Abstract
Microbial populations in indoor environments, where we live and eat, are important for public health. Various bacterial species reside in the kitchen, and refrigerators, the major means of food storage within kitchens, can be a direct source of food borne illness. Therefore, the monitoring of microbiota in the refrigerator is important for food safety. We investigated and compared bacterial communities that reside in the vegetable compartment of the refrigerator and on the seat of the toilet, which is recognized as highly colonized by microorganisms, in ten houses using high-throughput sequencing. Proteobacteria, Firmicutes, Actinobacteria, and Bacteroidetes were predominant in refrigerator and toilet samples. However, Proteobacteria was more abundant in the refrigerator, and Firmicutes was more abundant in the toilet. These household bacterial communities were compared with those of human skin and gut to identify potential sources of household bacteria. Bacterial communities from refrigerators and toilets shared more species in common with human skin than gut. Opportunistic pathogens, including Propionibacterium acnes, Bacteroides vulgatus, and Staphylococcus epidermidis, were identified as species shared with human skin and gut microbiota. This approach can provide a general background of the household microbiota and a potential method of source-tracking for public health purposes.
Electronic supplementary material
The online version of this article (doi:10.1007/s00284-013-0401-y) contains supplementary material, which is available to authorized users.
Introduction
Indoor microbes have been studied in the context of human health using culture-dependent and -independent techniques. Most studies focused on the bacterial contamination of surfaces in kitchens and restrooms, which are easily colonized by microbes [9, 10, 15, 22, 24]. Some pathogenic bacteria can survive on the surfaces in these environments for some time, and contamination of food by these pathogenic bacteria can cause illness. Microbial contaminations of refrigerators have been studied, because refrigerators are used to store food [2, 4, 7, 14]. Moisture and nutrients (food particles) in refrigerators provide favorable growth conditions for contaminating bacteria from unwashed raw foods, leaking packages, and hands. In particular, higher bacterial counts and temperatures in vegetable compartments could cause critical problems [4]. Recently, a German outbreak caused by Shiga-toxin producing Escherichia coli O104:H4 illustrated that unwashed vegetables could be a risk element [3]. Therefore, the study of bacterial contamination in the vegetable compartments of refrigerators is important for public health.
Most of the previously reported culture-dependent studies of kitchen and refrigerator microbes focused on pathogen detection [7, 14, 21, 22, 30]. The recent advent of next generation sequencing techniques provides unprecedented data on the microbial composition, and the ecology of various environments, including indoor spaces [9, 10, 12, 15]. Analyses of microbes in various environments by high-throughput sequencing can benefit various fields, including source-tracking. Identification of the sources of bacterial contamination in indoor environment is important for managing food safety. Human skin is a primary source of bacteria in indoor environments, and individuals can transmit bacterial pathogens by touching indoor spaces [9, 10]. Comparing various parts of the human microbiome with microbial communities in indoor environments can identify bacterial species commonly found in both environments and thereby suggest the source of contamination or transmission.
In this study, we characterized bacterial communities within vegetable compartments of refrigerators and on toilet seats by using pyrosequencing based on 16S rRNA genes. The comparison of bacterial communities analyzed in this study with published human microbiome data provides further insight into shared species and sources of bacteria on the surfaces of refrigerators and toilets. Opportunistic pathogens were shared between the human skin microbiome and microbial populations in refrigerators and toilets.
