Dear Editor,
The AMP-activated protein kinase (AMPK) is the key energy sensor in response to various stresses that decrease cellular ATP levels, including low glucose, hypoxia, ischemia and heat shock. Recently, evidence has suggested that AMPK also plays critical roles in coupling the cellular bio-energetic state to various physiological processes, such as tumor suppression, cell polarity, life span, circadian clock, gene transcription and autophagy. Thus, AMPK represents a potential drug target for metabolic disorders and cancer treatment1,2.
AMPK is a heterotrimeric protein kinase composed of one catalytic (α) and two regulatory (β and γ) subunits (Figure 1A). The α-subunit contains an N-terminal kinase domain, followed by an autoinhibitory domain (AID), a regulatory linker region (α-linker) and a C-terminal β/γ-subunit interacting domain (α-CTD). Phosphorylation of a conserved threonine in the kinase domain (Thr172 in rat α1) by upstream protein kinases is the prime activation step of AMPK, while the adjacent AID directly binds to the kinase domain and inhibits its catalytic activity3. The heterotrimeric core structures of the yeast and mammalian AMPKs, including the entire γ-subunit and the interacting fragments of the α- and β-subunits, have revealed that the C-terminal domain of the β-subunit (β-CTD) serves as a bridge between the α-CTD and the γ-subunit and that the γ-subunit adopts a pseudosymmetric conformation comprising four tandem cystathionine β-synthase (CBS1-4) motifs for adenine nucleotide binding4. Binding of AMP and ADP to the γ-subunit has activating effects in protecting AMPK against dephosphorylation and/or promoting its phosphorylation5,6. In addition, raising the cellular AMP level will allosterically enhance the activity of mammalian AMPK that has been primarily activated by upstream kinases. Therefore, a central issue pertaining to the regulation of AMPK lies in how AMP binding to the γ-subunit ultimately regulates the kinase activity in the α-subunit.
Figure 1.
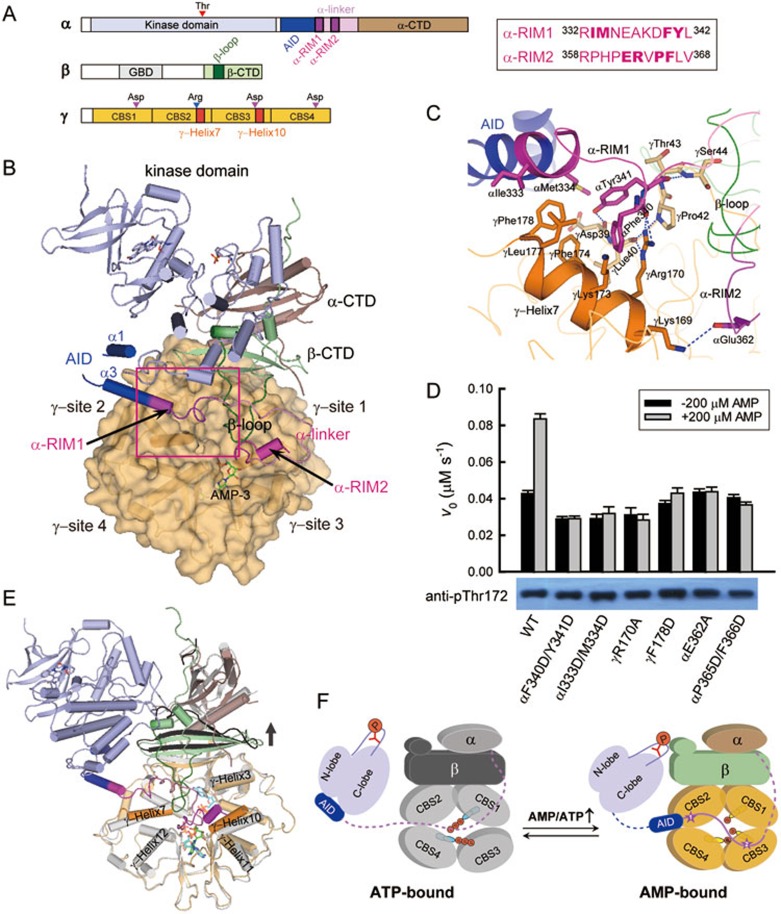
AMPK regulation via recruitment of α-RIM1 to γ-subunit site 2. (A) Schematic diagram of the αβγ-heterotrimeric AMPK. The color codes are as follows: kinase domain (light blue), AID (blue), α-linker (pink), α-RIM1/2 (magenta), α-CTD (brown), β-CTD (green), γ-subunit (light orange). The sequences for α-RIM1 and α-RIM2 in rat α1 are indicated on the right. (B) Graphical representations of the rebuilt active AMPK9. The color scheme follows the schematic diagram in A with AMP highlighted as green stick. The traceable helices α1 and α3 of AID are indicated. (C) Contacts between α-RIM1 and γ-site 2. The interacting residues were highlighted as sticks, and the hydrogen bonds were indicated by blue dashed lines. (D) Both α-RIM1 and α-RIM2 play significant roles in the allosteric activation of AMPK. The kinase assay for AMPK was performed in the presence of 7 nM AMPK (wild type or mutants), 200 μM SAMS, 1 mM ATP and 10 mM MgCl2, with or without the addition of 200 μM AMP (mean ± SD, n = 3). The bottom panel shows the phosphorylation levels of all AMPK proteins detected by western blotting using a specific antibody against phosphorylated AMPK (anti-pThr172). (E) Structural comparison of the rebuilt active AMPK9 with the ATP-bound core structure (PDB code: 4EAK)7. The color scheme for the ATP-bound core is as follows: α- and γ-subunits in grey, β-subunit in black and two ATP molecules at sites 1 and 4 in cyan. (F) Allosteric regulation of mammalian AMPK by AMP/ATP. The γ-subunit of AMPK adopts an inactive, ATP-bound conformation (left) when the cellular AMP concentration is low. The α-linker is likely flexible and extended (dotted line), and AID directly binds to the kinase domain and inhibits the AMPK activity. Under energy stress, the γ-subunit is bound by AMP (right), undergoes substantial conformational changes and recruits both α-RIM1 and α-RIM2. The constrained α-linker pulls AID away from the kinase domain, and the activity of AMPK is eventually stimulated.
