Abstract
Background
Predictive modeling of the biological effects of nanomaterials is critical for industry and policymakers to assess the potential hazards resulting from the application of engineered nanomaterials.
Methods
We generated an experimental dataset on the toxic effects experienced by embryonic zebrafish due to exposure to nanomaterials. Several nanomaterials were studied, such as metal nanoparticles, dendrimer, metal oxide, and polymeric materials. The embryonic zebrafish metric (EZ Metric) was used as a screening-level measurement representative of adverse effects. Using the dataset, we developed a data mining approach to model the toxic endpoints and the overall biological impact of nanomaterials. Data mining techniques, such as numerical prediction, can assist analysts in developing risk assessment models for nanomaterials.
Results
We found several important attributes that contribute to the 24 hours post-fertilization (hpf) mortality, such as dosage concentration, shell composition, and surface charge. These findings concur with previous studies on nanomaterial toxicity using embryonic zebrafish. We conducted case studies on modeling the overall effect/impact of nanomaterials and the specific toxic endpoints such as mortality, delayed development, and morphological malformations. The results show that we can achieve high prediction accuracy for certain biological effects, such as 24 hpf mortality, 120 hpf mortality, and 120 hpf heart malformation. The results also show that the weighting scheme for individual biological effects has a significant influence on modeling the overall impact of nanomaterials. Sample prediction models can be found at http://neiminer.i-a-i.com/nei_models.
Conclusion
The EZ Metric-based data mining approach has been shown to have predictive power. The results provide valuable insights into the modeling and understanding of nanomaterial exposure effects.
Keywords: nanomaterial exposure effects, biological response, toxicity, embryonic zebrafish, data mining, numerical prediction
Introduction
Nanobiotechnology is an interdisciplinary field at the intersection of nanoscience, biology, and engineering.1 There has been a rapid proliferation of different types of nanomaterials with novel properties, offering extraordinary opportunities in various applications such as in vivo cancer diagnosis,2 implantable devices,3 drug delivery systems,4,5 and gene delivery systems.6,7 Such rapid growth of nanobiotechnology will obviously result in increased exposure of humans and the environment to nanomaterials. Hence, there is a need to systematically investigate the potential biological and environmental impacts of newly emerging nanomaterials.
The nanotechnology community began to evaluate the effects of exposure to nanomaterials in the early 1990s.8 Many studies have focused on respiratory system exposure to assess the bioeffects of engineered nanomaterials such as metal oxide nanomaterials9,10 and carbon-based nanomaterials.11 The results suggest that the hazard potential depends on a combination of material properties such as particle shape, size, solubility, and crystalline phase.10 Other exposure routes that have been studied include gastrointestinal system, cardiovascular system, central nervous system, and skin.12 So far, nanomaterials have been mainly tested with in vitro toxicity assays that analyze cellular parameters such as cell viability, oxidative stress, genotoxicity, and inflammatory response.10 In comparison to animal models, in vitro studies allow for a more efficient determination of toxicity endpoints. However, the results from in vitro studies may mislead safety assessment efforts and will require further verification from in vivo animal experiments.10,12
The biological activity of nanomaterials depends on inherent physicochemical properties not routinely considered in toxicity studies. Nanomaterials can have distinct health effects compared with bulk materials of the same chemical composition.12,13 For example, studies have shown that nanocopper particles induce toxicological effects and injuries on the kidney, liver, and spleen of mice while micro-copper particles do not.13 Thus, the existing knowledge of safety evaluation for bulk materials may not apply to the safety assessment of nanomaterials.
Information currently being acquired on the bioeffects of nanomaterials is extremely diverse, including a multitude of widely varying nanomaterials that are being tested in a broad array of in vivo animal systems and in vitro assays.8–13 Knowledge of nanomaterial–biological interactions requires the inclusion and consideration of the entire body of data produced from global efforts in this research area. Compilation of this data will allow for the determination of nanomaterial structure–activity relationships. Such mathematical representations can be used to predict nanomaterial properties and activity in the absence of empirical data. Since nanotechnology fundamentally allows manipulation of matter at the atomic level, toxic interactions could potentially be minimized by creative design once we have sufficient knowledge of how nanomaterials interact with biological systems.
Our research goal is to organize and analyze data and compare results across research platforms in an effort to model robust structure–activity relationships. Instead of an ad hoc approach to building models, we leverage a nanomaterial environmental impact (NEI) modeling framework to guide the development of various models. This NEI modeling framework is based on the most recent innovation in NEI assessment and provides a foundation for the development of the key components of data collection, model discovery, and model composition.14,15
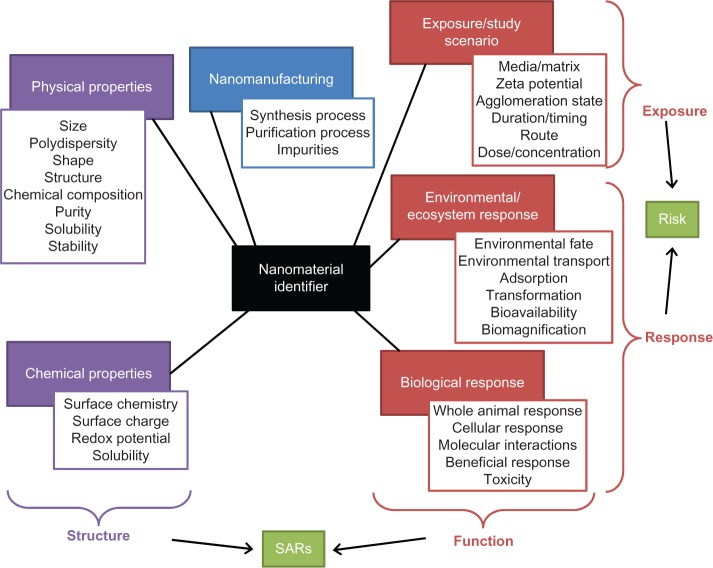
Figure 1 shows a schematic diagram of the NEI modeling framework. The scope covers physical and chemical properties of nanomaterials, exposure and study scenarios, environmental and ecosystem responses, biological responses, and their interactions. Physical and chemical properties describe the “structure” of the material, while exposure and study scenarios, environmental and ecosystem responses, and biological responses describe or are relevant to the “activity” of the material in different contexts. Structure–activity relationships are mathematical descriptions of the relationships between structural features (here we argue that complex descriptors such as 3D molecular simulation models could be used to describe structure) and the activity, which would represent diverse data on fate, transport, biological impacts, and ecosystem impacts. Exposure and study scenarios describe the “exposure” (the fate and update of the materials), while environmental and ecosystem, and biological responses describe the “response.” The interaction between exposure and response can be used to define risk. Because measured responses are related to nanomaterial structural properties, risk is also related to these properties.
Figure 1.
The scope of modeling nanomaterial environmental impact.
