Abstract
Fas-associated death domain-containing protein (FADD), a classical apoptotic signaling adaptor, participates in different nonapoptotic processes regulated by its phosphorylation. However, the influence of FADD on metabolism, especially glucose homeostasis, has not been evaluated to date. Here, using both two-dimensional electrophoresis and liquid chromatography linked to tandem mass spectrometry (LC/MS/MS), we found that glycogen synthesis, glycolysis, and gluconeogenesis were dysregulated because of FADD phosphorylation, both in MEFs and liver tissue of the mice bearing phosphorylation-mimicking mutation form of FADD (FADD-D). Further physiological studies showed that FADD-D mice exhibited lower blood glucose, enhanced glucose tolerance, and increased liver glycogen content without alterations in insulin sensitivity. Moreover, investigations on the molecular mechanisms revealed that, under basal conditions, FADD-D mice had elevated phosphorylation of Akt with alterations in its downstream signaling, leading to increased glycogen synthesis and decreased gluconeogenesis. Thus, we uncover a novel role of FADD in the regulation of glucose homeostasis by proteomic discovery and physiological validation.
Fas-associated death domain-containing protein (FADD)1, originally identified by the yeast two-hybrid system with the cytoplasmic domain of Fas (1, 2), is considered as an essential mediator of Fas-induced apoptosis (1–3). It is a 23-kDa protein consisting of a C-terminal death domain and an N-terminal death effector domain (4). Later studies have revealed that this adaptor molecule could transmit apoptotic signals of all known death domain receptors (5, 6). Apart from its critical role in apoptosis, FADD also participates in lymphocyte development and proliferation (7–10), cardiac embryonic development (11, 12), tumorigenesis (13), inflammation, and innate immunity (14–16). These non-apoptotic functions of FADD have been indicated to be partly attributable to its phosphorylation (serine 194 in human and serine 191 in mouse) (9, 10, 17, 18). This phosphorylation site locates at the C-terminal tail of FADD outside apoptotic domains and is regulated dependent on the cell cycle during T-cell activation (19). Mice bearing a mutation form of FADD mimicking constitutive phosphorylation at serine 191 (FADD-D) have been generated to investigate the physiological actions of FADD phosphorylation (8). It has been shown that FADD-D T cells are defective in cell cycle progression without impairment in apoptosis, suggesting an essential role of FADD phosphorylation in the regulation of growth and proliferation (8).
Glucose is an essential nutrient and major energy source in mammalian. Therefore, it is of great importance to maintain blood glucose. The glucose homeostasis is achieved by the balance of the storage of glucose as glycogen, the use of glucose through glycolysis, and the production of glucose via glycogenolysis and gluconeogenesis in liver (20). Glycogen synthesis and breakdown (glycogenolysis) are predominantly controlled by the coordination of two enzymes, glycogen synthase (GS) and glycogen phosphorylase (GP) (21). Glycolysis, initiated by the enzyme glucokinase (Gck), converts glucose into pyruvate and further produces a final production of ethanol or lactate by a serial of reactions (22). Gluconeogenesis is the opposite metabolic pathway of glycolysis, generating glucose from noncarbohydrate carbon substrates such as pyruvate, lactate, and so on (22, 23). Glucose-6-phosphatase (G6pc) and phosphoenolpyruvate carboxykinase (Pepck) are two rate-limiting enzymes for gluconeogenesis (20, 24). In addition, glucose metabolism is under the control of several hormones, such as insulin and glucagon (25). Insulin plays an important role in glucose homeostasis by activating protein kinase B/Akt, the key node in insulin signaling (26, 27). The activated Akt can phosphorylate and inactivate glycogen synthase kinase (GSK) 3β, a key negative regulator in glycogen synthesis (28, 29). Meanwhile, Akt phosphorylates the forkhead transcription factor (Foxo1), promoting nuclear to cytoplasmic translocation of Foxo1 and suppressing Foxo1- dependent transcription, such as G6pc and Pepck (30, 31).