Materials and Methods
Sampling and DNA Extraction
Swab samples were obtained from 5 × 5 cm surfaces of refrigerators (vegetable compartments) and toilets (seat part) at ten houses using an Easy swab kit (KOMED, Korea). Sampling was carried out in households with 4−5 family members. Samples were transported back to the laboratory under chilled conditions (4 °C) and processed within 6 h. To analyze culturable and unculturable bacterial communities, the genomic DNA on swab samples was extracted by two different methods. For culturable bacterial community, diluted swab samples (10−2) were inoculated on plate count agar (PCA; BD-Difco, Sparks, MD, USA) and nutrient agar (NA; BD-Difco) and incubated for 48 h at 30 °C. The surface of the cultured agar medium was washed and suspended in 1 mL of the extraction buffer from a FastDNA SPIN extraction kit (MP Biomedicals, Santa Ana, CA, USA) using a disposable spreader (SPL Life Sciences, Korea). Genomic DNA from the washed plates was then extracted using a FastDNA SPIN extraction kit. For unculturable bacterial community, metagenomic DNA in swab samples from refrigerators and toilets was extracted using a FastDNA SPIN extraction kit by following the manufacturer’s instructions.
Pyrosequencing
16S rRNA gene fragments corresponding to the V1−V3 regions were amplified from the genomic DNA of culture washing solutions and swab metagenomic samples using a previously described method [13]. For PCR, amplifications were performed in a final volume of 50 μL containing 10× Taq buffer, dNTP mixture (Takara, Shiga, Japan), 10 μM of each barcoded fusion primer (http://oklbb.ezbiocloud.net/content/1001), and 2 U of Taq polymerase (ExTaq, Takara) by a C1000 Touch thermal cycler (Bio-Rad, Hercules, CA, USA). After initial denaturation at 94 °C for 5 min, the product was amplified by 30 cycles of denaturation (30 s, 94 °C), primer annealing (30 s, 55 °C), and extension (30 s, 72 °C), with a final extension step of 7 min at 72 °C. The PCR product was confirmed by 2 % agarose gel electrophoresis and visualized under a Gel Doc system (Bio-Rad). The amplified products were purified with a QIAquick PCR purification kit (Qiagen, Valencia, CA, USA) and quantified using a PicoGreen dsDNA Assay kit (Invitrogen, Carlsbad, CA, USA). Equimolar concentrations of each amplicon from different samples were pooled and purified using an AMPure bead kit (Agencourt Bioscience, Beverly, MA, USA) and then amplified on sequencing beads by emulsion PCR. Recovered beads from emulsion PCR were deposited on a 454 Picotiter Plate and sequenced with a Roche/454 GS Junior system by following the manufacturer’s instructions.
Data Analysis
Raw sequence files were processed by (1) demultiplexing, (2) trimming primer sequence, (3) quality filtering, (4) sequencing error correction, (5) taxonomic assignment, and (6) detection of chimeras. Each sample was identified by a unique barcode in the demultiplexing step and low quality reads (average quality score <25 or read length <300 bp) were removed for further analysis. Pairwise sequence alignment and the hmm-search program of the HMMER 3.0 package [5] were used to trim primer sequences based on the profile of the 16S rRNA V1−V3 regions. To correct sequencing errors, representative sequences in clusters of trimmed sequences were chosen and considered for taxonomy identification (details in Supplementary Methods). Individual reads were assigned their taxonomic positions according to the highest pairwise similarity among the top five BLASTN hits against the EzTaxon-e database [16]. Chimera sequences were removed by UCHIME [6]. The read number in each sample was normalized by random subsampling. The diversity indices and species richness were calculated using three different methods: Cluster Database at High Identity with Tolerance (CD-HIT), Taxonomy-Based Clustering (TBC), and Taxonomy-Dependent Clustering (TDC)-TBC (details in Supplementary Methods). The compositions and proportions of bacterial species shared between two samples or sets of multiple samples were calculated using CLcommunity software (ChunLab, Inc., Korea). Similarity coefficients of Bray-Curtis, Jaccard and Sorenson abundance were calculated using Mothur [27], and the matrix of Fast UniFrac [11] was generated using CLcommunity. Principal coordinate analyses (PCoA) were used to represent the relationships between samples using calculated similarity coefficients. The significance of difference among bacterial communities was calculated by Libshuff analysis using Mothur. Pyrosequencing reads generated in this study are available at the EMBL SRA database under the study accession number ERP002164 (http://www.ebi.ac.uk/ena/data/view/ERP002164).