Abbreviation: SARs, structure–activity relationships.
The NEI modeling framework (shown in Figure 1) provides guidance for developing various models. In the present research, we focus on modeling potential biological impacts of engineered nanomaterials. Given the identification of the elements related to biological impact modeling, we have developed a data mining approach to parameterizing nanomaterial biological impact models based on experimental data. Data mining techniques, such as numerical prediction, can assist analysts in discovering risk assessment models for nanomaterials. These techniques provide an in-depth knowledge for each biological effect and facilitate quantitative structure–activity relationships modeling.
Several methods have been proposed in the literature to predict adverse effects of nanomaterials (the output) from chemical and biological information (the input). These methods use different ways to measure toxicological effects. Some focus on predicting cytotoxic effects examined in different human cell lines (eg, lung, liver, or kidney) employing different assays.16 Others focus on predicting rodent acute toxicity measured by median lethal dose (LD50) values.17 These methods are based on an ad hoc approach pertinent to specific applications only. They do not support comprehensive evaluation of nanomaterials at multiple levels of biological organization.
Compared with existing work on NEI modeling, our approach has several key advantages. First, our NEI modeling supports the evaluations for biocompatibility using a rapid in vivo system (embryonic zebrafish) to assess the biological activity and toxic potential of nanomaterials at multiple levels of biological organization (ie, molecular, cellular, systems, and organismal). Previous studies have shown that the embryonic zebrafish model is an efficient and powerful model in toxicity testing.18–22 Second, our modeling is integrated with the Nanomaterial–Biological Interactions (NBI) Knowledgebase,23 which provides comprehensive information on the effects of nanomaterial exposure on biological systems. In addition to the embryonic zebrafish experiments, the NBI Knowledgebase is expanding its data sources by including data from other animal systems and in vitro assays. Systematic analysis of disparate data on nanomaterial–biological interactions will provide capabilities to identify structure and design principles of high-performance, environmentally-benign nanomaterials. Third, our modeling leverages Intelligent Automation, Inc. (IAI) internal agent-based data mining framework (ABMiner, Intelligent Automation, Inc., Rockville, MD, USA)24,25 to build models for predicting toxicity or biological impact measures of interest. ABMiner incorporates hundreds of algorithms for both supervised and unsupervised learning. This wide range of algorithms will provide greater flexibility in meeting the needs of building different models for different exposure contexts. ABMiner also provides an optimization engine that can help find the best algorithm (eg, highest accuracy or lowest false positive rate) for a given problem based on optimization theory.
In this study, we investigate adverse effects based on a novel metric representative of adverse effects (toxicity) in embryonic zebrafish.26 The novelty of our approach compared to existing methods is the ability to systematically predict individual exposure effects (eg, mortality, delayed development, or organ malformations) and the overall exposure effect/impact of nanomaterials. The rest of the paper is organized as follows. We first describe materials and methods for modeling the effects of nanomaterial exposure on biological systems. Then we present results to demonstrate the analysis capability of our data-mining-based modeling framework. Next we discuss the results and their implications. Finally, we conclude the paper and outline future research.
Materials and methods
Materials
Nanomaterial exposure effects can be based on whole animal evaluations, cellular-level evaluations, or molecular-level evaluations. For example, whole animal evaluations could provide screening-level measurement using species of rat, mouse, zebrafish, and other animal models; cellular-level evaluations could have measures of different types of cell death; and molecular-level evaluations could include global gene expression, gene localization, and function.26 In the present study, we focus on whole animal evaluations using the embryonic zebrafish model. The embryonic zebrafish metric (EZ Metric) is a screening-level measurement representative of adverse effects (toxicity) in embryonic zebrafish.26,27 Specific biological effects, such as mortality, delayed development, or malformations, are measured after embryonic exposure to nanomaterials. The measure of a specific biological effect Ei is defined as:
| (1) |
where i is the index of a biological effect, Ni is the number of zebrafish embryos having effect i, and T is the total number of zebrafish embryos tested.
To measure the overall adverse effect, individual biological effects (toxic endpoints) are summed to generate a single additive EZ Metric score:
| (2) |
where i is the index of a biological effect, Ei is the measure of the ith biological effect, and n is the total number of biological effects.
To consider the potential impact of the different biological effects that are noted following exposure, individual endpoint frequencies are weighted against their likely biological importance and then summed up to represent a single weighted EZ Metric score:
| (3) |
where wi is the weight factor for the ith biological effect Ei. The weighting factors used for individual biological effects to calculate the overall weighted EZ Metric score are shown in Table 1.
Table 1.
Weight scheme for individual biological effects
| Time period | Abbreviation | Biological effect | Weight |
|---|---|---|---|
| 24 hpf | DP | Developmental progression (delayed development) | 0.3 |
| 24 hpf | M | Mortality | 1 |
| 24 hpf | N | Notochord malformation | 0.4 |
| 24 hpf | SM | Spontaneous movement | 0.2 |
| 120 hpf | A | Axis malformation | 0.4 |
| 120 hpf | B | Brain malformation | 0.6 |
| 120 hpf | C | Circulation | 0.2 |
| 120 hpf | CF | Caudal fin malformation | 0.1 |
| 120 hpf | E | Eye malformation | 0.2 |
| 120 hpf | H | Heart malformation | 0.6 |
| 120 hpf | J | Jaw malformation | 0.2 |
| 120 hpf | M | Mortality | 0.95 |
| 120 hpf | O | Otic malformation | 0.1 |
| 120 hpf | P | Pigmentation | 0.1 |
| 120 hpf | PF | Pectoral fin malformation | 0.1 |
| 120 hpf | SB | Swim bladder | 0.1 |
| 120 hpf | Sn | Snout malformation | 0.1 |
| 120 hpf | So | Somite malformation | 0.1 |
| 120 hpf | T | Trunk malformation | 0.3 |
| 120 hpf | TR | Touch response | 0.1 |
| 120 hpf | Y | Yolk sac edema | 0.5 |
Abbreviation: hpf, hours post-fertilization.
To explore the process of building risk assessment models for nanomaterials, we utilize the EZ Metric experimental data housed in Oregon State University’s NBI knowledge base23 to build prediction models. The dataset contains data fields on nanomaterial characterization (purity, size, shape, charge, composition, functionalization, and agglomeration state), synthesis methods, and nanomaterial–biological interactions (beneficial, benign, or deleterious) defined at multiple levels of biological organizations (molecular, cellular, and organismal). Currently, there are a total of 82 experiments conducted to test the biological interactions using nanomaterials with different properties and dosage concentrations. Based on the knowledge of domain experts, impact values are calculated for each experiment.