At present, proteomics coupled with bioinformatics analysis is increasingly used in biological research to fully understand the potential targets and signaling pathway networks (32). The proteomic platform provides a powerful tool for us to perform high-throughput studies allowing the detection of modulated proteins, which are regulated under abnormal conditions, and attempts to identify candidate pathways regulated by specific genes (33–35). In particular, the shotgun approach based on one-dimensional (1D) PAGE with liquid chromatography chip and Q-TOF MS/MS (HPLC-Chip/Q-TOF) allows the qualitative and quantitative analysis of a large number of proteins in complex samples, avoiding time-consuming two-dimensional electrophoresis (2-DE) and isotopic labeling (36). In this study, we first applied two-dimensional electrophoresis (2-DE)-proteomics approach to compare the different expression patterns of wild-type (WT) and FADD-D mouse embryonic fibroblasts (MEFs). Then, we further investigated WT and FADD-D livers by a label-free 1D-LC-MS/MS analysis using nanoLC coupled to a q-TOF mass spectrometer (HPLC-Chip/Q-TOF). Based on the information inherent in chromatographic data, MS spectra, and MS/MS-based peptide assignments, label-free quantitative strategies are attractive alternatives for quantitative LC/MS/MS-based proteomics because of their simplicity, affordability, and flexibility. Although suppressive ionization may affect the quantification, previous studies have demonstrated that spectral counting (peptide count), spectral ion intensity, or peak area of peptide ions correlates well with protein abundance in complex samples (37–39). Here, protein samples from two genotype livers were trypsinized before LC-MS analysis. Samples were analyzed in triplicates using 6550 nano HPLC-Chip/q-TOF mass spectrometer. The data were analyzed and semiquantified using Spectrum Mill bioinformatics tool. In total hundreds of proteins were found to be dysregulated because of FADD phosphorylation in both MEFs and livers. Using MetaCoreTM pathway analysis tools, we identified a number of differentially expressed proteins involved in glycogen metabolism, glycolysis, and gluconeogenesis, suggesting a novel role for FADD in glucose homeostasis. Therefore, we performed further physiological examinations on FADD-D mice to address this issue. Our data showed that FADD-D mice were hypoglycemic and had increased glucose tolerance. The hepatic glycogen accumulation in FADD-D mice was also enhanced. In addition, the phosphorylation form of Akt, a key regulator in glucose homeostasis (27, 40), was elevated in liver tissue of FADD-D mice. Collectively, these data suggest that FADD phosphorylation might have an impact on glucose homeostasis, revealing a novel function of FADD, which has not been evaluated to date.
EXPERIMENTAL PROCEDURES
Stable Cell Line Construction and Cell Culture
WT MEFs and FADD-D MEFs were all cultured in DMEM (Hyclone, Logan, UT) containing 10% FBS (Hyclone) with 50 U/ml penicillin/streptomycin. FADD mutant cell line FADD-D was constructed based on FADD−/− MEFs; the construction and validation of the cell lines were performed in Dr. Astar Winoto's Laboratory (UC Berkeley). Briefly, FADD-D mutant cDNAs were generated using PCR, as previously described (8), subcloned into the retroviral vector MSCV-Zeocin and transfected into Bosc packing cells. Supernatant was used to infect FADD knockout MEFs. Infected MEFs were selected with Zeocin for 1 month. The expression levels of FADD and FADD-D were examined using immunoblotting assay and were similar across different MEFs.
Two-dimensional Gel Electrophoresis and Matrix-assisted Laser Desorption Ionization-time-of-flight (MALDI-TOF) MS
MEFs (1 × 107) were all harvested when they reached 90% confluence, rinsed three times with ice-cold PBS and pelleted at 2000 × g for 5 min. Then, the pellets were lysed in a cell lysis solution containing 7 m urea, 2 m thiourea, 40 mm dithiotreitol, and 2% IPG buffer (pH 3–10 L, GE Healthcare) at a volume ratio of 1:10. After cyclic liquid nitrogen freezing-thawing treatment, the lysates were sonicated in short bursts to disrupt nucleic acids, and then were clarified by centrifugation at 40,000 × g at 4 °C for 60 min. The protein concentration in the supernatant was determined by the Bradford method. All procedures were performed on ice. The supernatant containing 120 μg proteins was first isoelectrically focused on a gel strip with 3–10 linear pH gradient, and then resolved on an SDS-polyacrylamide gel as previously reported (41). The separated proteins were visualized by silver diamine-staining as described before (42). After destaining with double distilled water, gels were scanned at 300 dpi resolution, and the images were processed using the Adobe Photoshop software (Adobe Systems) and analyzed using the Image Master PlatinumTM software (GE Healthcare) according to the manufacturer's procedures. Over a twofold change in protein concentration was considered to be significant (p < 0.01 by Mann-Whitney test). The differentially expressed proteins were cut and digested essentially as described by Yang et al. (43).
