Abstract
Background:
The Coxsackie- and Adenovirus Receptor (CAR) has been assigned two crucial attributes in carcinomas: (a) involvement in the regulation of growth and dissemination and (b) binding for potentially therapeutic adenoviruses. However, data on CAR expression in cancer types are conflicting and several entities have not been analysed to date.
Methods:
The expression of CAR was assessed by immunohistochemical staining of tissue microarrays (TMA) containing 3714 specimens derived from 100 malignancies and from 273 normal control tissues.
Results:
The expression of CAR was detected in all normal organs, except in the brain. Expression levels, however, displayed a broad range from being barely detectable (for example, in the thymus) to high abundance expression (for example, in the liver and gastric mucosa). In malignancies, a high degree of variability was notable also, ranging from significantly elevated CAR expression (for example, in early stages of malignant transformation and several tumours of the female reproductive system) to decreased CAR expression (for example, in colon and prostate cancer types).
Conclusion:
Our results provide a comprehensive insight into CAR expression in neoplasms and indicate that CAR may offer a valuable target for adenovirus-based therapy in a subset of carcinomas. Furthermore, these data suggest that CAR may contribute to carcinogenesis in an entity-dependent manner.
Keywords: Coxsackievirus and Adenovirus receptor, multitumour tissue microarray, differential expression
The Coxsackie- and Adenovirus Receptor (CAR), a transmembrane component of the tight junction complex, facilitates viral attachment onto the cellular surface, a crucial requirement for subsequent virus uptake (Bergelson et al, 1997; Cohen et al, 2001). Presence of CAR is therefore considered a critical determinant for the efficacy of therapeutic strategies employing adenoviruses. Hereby, attenuated adenoviruses, either replication-incompetent created to deliver therapeutic genes or viruses replicating restrictedly in certain cell types, may be used for the cancer treatment (Kasuya et al, 2007). In various human cancer types, however, particularly those displaying loss of differentiation, and advanced disease stages, reduced CAR presence has been documented (Heideman et al, 2001; Rauen et al, 2002; Sachs et al, 2002; Matsumoto et al, 2005; Korn et al, 2006; Anders et al, 2009; Wunder et al, 2012a, 2012b). In line with these observations, significant correlations between impaired CAR expression and a poor clinical outcome for gastric and bladder cancer patients were found (Matsumoto et al, 2005; Anders et al, 2009). Regulation of declined CAR expression in cancers has been attributed to activation of the Raf/MEK/ERK pathway and the TGF-β signalling, as well as hypoxia, epithelial–mesenchymal transdifferentiation and histone deacetylation of the CAR gene promoter (Brüning and Runnebaum, 2003; Pong et al, 2003; Anders et al, 2003a; Lacher et al, 2006; Küster et al, 2010a, 2010b; Lacher et al, 2011).
In contrast, CAR upregulation was found in cancers of the endometrium, ovary, cervix, breast and lung, as well as neuroblastomas and medulloblastomas (Martino et al, 2000; Martin et al, 2005; Persson et al, 2006; Wang et al, 2006; Reimer et al, 2007; Giaginis et al, 2008; Dietel et al, 2011). In breast and lung cancer types, high CAR expression has been linked to poor overall survival and shorter disease-free survival, respectively (Martin et al, 2005; Wunder et al, 2012a). Contrary to the loss of CAR in neoplasms, little is known about the molecular basis of CAR upregulation: in oesophageal squamous cell carcinoma cell lines, CAR expression was induced through the MAPK/ERK1/2 signalling, a pathway that also has been linked to CAR downregulation as described above (Ma et al, 2012). Furthermore, disruption of cellular organisation has been found to upregulate CAR in early breast cancer (Anders et al, 2003b).
Currently, it remains unclear whether these diverse results reflect entity-depending differences in CAR expression or might solely be caused by methodical differences. Nevertheless, given that differential CAR expression may indicate a progression step during malignant transformation, these previous findings might reflect the possible complex function of CAR. On one hand, loss of CAR has been suggested to decrease intercellular adhesion, promote proliferation, migration, invasion and metastatic potential of cancers, leading to the hypothesis of a tumour-suppressive role of CAR (Okegawa et al, 2000, 2001; Brüning and Runnebaum, 2004; Huang et al, 2005; Wang et al, 2005; Raschperger et al, 2006; Anders et al, 2009; Stecker et al, 2011). On the other hand, CAR has been implied to promote carcinogensesis, as increased CAR levels were found in early-stage breast cancer and breast cancer precursor cell lines (Anders et al, 2003b; Brüning et al, 2005).
Intrigued by these findings, we performed an immunohistochemical determination of CAR expression in a broad range of malignancies, corresponding precursor lesions as well as healthy controls employing tissue microarrays. Usage of this uniform methodical platform was chosen to generate data allowing for direct comparison between different organs, and hereby to identify neoplasms in which CAR expression might be of importance during malignant progression and the ones where it is not. By doing so, potential targets for adenovirus-mediated therapies based on CAR expression can be identified as well.
Materials and methods
Tissue microarrays and immunohistological investigations
The expression of CAR protein was assessed with immunohistochemical staining of tissue microarrays (TMA) containing a total of 3714 formalin-fixed, paraffin-embedded archival samples (diameter 0.6 mm) from a total of 100 different human tumours and preneoplastic lesions, as well as 273 corresponding controls derived from normal tissues (Simon and Sauter, 2002). All these samples (provided by RS and GS) were taken from tissues acquired for routine diagnostic purposes at the Department of Pathology, University Medical Center Hamburg-Eppendorf, in accordance with the principles of the ‘Ethik-Kommission der Ärztekammer, Hamburg'. The collection and TMA-based screenings of human tumour samples were in compliance with the ethical principles for medical research issued by the World Medical Association's Declaration of Helsinki.
For the subsequent immunohistochemical study, TMA sections were deparaffinized and dehydrated, employing standard procedures using rotihistol, isopropanol and ethanol. Following common antigen-retrieval methods including trypsin and microwave treatments in 10 mM citrate buffer (pH 6.0), tissues were blocked in milk and incubated with a primary polyclonal antibody against CAR (1:50, H-300: sc-15405, Biotechnology Inc., Santa Cruz, CA, USA) for 16 h at 4 °C. Subsequently, sections were incubated in biotinylated goat anti-rabbit immunoglobulin (1:400; Vector Laboratories, Burlingame, CA, USA), followed by treatment with the streptavidin-biotinylated horseradish peroxidase complex (Vectastain Elite ABC kit, Vector Laboratories). Using diaminobenzidine tetrahydrochloride (Sigma-Aldrich, Munich, Germany), sections were developed in hydrogen peroxide/PBS and counterstained with haemalaun. Immunostainings of CAR were analysed by two pathologists (DG and MV) blinded to clinicopathological data and scored according to (a) percentage of CAR-immunopositive cells (‘0': 0%, ‘1': <10%, ‘2': 11–50%, ‘3': 51–80 %, ‘4': 81–100%) and (b) staining intensity (‘0': no specific signal, ‘1': weak, ‘2': medium, ‘3' strong). On the basis of these data, the immunoreactive score (IRS) was calculated by percentage of positive cells × staining intensity score. For further evaluation, an IRS from 0 to 3 was considered CAR-negative, whereas 4–12 was regarded as CAR-positive.
Statistical methods
Statistical calculations using (Fisher's exact probability test or χ2 test, respectively) were performed using the SPSS software (version 11.5; SPSS Inc, Chicago, IL, USA).