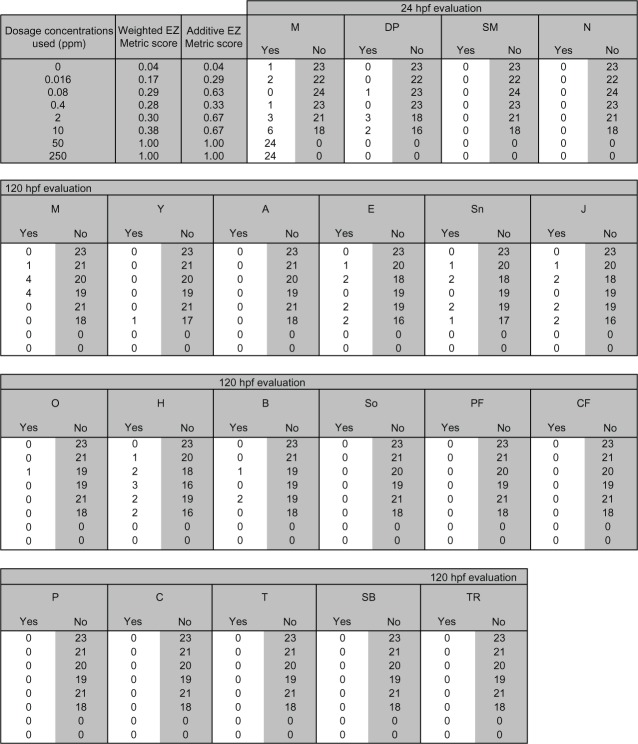
Figure 2 shows a sample experimental dataset with 24 zebrafish embryos exposed to the nanomaterial STARBURST® poly(amido amine) (PAMAM) Dendrimer DNT-107 in the concentration range of 0–250 parts per million (ppm). The primary exposure route is dermal, and the primary exposure delivery is waterborne. The 24 hours post-fertilization (hpf) evaluations include mortality, development progression/delayed development, spontaneous movement, and notochord malformation. The 120 hpf evaluations include mortality, yolk sac edema, axis malformation, eye malformation, snout malformation, jaw malformation, otic malformation, heart malformation, brain malformation, somite malformation, pectoral fin malformation, caudal fin malformation, pigmentation, circulation, trunk malformation, swim bladder, and touch response. For every dosage, the 24 hpf and 120 hpf toxicity evaluations are aggregated to calculate the additive and weighted EZ Metric scores using Equations 2 and 3.
Figure 2.
Sample nanomaterial–biological interaction data.
Abbreviations: A, axis malformation; B, brain malformation; C, circulation; CF, caudal fin malformation; DP, developmental progression; E, eye malformation; EZ Metric, embryonic zebrafish metric; H, heart malformation; hpf, hours post-fertilization; J, jaw malformation; M, mortality; N, notochord malformation; O, otic malformation; P, pigmentation; PF, pectoral fin malformation; SB, swim bladder; SM, spontaneous movement; Sn, snout malformation; So, somite malformation; T, trunk malformation; TR, touch response; Y, yolk sac edema.
Methods
Data mining approach to NEI modeling
Data mining techniques allow us to model important parameters related to NEI and to build predictive models for various biological effects of nanomaterials. Data mining can be addressed from different perspectives, such as classification and numerical prediction. In a classification problem, a model or classifier is constructed to predict categorical labels, such as “Yes” or “No,” and “safe” or “risky.” A numerical prediction problem is similar to a classification problem. Both of them model the relationship between multiple independent variables (the input) and one dependent variable (the output). Instead of having a nominal dependent variable, a numerical prediction problem has a numerical dependent variable. Both classification and numerical prediction are two-step processes. In the first step, training data are analyzed by a machine learning algorithm to build a model. In the second step, test data are used to estimate the accuracy or performance of the model.
In this study, we treat NEI modeling as a numerical prediction problem, where the input variables are nanomaterial properties (eg, material type, particle size distribution, shape, structure, surface charge, water solubility, etc) and exposure scenarios (eg, duration, continuity, exposure route, number of nanoparticles, etc) and the output variable is a numerical measure of a target biological effect. We can build prediction models to predict specific biological effects using the individual measured endpoints or we can build prediction models to predict the overall biological effect/impact using the additive or weighted EZ Metric scores.
Data mining is a generic approach and can be applied to different types of data on nanomaterial–biological interactions, as long as the input and output variables are appropriately defined. In addition to processing embryonic zebrafish experimental data, our modeling approach can be expanded to other types of biological systems as well. For example, we can build prediction models to predict the toxicological effects of cobalt nanoparticles in different human cell lines analyzed by in vitro assays.16 We can also predict the toxicity of copper nanoparticles in vivo measured by the median lethal dose (LD50) of mice.13 The information on predicting toxicity across data sources and organisms is also described in the Discussion section.
NEI model discovery
We developed a data mining solution for NEI modeling by leveraging IAI’s data mining tool ABMiner.24,25 ABMiner supports the full data mining cycle, including data set preparation, model discovery, and model deployment. For data set preparation, ABMiner provides an interface which helps the user to load data from various sources (eg, CVS files, and relational databases). For model discovery, ABMiner provides more than 400 machine learning algorithms (eg, classification and clustering algorithms) aggregated from IAI’s machine learning projects and open sources libraries such as Weka.28 These algorithms allow users to build various prediction models (eg, classification and numerical prediction models). ABMiner provides a dynamical visual representation of the model building process and the performance (eg, accuracy) of each model. Figure 3 shows a screenshot of ABMiner.
Figure 3.
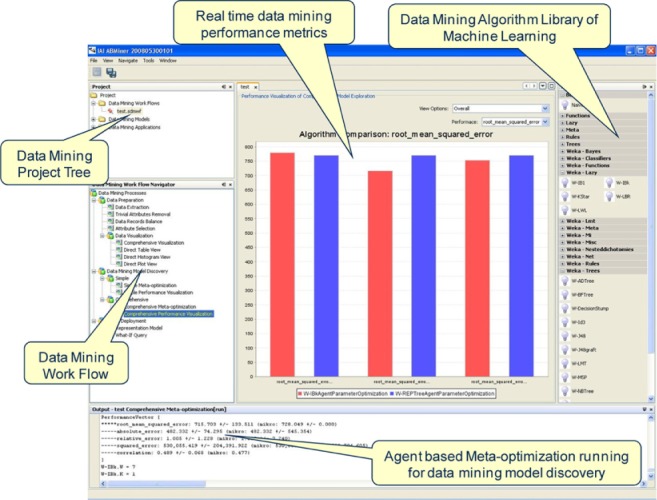
ABMiner screenshot.