MALDI samples were prepared according to a thin layer method described before (43, 44). Mass spectra were recorded on an Ultraflex II MALDI-TOF-TOF mass spectrometer (Bruker Daltonics GmbH, Bremen, Germany) under the control of FlexControlTM 3.0 software (Bruker Daltonik). MALDI-TOF spectra were recorded in the positive ion reflector mode in a mass range from 700–4000 Da, and the ion acceleration voltage was 25 kV. Acquired mass spectra were processed using the software FlexAnalysisTM 3.0 (Bruker Daltonics): Peak detection algorithm: SNAP (Sort, Neaten, Assign, and Place); signal-to-noise (S/N) threshold: 3; Quality Factor Threshold: 50. The tryptic auto-digestion ion picks (trypsin_[108–115], MH+842.509, trypsin_[58–77], MH+2211.104) were used as internal standards. Matrix and/or auto-proteolytic trypsin fragments or known contaminant ions (keratins) were excluded. The resulting peptide mass lists were used to search the IPI mouse database 3.29 (53981 sequences, 25507684 residues) with Mascot (v2.3.02) in automated mode. The following criteria were used for search parameters: significant protein MOWSE score at p < 0.05, minimum mass accuracy 120 ppm, trypsin as enzyme, 1 missed cleavage site allowed, alkylation of cysteine by carbamidomethylation as fixed modification and oxidation of methionine as variable modification. In addition, the Mascot Score and expectation of the first nonhomologous protein to the highest ranked hit were checked.
Animals
All studies involving mice with high standard animal welfare were approved by Nanjing University Animal Care and Use Committee. Animals were maintained in a specific pathogen-free animal facility on a 12-h light-dark cycle at an ambient temperature of 21 °C. They were given free access to water and food. Experiments were performed on mice aged between 8–10 weeks. Details of generating FADD-D mice have been described previously (8).
Protein Extraction, Separation, and Digestion
Proteins were extracted from fresh livers as described previously with some modifications (45). Briefly, minced livers were homogenized with 5 volumes of ice-cold lysis solution using a loose fitting Dounce homogenizer. After filtering through four layers of nylon gauze, the homogenate was centrifuged at 12,000 × g for 10 min, and then the supernatant was collected and the protein concentration was determined using the Bradford protein assay kit (Bio-Rad, Hercules, CA). After being separated by SDS-PAGE, the PAGE was stained with Coomassie brilliant blue G-250 and cut into slices. Before MS analysis, the gel was destained with 100 mm ammonium bicarbonate (ABC)/50% acetonitrile (ACN) (1:1, v/v), and ACN was added to dehydrate. Subsequently, trypsin (10 ng/μl) was added to the gel pieces, and then ABC/ACN (9:1, v/v) of 40 mm was added to cover, and digested overnight. Then the digest pieces were extracted with a solution containing 50% ACN and 5% formic acid (FA). The digested peptides were concentrated by freeze drier and stored at −80 °C for further MS analysis.
LC-CHIP Q-TOF MS/MS Analysis
The LC-CHIP Q-TOF MS/MS analysis was conducted as described previously (46). The HPLC-Chip is a microfluidic device that integrates sample preparation and analysis on a single chip, which includes a sample enrichment column, an analytical column, a nano-spray tip, and all connections between them. It inserts into the HPLC-Chip/MS interface, which includes the nano-spray source, connections to the LC pumps and autosampler, and the microvalve for column switching. The chip is hydraulically interfaced to a nano-HPLC pump and an autosampler through a face seal rotaryvalve, thus eliminating conventional LC fittings and connections. Briefly, the lyophilized peptides were resuspended in 0.1% FA. The resuspended peptide solution was enriched and fractionated with HPLC-Chip (Agilent 1200 Series HPLC systems). The software HPLC-Chip Cube MS interface was used to control the whole analytical process. A total of 1.0 μl sample (200 ng) was injected into the enrichment column to desalt and analyze online through MSn after isocratic elution and gradient elution by enrichment and separation columns, respectively. The peptides were loaded into the enrichment column before analytical separation. A Agilent 6550 ESI Q-TOF Mass Spectrometer adopted Chip cube was used as an ion source. Precursor selection selected three max precursors per cycle, active exclusion by enabled mode, excluded after 1 spectrum, and released after 0.25 min and the MS or MS/MS scanning range of m/z 300 to 3000 or 100 to 3000.
Database Search for Protein Identification
Database search was applied as previously reported (46). Briefly, the data of MS and MS/MS were analyzed using the Agilent G2721AA Spectrum Mill MS Proteomics Workbench (Rev A.03.03.078) against the database of UniProtKB/SWISSProt, species Mus musculus (mouse). The value of peptide spectral intensity was obtained from the analyzed data of MS and MS/MS by the spectrum mill proteomics workbench. The Spectrum Mill Data Extractor program prepared MS/MS data files for processing. Autovalidation was carried out after searching by calculated reversed database scores to rule out false positives.
Bioinformatics Analysis
The data set with a list of regulated proteins identified by proteomics was analyzed further by pathway analysis using the network building tool MetaCoreTM version 5.4 (GeneGo, St. Joseph, MI). MetaCoreTM is an integrated software suite for functional analysis of experimental data. It is based on a proprietary manually curated database of protein–protein, protein–DNA and protein compound interactions, metabolic and signaling pathways and the effects of bioactive molecules in gene expression. The networks can be visualized graphically as nodes (proteins) and edges (the relationship between proteins) alongside the empirical expression pattern. Differentially expressed proteins were converted into gene symbols and uploaded into MetaCoreTM for analysis. For enrichment analysis, gene IDs of the uploaded files were matched with gene IDs in GeneGo ontologies in MetaCoreTM that includes GeneGo Pathway Maps.