Results
Expression of CAR in carcinomas and corresponding normal tissues
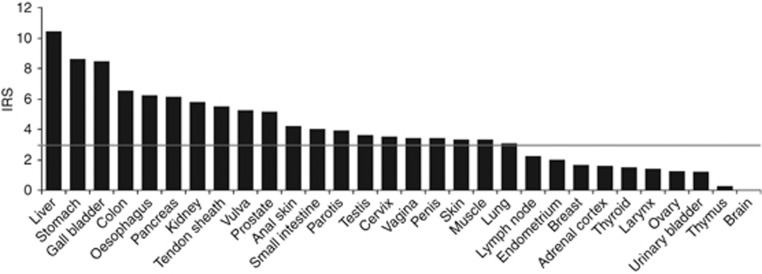
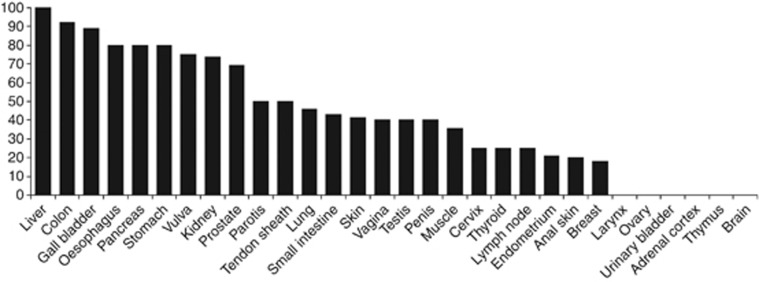
Immunopositivity of CAR was found in normal samples of all entities, except of the brain. Expression levels, however, displayed a high variability ranging from abundant presence in the liver, stomach and gall bladder to barely detectable, such as in the thymus (Figure 1). Highest percentages of CAR-positive tissues (IRS >3) were seen in the liver, colon, gall bladder, oesophagus, pancreas, stomach and vulva. On the other hand, low counts of CAR-positive cases (maximum of 25%) were noted in the cervix, thyroid, lymph nodes, endometrium, anal skin and breast. No cases with an IRS >3 were observed in the larynx, ovary, urinary bladder, adrenal cortex, thymus and brain (Figure 2).
Figure 1.
CAR expression in normal samples: CAR protein expression was determined with immunohistochemical staining. On the basis of the immunoreactive score, entities were considered CAR-negative (IRS 0–3) or CAR-positive (IRS 4–12) (grey line).
Figure 2.
Percentage of CAR-positive cases in normal samples: Portion of CAR-positive cases was calculated on the basis of the IRS (see Figure 1) with 25% or less being considered low CAR-expressing entities.
In neoplasias, a great degree of diversity of CAR expression was notable as well, with ubiquitous CAR expression in several early stages of malignant transformation such as non-invasive urinary bladder cancer, Warthin Tumours, thyroid adenoma and basalioma. In advanced stages, high CAR expression levels were detected, for instance, in hepatocellular and endometroid carcinomas. In contrast, low CAR expression levels were found in prostate cancer, various subtypes of breast cancer, Merkel cell carcinoma and desmoid tumours (Table 1).
Table 1. CAR immunopositivity in neoplasms and controls.
| Tissue type | n | Intensity (mean) | Pos. cells (mean) | IRS (mean) | CAR positive n | % | P-value |
|---|---|---|---|---|---|---|---|
|
Respiratory tract tumours | |||||||
|
Larynx | |||||||
| Larynx. normal | 5 | 1.2 | 1.2 | 1.4 | 0 | 0 | |
| Larynx. carcinoma |
55 |
1.96 |
2.2 |
4.8 |
35 |
63.6 |
0.006 |
|
Lung | |||||||
| Lung. normal | 24 | 1.54 | 1.83 | 3.04 | 11 | 45.8 | |
| Lung cancer. adenocarcinoma | 68 | 2.13 | 2.6 | 5.68 | 56 | 82.4 | 0.001 |
| Lung cancer. bronchioalveolary carcinoma | 13 | 1.85 | 2.62 | 5.31 | 9 | 69.2 | 0.173 |
| Lung cancer. large cell cancer | 45 | 1.78 | 2.22 | 4.36 | 25 | 55.6 | 0.441 |
| Lung cancer. NSCLC | 10 | 1.3 | 1.9 | 3 | 4 | 40 | 0.755 |
| Lung cancer. small cell cancer | 13 | 1.54 | 1.77 | 3.23 | 6 | 46.2 | 0.985 |
| Lung cancer. SQCC | 57 | 2.19 | 2.84 | 6.26 | 48 | 84.2 | < 0.0001 |
| Lymphoepithelial tumour | 5 | 0.8 | 1.2 | 1.8 | 1 | 20 | n.a. |
| Malignant mesothelioma | 24 | 2.08 | 2.25 | 5.17 | 14 | 58.3 | n.a. |
| Mucoepidermoid carcinoma | 17 | 2.12 | 2.59 | 5.88 | 14 | 82.4 | n.a. |
| Oral cavity. carcinoma |
53 |
1.81 |
2.34 |
4.72 |
29 |
54.7 |
n.a. |
|
Parotis | |||||||
| Parotis. normal | 10 | 1.8 | 1.9 | 3.9 | 5 | 50 | |
| Parotis. pleomorphic adenoma | 60 | 1.02 | 1.18 | 1.57 | 7 | 11.7 | 0.003 |
| Warthin's tumour |
54 |
2.76 |
3.13 |
9.2 |
46 |
85.2 |
0.011 |
|
Gastrointestinal tumours | |||||||
|
Colon | |||||||
| Colon. normal | 14 | 2.38 | 2.69 | 6.54 | 12 | 92.3 | |
| Colon adenoma. low grade | 45 | 2.49 | 2.27 | 5.82 | 36 | 80 | 0.301 |
| Colon adenoma. high grade | 30 | 2.77 | 2 | 5.63 | 21 | 70 | 0.112 |
| Colon cancer |
59 |
2.12 |
1.67 |
3.98 |
30 |
50 |
0.005 |
|
Esophagus | |||||||
| Esophagus. normal | 5 | 1.8 | 3.2 | 6.2 | 4 | 80 | |
| Esophageal carcinoma. adenocarcinoma | 59 | 2.29 | 2.24 | 5.19 | 40 | 67.8 | 0.572 |
| Esophageal carcinoma. SQCC |
56 |
2.41 |
2.48 |
6.09 |
48 |
85.7 |
0.73 |
|
Gall bladder | |||||||
| Gall bladder. normal | 9 | 2.56 | 3.11 | 8.44 | 8 | 88.9 | |
| Gall bladder carcinoma | 25 | 2.08 | 2.12 | 4.68 | 15 | 60 | 0.112 |
| Gastrointestinal stroma tumour (GIST) |
46 |
1.59 |
2.37 |
4.09 |
21 |
45.7 |
n.a. |
|
Liver | |||||||
| Liver. normal | 5 | 2.6 | 4 | 10.4 | 5 | 100 | |
| Hepatocellular carcinoma |
53 |
2.43 |
3 |
7.77 |
45 |
84.9 |
0.349 |
|
Pancreas | |||||||
| Pancreas. normal | 10 | 2.3 | 2.5 | 6.1 | 8 | 80 | |
| Pancreatic cancer. ductal adenocarcinoma | 53 | 2.11 | 2.13 | 4.72 | 40 | 75.5 | 0.758 |
| Pancreatic cancer. neuroendocrine | 18 | 1.89 | 2.61 | 5.11 | 13 | 72.2 | 0.649 |
| Pancreatic cancer. papilla. adeno |
28 |
2.36 |
2.32 |
5.71 |
23 |
82.1 |
0.881 |
|
Small intestine | |||||||