Although the γ-subunit contains four potential nucleotide binding sites (1-4), site 2 in mammalian AMPKs possesses a key Asp-to-Arg substitution and remains unoccupied even in the presence of high concentrations of AMP or ATP4,7. The other three sites (1, 3 and 4) on the γ-subunit appear to reversibly bind three AMP or two ATP molecules, which results in substantial conformational changes and distinct activities of AMPK7. Our structure-based mutagenesis studies have demonstrated that two of the three nucleotide-binding sites, γ-site 3 and γ-site 4, are important for AMPK allosteric activation. Xiao et al.8 reported the structure of an active AMPK heterotrimer, comprising the kinase domain, AID and the regulatory α-linker of the α-subunit in addition to the core fragment, which revealed additional inter-subunit interactions involving the activation loop of the kinase domain and a small segment within the α-linker. However, in the same study, it was concluded that mammalian AMPK does not contain an AID domain, inconsistent with our results3. We found that the amino acid sequence of the AID and α-linker region in that AMPK structure was incorrectly assigned due to the poor electron density9. We have thus rebuilt the model and determined the structures of two isolated mammalian AID domains, which affirms the universal presence of AID (Figure 1B and Supplementary information, Figure S1). More importantly, we identified the α-RIM (regulatory subunit interacting motif) sandwiched in between γ-site 3 and the induced β-loop (Supplementary information, Figure S2), and we also determined the essential role of α-RIM in AMPK allosteric regulation upon AMP binding9.
Intriguingly, the α-linker segment prior to the α-RIM reaches the other side of the induced β-loop and then packs against the non-bound site 2 on the γ-subunit (Figure 1B). This α- and γ-subunit interface is mediated by a novel α-subunit motif containing the last turn of AID helix α3 and the following flexible loop (rat α1 332-342). According to the linear sequence of the α-subunit, we hereafter refer to this new segment as α-RIM1 and rename the previously identified α-RIM as α-RIM2. At the interface between α-RIM1 and γ-site 2, the aromatic residues αPhe340 and αTyr341 interact with γLys173, γPhe174 and γLeu177 from the γ-Helix7; in turn, the hydrophobic γPhe178 inserts between αIle333 and αMet334 (Figure 1C). In addition, the highly conserved γArg170, which is believed to be the main determinant of the non-bound site 2, further strengthens this interface by participating in multiple hydrophilic interactions with the main chains of αPhe340 in α-RIM1 and γLeu40 and γPro42 from an N-terminal segment of the γ-subunit, while the main chains of γAsp39, γThr43 and γSer44 from this γ-subunit segment form hydrogen bonds with both the side and main chains of αTyr341 in α-RIM1. Therefore, both the γ-Helix7 at site 2 and the spatially adjacent γ-subunit segment (rat γ1 39-44) play important roles in recruiting the newly identified α-RIM1. On close inspection, we also found additional hydrophobic interactions involving the previous α-RIM2 (Supplementary information, Figure S3). The αPro365 and αPhe366, which follow the two important charged residues αGlu362 and αArg3639, nestle into a hydrophobic pocket formed by six residues at γ-site 3 (γPhe243, γIle246, γAsn247, γAla250, γHis267 and γVal64). Notably, the aromatic ring of γPhe243, the residue preceding the indispensable and highly conserved γAsp244, also stabilizes the ribose ring of AMP bound at site 3. Taken together, two tandem α-subunit motifs, the α-RIM1 and the previously defined α-RIM2, bind to site 2 and site 3 on the γ-subunit, respectively.