Metabolic Analysis
Mice were fasted overnight. Blood glucose concentrations were measured using a glucometer (Roche, Mannheim, Germany). Serum insulin was determined using Rat/Mouse Insulin ELISA (Millipore, Billerica, MA). Serum glucagon was determined by Bio-Plex kit (Bio-Rad Laboratories, Hercules, CA).
Intraperitoneal Glucose and Insulin Tolerance Test
For intraperitoneal glucose tolerance test (IPGTT), we subjected mice to an overnight fast followed by intraperitoneal glucose injection (1.0 g/kg). Blood glucose was measured at 0, 15, 30, 60, and 90 min after the injection. For intraperitoneal insulin tolerance test (IPITT), mice were fasted for 6 h, followed by intraperitoneal insulin injection (0.5 U/kg). Blood glucose levels were then measured at 0, 15, 30, 60, and 90 min after the injection.
Glycogen Content
The glycogen content was determined according to the method described previously (47). Briefly, mice were fasted overnight and sacrificed. Liver samples (30–90 mg) were weighted and digested in 300 μl of 30% (w/v) KOH at the boiling water bath for 30 min. After the solution cooled to room temperature, 100 μl 1 m Na2SO4 and 800 μl ethanol were added and boiled for 5 min to precipitate glycogen. The samples were centrifuged for 5 min at 10,000 rpm and the pellets were washed with 95% ethanol and dried on air. Pellets were reconstituted in 200 μl 0.2 m Na-acetate buffer (pH 4.8) and hydrolyzed with 1.25 mg/ml amyloglucosidase (Sigma-Aldrich, St. Louis, MO) at 55 °C for 2 h. Glucose concentration was determined using Glucose (GO) Assay Kit (Sigma-Aldrich, St. Louis, MO).
Histology
Livers collected from euthanized animals were fixed in 10% phosphate-buffered formalin and embedded in paraffin, then, 5-μm-thick sections were stained with hematoxylin and eosin (H&E) for routine histology. Liver samples were fixed in Carnoy solution (60% ethanol, 30% chloroform, 10% glacial acetic acid) for glycogen detection and stained with Periodic Acid-Schiff (PAS) Kit (Sigma-Aldrich, St. Louis, MO).
For immunohistochemical staining, liver sections were immunostained with anti-GS antibody (Cell Signaling Technology, Beverly, MA) and visualized with DAB. The localization of Foxo1 in liver was detected by staining liver sections with anti-Foxo1 antibody (Cell Signaling Technology, Beverly, MA) and Alexa Fluor 488-conjugated anti-rabbit IgG antibody (Invitrogen, Carlsbad, CA) was used as the secondary antibody. Nuclei were stained with Hoechst. The staining was monitored with an Axiophot2 microscope (Zeiss, Jena, Germany).
For electron microscopic examination, livers were fixed with 2.5% glutaraldehyde in 0.1 m phosphate buffer (pH 7.4). The tissues were postfixed in 2% osmium tetroxide solution and embedded in epoxy resin. Sections were examined with electron microscope (Zeiss, Jena, Germany).
Western Blotting
Mice fasted overnight were injected intraperitoneally with 1 U/kg insulin. After 15 min, mice were sacrificed and tissues were collected. Livers were homogenized in lysis buffer. After centrifugation at 12,000 rpm, the supernatant was collected, and protein concentration was quantified by Bradford assays.
For Western blotting, equal amounts of protein (50 μg) were electrophoresed by 6–12% SDS-PAGE and then transferred onto a PVDF membrane. Blots were probed with antibodies for Akt, phospho-Akt (Ser473), phospho-GSK3β (Ser9), GS (Cell Signaling Technology, Beverly, MA, USA), GSK3β (BD Biosciences, Franklin Lakes, NJ), Ppp1ca, Ldha, Pepck (Santa Cruz Biotechnology, Inc., Santa Cruz, CA), Glucokinase (Bioworld Technology, Inc., MN), Pgam1, Aldoa, Tpi1, Kpyr, and G6pc (Bioss, Beijing Biosynthesis Biotechnology, Inc., Beijing, China).
RNA Extraction and Quantitative Real-time PCR
Total RNA was extracted from tissues using a TRIzol reagent (Invitrogen, Carlsbad, CA) following the manufacturer's instructions and was used to prepare cDNA by PrimeScript RT reagent kit (Takara, Otsu, Shiga, Japan). Quantitative real-time PCR (qRT-PCR) was performed on an ABI system (Applied Biosystems, Foster City. CA). The primers used in the qRT-PCR were listed in supplemental Table S1. All data were means of fold change of triplicate analyses and normalized with those of GAPDH.