| Small intestine. normal | 7 | 1.57 | 2.29 | 4 | 3 | 42.9 | |
| Small intestine carcinoma |
22 |
1.86 |
1.82 |
3.45 |
11 |
50 |
0.742 |
|
Stomach | |||||||
| Stomach. normal | 5 | 2.6 | 3.2 | 8.6 | 4 | 80 | |
| Stomach cancer. diffuse type | 54 | 1.54 | 1.85 | 3.15 | 20 | 37 | 0.061 |
| Stomach cancer. intestinal type | 56 | 1.55 | 2.23 | 3.52 | 22 | 39.3 | 0.078 |
| Oncocytoma |
62 |
2.48 |
2.94 |
7.71 |
54 |
87.1 |
0.654 |
|
Anal skin | |||||||
| Anal skin. normal | 5 | 2 | 2 | 4.2 | 1 | 20 | |
| Anal cancer |
15 |
1.8 |
2.13 |
3.93 |
7 |
46.7 |
0.292 |
|
Gynecological tumours | |||||||
|
Breast | |||||||
| Breast. normal | 11 | 1.18 | 1.18 | 1.64 | 2 | 18.2 | |
| Breast cancer. apocrine carcinoma | 14 | 1 | 2 | 2.57 | 3 | 21.4 | 0.84 |
| Breast cancer. ductal carcinoma | 60 | 0.75 | 0.97 | 1.18 | 5 | 8.3 | 0.314 |
| Breast cancer. kribriform carcinoma | 24 | 0.79 | 0.92 | 1.29 | 2 | 8.3 | 0.395 |
| Breast cancer. lobulary carcinoma | 64 | 0.42 | 0.5 | 0.59 | 3 | 4.7 | 0.097 |
| Breast cancer. medullary carcinoma | 63 | 1.17 | 1.62 | 2.32 | 14 | 22.2 | 0.764 |
| Breast cancer. mucinous carcinoma | 59 | 1.03 | 1.42 | 2.05 | 12 | 20.3 | 0.87 |
| Breast cancer. phylloid carcinoma | 47 | 0.62 | 0.7 | 0.83 | 2 | 4.3 | 0.101 |
| Breast cancer. tubulary carcinoma |
58 |
1.21 |
1.36 |
2.45 |
14 |
24.1 |
0.668 |
|
Cervix | |||||||
| Cervix. normal | 4 | 1.5 | 2 | 3.5 | 1 | 25 | |
| Cervical cancer. adenocarcinoma | 42 | 2.4 | 2.48 | 6.64 | 33 | 78.6 | 0.02 |
| Cervical cancer. adenosquamous carcinoma | 2 | 2 | 1.5 | 3.5 | 1 | 50 | 0.54 |
| Cervical cancer. SQCC |
63 |
2.13 |
2.83 |
6.29 |
52 |
82.5 |
0.006 |
|
Endometrium | |||||||
| Endometrium. normal | 19 | 1.26 | 1.32 | 2 | 4 | 21.1 | |
| Endometrial cancer. endometroid carcinoma | 60 | 2.62 | 2.9 | 7.75 | 55 | 91.7 | < 0.0001 |
| Endometrial cancer. serous carcinoma |
53 |
2.13 |
2.4 |
5.51 |
40 |
75.5 |
< 0.0001 |
|
Ovary | |||||||
| Ovar. normal | 4 | 1.25 | 0.75 | 1.25 | 0 | 0 | |
| Ovarian cancer. brenner tumour | 40 | 2.05 | 2.38 | 5.53 | 22 | 55 | 0.036 |
| Ovarian cancer. endometroid carcinoma | 22 | 2.9 | 3 | 8.81 | 21 | 100 | < 0.0001 |
| Ovarian cancer. mucinous carcinoma | 44 | 2.25 | 3.07 | 7.09 | 44 | 90 | < 0.0001 |
| Ovarian cancer. serous carcinoma | 63 | 1.87 | 2.87 | 5.62 | 47 | 74.6 | 0.002 |
| Teratoma |
57 |
1.37 |
1.33 |
2.74 |
18 |
31.6 |
0.181 |
|
Vagina | |||||||
| Vagina. normal | 5 | 1.8 | 1.4 | 3.4 | 2 | 40 | |
| Vagina carcinoma. SQCC |
20 |
2.05 |
2.3 |
5 |
14 |
70 |
0.211 |
|
Vulva | |||||||
| Vulva. normal | 4 | 2.25 | 2.5 | 5.25 | 3 | 75 | |
| Vulva carcinoma. SQCC |
60 |
2.02 |
3.1 |
6.32 |
54 |
90 |
0.352 |
|
Genitourinary tract tumours | |||||||
|
Testis | |||||||
| Testis. normal | 5 | 1.8 | 1.8 | 3.6 | 2 | 40 | |
| Testis. non-seminoma | 44 | 1.18 | 1.77 | 2.75 | 13 | 29.5 | 0.631 |
| Testis. seminoma |
92 |
1.27 |
2.02 |
2.71 |
24 |
26.1 |
0.494 |
|
Penis | |||||||
| Penis. normal | 5 | 1.4 | 2.2 | 3.4 | 2 | 40 | |
| Penile carcinoma |
46 |
1.5 |
2.28 |
3.8 |
21 |
45.7 |
0.809 |
|
Prostate | |||||||
| Prostate. normal | 26 | 1.88 | 2.54 | 5.15 | 18 | 69.2 | |
| Prostate cancer |
63 |
0.86 |
1.1 |
1.33 |
5 |
7.9 |
< 0.0001 |
|
Renal cell cancer | |||||||
| Kidney. normal | 19 | 1.95 | 2.68 | 5.79 | 14 | 73.7 | |
| Renal cell cancer. chromophobic | 56 | 1.95 | 2.98 | 5.95 | 44 | 78.6 | 0.66 |
| Renal cell cancer. clear cell | 68 | 1 | 1.38 | 1.62 | 4 | 5.9 | <0.0001 |
| Renal cell cancer. papillary | 31 | 1.68 | 2.58 | 4.58 | 18 | 58.1 | 0.264 |
| Renal cell cancer. colibri |
9 |
0.89 |
2.22 |
2.56 |
3 |
33.3 |
0.041 |
|
Urinary bladder | |||||||
| Urinary bladder. normal | 5 | 1 | 1 | 1.2 | 0 | 0 | |
| Urinary bladder cancer. non-invasive (pTa) | 60 | 2.75 | 3.35 | 9.67 | 53 | 88.3 | <0.0001 |
| Urinary bladder cancer. invasive (pT2-4) | 60 | 1.88 | 2.2 | 4.42 | 34 | 56.7 | 0.015 |
| Urinary bladder cancer. colibri |
10 |
1.4 |
2.1 |
2.9 |
3 |
30 |
0.171 |
|
Neuroendocrine tumours | |||||||
|
Adrenal cortex | |||||||
| Adrenal cortex. normal | 5 | 1 | 1.6 | 1.6 | 0 | 0 | |
| Adrenal cortex. adenoma | 21 | 1.52 | 2.38 | 3.62 | 8 | 38.1 | 0.097 |
| Adrenal cortex. carcinoma | 8 | 1.63 | 2.63 | 4.63 | 3 | 37.5 | 0.118 |
| Carcinoid | 38 | 1.92 | 2.26 | 4.47 | 23 | 60.5 | n.a. |
| Paraganglioma | 34 | 1.88 | 2.41 | 4.82 | 19 | 55.9 | n.a. |
| Phaeochromocytoma |
65 |
1.16 |
1.73 |
2.5 |
16 |
25 |
0.202 |
|
Thyroid | |||||||
| Thyroid. normal | 4 | 1 | 0.75 | 1.5 | 1 | 25 | |
| Thyroid carcinoma. anaplastic | 3 | 1.67 | 1 | 1.67 | 0 | 0 | 0.35 |