Results and Discussion
Comparison of Bacterial Communities Originated from Surfaces of Refrigerators and Toilets
The bacterial communities in swab samples were analyzed using high-throughput 16S rRNA gene pyrosequencing. Diversity indices calculated by three different methods are presented in Supplementary Table S1. In refrigerator and toilet samples, the richness and diversity of the communities obtained from metagenomic DNAs were higher than those obtained from plate washed DNA. Although the values calculated by the TBC method were higher than those calculated by the CD-HIT and TDC-TBC methods, the diversity trends in each sample were similar among the three methods. Four phyla, namely Proteobacteria, Firmicutes, Bacteroidetes, and Actinobacteria, were dominant (over 98 % of total reads from each sample) in the mean bacterial community, which was obtained by pooling the culture-independent results from the refrigerator and toilet surfaces of ten households (Fig. 1a). These major phyla were also identified in previous indoor studies [1, 10, 15]. Although the compositions of dominant phyla were similar in surfaces of refrigerators and toilets, the proportions of the phyla varied. Proteobacteria was the most prevalent phylum in refrigerator samples (63.6 % of total reads) and toilet samples (42.2 %). The relative abundance of Firmicutes in toilet samples (36.2 % of total reads) was higher than the refrigerator samples (15.7 %). A total of 30 phyla were detected in refrigerator samples, while 16 phyla were obtained from toilet samples. This could be due to differences in survivability that depend on the moisture or temperature of surfaces and the frequency of transmission.
Fig. 1.
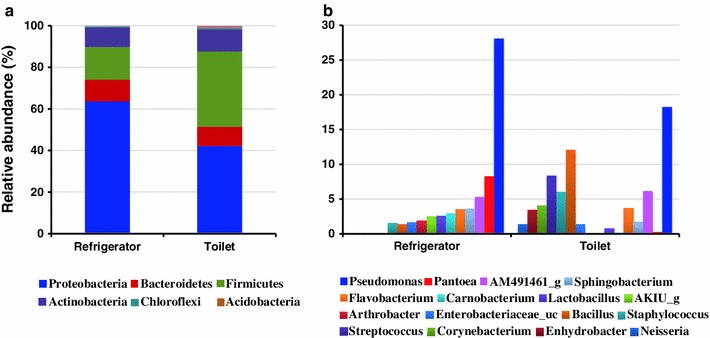
The average compositions of bacterial communities obtained from the vegetable compartments of refrigerators and from toilets using culture-independent method were analyzed and compared. a The compositions of phyla detected in refrigerators and toilet samples were compared. The phylum represented by each color is defined below figure. b The compositions of the top ten genera detected in each sample were compared. The names of the genera appear below the figure. The nomenclatures of phylotypes are based on the EzTaxon-e database (Kim et al., 2012; http://eztaxon-e.ezbiocloud.net/)
The compositions of the top ten most prevalent genera in each sample showed clear differences between bacterial communities of refrigerators and toilets (Fig. 1b). Pseudomonas and Pantoea within Gammaproteobacteria were identified as the dominant genera in refrigerator samples. Although the genus Pseudomonas was also dominant in toilet samples, the proportion of Pantoea was relatively low and Bacillus, Staphylococcus, and Streptococcus within Firmicutes were dominant genera. The bacterial communities present in the individual samples obtained from each house are presented in Supplementary Fig. S1. The number of toilet samples was smaller than that of refrigerator samples because, sufficient DNA was not always isolated from swab samples of toilet seat surfaces. This is probably because toilet surfaces are cleaned more frequently than the vegetable compartments of refrigerators in general households. The compositions of bacterial communities in refrigerators of most houses obtained by plate washing method were similar to those obtained by culture-independent methods, except #6 house. However, only 5 out of 30 phyla were detected in the plate washing results, and the proportions of each member in bacterial communities were different between two methods. The differences between culture-based plate washing and culture-independent surveys were significant in toilet samples obtained from identical houses (#1 and 3): Firmicutes and Actinobacteria were more abundant in culture-based plate washing results. This difference could be due to the selectivity of PCA or NA media for cultured bacteria found in the bacterial community on toilet seats. The genus Staphylococcus was the most dominant bacteria obtained by culture-based plate washing method in toilet samples (average 45.9 % of total reads). The phylum and genus compositions in the refrigerator and toilet samples were unique because the people and their behaviors (e.g., frequency of cleaning, cleaning products used, kinds of refrigerators and toilets, and usage patterns) varied in each household.