Statistical Analysis
Data were presented as means ± S.E. The difference between the two genotype groups was analyzed by two-tailed Student's t test using Prism software (GraphPad, San Diego, CA). Values were considered statistically significant at p < 0.05.
RESULTS
2-DE-Based Proteomics Analysis
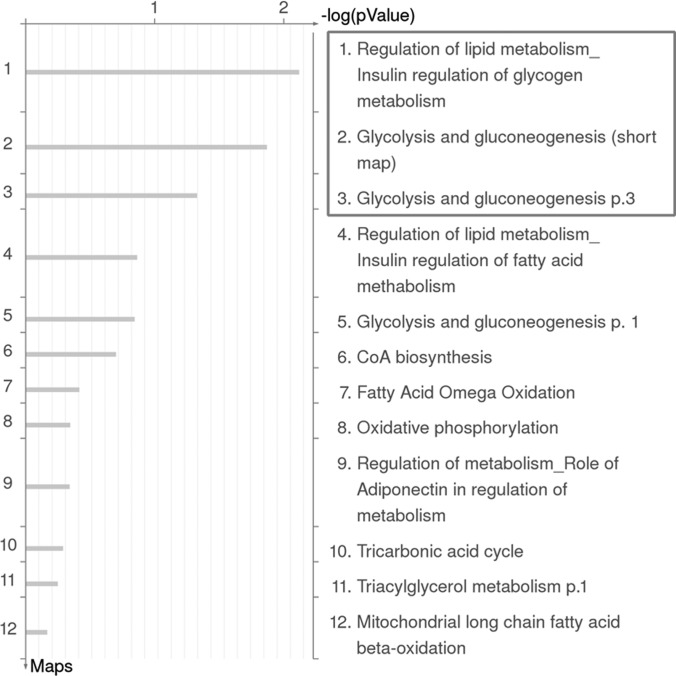
Considering the significance of serine 191 phosphorylation at FADD C-terminal region, we sought to reveal the molecular pathways directly or indirectly controlled by this phosphorylation and underscore its critical roles in nonapoptotic processes by the high throughput proteomic approaches. The confirmation of FADD-D MEF cell line was validated by Western blotting as shown in supplemental Fig. S1A. Proteomic profiling of FADD-D and wild type (WT) control MEFs was performed in parallel by two-dimensional electrophoresis analysis. supplemental Fig. S2 showed the typical 2-D gel images of the two cell lines respectively. A total of 35 proteins with their identified specific MS/MS sequences, were identified after in-gel digestion, MALDI-TOF/TOF analysis, and IPI mouse database search. To further interpret the likely roles of differentially expressed proteins induced by FADD phosphorylation, we used the MetaCoreTM pathway mapping tool to analyze and build the biological networks related to these proteins. GeneGo Map Folder analysis was applied to show that energy metabolism and its regulation have the highest significance. Fig. 1 showed detailed GeneGo Maps in Fold “energy metabolism and its regulation,” among which insulin regulation of glycogen metabolism, glycolysis and gluconeogenesis were the most significant pathways. Six of the changed proteins were involved in these pathways (Table I and supplemental Fig. S3). supplemental Fig. S4A–S4C further displayed the pathway maps associated with glycogen metabolism, glycolysis, and gluconeogenesis. These results indicated that glucose metabolism may be dysregulated because of FADD phosphorylation in MEFs.
Fig. 1.
Representation of ontological category of differentially expressed proteins in control and FADD-D cell lines by GeneGo Map Folders analysis. The results were ordered by -log10 of the p value of the hypergeometric distribution. Detailed GeneGo Maps in Fold “energy metabolism and its regulation” were shown.
Table I. Deregulated proteins involved in glycolysis and gluconeogenesis in FADD-D cells compared to FADD cells. The identification information for each protein is documented in supplemental File S1.