| Thyroid carcinoma. follicular | 46 | 2.5 | 2.63 | 7.13 | 37 | 80.4 | 0.013 |
| Thyroid carcinoma. medullary | 25 | 2.24 | 2.08 | 4.6 | 15 | 60 | 0.191 |
| Thyroid carcinoma. papillary | 47 | 2.51 | 2.62 | 7.13 | 40 | 85.1 | 0.004 |
| Thyroid. adenoma |
62 |
2.82 |
2.89 |
8.3 |
56 |
90.3 |
< 0.0001 |
|
Hematological neoplasias | |||||||
|
Lymph node | |||||||
| Lymph node. normal | 20 | 1.1 | 1.25 | 2.2 | 5 | 25 | |
| Hodgkin's lymphoma | 38 | 1.29 | 1.5 | 1.95 | 5 | 13.2 | 0.256 |
| Non-Hodgkin's lymphoma |
8 |
1.38 |
1.25 |
2.13 |
1 |
12.5 |
0.466 |
|
Thymus | |||||||
| Thymus. normal | 4 | 0.25 | 0.25 | 0.25 | 0 | 0 | |
| Thymoma |
55 |
1.24 |
1.76 |
2.84 |
20 |
36.4 |
0.138 |
|
Neuronal tumours | |||||||
|
Brain | |||||||
| Brain. normal | 5 | 0 | 0 | 0 | 0 | 0 | |
| Astrocytoma | 37 | 1.11 | 1.68 | 2.46 | 11 | 29.7 | 0.156 |
| Ependymoma | 10 | 1.1 | 1.1 | 1.4 | 1 | 10 | 0.464 |
| Medulloblastoma | 4 | 1.75 | 1.5 | 2.75 | 2 | 50 | 0.073 |
| Oligodendroglioma | 23 | 1.39 | 1.52 | 2.74 | 7 | 30.4 | 0.154 |
| Neuroblastoma |
48 |
1.88 |
1.9 |
4.17 |
26 |
54.2 |
0.021 |
|
Soft tissue tumours | |||||||
|
Muscle | |||||||
| Muscle. normal | 14 | 1.64 | 1.79 | 3.29 | 5 | 35.7 | |
| Angiosarcoma | 7 | 1.57 | 1.43 | 2.43 | 2 | 28.6 | 0.743 |
| Dermatofibrosarcoma protuberans | 5 | 0.8 | 1 | 1.4 | 1 | 20 | 0.516 |
| Carcinosarcoma | 36 | 0.94 | 1.58 | 1.92 | 7 | 19.4 | 0.226 |
| Desmoid tumour |
9 |
0.44 |
0.67 |
0.67 |
0 |
0 |
0.043 |
|
Tendon sheat | |||||||
| Tendon sheat. normal | 2 | 2 | 2.5 | 5.5 | 1 | 50 | |
| Giant cell tumour of the tendon sheat | 33 | 2.03 | 2.3 | 4.73 | 24 | 72.7 | 0.49 |
| Granular cell cancer | 7 | 1.29 | 1.71 | 2.43 | 2 | 28.6 | n.a. |
| Haemangiopericytoma | 7 | 1.29 | 1.57 | 2.43 | 2 | 28.6 | n.a. |
| Leiomyoma | 24 | 1.29 | 1.96 | 3.17 | 11 | 45.8 | 0.542 |
| Leiomyosarcoma | 28 | 1.36 | 2.25 | 3.61 | 12 | 42.9 | 0.657 |
| Liposarcoma | 16 | 0.81 | 1.38 | 1.5 | 2 | 12.5 | n.a. |
| Malignant fibrous histiocytoma | 24 | 1.08 | 1.42 | 2.21 | 4 | 16.7 | n.a. |
| Malignant schwannoma | 14 | 1.14 | 1.14 | 2 | 2 | 14.3 | n.a. |
| Neurofibroma | 49 | 1.02 | 1.12 | 2.04 | 10 | 20.4 | n.a. |
| Stroma sarcoma |
11 |
1 |
1.64 |
2.09 |
3 |
27.3 |
0.653 |
|
Bone tumours | |||||||
| Chondrosarcoma |
4 |
1.75 |
2 |
4.25 |
2 |
50 |
n.a. |
|
Skin tumours | |||||||
|
Skin | |||||||
| Skin. normal | 17 | 1.65 | 1.76 | 3.29 | 7 | 41.2 | |
| Basal cell adenoma | 34 | 1.56 | 1.59 | 2.74 | 14 | 41.2 | 1 |
| Basalioma | 46 | 2.54 | 3.04 | 7.91 | 42 | 91.3 | < 0.0001 |
| Malignant melanoma | 30 | 1.43 | 1.83 | 2.9 | 9 | 30 | 0.437 |
| Merkel cell cancer |
2 |
1 |
1 |
1 |
0 |
0 |
0.253 |
|
Naevus | |||||||
| Naevus. benign | 40 | 2.08 | 2.08 | 4.55 | 24 | 60 | 0.192 |
| Pilomatrixoma | 38 | 1.39 | 1.55 | 2.82 | 16 | 41.2 | 0.949 |
| Skin cancer. SQCC | 45 | 1.6 | 2.36 | 3.82 | 22 | 48.9 | 0.587 |
NOTE. All tissue samples were derived from surgically removed specimens. Results were calculated either by Fisheŕs exact test or by Chi-square test when applicable. Significant results for differential presence compared with available normal controls are shown in bold.
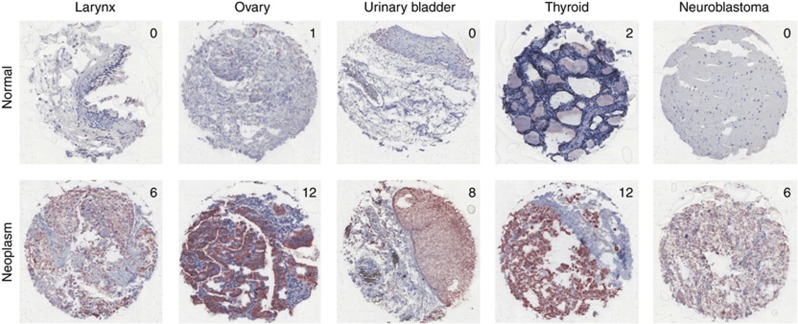
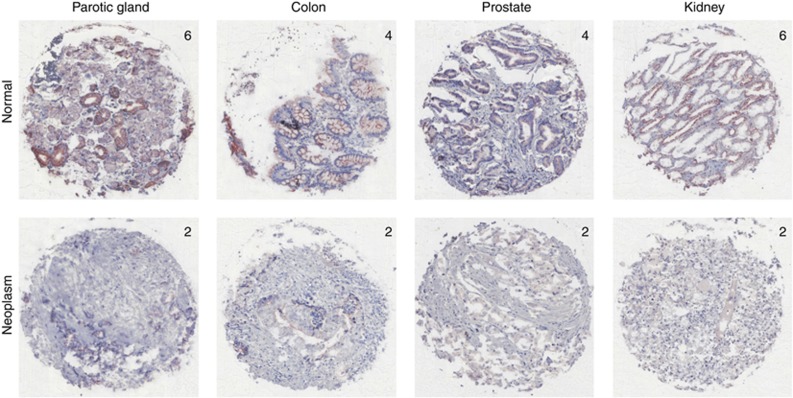
In comparison with healthy controls, significantly increased numbers of CAR-positive cases were found in basalioma, larynx carcinoma, Warthin's tumour, lung cancer, cervical cancer, endometrial cancer, ovarian cancer, urinary bladder cancer, thyroid adenoma and carcinoma, as well as in neuroblastoma (Table 1; Figure 3). On the other hand, significantly lower CAR expression levels were seen in pleomorphic adenoma of the parotid gland, colon cancers, prostate cancers, as well as subtypes of renal cell cancers (Table 1; Figure 4). To assess whether CAR presence correlates with clinicopathological parameters, we compared our findings for CAR immunopositivity with tumour grade (G), local tumour growth (T-category) and nodal status (N-category) where applicable, revealing the loss of CAR in locally advanced colon cancers (Table 2).
Figure 3.
CAR overexpression in neoplasms: Representative examples of entities displaying elevated CAR protein expression compared with respective normal controls (numbers = IRS of the individual specimen). Ovary: endometroid carcinoma; urinary bladder cancer: non-invasive/pTa; thyroid: papillary carcinoma.
Figure 4.
Loss of CAR expression in neoplasms: Typical sites with significant down regulation of CAR protein expression (numbers = IRS of the individual specimen). Kidney: clear cell renal cancer.