Given a dataset on nanomaterial–biological interactions (eg, embryonic zebrafish experimental dataset), we apply numerical prediction algorithms to the data to derive the mathematical models that can predict biological effects of interest from nanomaterial properties and exposure scenarios. Sample numerical prediction algorithms in ABMiner include:
IBK – IBK is a K-nearest neighbor predictor. It assigns an input (eg, a nanomaterial and a corresponding exposure scenario) to the output label (eg, toxicity) most common amongst its K nearest neighbors. The predictor can select the appropriate number of K based on cross-validation. When K is selected by cross-validation for numeric class attributes (labels), it minimizes mean squared error or mean absolute error.29
Bagging – Bagging is a meta-learning algorithm for generating multiple versions of a predictor and using these to get an aggregated predictor.30 The multiple versions are formed by making bootstrap replicates of the dataset and using these as new learning sets. The default base predictor is REPTree,31 a fast decision tree learner.
M5P – M5P is a tree algorithm that implements base routines for generating M5 Model trees and rules. The original algorithm, M5, was invented by Quinlan32 and Wang and Witten made improvements.33
KStar – KStar is an instance-based classifier where the class of a test instance is based upon the class of those training instances similar to it.34 KStar differs from other instance-based learners in that it uses an entropy-based distance function.
ABMiner employs a k-fold cross validation method to evaluate models. For numerical prediction, common evaluation criteria include root mean squared error, absolute error, relative error, and accuracy measured by the correlation between the actual label and predicted label. In k-fold cross validation, the data set is divided into k subsets. Each time, one of the k subsets is used as the test set and the other k − 1 subsets are put together to form a training set. Then the average performance across all k trials is computed. The advantage of this method is that it matters less how the data gets divided. Every data point is included in a test set exactly once, and is included in a training set k − 1 times. The variance of the resulting estimate is reduced as k is increased. Based on the cross validation results, users can compare the models and select the best model (eg, the model with the highest accuracy) for deployment.
NEI model deployment
ABMiner provides a model base component25 that enables a well-trained NEI model, together with its underlying data set and performance metrics, to be wrapped up and accessible across various platforms. There are two unique features of the model base. First, the model base is able to host the datasets and knowledge models from data mining activities, which are obtained by using IAI ABMiner integrated development environment. Second, the model base is able to expose these data mining outcomes (ie, predictive models) to the community through a model deployment interface. By accessing the model base, users can browse NEI knowledge models, and view the dataset associated with a knowledge model and query the knowledge model itself. This will offer industry, academia, and regulatory agencies a tool to rationally inquire about nanomaterial exposure effects in biological systems.
Results
Data preparation
EZ Metric impact measurements are located in separate datasets within the NBI. To get a comprehensive dataset for modeling biological effects, we developed an algorithm to integrate all the experimental data (spreadsheet files) into a dataset with all the nanomaterial properties, experimental design parameters, dosage, and impact measurements. The dataset contains test results on 82 nanomaterials including metal nanoparticles, dendrimer, metal oxide, polymeric, and others. Each nanomaterial is tested at eight different dosage concentrations. Therefore, there are 656 test results (samples) in the data.
We performed data cleaning to remove trivial attributes (those that have the same values for all experiments) in the data. After this step, we had 21 attributes: 20 input attributes and one output attribute for modeling. The 20 input attributes included: material type, synthesis precursors, purity, primary particle size: average (nm), primary particle size: minimum (nm), primary particle size: maximum (nm), core shape, core structure, core atomic composition, number of core atoms, mass core atoms (ng), shell composition, outermost surface functional groups, minimum number of ligands, surface charge: (positive, negative, or neutral), solubility/dispersity medium, primary exposure route, material zeta potential in media (mV), stable average agglomerate size in media (nm), and dosage concentrations used (ppm). The output attribute is the prediction label: it can be the measure of specific biological effects (eg, 24 hpf mortality Metric score) or the measure of aggregated overall effect/impact (eg, additive EZ Metric score).
Specific effect modeling
This modeling focuses on the impact of nanomaterials measured by specific responses or effects, including the 24 hpf evaluation such as mortality, delayed development, spontaneous movement, notochord malformation, and the 120 hpf evaluation such as mortality, yolk sac edema, axis malformation, eye malformation, and many other effects. This kind of modeling allows us to examine biological effects at a high resolution.
To model a specific effect, we first load the corresponding data into ABMiner. For example, Figure 4 shows the metadata of the 24 hpf mortality data. The data has one prediction label (relative 24 hpf M) and 20 input attributes. We can evaluate the worth of an input attribute using an algorithm called RELIEF.35 The key idea of RELIEF is to estimate attributes according to how well their values distinguish among the instances that are near each other. The rationale is that good attributes should differentiate between instances from different classes and should have the same value for instances from the same class. Specifically, the weight for each attribute Wa is defined as:
Figure 4.
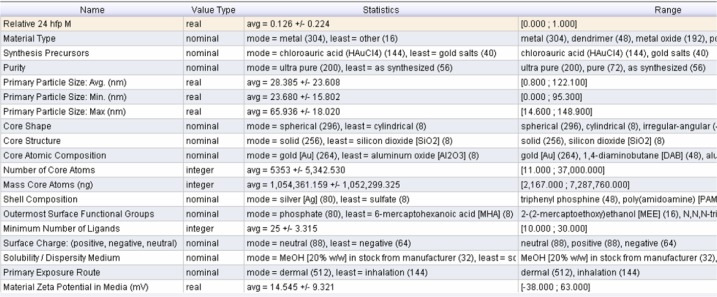
Metadata of the integrated data set.
Abbreviations: Avg, average; Max, maximum; Min, minimum.
| (4) |
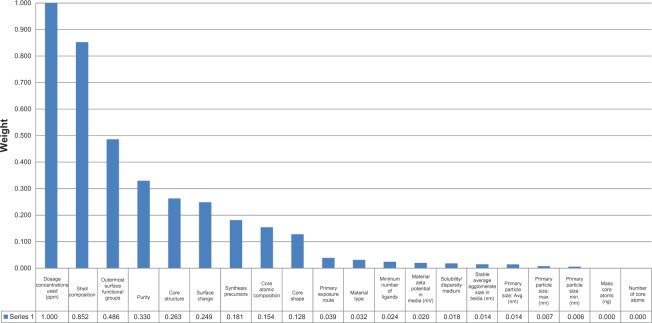
where a is an attribute and P is the probability. RELIEF can operate on both discrete and continuous class labels. Figure 5 shows the attribute weights calculated by RELIEF for the 24 hpf mortality data. In this case, dosage, shell composition, outermost surface functional groups, purity, core structure, and surface charge are significant attributes for predicting the 24 hpf mortality. In high-dimensional data mining, we can select a small number of attributes with relatively high weights. Here, the embryonic zebrafish data has 20 input attributes and 656 examples. Since the number of attributes is not large, we use all 20 attributes for model building.
Figure 5.
Attribute weights with respect to the 24 hpf mortality.
Abbreviation: hpf, hours post-fertilization.