| Spot No. | Ratio of FADD-D to FADD | Identified protein | Network objects | NCBI accession No. | Coverage matched peptides | MowseScore |
|---|---|---|---|---|---|---|
| 1 | -2.45 | Phosphoenolpyruvate carboxykinase, mitochondrial precursor | PPCKM | GI:52783203 | 31% | 125 |
| 2 | 3.13 | Glucokinase | HXK4 | GI:1708365 | 33% | 110 |
| 3 | -2.78 | Glucose-6-phosphatase | G6PT | GI:341940721 | 41% | 148 |
| 4 | 3.79 | Phosphoglycerate mutase 1 | PGAM1 | GI:20178035 | 67% | 154 |
| 5 | 2.88 | l-lactate dehydrogenase A chain | LDHA | GI:126048 | 36% | 159 |
| 6 | 2.86 | Serine/threonine protein phosphatase PP1-alpha catalytic subunit (PP1 cat alpha) | PP1-cat | GI:49065812 | 30% | 85 |
HPLC-Chip/Q-TOF Based Proteomics Analysis
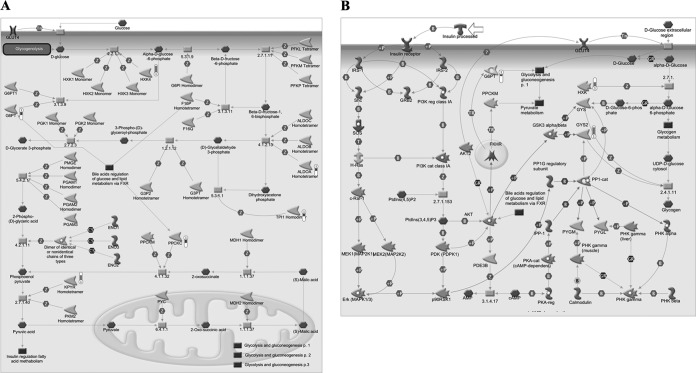
To test whether FADD phosphorylation has a critical role on glucose metabolism, we generated FADD-D mice, an animal model containing FADD phosphorylation-mimicking mutation of serine residue at 191 on the background of FADD−/− (8). The expression levels of FADD and its phosphorylation form in livers of FADD-D and its littermate control mice were confirmed by Western blotting as shown in supplemental Fig. S1B. HPLC-Chip/Q-TOF technology was applied to quantitatively determine differentially expressed proteins between control and FADD-D mice livers. A t test identified 122 differential proteins. MetaCoreTM pathway mapping tool revealed that seven proteins among them were involved in glycogen metabolism, glycolysis, and gluconeogenesis processes (Fig. 2 and Table II), which was partially consistent with the above cellular proteomics results. These data suggest a crucial role of FADD in glucose metabolism.
Fig. 2.
GeneGO pathway showing changes in expression of liver proteins involved in glycolysis and gluconeogenesis (A), insulin regulation of glycogen metabolism (B) upon FADD phosphorylation. The various proteins on this map are represented by different symbols (representing the functional class of the protein). Thermometers with blue or red shading next to symbols depict proteins identified in the present study: blue color represents the proteins that were down-regulated in FADD-D mice liver relative to control group; red color represents the proteins that were up-regulated in FADD-D mice liver relative to control group.
Table II. Deregulated proteins involved in glycolysis and gluconeogenesis in FADD-D mice liver compared to control mice liver. The identification information for each protein is documented in supplemental File S2.
| No. | Identified protein | Network objects | Swiss-Prot No. | Molecular weight | Ratio of FADD-D to control |
|---|---|---|---|---|---|
| 1 | Glucose-6-phosphatase, catalytic | G6PT | P35576 | 40,473 | -1.97 |
| 2 | Glucokinase (hexokinase 4) | HXK4 | P52792 | 52,089 | 4.65 |
| 3 | Phosphoenolpyruvate carboxykinase 1, cytosolic | PPCKC | Q9Z2V4 | 69,355 | -2.34 |
| 4 | Triosephosphate isomerase 1 | TPI1 | P17751 | 32,192 | -1.76 |
| 5 | Pyruvate kinase isozymes R/L | KPYR | P53657 | 62,309 | 3.42 |
| 6 | Fructose-bisphosphate aldolase A | ALDOA | P05064 | 39,356 | -1.68 |
| 7 | Glycogen synthase, liver | GYS2 | Q8VCB3 | 80,871 | 8.56 |