To assess the importance of the aforementioned inter-subunit interactions, we generated a series of point mutations on the rat α1β1γ1-holoenzyme and examined their effects on AMPK allosteric activation. In the presence of 200 μM AMP, the activity of the wild-type AMPK was stimulated by ∼2-fold (Figure 1D). In contrast, mutating the key α-RIM1 residues largely abolished the AMP dependence as observed in the mutants in which the hydrophobic pairs of interacting residues were replaced with charged residues (αF340D/Y341D and αI333D/M334D). Similarly, the γ-site 2 mutants (γR170A and γF178D) no longer responded to the increase in AMP concentration. We also simultaneously substituted the two hydrophobic residues in the α-RIM2, and the AMP dependence of the αP365D/F366D mutant was completely abolished as previously observed in other α-RIM2 mutants, including αE362A9. The independence of these mutant AMPKs on changes in AMP concentration clearly demonstrates that both α-RIM1 and α-RIM2 play important roles in the allosteric regulation of AMPK activity.
Significantly, in the vertebrate AMPK homologs, 7 out of the 8 important interacting residues in the new α-RIM1 and previous α-RIM2 are identical, and the remaining one is highly conserved (Supplementary information, Figure S1). The interacting γ-subunit residues are also strictly conserved across vertebrates (Supplementary information, Figure S4). Therefore, various AMPK αβγ-heterotrimers may adopt similar inter-subunit interactions and be activated via a conserved molecular mechanism. By contrast, some of these residues in the α-RIM1/2 and γ-sites 2 and 3 are not conserved in invertebrate AMPKs, such as those from Caenorhabditis elegans, Arabidopsis thaliana, Saccharomyces cerevisiae and Schizosaccharomyces pombe. This provides a possible explanation for why only vertebrate AMPKs can be allosterically activated by AMP.
The next question is how AMPK senses the change in the cellular AMP/ATP levels. We have shown that the mammalian AMPK core structures in complex with ATP or AMP adopt distinct conformations, in that the γ-site 3 is free of any nucleotide in the ATP-bound state but bound by AMP in the AMP-bound state7. This active AMPK structure in complex with two AMP molecules at γ-sites 3 and 4 adopts an almost identical conformation to the AMP-bound core structure (Supplementary information, Figure S2), but it exhibits substantial conformational changes around γ-site 3 compared to the ATP-bound structure (Figure 1E). In the ATP-bound structure, the α-CTD and β-CTD, as well as the first β-strand of the γ-subunit that forms an inter-subunit β-sheet with the β-CTD, collectively move upwards, which would abolish the multiple hydrogen bonds between the main chains of the γ-subunit N-terminal segment and the αTyr341 of α-RIM1 (Figure 1E and Supplementary information, Figure S5A). The side chains of γArg170 and γLys173 at γ-site 2 also shift away from α-RIM1, which may disrupt both the hydrophilic and hydrophobic interactions with αPhe340. In addition, the γ-Helix7 tilts ∼5° and forms an additional turn at the C-terminus, and the aromatic ring of γPhe178 thereby rotates almost 180° and moves away from the hydrophobic side chains on the AID helix α3. Hence, when the γ-subunit is bound by ATP, most of the interactions between the α-RIM1 and the γ-subunit would also be disrupted. Upon ATP binding, the most significant shift (up to 6 Å) occurs in γ-Helix10, which contains the highly conserved Asp244 as well as four of the six residues accommodating the hydrophobic αPro365 and αPhe366 in α-RIM2 (Supplementary information, Figures S5B and S3). The spatially adjacent γ-Helix11 and γ-Helix3 also move away from α-RIM2; specifically, the imidazole side chain of γHis267 rotates ∼180° and shifts > 5 Å. In addition, the ε-amino group of γLys169 is pushed away from the key αGlu362 by the dramatically changed γHis297. Therefore, the γ-site 3 in the ATP-bound state is distorted and unable to recruit α-RIM2. In contrast, AMP binding to the γ-subunit induces substantial changes and enables recruitment of both α-RIM1 and α-RIM2 to the γ-subunit.