Identification of Bacterial Species Shared with Human Microbiome
Several studies have reported that most indoor bacteria could be of human origin, particularly from human skin such as hands [9, 10, 24]. To identify bacterial species present on human skin and in the two indoor environments, bacterial communities obtained in this study were compared with microbiota from human skin and fecal samples. Data on the human microbiome were downloaded from the human microbiome Project [19]. Skin and gut microbiome data were selected because of the possibility of direct contact with the surfaces of refrigerators and toilets. On an average, 15.6 % of the bacterial species obtained from human skin and 4.9 % of the species obtained from human gut samples were shared with the bacterial communities in refrigerators (Fig. 2). The proportion of species shared by bacterial communities from toilets and human skin samples (51.6 %) was higher than the proportion of species shared by those from toilets and the human gut microbiome (15.4 %). This result indicates that the human skin microbiome could be a significant source of bacterial transmission by touch or exposure even on the surface of the toilet. This is similar to the results of public restrooms, where human skin was identified as the principal source of bacteria [9]. The proportion of bacteria shared by human skin and the surface of the toilet was higher than that shared by human skin and the refrigerator because of the higher frequency of human contact with toilets. The species shared between human skin and refrigerators were similar to those shared between human skin and toilet surfaces. These results support the previous findings that most indoor bacteria could originate from human skin and indicate that particular bacteria could be attached to and survive for long periods on indoor surfaces [9, 10]. Of the shared species found in the gut microbiome, Bacteroides vulgatus was the most abundant on the surfaces of refrigerators and toilets, but the composition of shared species was different on the two surfaces (Fig. 2). This could be due to direct or indirect exposure of fecal bacteria to the surfaces of refrigerators or toilets. Propionibacterium acnes was the most abundant species shared between human skin and the surfaces of refrigerators and toilets. This species is a member of the normal flora of the skin, oral cavity, large intestine, and other human body sites. It mainly plays a role in acne, and it can cause postoperative and device-related infections as an opportunistic pathogen [18, 23]. Staphylococcus epidermidis and Staphylococcus hominis are commensal bacteria in human skin; they inhibit virulent bacteria such as Staphylococcus aureus. However, they are also opportunistic pathogens that cause nosocomial infections by dwelling inside medical devices [8, 25]. Bacteroides vulgatus is the most abundant of the species shared between the human gut microbiome and the surfaces of refrigerators or toilets. Although this bacterium is one of the predominant bacteria in the gut of a healthy person, it was isolated from a patient with Crohn’s disease and identified as an antibiotic-resistant pathogen [17, 26]. The distribution patterns of these opportunistic pathogens pose considerable issues for explaining potential contamination of foods or residential environments. Bacterial communities on the surfaces of refrigerator vegetable compartments could be transferred to the vegetables and cause food borne illness, such as the German outbreak of E. coli in 2011 [3].
Fig. 2.