Low Blood Glucose Level in FADD-D Mice
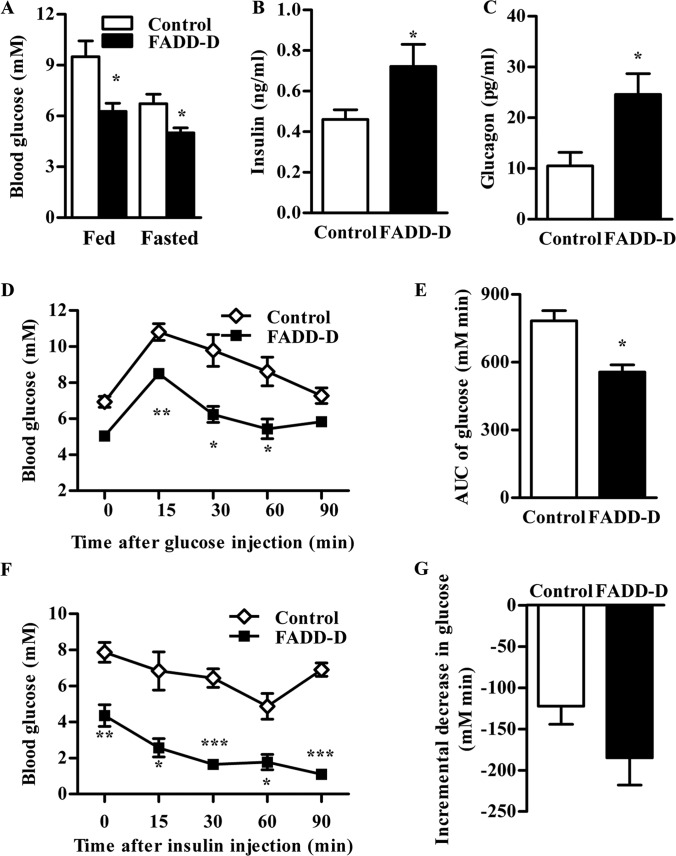
To clarify the specific role of FADD phosphorylation on glucose metabolism physiologically, we examined blood glucose level in FADD-D mice, and found that FADD-D mice displayed notably lower fed and fasting blood glucose levels (Fig. 3A). Because glucose homeostasis is mainly regulated by insulin and glucagon (26, 48), we also detected their concentrations in serum. Fasting insulin levels were higher in FADD-D mice (Fig. 3B). In addition, glucagon, the counter-regulatory hormone of insulin, was about three times the level of control mice in FADD-D mice (Fig. 3C).
Fig. 3.
Low blood glucose and increased glucose tolerance in FADD-D mice. A, Fed and fasted blood glucose level in mice (n = 4–9 for each group). B and C, Fasted serum insulin (B) and glucagon (C) levels in mice (n = 6–12 for each group). D, IPGTT of FADD-D (black squares) and control mice (white diamonds). Mice were injected intraperitoneally with 1.0 g/kg glucose, and blood glucose levels were monitored at the intervals indicated (n = 3). E, The histogram represents the cumulative increase in blood glucose from basal level after the injection of glucose during IPGTT. F, IPITT of FADD-D (black squares) and control mice (white diamonds). Mice were injected intraperitoneally with 0.5 U/kg insulin, and blood glucose levels were monitored at the intervals indicated (n = 3). G, The histogram represents the cumulative decrease in blood glucose from the basal level after the injection of insulin during IPITT. Data are presented as means ± S.E. Statistical significance was assessed by two-tailed Student's t test, *p < 0.05, **p < 0.01, ***p < 0.001.
Improved Glucose Tolerance in FADD-D Mice
To further explore the effects of FADD-D on glucose homeostasis in vivo, we performed an intraperitoneal glucose tolerance test (IPGTT). In an IPGTT assay, the glucose levels were significantly decreased in FADD-D mice (Fig. 3D) and the incremental area under the blood glucose curve of FADD-D mice was only about two-thirds of that in control mice (Fig. 3E), indicating an improved glucose clearance in FADD-D mice after a glucose challenge.
The Intraperitoneal insulin tolerance test (IPITT) was also conducted to evaluate whether insulin sensitivity was changed in FADD-D mice. After insulin injection, the blood glucose of FADD-D mice decreased to the level dramatically lower than that observed in littermates (Fig. 3F). But there was no significant difference in the relative decreased glucose from basal levels between FADD-D and control mice (Fig. 3G), suggesting that insulin sensitivity in FADD-D was unaltered.
Increased Glycogen Accumulation in Liver of FADD-D Mice
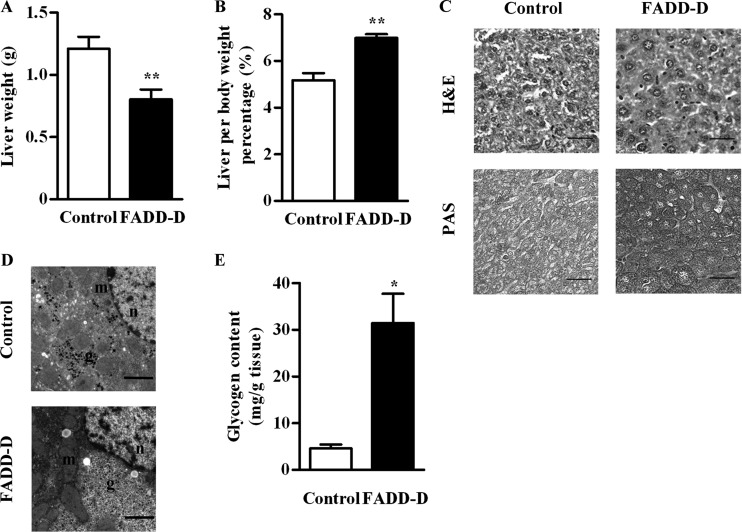
Blood glucose homeostasis is maintained by a balance between glucose production, uptake, and storage, which mainly take place in liver (20). So, we next examined the liver of these mice. Liver weights of FADD-D mice were decreased compared with their littermate controls (Fig. 4A). But when normalized to the body weight, the relative liver weight was significantly higher in FADD-D mice (Fig. 4B).