Then, why and how would the binding of two RIM motifs ultimately enhance AMPK activity in the α-subunit? We suggested previously that the γ-sites 3 and 4 of mammalian AMPKs are indispensable for AMP-induced allosteric regulation and that the γ-site 1 has little effect7. It is natural that α-RIM2 plays an important role in the AMP regulation of AMPK activity because α-RIM2 packs against the essential γ-site 3 in its AMP-bound state9. Here we demonstrate that α-RIM1, though surprisingly bound to the nucleotide-free γ-site 2, has a significant effect on AMPK regulation. As α-RIM1 partially overlaps with the AID domain, binding of α-RIM1 to the γ-site 2 would recruit the AID domain to the proximity of the γ-subunit. More importantly, all the AID side chains that were shown to interact with the kinase domain in the autoinhibited AMPK structure3 are facing the γ-subunit, and binding of the kinase domain to AID in the active AMPK conformation would thus cause severe steric clash between the kinase domain and the γ-subunit (Supplementary information, Figure S6). As revealed in our rebuilt structure of the active AMPK, binding of α-RIM1 and α-RIM2 to the AMP-bound γ-subunit forces the kinase domain to dissociate from AID and pack against the β-CTD, thereby relieving the inhibitory function of AID.
A full understanding of the regulation of AMPK upon energy stress may facilitate the design of novel therapeutics for metabolic diseases. Together with our previous studies, the structural and mutational analyses demonstrate that the three regulatory elements in the α-subunit, AID, α-RIM1 and α-RIM2, are indispensable for the allosteric activation of AMPK, leading to a more complete understanding of the allosteric regulation of mammalian AMPK (Figure 1F). When the cellular AMP concentration is low, α-RIM1 and α-RIM2 do not bind to the ATP-bound γ-subunit. AID directly binds to the kinase domain, thus leading to the autoinhibition of AMPK activity. Under energy stress (increased AMP levels), the γ-subunit is bound by AMP and undergoes substantial conformational changes. The α-RIM1 and α-RIM2 are recruited to γ-site 2 and γ-site 3, respectively. The AID domain is thus pulled into the proximity of γ-site 2 and dissociates from the kinase domain. The free kinase domain then binds to the β-CTD, and the heterotrimeric AMPK holoenzyme adopts a more compact, fully activated conformation10. Following this model, molecules that can stabilize the interactions between α-RIM1/2 and γ-sites 2/3 or disrupt those between AID and the kinase domain would retain AMPK in a stimulated state.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (31130062), the National Key Basic Research Program of China (2013CB530603) and Tsinghua University (20121080028).
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Information
Sequence alignment of the α-subunit regulatory region from representative AMPK homologues generated by ClustalW.
Structural comparison of the rebuilt active AMPK2 with the AMP-bound core structure (PDB code: 4EAI)4.
Stereo view of interactions of the α-RIM2 with the γ-subunit site 3 and the newly formed β-loop.
Sequence alignment of AMPK γ-subunits.
Close-up views of the substantial changes between the rebuilt active AMPK2 and the ATP-bound core structure (PDB code: 4EAK)4.
Superposition of the two AID domains in the S. pombe KD-AID structure (left) and the active AMPK trimer (right).
Materials and methods
References
- Steinberg GR, Kemp BE. Physiol Rev. 2009. pp. 1025–1078. [DOI] [PubMed]
- Hardie DG. Biochem Soc Trans. 2011. pp. 1–13. [DOI] [PubMed]
- Chen L, Jiao ZH, Zheng LS, et al. Nature. 2009. pp. 1146–1149. [DOI] [PubMed]
- Kemp BE, Oakhill JS, Scott JW. Structure. 2007. pp. 1161–1163. [DOI] [PubMed]
- Hardie DG, Carling D, Gamblin SJ. Trends Biochem Sci. 2011. pp. 470–477. [DOI] [PubMed]
- Oakhill JS, Scott JW, Kemp BE. Trends Endocrinol Metab. 2012. pp. 125–132. [DOI] [PubMed]
- Chen L, Wang J, Zhang YY, et al. Nat Struct Mol Biol. 2012. pp. 716–718. [DOI] [PubMed]
- Xiao B, Sanders MJ, Underwood E, et al. Nature. 2011. pp. 230–233. [DOI] [PMC free article] [PubMed]
- Chen L, Xin FJ, Wang J, et al. Nature. 2013. pp. E8–E10. [DOI] [PubMed]
- Zhu L, Chen L, Zhou XM, et al. Structure. 2011. pp. 515–522. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequence alignment of the α-subunit regulatory region from representative AMPK homologues generated by ClustalW.
Structural comparison of the rebuilt active AMPK2 with the AMP-bound core structure (PDB code: 4EAI)4.
Stereo view of interactions of the α-RIM2 with the γ-subunit site 3 and the newly formed β-loop.
Sequence alignment of AMPK γ-subunits.
Close-up views of the substantial changes between the rebuilt active AMPK2 and the ATP-bound core structure (PDB code: 4EAK)4.
Superposition of the two AID domains in the S. pombe KD-AID structure (left) and the active AMPK trimer (right).
Materials and methods