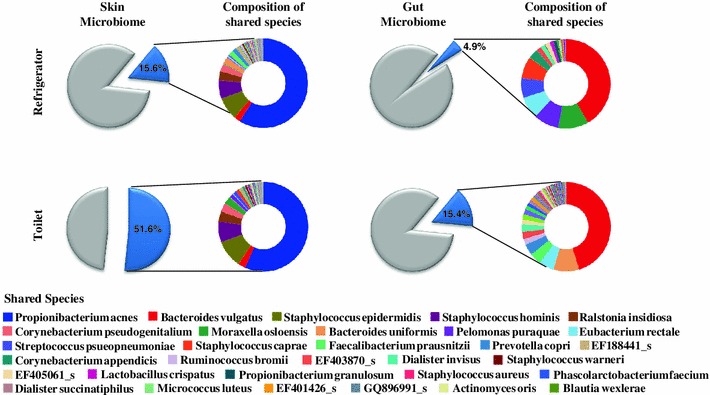
The proportion of species within the human skin and gut microbiomes shared with bacteria obtained from refrigerator and toilet samples is indicated by the blue piece of pie. The compositions of the shared species are presented in the colored pie chart. The largest piece of pie indicates the species of highest abundance in skin or fecal microbiome samples
Comparison of PCoA Plots Based on Four Different Statistical Calculations of Community Distance
PCoA plots based on four different statistical calculations of distance were compared to analyze the relationships among the samples (Fig. 3). Although there were variations in the bacterial communities obtained from refrigerator, toilet, and skin, these communities were more related to each other than to communities from fecal samples in PCoA plots using the Fast UniFrac distance (Fig. 3a). Bacterial communities obtained from refrigerators and toilets were similar in PCoA plots using the Bray–Curtis and Sorenson abundance coefficients (Fig. 3b, d). This might be due to the fact that samples obtained from refrigerators and toilets in the same house were exposed to the same people. Bacterial communities of human skin were more similar to those from refrigerator or toilet samples than human fecal samples in Fast UniFrac, Bray–Curtis, and Sorenson methods, which is consistent with the results based on the calculation of shared species between the samples (Fig. 2). However, bacterial communities from fecal samples were more similar to bacterial communities from toilet and refrigerator samples in the Jaccard abundance analysis (Fig. 3c). The bacterial communities obtained from toilets were more similar to those of fecal samples than to the bacterial communities of other samples. These analyses also showed that microbes within the human body could be a source of bacteria in indoor environments. The Libshuff analyses showed the significant differences among bacterial communities (P < 0.05).
Fig. 3.
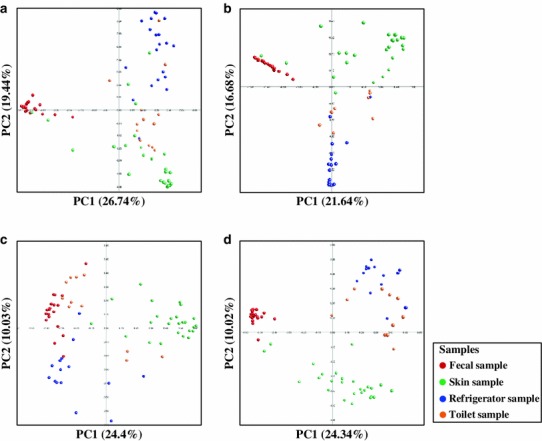
Similarities between bacterial communities that originated from refrigerator, toilet, human skin, and gut samples were analyzed and compared by PCoA. Similarities between communities were calculated by a Fast UniFrac, b Bray-Curtis, c Jaccard abundance, and d Sorenson abundance similarity coefficient using the Mothur program [27]
Conclusion
The initiation of food borne illness has been reported to occur more frequently in private homes than in commercial operations [28, 29]. Refrigerators in kitchens could be colonized by bacteria, and these bacteria might contaminate other stored foods or attach to and survive on the internal surface of the refrigerator, thereby posing risks of indirect, long-term contamination during subsequent food preparation activities [20–22, 30]. In this study, most bacteria detected were probably not pathogens or opportunistic pathogens, and genera belonging to common pathogens were detected in only a very small fraction of communities on the surfaces of refrigerators and toilets. However, their presence could influence other microorganisms, since they survive on and are transmitted to the surfaces of indoor environments. This potential risk can be prevented by wrapping stored foods and regularly cleaning indoor environments, including refrigerators. The expansion of studies on indoor microbial communities using high-throughput molecular methods will advance our understanding of microorganisms in indoor environments and improve preventive measures for public health.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Acknowledgments
This study was supported by Industrial Strategic Technology Development Program (10040176) funded by the Korean Ministry of Knowledge Economy (MKE).