Table 2. CAR presence and clinico-pathological parameters.
| |
pT |
|
pN |
|
G |
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Tissue type | 1 | 2 | 3 | 4 | P-value | 0 | 1 | 2 | 3 | P-value | 1 | 2 | 3 | 4 | P-value |
| Larynx. carcinoma |
|
|
|
|
0.283 |
|
|
|
|
0.749 |
|
|
|
|
0.302 |
| CAR− | 0 | 2 | 2 | 4 | 5 | 1 | 0 | 0 | 1 | 8 | 1 | ||||
| CAR+ |
|
3 |
8 |
6 |
4 |
|
9 |
2 |
2 |
1 |
|
0 |
20 |
3 |
|
| Lung cancer. adenocarcinoma |
|
|
|
|
0.581 |
|
|
|
|
0.073 |
|
|
|
|
0.452 |
| CAR− | 2 | 5 | 0 | 0 | 0 | 5 | 1 | 0 | 3 | 4 | |||||
| CAR+ |
9 |
13 |
3 |
4 |
|
11 |
6 |
6 |
1 |
|
|
13 |
9 |
|
|
| Lung cancer. bronchioalveolary carcinoma |
|
|
|
|
0.956 |
|
|
|
|
0.307 |
|
|
|
|
0.018 |
| CAR− | 1 | 2 | 3 | 0 | 0 | 2 | 0 | 0 | |||||||
| CAR+ |
2 |
4 |
|
|
|
2 |
2 |
1 |
|
|
0 |
4 |
2 |
|
|
| Lung cancer. large cell cancer |
|
|
|
|
0.274 |
|
|
|
|
0.222 |
|
|
|
|
0.147 |
| CAR− | 0 | 2 | 1 | 0 | 1 | 1 | 1 | 3 | 0 | ||||||
| CAR+ |
1 |
6 |
2 |
2 |
|
4 |
4 |
3 |
|
|
|
|
2 |
2 |
|
| Lung cancer. NSCLC |
|
|
|
|
0.664 |
|
|
|
|
0.444 |
|
|
|
|
0.816 |
| CAR - | 3 | 3 | 1 | 1 | 5 | 1 | 0 | 0 | 6 | 2 | |||||
| CAR + |
7 |
23 |
6 |
3 |
|
15 |
14 |
6 |
1 |
|
|
26 |
7 |
|
|
| Colon cancer |
|
|
|
|
0.010 |
|
|
|
|
|
|
|
|
|
|
| CAR− | 0 | 3 | 14 | 12 | |||||||||||
| CAR+ |
2 |
8 |
8 |
5 |
|
|
|
|
|
|
|
|
|
|
|
| Cervical cancer. adenocarcinoma |
|
|
|
|
0.513 |
|
|
|
|
0.457 |
|
|
|
|
0.112 |
| CAR− | 5 | 0 | 3 | 0 | 2 | 0 | 1 | ||||||||
| CAR+ |
18 |
4 |
|
|
|
10 |
5 |
|
|
|
2 |
7 |
5 |
|
|
| Cervical cancer. SQCC |
|
|
|
|
0.248 |
|
|
|
|
0.966 |
|
|
|
|
0.662 |
| CAR− | 10 | 1 | 0 | 7 | 2 | 4 | 7 | ||||||||
| CAR+ |
31 |
12 |
|
3 |
|
31 |
11 |
|
|
|
|
13 |
31 |
|
|
| Endometrial cancer. endometroid |
|
|
|
|
0.646 |
|
|
|
|
0.535 |
|
|
|
|
0.922 |
| CAR− | 5 | 0 | 0 | 1 | 1 | 3 | 1 | 1 | |||||||
| CAR+ |
41 |
7 |
5 |
|
|
20 |
4 |
|
|
|
29 |
15 |
9 |
|
|
| Ovarian cancer. mucinous |
|
|
|
|
0.438 |
|
|
|
|
0.803 |
|
|
|
|
0.275 |
| CAR− | 0 | 0 | 0 | 0 | 0 | 2 | 0 | ||||||||
| CAR+ |
11 |
1 |
|
|
|
3 |
1 |
|
|
|
9 |
9 |
4 |
|
|
| Ovarian cancer. serous |
|
|
|
|
0.312 |
|
|
|
|
0.876 |
|
|
|
|
0.120 |
| CAR− | 1 | 0 | 3 | 3 | 5 | 1 | 2 | 12 | |||||||
| CAR+ |
1 |
3 |
17 |
|
|
11 |
12 |
|
|
|
2 |
20 |
25 |
|
|
| Renal cell cancer. chromophobic |
|
|
|
|
0.523 |
|
|
|
|
|
|
|
|
|
0.553 |
| CAR− | 3 | 2 | 1 | 0 | 5 | 1 | 0 | ||||||||
| CAR+ |
16 |
10 |
6 |
|
|
|
|
|
|
|
5 |
23 |
2 |
2 |
|
| Renal cell cancer. colibri |
|
|
|
|
0.337 |
|
|
|
|
0.392 |
|
|
|
|
0.809 |
| CAR− | 2 | 1 | 3 | 2 | 1 | 1 | 3 | 1 | |||||||
| CAR + |
0 |
1 |
1 |
|
|
0 |
2 |
0 |
|
|
|
2 |
1 |
|
|
| Renal cell cancer. clear cell |
|
|
|
|
0.949 |
|
|
|
|
0.465 |
|
|
|
|
0.635 |
| CAR− | 46 | 3 | 13 | 16 | 2 | 8 | 45 | 8 | |||||||
| CAR+ |
3 |
0 |
1 |
|
|
0 |
0 |
|
|
|
0 |
3 |
1 |
|
|
| Renal cell cancer. papillary |
|
|
|
|
0.123 |
|
|
|
|
0.100 |
|
|
|
|
0.495 |
| CAR− | 9 | 2 | 2 | 1 | 2 | 3 | 9 | 1 | |||||||
| CAR+ |
12 |
2 |
0 |
|
|
0 |
0 |
|
|
|
5 |
11 |
0 |
|
|
| Urinary bladder cancer. colibri |
|
|
|
|
0.724 |
|
|
|
|
0.665 |
|
|
|
|
|
| CAR− | 1 | 1 | 2 | 2 | 5 | 1 | 1 | ||||||||
| CAR+ |
0 |
1 |
1 |
0 |
|
2 |
0 |
1 |
|
|
|
|
|
|
|
| Urinary bladder cancer |
|
|
|
|
0.622 |
|
|
|
|
0.101 |
|
|
|
|
0.234 |
| CAR− | 21 | 5 | 0 | 1 | 2 | 0 | 3 | 23 | |||||||
| CAR+ |
|
28 |
5 |
1 |
|
0 |
0 |
3 |
|
|
|
8 |
26 |
|
|
| Thyroid carcinoma. follicular |
|
|
|
|
0.468 |
|
|
|
|
|
|
|
|
|
|
| CAR− | 7 | 2 | 0 | 0 | |||||||||||
| CAR + |
29 |
3 |
4 |
1 |
|
|
|
|
|
|
|
|
|
|
|
| Thyroid carcinoma. medullary |
|
|
|
|
0.551 |
|
|
|
|
0.672 |
|
|
|
|
|
| CAR− | 0 | 2 | 1 | 0 | 0 | 1 | |||||||||
| CAR+ |
1 |
4 |
0 |
1 |
|
1 |
2 |
|
|
|
|
|
|
|
|
| Thyroid carcinoma. papillary |
|
|
|
|
0.383 |
|
|
|
|
0.673 |
|
|
|
|
0.386 |
| CAR− | 0 | 4 | 1 | 0 | 0 | 1 | 1 | 0 | |||||||
| CAR+ | 8 | 16 | 4 | 7 | 4 | 6 | 1 | 1 | |||||||
NOTE. Results were calculated by Chi-square test for neoplasms showing differential CAR presence compared with respective controls. Significant results are shown in bold.