ABMiner lists a set of applicable algorithms for numerical prediction. We can conveniently select algorithms and start building models using the selected algorithms. In this scenario, four algorithms are selected for model building, including IBK, Bagging, M5P, and KStar. Using ABMiner, we compare the performance of different learning algorithms for modeling each specific effect measured by EZ Metric score. Table 2 shows the best performance algorithm for modeling each specific effect. Here the best performance algorithm means the algorithm which generates the highest correlation between the actual and predicted EZ Metric score.
Table 2.
Best algorithm for each biological effect
| Biological effect | Best algorithm | Pearson correlation between the actual and predicted score |
|---|---|---|
| 24 hpf M | IBK | 0.837 |
| 120 hpf M | IBK | 0.765 |
| 120 hpf H | IBK | 0.725 |
| 120 hpf J | IBK | 0.667 |
| 120 hpf T | IBK | 0.657 |
| 24 hpf DP | IBK | 0.591 |
| 120 hpf P | IBK | 0.565 |
| 120 hpf E | IBK | 0.544 |
| 120 hpf Sn | IBK | 0.486 |
| 120 hpf TR | IBK | 0.476 |
| 120 hpf CF | IBK | 0.441 |
| 120 hpf Y | Bagging | 0.439 |
| 120 hpf PF | IBK | 0.387 |
| 120 hpf SB | M5P | 0.380 |
| 120 hpf C | IBK | 0.368 |
| 120 hpf O | IBK | 0.331 |
| 120 hpf B | IBK | 0.297 |
| 120 hpf A | IBK | 0.294 |
| 120 hpf So | Bagging | 0.262 |
| 24 hpf N | M5P | 0.125 |
| 24 hpf SM | Kstar | −0.003 |
Abbreviations: A, axis malformation; B, brain malformation; C, circulation; CF, caudal fin malformation; DP, developmental progression; E, eye malformation; H, heart malformation; hpf, hours post-fertilization; J, jaw malformation; M, mortality; N, notochord malformation; O, otic malformation; P, pigmentation; PF, pectoral fn malformation; SB, swim bladder; SM, spontaneous movement; Sn, snout malformation; So, somite malformation; T, trunk malformation; TR, touch response; Y, yolk sac edema.
The results show that we have different modeling accuracies for different biological effects. For example, we achieve a high correlation of 0.837 for modeling 24 hpf mortality, while we only achieve a correlation of −0.003 for modeling 24 hpf spontaneous movement. In most cases, IBK achieves the highest correlation among all algorithms. However, IBK is not the best algorithm for modeling certain effects such as 24 hpf spontaneous movement, 24 hpf notochord malformation, 120 hpf yolk sac edema, 120 hpf somite malformation, and 120 hpf swim bladder.
Overall effect modeling
This modeling focuses on the general impact of exposure to nanomaterials on biological responses measured by the additive EZ Metric score and the weighted EZ Metric score. Using ABMiner, we compare the performance of different algorithms for modeling the overall impact.
Table 3 shows the prediction results. The algorithm of Bagging offers the best performance with a correlation of 0.653 between the actual and predicted additive EZ Metric score. IBK and Bagging both have the best performance with a correlation of 0.792 between the actual and predicted weighted EZ Metric score. Figure 6 shows a sample correlation map for the weighted EZ Metric score case by the Bagging algorithm. In general, the results show that using the weighted EZ Metric score achieves better prediction performance than using the additive EZ Metric score.
Table 3.
Prediction results of overall effect modeling
| Algorithm | Pearson correlation between the actual and predicted overall effect
|
|
|---|---|---|
| Overall effect measured by the “additive EZ Metric score” | Overall effect measured by the “weighted EZ Metric score” | |
| IBK | 0.640 | 0.792 |
| Bagging | 0.653 | 0.792 |
| M5P | 0.542 | 0.766 |
| KStar | 0.548 | 0.685 |
Abbreviation: EZ Metric, embryonic zebrafish metric.
Figure 6.
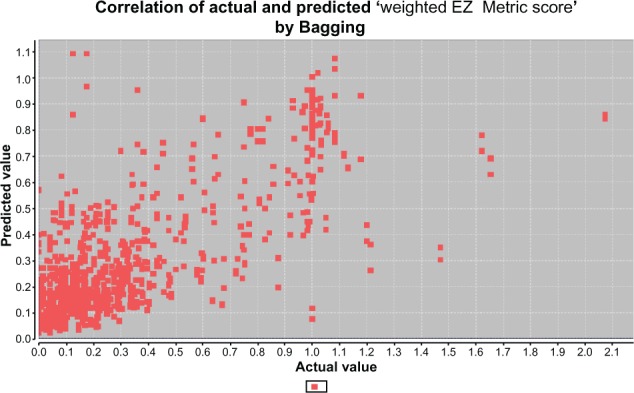
Correlation result of algorithm Bagging.
Abbreviation: EZ Metric, embryonic zebrafish metric.
Model base
To make the prediction models more generally available, we implemented a model base of NEI prediction models. The model base is able to host and manage the datasets and NEI models built by different algorithms. Users can gain information from the NEI models and datasets to inform the nanomaterial design process in creating benign materials and processes.
To illustrate the concept, we have trained several NEI models and deposited them into a sample Model Base at http://neiminer.i-a-i.com/nei_models. The model base interface allows the user to select a model from the list, view and edit the model description, visualize the dataset used to train the model, and query the model for predictive information about the target biological effect.
Model query
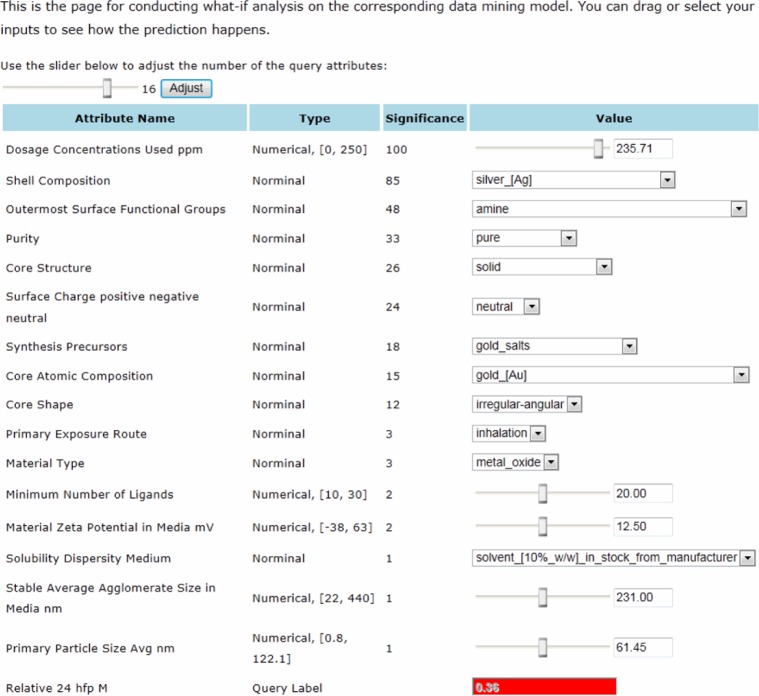
To demonstrate the utility of data-mining-based predictive modeling, we built an interface for biological effect assessment by invoking the underlying prediction models. Figure 7 shows the interface for querying the prediction model of 24 hpf mortality built using the M5P algorithm. Users can make “what if” queries on the model by varying the values of the input attributes. For example, we can simulate a scenario by setting the desired values of the nanomaterial properties (eg, shell composition, outermost surface function groups, or core structure) and exposure conditions (eg, dosage concentrations used or primary exposure route). The interface will automatically display the predicted EZ Metric score of 24 hpf mortality for the simulated scenario. The predicted EZ Metric score provides valuable information on the toxic potential of the target nanomaterial. For example, if the EZ Metric score is smaller than the lower-level threshold of acceptable mortality rate (eg, 0.2), the material can be interpreted as “likely benign;” if the EZ Metric score is larger than the upper-level threshold of unacceptable mortality rate (eg, 0.625), the material can be interpreted as having high toxic potential and requires further testing; if the EZ Metric score is between the lower-level and upper-level thresholds, the material can be interpreted as suspect nanomaterial having moderate toxic potential.26
Figure 7.
