010;27(10):107801. [Google Scholar]
- Wen J. Detection of protein microarrays by oblique-incidence reflectivity difference technique. Sci. Chi. Phys. Mech. Astronomy. 2010;53(2):306. [Google Scholar]
- Xie Z., Hu S., Blackshaw S., Zhu H., Qian J. hPDI: a database of experimental human protein-DNA interactions. Bioinformatics. 2010;26:287–289. doi: 10.1093/bioinformatics/btp631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H., Park S.H., Finkelstein G., Reif J.H., LaBean T.H. DNA-templated self-assembly of protein arrays and highly conductive nanowires. Science. 2003;301:1882–1884. doi: 10.1126/science.1089389. [DOI] [PubMed] [Google Scholar]
- Zajac A., Song D., Qian W., Zhukov T. Protein microarrays and quantum dot probes for early cancer detection. Colloids Surf. B. Biointerfaces. 2007;58:309–314. doi: 10.1016/j.colsurfb.2007.02.019. [DOI] [PubMed] [Google Scholar]
- Zhang K., Diehl M.R., Tirrell D.A. Artificial polypeptide scaffold for protein immobilization. J. Am. Chem. Soc. 2005;127:10136–10137. doi: 10.1021/ja051457h. [DOI] [PubMed] [Google Scholar]
- Zhao S. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327:1000–1004. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H. Two-color, rolling-circle amplification on antibody microarrays for sensitive, multiplexed serum-protein measurements. Genome Biol. 2004;5:R28. doi: 10.1186/gb-2004-5-4-r28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H. Analysis of yeast protein kinases using protein chips. Nat. Genet. 2000;26:283–289. doi: 10.1038/81576. [DOI] [PubMed] [Google Scholar]
- Zhu H. Global analysis of protein activities using proteome chips. Science. 2001;293:2101–2105. doi: 10.1126/science.1062191. [DOI] [PubMed] [Google Scholar]
- Zhu H. Severe acute respiratory syndrome diagnostics using a coronavirus protein microarray. Proc. Natl. Acad. Sci. U. S. A. 2006;103:4011–4016. doi: 10.1073/pnas.0510921103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J. RNA-binding proteins that inhibit RNA virus infection. Proc. Natl. Acad. Sci. U. S. A. 2007;104:3129–3134. doi: 10.1073/pnas.0611617104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J. Protein array identification of substrates of the Epstein-Barr Virus protein kinase BGLF4. J. Virol. 2009;83:5219–5231. doi: 10.1128/JVI.02378-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziauddin J., Sabatini D.M. Microarrays of cells expressing defined cDNAs. Nature. 2001;411:107–110. doi: 10.1038/35075114. [DOI] [PubMed] [Google Scholar]