References
- 1.Aydogdu H, Asan A, Otkun MT. Indoor and outdoor airborne bacteria in child day-care centers in Edirne City (Turkey), seasonal distribution and influence of meteorological factors. Environ Monit Assess. 2010;164(1–4):53–66. doi: 10.1007/s10661-009-0874-0. [DOI] [PubMed] [Google Scholar]
- 2.Barker J, Bloomfield SF. Survival of Salmonella in bathrooms and toilets in domestic homes following salmonellosis. J Appl Microbiol. 2000;89(1):137–144. doi: 10.1046/j.1365-2672.2000.01091.x. [DOI] [PubMed] [Google Scholar]
- 3.Buchholz U, Bernard H, Werber D, Bohmer MM, Remschmidt C, Wilking H, Delere Y, an der Heiden M, Adlhoch C, Dreesman J, et al. German outbreak of Escherichia coli O104:H4 associated with sprouts. New Engl J Med. 2011;365(19):1763–1770. doi: 10.1056/NEJMoa1106482. [DOI] [PubMed] [Google Scholar]
- 4.Carpentier B, Lagendijk E, Chassaing D, Rosset P, Morelli E, Noel V. Factors impacting microbial load of food refrigeration equipment. Food Control. 2012;25(1):254–259. doi: 10.1016/j.foodcont.2011.10.051. [DOI] [Google Scholar]
- 5.Eddy SR. Accelerated profile HMM searches. PLoS Comput Biol. 2011;7(10):e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27(16):2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Evans JA, Russell SL, James C, Corry JEL. Microbial contamination of food refrigeration equipment. J Food Eng. 2004;62(3):225–232. doi: 10.1016/S0260-8774(03)00235-8. [DOI] [Google Scholar]
- 8.Fey PD, Olson ME. Current concepts in biofilm formation of Staphylococcus epidermidis. Future Microbiol. 2010;5(6):917–933. doi: 10.2217/fmb.10.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flores GE, Bates ST, Knights D, Lauber CL, Stombaugh J, Knight R, Fierer N. Microbial biogeography of public restroom surfaces. PLoS One. 2011;6(11):e28132. doi: 10.1371/journal.pone.0028132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flores GE, Bates ST, Caporaso JG, Lauber CL, Leff JW, Knight R, Fierer N. Diversity, distribution and sources of bacteria in residential kitchens. Environ Microbiol. 2013;15(2):588–596. doi: 10.1111/1462-2920.12036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hamady M, Lozupone C, Knight R. Fast UniFrac: facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J. 2010;4(1):17–27. doi: 10.1038/ismej.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hewitt KM, Gerba CP, Maxwell SL, Kelley ST. Office space bacterial abundance and diversity in three metropolitan areas. PLoS One. 2012;7(5):e37849. doi: 10.1371/journal.pone.0037849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hur M, Kim Y, Song HR, Kim JM, Choi YI, Yi H. Effect of genetically modified poplars on soil microbial communities during the phytoremediation of waste mine tailings. Appl Environ Microbiol. 2011;77(21):7611–7619. doi: 10.1128/AEM.06102-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jackson V, Blair IS, McDowell DA, Kennedy J, Bolton DJ. The incidence of significant food borne pathogens in domestic refrigerators. Food Control. 2007;18(4):346–351. doi: 10.1016/j.foodcont.2005.10.018. [DOI] [Google Scholar]
- 15.Kembel SW, Jones E, Kline J, Northcutt D, Stenson J, Womack AM, Bohannan BJM, Brown GZ, Green JL. Architectural design influences the diversity and structure of the built environment microbiome. ISME J. 2012;6(8):1469–1479. doi: 10.1038/ismej.2011.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, Park SC, Jeon YS, Lee JH, Yi H, et al. Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol. 2012;62(Pt 3):716–721. doi: 10.1099/ijs.0.038075-0. [DOI] [PubMed] [Google Scholar]