Query interface for 24 hpf mortality.
Notes: The underlying model is built using the M5P algorithm. The model can dynamically predict the 24 hpf mortality value for different combinations of attribute values.
Abbreviations: Avg, average; hpf, hours post-fertilization; M, mortality.
Discussion
Our present study focuses on the evaluations for biocompatibility using the embryonic zebrafish system to assess the biological activity and toxic potential of nanomaterials. Zebrafish exhibit remarkable similarity to other high-order vertebrates including humans in their molecular signaling pathways, cellular structure, and physiology.18 The embryonic zebrafish assay is increasingly being used in studies of toxicity of nanomaterials. Examples include the use of zebrafish embryos for testing zinc oxide,19 silver nanoparticles,20 gold nanoparticles,21 and carbon fullerenes.22 Most of these studies focus on the evaluation of one type of nanomaterial. To enable systematic assessment of the embryonic zebrafish as a useful predictive model in nanomaterial toxicity, we aim to quantify the adverse effects in zebrafish embryos on exposure to different kinds of nanomaterials (eg, metal nanoparticles, dendrimer, metal oxide, and polymeric).
Specifically, we developed a generic data mining approach to build predictive models for assessing the unintended biological effects of different types of nanomaterials. Data mining supports different perspectives of modeling, such as classification and numerical prediction. When the biological effect is measured by nominal values, classification models can be built to predict the nominal labels. When the biological effect is measured by numerical values, numerical prediction models can be built to predict the numerical labels. Here, we focus on predicting the biological effects measured by numerical EZ Metric scores. Specific biological effects, such as mortality, delayed development, or malformations, can be predicted by models built using different learning algorithms, such as a nearest neighbor algorithm and a tree algorithm. These algorithms can also be used to build models that predict the overall biological effect measured by the additive or weighted EZ Metric scores.
The dataset used for data mining included 20 input attributes that described the properties of nanomaterials and exposure scenarios. The output is either a specific biological effect or the overall biological effect. We used a feature evaluation algorithm called RELIEF to evaluate the weight of an input attribute with respect to the output. We found several important attributes that contribute to the 24 hpf mortality, including dosage concentration, shell composition, outermost surface functional groups, purity, core structure, and surface charge. Previous studies also showed that these attributes are strong indicators of mortality and malformations. For example, one study reported a concentration-dependent increase in mortality in silver nanoparticle treated zebrafish embryos.20 Another study showed that nanoparticle surface charge significantly influences mortality and malformations in zebrafish embryos exposed to gold nanoparticles.21
The results of specific biological effect modeling show that we can achieve high prediction accuracy for certain biological effects (refer to Table 2). For example, we achieved a correlation of 0.837 for modeling 24 hpf mortality, a correlation of 0.765 for modeling 120 hpf mortality, and a correlation of 0.725 for modeling 120 hpf heart malformation. However, some biological effects cannot be accurately predicted. For example, we only achieved a correlation of −0.003 for modeling 24 hpf spontaneous movement. The results suggest that different biological effects may have different modeling accuracy given the same set of algorithms. We need to further explore the best algorithm for modeling each specific effect.
Our results on overall impact modeling show that using the weighted EZ Metric score generally achieves better prediction performance than using the additive EZ Metric score (refer to Table 3). For example, the M5P algorithm only generated a correlation of 0.542 between the actual and predicted unweighted overall scores, but it generated a much higher correlation of 0.766 between the actual and predicted overall scores using the initial weighting scheme. The results imply that the weighting scheme for individual biological effects does have an influence on the performance of predicting the general exposure impact.
To demonstrate the utility of prediction models, we implemented a model base with a model deployment or query interface. The model base is a host of NEI models and associated datasets. The model query interface utilizes predictive information gained from the prediction models to provide valuable information on the toxic potential of the target nanomaterial. The user can adjust the input attributes (nanomaterial properties, exposure scenarios) to predict potential biological impacts of unsynthesized nanomaterials.
The current dataset to which the NEI model was applied is comprised of multiple morphological, developmental, and behavioral endpoints; which means that expanding the model to include additional endpoints is easily achievable, and in fact, is the next step in model development. The goal of the NEI modeling is to be inclusive of different types of hazard data relative to nanomaterials with the aim to integrate toxicological data across disparate organisms, under varied exposure scenarios, and assimilate weight-of-the-evidence hazard rankings that are more robust than traditional individual metrics. It is envisioned that consensus modeling36 can be implemented within this model framework where endpoints are found to be representative of similar effects (eg, mortality in zebrafish embryos, cytotoxicity measures in various cell types). Where significant variance is found between/among comparable measures, data mining allows for more detailed analysis with statistical validity. In summary, NEI modeling allows for differential inclusion of data within the framework and consensus modeling approaches allow cross-comparison of the hazard ranking across laboratories and organisms.
In addition to building exposure effect/toxicity models, data mining techniques (eg, classification, clustering, or numerical prediction) support the development of other types of models related to the source, transport, and exposure routes of nanomaterials. Sample models include air quality models, hazardous air pollutant exposure models, consensus transport models, bioavailability, biomagnifications, and exposure assessment models.37 We can apply model composition techniques14 to form more comprehensive models and predictability.
Conclusion
Nanobiotechnology is most likely to gain acceptance if environmental and human health considerations are investigated systematically. Modeling NEI is critical to the understanding of the biological effects and health impact of exposure to nanomaterials. To address this critical need, we have developed a data mining approach to NEI modeling. Specifically, we treated NEI modeling as a numerical prediction problem, where the inputs are nanomaterial properties and exposure scenarios, and the outputs are numerical measures of biological effects. We utilized learning algorithms, such as IBK (a nearest neighbor predictor), M5P (a decision tree predictor), and Bagging (a bootstrapping meta-learning algorithm), to train biological effect prediction models from labeled data.