Table 3. Findings of differential CAR presence in human neoplasms compared with previous publications.
|
CAR upregulation | ||
| Basalioma | N | — |
| Thyroid adenoma | N | — |
| Warthin's tumours | N | — |
| Laryngeal cancer | N | — |
| Thyroid carcinoma | A | (Marsee et al, 2005; Giaginis et al, 2010) |
| Lung cancer | A | (Wang et al, 2006; Chen et al 2013) |
| Neuroblastomas | A | (Persson et al, 2006) |
| Medulloblastomas | A | (Persson et al, 2006) |
| Endometrial cancer | A | (Giaginis et al, 2008) |
| Ovarian cancer | A | (Reimer et al, 2007) |
| Cervical carcinoma | A | (Dietel et al, 2011) |
| Non-invasive urinary bladder cancer |
D |
(Sachs et al, 2002; Matsumoto et al, 2005; Buscarini et al, 2007) |
|
CAR downregulation | ||
| Pleomorphic adenoma (parotid gland) | N | — |
| Colon | A | (Korn et al, 2006; Zhang et al, 2008; Stecker et al, 2011) |
| Prostate | A | (Rauen et al, 2002) |
| Kidney | A | (Okegawa et al, 2001) |
Abbreviations: A=agreement; D=disagreement; N=new finding.
Discussion
Our data provide insight into CAR expression levels in a broad range of neoplasias and their corresponding normal tissues, including several that have not been investigated before. As the samples in our analysis were all stained in one procedure, the results allow for a direct comparison between different entities for the first time. It reveals considerable differences in CAR expression levels and confirms the hypothesis of entity-specific expression pattern. Hereby, our data may provide a basis to gain further insight into the complex and potentially organ-site-specific function and regulation of CAR during carcinogenesis. Furthermore, entities with high CAR presence identified by our study may pose promising targets for therapeutic adenoviruses.
In normal tissues, our observations of high CAR expression level within the liver, stomach, colon and pancreas are in agreement with previous reports (Korn et al, 2006; Anders et al, 2009; Stecker et al, 2011). Our finding of profuse immunopositivity within the gall bladder marks the first description of this phenomenon to our best knowledge. These data suggest a particular impact of CAR within the gastrointestinal tract. In line with this hypothesis, functional CAR knockout in a murine model led to a dilated intestinal tract (Pazirandeh et al, 2011). Despite abundant CAR presence, symptomatic infections of these organs by Adeno- and Coxsackieviruses are rare because of the limited access to CAR and acquired immunity. Nevertheless, high CAR expression within the liver may lead to substantial unwanted sequestering of systemically administered therapeutic adenoviruses (Arnberg, 2012). In contrast, we found no detectable CAR immunopositivity within the brain, in line with previous studies showing the white matter being CAR-negative and scattered CAR-positive neurons within the neocortex and in ependymal cells only (Johansson et al, 1999; Persson et al, 2006).
In several early neoplasms, we did note significantly elevated CAR expression levels. Hereby, our finding of increased CAR expression in basaliomas, thyroid adenomas and Warthin's tumours – benign neoplasms of the salivary glands – are the first description of this fact to our best knowledge. The later might be of particular interest, as we did note a significant impairment of CAR in pleomorphic adenoma of the parotid gland also. The functional impact of this result, however, remains to be elucidated.
In cancer types, our finding of significant CAR increase in laryngeal carcinoma marks the first description of this phenomenon to our best knowledge. Therefore, our data may initiate further studies in this entity. In line with prior reports, we noted abundant CAR presence in several subtypes of thyroid carcinoma (Marsee et al, 2005; Giaginis et al, 2010), in lung cancer (Wang et al, 2006; Chen et al, 2013), as well as in neuro- and medulloblastomas (Persson et al, 2006). Moreover, in agreement with previous reports we found significantly hightened CAR presence in cancers of the endometrium (Giaginis et al, 2008), ovary (Reimer et al, 2007) and cervix (Dietel et al, 2011). These data suggest that CAR overexpression occurs preferentially in cancers of the female reproductive system, contrary to reduced CAR presence in neoplasms of the testis and prostate. The reason for this phenomenon remains unclear, yet hormone-driven effects might be of particular interest. Previously, an increased CAR expression by treatment with estradiol was found in hormone receptor-positive breast cancer and ovarian cancer cell lines (You et al, 2001; Auer et al, 2009). Furthermore, our data imply that CAR may have little impact on breast cancer as we did not observe distinct expression changes in breast epithelium in contrast to a previous study, describing elevated transcriptional CAR expression in breast cancers (Martin et al, 2005).
In disagreement with previous reports we noted a significantly increased CAR presence in non-invasive urinary bladder cancers because of the low presence of CAR in normal urinary bladder samples (Sachs et al, 2002; Matsumoto et al, 2005; Buscarini et al, 2007). These differences might be caused by methodical differences such as the use of different antibodies, yet may be explained by the limited number of healthy cases in our study also.
Concerning CAR downregulation, our finding in cancer types is in agreement with previous studies for the colon (Korn et al, 2006; Zhang et al, 2008; Stecker et al, 2011), prostate (Rauen et al, 2002) and kidney (Okegawa et al, 2001).
For a subset of entities, access to clinicopathological data allowed for further analysis of potential relations to CAR presence. Our finding of CAR downregulation in locally advanced colon cancers underlines the concept of CAR's tumour-suppressive role in this entity (Stecker et al, 2011). However, as our study aims for a comprehensive evaluation of neoplasms, it is limited although concerning sample numbers and clinicopathological data for individual entities. Therefore, our study potentially underestimates associations between CAR and clinicopathological properties.
In conclusion, our data suggest that differential expression of CAR in cancer types represents an entity-specific phenomenon with CAR upregulation happening more frequently than its downregulation. These findings shed a new light on CAR regulation in cancer types also. To date, mainly CAR downregulation in cancer types has been investigated. Hereby, activation of the Raf/MEK/ERK pathway and TGF-β signalling, hypoxia, epithelial–mesenchymal transdifferentiation and histone deacetylation of the CAR gene promoter were identified as regulators of CAR expression (Brüning and Runnebaum, 2003; Pong et al, 2003; Anders et al, 2003a; Lacher et al, 2006; Küster et al, 2010a, 2010b; Lacher et al, 2011). In contrast, few studies have investigated the mechanism of CAR upregulation. Therefore, it remains to be elucidated whether the MAPK/ERK1/2 signalling induces CAR expression in other entities than oesophageal squamous cell carcinomas (Ma et al, 2012), and, for instance, whether hormones influence CAR levels in cancer types as discussed above. Furthermore, our findings have potential implications for the understanding of the function of CAR in cancer types. To date, CAR has been mainly attributed cancer-suppressive properties. Previous studies on CAR function, however, have been performed in models of advanced cancer types of the colon, prostate and kidney. However, all these entities do belong to the limited number of sites showing significant CAR downregulation in our study. Upregulation of CAR on the other hand might be suggestive of a tumour-promoting function of CAR in several other organs. In line with this hypothesis, an association has been found between high CAR expression and increased proliferation and/or invasion in endometrial, ovarian, and cervical cancers as well as in lung cancer (Brüning et al, 2005; Giaginis et al, 2008; Dietel et al, 2011; Chen et al, 2013). Furthermore, CAR has been shown to foster early carcinogenesis in ovarian and cervical cancers, with CAR-expressing cell lines displaying less sensitivity towards apoptotic stimuli (Brüning et al, 2005). On the other hand, migration and metastatic phenotypes are being suppressed by CAR overexpression in cell lines derived from the same entities (Brüning and Runnebaum, 2004; Wang et al, 2005). These results underline that off course CAR expression levels per se do not allow for prediction of functional impact. Nevertheless, our findings may serve as a guide to neoplasms potentially influenced by CAR.
Acknowledgments
We thank Jennifer Schröder-Schwarz for technical assistance. This study was supported by a grant from the Deutsche Forschungsgemeinschaft to MA.