- 17.Kumar V, Sivaraman J. Structural characterization of BVU_3255, a methyltransferase from human intestine antibiotic resistant pathogen Bacteroides vulgatus. J Struct Biol. 2011;176(3):409–413. doi: 10.1016/j.jsb.2011.08.007. [DOI] [PubMed] [Google Scholar]
- 18.Marinelli LJ, Fitz-Gibbon S, Hayes C, Bowman C, Inkeles M, Loncaric A, Russell DA, Jacobs-Sera D, Cokus S, Pellegrini M, et al. Propionibacterium acnes bacteriophages display limited genetic diversity and broad killing activity against bacterial skin isolates. mBio. 2012;3(5):00279. doi: 10.1128/mBio.00279-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Methe BA, Nelson KE, Pop M, Creasy HH, Giglio MG, Huttenhower C, Gevers D, Petrosino JF, Abubucker S, Badger JH, et al. A framework for human microbiome research. Nature. 2012;486(7402):215–221. doi: 10.1038/nature11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Michaels B, Ayers T, Celis M, Gangar V. Inactivation of refrigerator biofilm bacteria for application in the food service environment. Food Service Technol. 2001;1:169–179. doi: 10.1046/j.1471-5740.2001.00022.x. [DOI] [Google Scholar]
- 21.Ojima M, Toshima Y, Koya E, Ara K, Kawai S, Ueda N. Bacterial contamination of Japanese households and related concern about sanitation. Int J Environ Heal R. 2002;12(1):41–52. doi: 10.1080/09603120120110040. [DOI] [PubMed] [Google Scholar]
- 22.Ojima M, Toshima Y, Koya E, Ara K, Tokuda H, Kawai S, Kasuga F, Ueda N. Hygiene measures considering actual distributions of microorganisms in Japanese households. J Appl Microbiol. 2002;93(5):800–809. doi: 10.1046/j.1365-2672.2002.01746.x. [DOI] [PubMed] [Google Scholar]
- 23.Perry A, Lambert P. Propionibacterium acnes: infection beyond the skin. Expert Rev Anti-Infe. 2011;9(12):1149–1156. doi: 10.1586/eri.11.137. [DOI] [PubMed] [Google Scholar]
- 24.Rintala H, Pitkaeranta M, Toivola M, Paulin L, Nevalainen A. Diversity and seasonal dynamics of bacterial community in indoor environment. BMC Microbiol. 2008;8:56. doi: 10.1186/1471-2180-8-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rogers KL, Fey PD, Rupp ME. Coagulase-negative Staphylococcal infections. Infect Dis Clin N Am. 2009;23(1):73–98. doi: 10.1016/j.idc.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 26.Ruselervanembden JGH, Vanderhelm R, Vanlieshout LMC. Degradation of intestinal glycoproteins by Bacteroides-Vulgatus. FEMS Microbiol Lett. 1989;58(1):37–41. doi: 10.1111/j.1574-6968.1989.tb03014.x. [DOI] [PubMed] [Google Scholar]
- 27.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75(23):7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scott E. Food borne disease and other hygiene issues in the home. J Appl Bacteriol. 1996;80(1):5–9. doi: 10.1111/j.1365-2672.1996.tb03181.x. [DOI] [PubMed] [Google Scholar]
- 29.Scuderi G, Fantasia M, Filetici E, Anastasio MP. Food borne outbreaks caused by Salmonella in Italy, 1991–4. Epidemiol Infect. 1996;116(3):257–265. doi: 10.1017/S0950268800052559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sinclair RG, Gerba CP. Microbial contamination in kitchens and bathrooms of rural Cambodian village households. Lett Appl Microbiol. 2011;52(2):144–149. doi: 10.1111/j.1472-765X.2010.02978.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