We generated an embryonic zebrafish experimental dataset with 656 test results (samples) across nanomaterials at different dosage concentrations. We evaluated the weights of input attributes (nanomaterial properties and exposure scenarios) with respect to the output (toxic endpoints). We found several important indicators of the 24 hpf mortality, including dosage concentration, shell composition, outermost surface functional groups, purity, core structure, and surface charge. These findings are consistent with the results of previous studies on nanomaterial toxicity using embryonic zebrafish.21,22,27,39,40
Using the experimental data set, we conducted case studies of specific biological effect modeling, as well as overall impact modeling. Our study clearly demonstrated that we can achieve high prediction accuracy for certain biological effects such as 24 hpf mortality, 120 hpf mortality, and 120 hpf heart malformation. Our study also showed that the weighting scheme for different biological effects is an important factor for predicting the overall effect/impact of nanomaterials. This demonstrates the predictive power of our EZ Metric-based data mining approach.
To make the prediction models more generally available, we developed a model base component. The model base is able to host and manage NEI models and associated datasets. It provides a query interface to inform the nanomaterial design process for benign materials and processes. This will offer a tool to industry, academia, and regulatory agencies to rationally investigate the adverse effects of nanomaterials in biological systems.
Taken together, our modeling results provide insights into the understanding and modeling of the effects of nanomaterial exposure on biological systems. For future work, we will enhance the NEI modeling framework by collaborating with domain experts in the following areas: (1) acquiring more data to refine and expand the impact prediction models; (2) developing models on nanomaterial structure–activity relationships using large-scale experimental databases of reasonable diversity (eg, heterogeneous nanoparticles and biological receptors); (3) expanding the scope of nanomaterial biological impact to include various elements in four different layers of the FRAMES framework: source, transport, food chain, and exposure/risk; (4) developing model composition techniques to form more comprehensive models and predictability; and (5) adopting ideas from the recent NanoInformatics 2020 Roadmap38 to develop innovative nanoinformatics applications.
Acknowledgments
We wish to thank US Army Corps of Engineers for their support under research contract numbers W912HZ-11-P-0009 and W912HZ-12-C-0004; NIEHS grants ES017552-01A2, ES016896-01, and P30 ES03850; and AFRL FA8650-05-1-5041. We thank the reviewers for their valuable comments. We also thank Ms Nalini Krishnankutty for language copy editing.
Footnotes
Disclosure
An earlier version of this paper was presented at the IEEE Workshop on Nanoinformatics for Biomedicine, in conjunction with the 2012 IEEE International Conference on Bioinformatics and Biomedicine (BIBM 2012). The authors report no conflicts of interest in this work.
References
- 1.Niemeyer CM, Mirkin CA. Nanobiotechnology: Concepts, Applications and Perspectives. Hoboken: Wiley-VCH Weinheim; 2004. [Google Scholar]
- 2.Jain KK. The Handbook of Nanomedicine. Totowa, NJ: Humana/Springer; 2008. [Google Scholar]
- 3.Elbaz J, Willner I. DNA origami: nanorobots grab cellular control. Nat Mater. 2012;11(4):276–277. doi: 10.1038/nmat3287. [DOI] [PubMed] [Google Scholar]
- 4.Subramani KA, Pathak SA, Hosseinkhani H. Recent trends in diabetes treatment using nanotechnology. Dig J Nanomat Biostructures. 2012;7(1):85–95. [Google Scholar]
- 5.Hosseinkhani H, Hosseinkhani M. Biodegradable polymer-metal complexes for gene and drug delivery. Curr Drug Saf. 2009;4(1):79–83. doi: 10.2174/157488609787354477. [DOI] [PubMed] [Google Scholar]
- 6.Hosseinkhani H. DNA nanoparticles for gene delivery to cells and tissue. Int J Nanotechnol. 2006;3:416–461. [Google Scholar]
- 7.Hosseinkhani H, Tabata Y. Self assembly of DNA nanoparticles with polycations for the delivery of genetic materials into cells. J Nanosci Nanotechnol. 2006;6(8):2320–2328. doi: 10.1166/jnn.2006.507. [DOI] [PubMed] [Google Scholar]
- 8.Oberdörster G, Ferin J, Gelein R, Soderholm SC, Finkelstein J. Role of the alveolar macrophage in lung injury: studies with ultrafine particles. Environ Health Perspect. 1992;97:193–199. doi: 10.1289/ehp.97-1519541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Donaldson K, Borm PJ, Oberdorster G, Pinkerton KE, Stone V, Tran CL. Concordance between in vitro and in vivo dosimetry in the proinflammatory effects of low-toxicity, low-solubility particles: the key role of the proximal alveolar region. Inhal Toxicol. 2008;20(1):53–62. doi: 10.1080/08958370701758742. [DOI] [PubMed] [Google Scholar]
- 10.Landsiedel R, Ma-Hock L, Kroll A, et al. Testing metal-oxide nanomaterials for human safety. Adv Mater. 2010;22(24):2601–2627. doi: 10.1002/adma.200902658. [DOI] [PubMed] [Google Scholar]
- 11.Lam CW, James JT, McCluskey R, Arepalli S, Hunter RL. A review of carbon nanotube toxicity and assessment of potential occupational and environmental health risks. Crit Rev Toxicol. 2006;36(3):189–217. doi: 10.1080/10408440600570233. [DOI] [PubMed] [Google Scholar]
- 12.Zhao Y, Wang B, Feng W, Bai C. Encyclopedia of Life Support Systems (EOLSS): Nanoscience and Nanotechnology. Oxford: Eolss Publishers; 2009. Nanotoxicology: Toxicological and biological activities of nanomaterials; pp. 1–68. [Google Scholar]
- 13.Chen Z, Meng H, Xing G, et al. Acute toxicological effects of copper nanoparticles in vivo. Toxicol Lett. 2006;163(2):109–120. doi: 10.1016/j.toxlet.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 14.Tang K, Liu X, Harper S, Steevens J, Xu R. NEIMiner: a model driven data mining system for studying environmental impact of nanomaterials; Bioinformatics and Biomedicine Workshops (BIBMW), 2012 IEEE International Conference. Proceedings of the 2012 IEEE International Conference on Bioinformatics and Biomedicine Workshops (BIBMW); October 4–7, 2012; Philadelphia, PA, USA. IEEE Xplore. 2012. pp. 895–902. [Google Scholar]