Footnotes
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
References
- Anders M, Christian C, McMahon M, McCormick F, Korn WM. Inhibition of the Raf/MEK/ERK pathway up-regulates expression of the coxsackievirus and adenovirus receptor in cancer cells. Cancer Res. 2003a;63:2088–2095. [PubMed] [Google Scholar]
- Anders M, Hansen R, Ding R-X, Rauen KA, Bissell MJ, Korn WM. Disruption of 3D tissue integrity facilitates adenovirus infection by deregulating the coxsackievirus and adenovirus receptor. Proc Natl Acad Sci USA. 2003b;100:1943–1948. doi: 10.1073/pnas.0337599100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders M, Vieth M, Röcken C, Ebert M, Pross M, Gretschel S, Schlag PM, Wiedenmann B, Kemmner W, Höcker M. Loss of the coxsackie and adenovirus receptor contributes to gastric cancer progression. Br J Cancer. 2009;100:352–359. doi: 10.1038/sj.bjc.6604876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnberg N. Adenovirus receptors: implications for targeting of viral vectors. Trends Pharmacol Sci. 2012;33:442–448. doi: 10.1016/j.tips.2012.04.005. [DOI] [PubMed] [Google Scholar]
- Auer D, Reimer D, Porto V, Fleischer M, Roessler J, Wiedemair A, Marth C, Müller-Holzner E, Daxenbichler G, Zeimet AG. Expression of coxsackie-adenovirus receptor is related to estrogen sensitivity in breast cancer. Breast Cancer Res Treat. 2009;116:103–111. doi: 10.1007/s10549-008-0108-0. [DOI] [PubMed] [Google Scholar]
- Bergelson JM, Cunningham JA, Droguett G, Kurt-Jones EA, Krithivas A, Hong JS, Horwitz MS, Crowell RL, Finberg RW. Isolation of a common receptor for Coxsackie B viruses and adenoviruses 2 and 5. Science. 1997;275:1320–1323. doi: 10.1126/science.275.5304.1320. [DOI] [PubMed] [Google Scholar]
- Brüning A, Runnebaum IB. CAR is a cell-cell adhesion protein in human cancer cells and is expressionally modulated by dexamethasone, TNFalpha, and TGFbeta. Gene Ther. 2003;10:198–205. doi: 10.1038/sj.gt.3301887. [DOI] [PubMed] [Google Scholar]
- Brüning A, Runnebaum IB. The coxsackie adenovirus receptor inhibits cancer cell migration. Exp Cell Res. 2004;298:624–631. doi: 10.1016/j.yexcr.2004.05.001. [DOI] [PubMed] [Google Scholar]
- Brüning A, Stickeler E, Diederich D, Walz L, Rohleder H, Friese K, Runnebaum IB. Coxsackie and adenovirus receptor promotes adenocarcinoma cell survival and is expressionally activated after transition from preneoplastic precursor lesions to invasive adenocarcinomas. Clin Cancer Res. 2005;11:4316–4320. doi: 10.1158/1078-0432.CCR-04-2370. [DOI] [PubMed] [Google Scholar]
- Buscarini M, Quek ML, Gilliam-Hegarich S, Kasahara N, Bochner B. Adenoviral receptor expression of normal bladder and transitional cell carcinoma of the bladder. Urol Int. 2007;78:160–166. doi: 10.1159/000098076. [DOI] [PubMed] [Google Scholar]
- Chen Z, Wang Q, Sun J, Gu A, Jin M, Shen Z, Qiu Z, Wang J, Wang X, Zhan Z, Li J-W. Expression of the coxsackie and adenovirus receptor in human lung cancers. Tumour Biol. 2013;34:17–24. doi: 10.1007/s13277-012-0342-2. [DOI] [PubMed] [Google Scholar]
- Cohen CJ, Shieh JT, Pickles RJ, Okegawa T, Hsieh JT, Bergelson JM. The coxsackievirus and adenovirus receptor is a transmembrane component of the tight junction. Proc Natl Acad Sci USA. 2001;98:15191–15196. doi: 10.1073/pnas.261452898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietel M, Häfner N, Jansen L, Dürst M, Runnebaum IB. Novel splice variant CAR 4/6 of the coxsackie adenovirus receptor is differentially expressed in cervical carcinogenesis. J Mol Med. 2011;89:621–630. doi: 10.1007/s00109-011-0742-6. [DOI] [PubMed] [Google Scholar]
- Giaginis C, Zarros A, Alexandrou P, Klijanienko J, Delladetsima I, Theocharis S. Evaluation of coxsackievirus and adenovirus receptor expression in human benign and malignant thyroid lesions. Apmis. 2010;118:210–221. doi: 10.1111/j.1600-0463.2009.02582.x. [DOI] [PubMed] [Google Scholar]
- Giaginis CT, Zarros AC, Papaefthymiou MA, Papadopouli AE, Sfiniadakis IK, Theocharis SE. Coxsackievirus and adenovirus receptor expression in human endometrial adenocarcinoma: possible clinical implications. World J Surg Oncol. 2008;6:59. doi: 10.1186/1477-7819-6-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heideman DA, Snijders PJ, Craanen ME, Bloemena E, Meijer CJ, Meuwissen SG, van Beusechem VW, Pinedo HM, Curiel DT, Haisma HJ, Gerritsen WR. Selective gene delivery toward gastric and oesophageal adenocarcinoma cells via EpCAM-targeted adenoviral vectors. Cancer Gene Ther. 2001;8:342–351. doi: 10.1038/sj.cgt.7700313. [DOI] [PubMed] [Google Scholar]
- Huang K-C, Altinoz M, Wosik K, Larochelle N, Koty Z, Zhu L, Holland PC, Nalbantoglu J. Impact of the coxsackie and adenovirus receptor (CAR) on glioma cell growth and invasion: requirement for the C-terminal domain. Int J Cancer. 2005;113:738–745. doi: 10.1002/ijc.20623. [DOI] [PubMed] [Google Scholar]
- Johansson CB, Momma S, Clarke DL, Risling M, Lendahl U, Frisén J. Identification of a neural stem cell in the adult mammalian central nervous system. Cell. 1999;96:25–34. doi: 10.1016/s0092-8674(00)80956-3. [DOI] [PubMed] [Google Scholar]
- Kasuya H, Takeda S, Shimoyama S, Shikano T, Nomura N, Kanazumi N, Nomoto S, Sugimoto H, Nakao A. Oncolytic virus therapy—foreword. Curr Cancer Drug Targets. 2007;7:123–125. doi: 10.2174/156800907780058826. [DOI] [PubMed] [Google Scholar]
- Korn WM, Macal M, Christian C, Lacher MD, McMillan A, Rauen KA, Warren RS, Ferrell L. Expression of the coxsackievirus- and adenovirus receptor in gastrointestinal cancer correlates with tumour differentiation. Cancer Gene Ther. 2006;13:792–797. doi: 10.1038/sj.cgt.7700947. [DOI] [PubMed] [Google Scholar]
- Küster K, Grötzinger C, Koschel A, Fischer A, Wiedenmann B, Anders M. Sodium butyrate increases expression of the coxsackie and adenovirus receptor in colon cancer cells. Cancer Invest. 2010a;28:268–274. doi: 10.3109/07357900902783195. [DOI] [PubMed] [Google Scholar]
- Küster K, Koschel A, Rohwer N, Fischer A, Wiedenmann B, Anders M. Downregulation of the coxsackie and adenovirus receptor in cancer cells by hypoxia depends on HIF-1alpha. Cancer Gene Ther. 2010b;17:141–146. doi: 10.1038/cgt.2009.49. [DOI] [PubMed] [Google Scholar]
- Lacher MD, Shiina M, Chang P, Keller D, Tiirikainen MI, Korn WM. ZEB1 limits adenoviral infectability by transcriptionally repressing the coxsackie virus and adenovirus receptor. Mol Cancer. 2011;10:91. doi: 10.1186/1476-4598-10-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacher MD, Tiirikainen MI, Saunier EF, Christian C, Anders M, Oft M, Balmain A, Akhurst RJ, Korn WM. Transforming growth factor-beta receptor inhibition enhances adenoviral infectability of carcinoma cells via up-regulation of Coxsackie and Adenovirus Receptor in conjunction with reversal of epithelial-mesenchymal transition. Cancer Res. 2006;66:1648–1657. doi: 10.1158/0008-5472.CAN-05-2328. [DOI] [PubMed] [Google Scholar]