- 15.Liu X, Tang K, Harper S, Harper B, Steevens J, Xu R. Predictive modeling of nanomaterial biological effects; Bioinformatics and Biomedicine Workshops (BIBMW), 2012 IEEE International Conference. Proceedings of the 2012 IEEE International Conference on Bioinformatics and Biomedicine Workshops (BIBMW); October 4–7, 2012; Philadelphia, PA, USA. IEEE Xplore. 2012. pp. 859–863. [Google Scholar]
- 16.Horev-Azaria L, Kirkpatrick CJ, Korenstein R, et al. Predictive toxicology of cobalt nanoparticles and ions: comparative in vitro study of different cellular models using methods of knowledge discovery from data. Toxicol Sci. 2011;122(2):489–501. doi: 10.1093/toxsci/kfr124. [DOI] [PubMed] [Google Scholar]
- 17.Zhu H, Ye L, Richard A, et al. A novel two-step hierarchical quantitative structure-activity relationship modeling work fow for predicting acute toxicity of chemicals in rodents. Environ Health Perspect. 2009;117(8):1257–1264. doi: 10.1289/ehp.0800471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hill AJ, Teraoka H, Heideman W, Peterson RE. Zebrafish as a model vertebrate for investigating chemical toxicity. Toxicol Sci. 2005;86(1):6–19. doi: 10.1093/toxsci/kfi110. [DOI] [PubMed] [Google Scholar]
- 19.Bai W, Zhang Z, Tian W, et al. Toxicity of zinc oxide nanoparticles to zebrafish embryo: a physicochemical study of toxicity mechanism. J Nanopart Res. 2010;12(5):1645–1654. [Google Scholar]
- 20.Asharani PV, Wu YL, Gong Z, Valiyaveettil S. Toxicity of silver nanoparticles in zebrafish models. Nanotechnology. 2008;19(25):1–8. doi: 10.1088/0957-4484/19/25/255102. [DOI] [PubMed] [Google Scholar]
- 21.Harper SL, Carriere JL, Miller JM, Hutchison JE, Maddux BL, Tanguay RL. Systematic evaluation of nanomaterial toxicity: utility of standardized materials and rapid assays. ACS Nano. 2011;5(6):4688–4697. doi: 10.1021/nn200546k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Usenko CY, Harper SL, Tanguay RL. In vivo evaluation of carbon fullerene toxicity using embryonic zebrafish. Carbon N Y. 2007;45(9):1891–1898. doi: 10.1016/j.carbon.2007.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nanomaterial-Biological Interactions Knowledgebase [homepage on the Internet] Corvallis, OR: Nanomaterial-Biological Interactions Knowledgebase; Available from: http://oregonstate.edu/nbiAccessed November 25, 2012 [Google Scholar]
- 24.Liu X, Tang K, Buhrman JR, Cheng H. An agent-based framework for collaborative data mining optimization; Collaborative Technologies and Systems (CTS), 2010 International Symposium. Proceedings of the IEEE International Symposium on Collaborative Technologies and Systems (CTS); May 17–21, 2010; Chicago, IL, USA. IEEE Xplore. 2010. pp. 295–301. [Google Scholar]
- 25.Tang K, Liu X, Tang Y, Manikonda V, Buhrman JR, Cheng H. ABMiner: A scalable data mining framework to support human performance analysis. In: Duffy VG, editor. Advances in Applied Digital Human Modeling. Boca Raton, FL: CRC Press; 2010. pp. 639–648. [Google Scholar]
- 26.Harper SL. An EZ Metric for Defining the A in nanoSARs. Nano WG Meeting organized by NCI. July2011 [Google Scholar]
- 27.Harper SL, Dahl JA, Maddux BLS, Tanguay RL, Hutchison JE. Proactively designing nanomaterials to enhance performance and minimise hazard. Int J Nanotechnol. 2008;5(1):124–142. [Google Scholar]
- 28.Hall M, Frank E, Holmes G, Pfahringer B, Reutemann P, Witten IH. The WEKA data mining software: an update. SIGKDD Explor. 2009;11(1):10–18. [Google Scholar]
- 29.Aha D, Kibler D, Albert MK. Instance-based learning algorithms. Machine Learning. 1991;6(1):37–66. [Google Scholar]
- 30.Breiman L. Bagging predictors. Machine Learning. 1996;24(2):123–140. [Google Scholar]
- 31.Witten IH, Frank E. Data Mining: Practical Machine Learning Tools and Techniques. 2nd ed. San Francisco: Morgan Kaufmann; 2005. [Google Scholar]
- 32.Quinlan RJ. In: Adams, Sterling, editors. Learning with continuous classes; Proceedings AI’92: Proceedings of the 5th Australian Joint Conference on Artificial Intelligence; November 16–18, 1992; Singapore. 1992. pp. 343–348. [Google Scholar]
- 33.Wang Y, Witten IH. Induction of model trees for predicting continuous classes; Proceedings of the 9th European Conference on Machine Learning; April 23–25, 1997; Prague, Czech Republic. [Google Scholar]
- 34.Cleary JG, Trigg LE. In: Prieditis A, Russel SJ, editors. K*: An instance-based learner using an entropic distance measure; Machine Learning: Proceedings of the 12th International Conference on Machine Learning; July 9–12, 1995; Tahoe City, CA, USA. 1995. pp. 108–114. [Google Scholar]
- 35.Kira K, Rendell LA. A practical approach to feature selection; Proceedings of the 9th International Workshop on Machine Learning; 1992; Aberdeen, Scotland. San Francisco: Morgan Kaufmann Publishers Inc. 1992. pp. 249–256. [Google Scholar]
- 36.Tong W, Fang H, Xie Q, et al. Gaining confidence on molecular classification through consensus modeling and validation. Toxicol Mech Methods. 2006;16(2–3):59–68. doi: 10.1080/15376520600558259. [DOI] [PubMed] [Google Scholar]
- 37.Whelan G, Castleton KJ, Buck JW, et al. Concepts of a Framework for Risk Analysis in Multimedia Environmental Systems Richland, WA: Pacific Northwest National Laboratory; 1997Available from: http://mepas.pnnl.gov/framesv1/documents/PNNL11748-frames_doc.pdfAccessed November 25, 2012 [Google Scholar]
- 38.Nanoinformatics 2020 Roadmap [website on the Internet] National Nanomanufacturing Network; 2011Available from: http://www.internano.org/content/view/510/251/Accessed November 25, 2012 [Google Scholar]
- 39.Harper SL, Usenko CY, Hutchison J, Maddux BLS, Tanguay RL. In vivo biodistribution and toxicity depends on nanomaterial composition, size, surface functionalization and route of exposure. Journal of Experimental Nanoscience. 2008;3:195–206. [Google Scholar]
- 40.Usenko CY, Harper SL, Tanguay RL. Exposure to C60 elicits an oxi-dative stress response in embryonic zebrafish. Toxicology and Applied Pharmacology. 2008;229:44–55. doi: 10.1016/j.taap.2007.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]