- Ma J, Zhao J, Lu J, Jiang Y, Yang H, Li P, Zhao M, Liu K, Dong Z. Coxsackievirus and adenovirus receptor promotes antitumor activity of oncolytic adenovirus H101 in oesophageal cancer. Int J Mol Med. 2012;30:1403–1409. doi: 10.3892/ijmm.2012.1133. [DOI] [PubMed] [Google Scholar]
- Marsee DK, Vadysirisack DD, Morrison CD, Prasad ML, Eng C, Duh Q-Y, Rauen KA, Kloos RT, Jhiang SM. Variable expression of coxsackie-adenovirus receptor in thyroid tumors: implications for adenoviral gene therapy. Thyroid. 2005;15:977–987. doi: 10.1089/thy.2005.15.977. [DOI] [PubMed] [Google Scholar]
- Martin TA, Watkins G, Jiang WG. The Coxsackie-adenovirus receptor has elevated expression in human breast cancer. Clin Exp Med. 2005;5:122–128. doi: 10.1007/s10238-005-0076-1. [DOI] [PubMed] [Google Scholar]
- Martino TA, Petric M, Weingartl H, Bergelson JM, Opavsky MA, Richardson CD, Modlin JF, Finberg RW, Kain KC, Willis N, Gauntt CJ, Liu PP. The coxsackie-adenovirus receptor (CAR) is used by reference strains and clinical isolates representing all six serotypes of coxsackievirus group B and by swine vesicular disease virus. Virology. 2000;271:99–108. doi: 10.1006/viro.2000.0324. [DOI] [PubMed] [Google Scholar]
- Matsumoto K, Shariat SF, Ayala GE, Rauen KA, Lerner SP. Loss of coxsackie and adenovirus receptor expression is associated with features of aggressive bladder cancer. Urology. 2005;66:441–446. doi: 10.1016/j.urology.2005.02.033. [DOI] [PubMed] [Google Scholar]
- Okegawa T, Li Y, Pong RC, Bergelson JM, Zhou J, Hsieh JT. The dual impact of coxsackie and adenovirus receptor expression on human prostate cancer gene therapy. Cancer Res. 2000;60:5031–5036. [PubMed] [Google Scholar]
- Okegawa T, Pong RC, Li Y, Bergelson JM, Sagalowsky AI, Hsieh JT. The mechanism of the growth-inhibitory effect of coxsackie and adenovirus receptor (CAR) on human bladder cancer: a functional analysis of car protein structure. Cancer Res. 2001;61:6592–6600. [PubMed] [Google Scholar]
- Pazirandeh A, Sultana T, Mirza M, Rozell B, Hultenby K, Wallis K, Vennström B, Davis B, Arner A, Heuchel R, Löhr M, Philipson L, Sollerbrant K. Multiple phenotypes in adult mice following inactivation of the Coxsackievirus and Adenovirus Receptor (Car) gene. PLoS One. 2011;6:e20203. doi: 10.1371/journal.pone.0020203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson A, Fan X, Widegren B, Englund E. Cell type- and region-dependent coxsackie adenovirus receptor expression in the central nervous system. J Neurooncol. 2006;78:1–6. doi: 10.1007/s11060-005-9055-3. [DOI] [PubMed] [Google Scholar]
- Pong R-C, Lai Y-J, Chen H, Okegawa T, Frenkel E, Sagalowsky A, Hsieh J-T. Epigenetic regulation of coxsackie and adenovirus receptor (CAR) gene promoter in urogenital cancer cells. Cancer Res. 2003;63:8680–8686. [PubMed] [Google Scholar]
- Raschperger E, Thyberg J, Pettersson S, Philipson L, Fuxe J, Pettersson RF. The coxsackie- and adenovirus receptor (CAR) is an in vivo marker for epithelial tight junctions, with a potential role in regulating permeability and tissue homeostasis. Exp Cell Res. 2006;312:1566–1580. doi: 10.1016/j.yexcr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- Rauen KA, Sudilovsky D, Le JL, Chew KL, Hann B, Weinberg V, Schmitt LD, McCormick F. Expression of the coxsackie adenovirus receptor in normal prostate and in primary and metastatic prostate carcinoma: potential relevance to gene therapy. Cancer Res. 2002;62:3812–3818. [PubMed] [Google Scholar]
- Reimer D, Steppan I, Wiedemair A, Concin N, Hofstetter G, Marth C, Müller-Holzner E, Zeimet AG. Soluble isoforms but not the transmembrane form of coxsackie-adenovirus receptor are of clinical relevance in epithelial ovarian cancer. Int J Cancer. 2007;120:2568–2575. doi: 10.1002/ijc.22580. [DOI] [PubMed] [Google Scholar]
- Sachs MD, Rauen KA, Ramamurthy M, Dodson JL, De Marzo AM, Putzi MJ, Schoenberg MP, Rodriguez R. Integrin alpha(v) and coxsackie adenovirus receptor expression in clinical bladder cancer. Urology. 2002;60:531–536. doi: 10.1016/s0090-4295(02)01748-x. [DOI] [PubMed] [Google Scholar]
- Simon R, Sauter G. Tissue microarrays for miniaturized high-throughput molecular profiling of tumors. Exp Hematol. 2002;30:1365–1372. doi: 10.1016/s0301-472x(02)00965-7. [DOI] [PubMed] [Google Scholar]
- Stecker K, Vieth M, Koschel A, Wiedenmann B, Röcken C, Anders M. Impact of the coxsackievirus and adenovirus receptor on the adenoma-carcinoma sequence of colon cancer. Br J Cancer. 2011;104:1426–1433. doi: 10.1038/bjc.2011.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Chen G, Li F, Zhou J, Lu Y, Ma D.2005Inhibitory effect of coxsackie adenovirus receptor on invasion and metastasis phenotype of ovarian cancer cell line SKOV3 J Huazhong Univ Sci Technol Med Sci 2585–87.93. [DOI] [PubMed] [Google Scholar]
- Wang Y, Wang S, Bao Y, Ni C, Guan N, Zhao J, Salford LG, Widegren B, Fan X. Coxsackievirus and adenovirus receptor expression in non-malignant lung tissues and clinical lung cancers. J Mol Histol. 2006;37:153–160. doi: 10.1007/s10735-006-9055-4. [DOI] [PubMed] [Google Scholar]
- Wunder T, Schmid K, Wicklein D, Groitl P, Dobner T, Lange T, Anders M, Schumacher U. Expression of the coxsackie adenovirus receptor in neuroendocrine lung cancers and its implications for oncolytic adenoviral infection. Cancer Gene Ther. 2012a;20 (1:25–32. doi: 10.1038/cgt.2012.80. [DOI] [PubMed] [Google Scholar]
- Wunder T, Schumacher U, Friedrich RE. Coxsackie adenovirus receptor expression in carcinomas of the head and neck. Anticancer Res. 2012b;32:1057–1062. [PubMed] [Google Scholar]
- You Z, Fischer DC, Tong X, Hasenburg A, Aguilar-Cordova E, Kieback DG. Coxsackievirus-adenovirus receptor expression in ovarian cancer cell lines is associated with increased adenovirus transduction efficiency and transgene expression. Cancer Gene Ther. 2001;8:168–175. doi: 10.1038/sj.cgt.7700284. [DOI] [PubMed] [Google Scholar]
- Zhang N-H, Song L-B, Wu X-J, Li R-P, Zeng M-S, Zhu X-F, Wan D-S, Liu Q, Zeng Y-X, Zhang X-S. Proteasome inhibitor MG-132 modifies coxsackie and adenovirus receptor expression in colon cancer cell line lovo. Cell Cycle. 2008;7:925–933. doi: 10.4161/cc.7.7.5621. [DOI] [PubMed] [Google Scholar]