Fig. 4.
Increased hepatic glycogen content in FADD-D mice. A, Liver weights of FADD-D and control mice (n = 4). B, Weights of liver normalized by body weight for FADD-D and control mice (n = 4). C, H&E staining and PAS staining for glycogen in the liver sections of FADD-D and control mice. Representative images are shown. Scale bar, 20 μm. D, The representative images of electron microscopic analysis of liver. Scale bar, 2 μm. “g” indicates glycogen, “m” indicates mitochondria, and “n” indicates nucleus. E, Hepatic glycogen content determined by glucose assay kit (n = 6). Data are presented as means ± S.E. Statistical significance was assessed by two-tailed Student's t test, *p < 0.05, **p < 0.01.
Histology analysis of liver showed there was no notable difference between FADD-D and control mice on Hematoxylin and Eosin (H&E) stains. Periodic acid-Schiff (PAS) staining revealed a remarkable increased hepatic glycogen accumulation in FADD-D mice (Fig. 4C), which was further confirmed by electron microscopy (Fig. 4D). In addition, the biochemical assay demonstrated that there was an approximate sixfold increase in hepatic glycogen content in FADD-D mice compared with littermate controls (Fig. 4E).
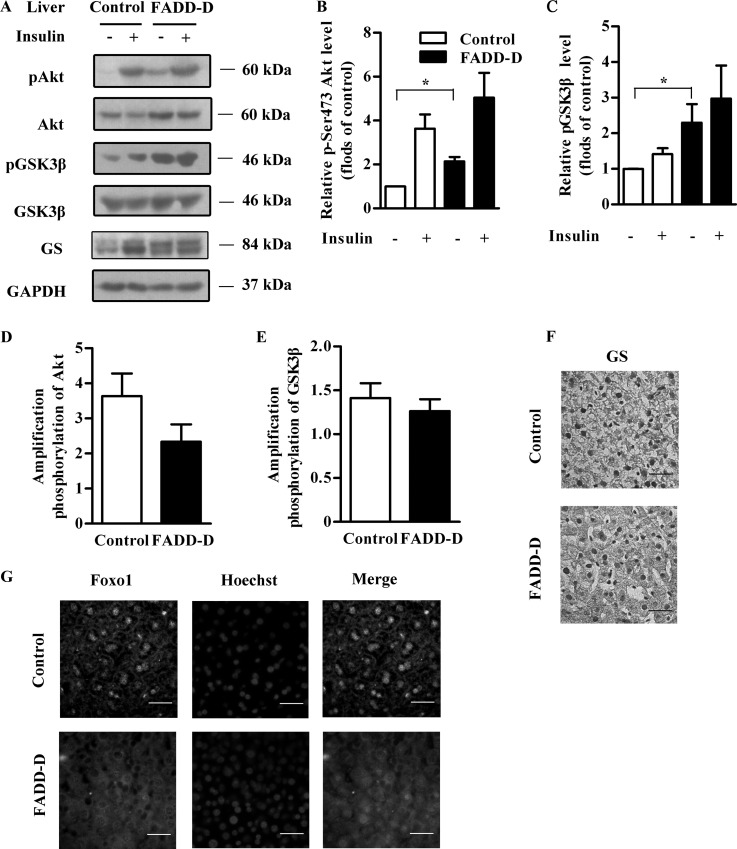
Enhanced Akt Phosphorylation in FADD-D Mice
To investigate the molecular mechanisms underlying the hypoglycemia and hepatic glycogen accumulation in FADD-D mice, we studied the insulin signaling, which plays a critical role in glucose homeostasis (26, 27). Under resting conditions, there was a significant increase in the phosphorylation of Akt in FADD-D mice liver (Figs. 5A and 5B). After insulin stimulation, FADD-D and control mice both had increased Akt phosphorylation. But, consistent with the above IPITT results, the amplification of Akt phosphorylation after insulin stimulation showed no significant difference between FADD-D and control mice (Fig. 5D), indicating unaltered insulin sensitivity in FADD-D mice.
Fig. 5.
Enhanced Akt phosphorylation in the liver of FADD-D mice. A, Western blots of liver tissue lysates from FADD-D and control mice were performed after insulin treatment to assess phospho-Akt, phospho-GSK3β, as well as total Akt, GSK3β, GS and GAPDH expression levels (n = 3 for each group). B and C, Densitometric analysis of phospho-Akt (B) and phospho-GSK3β levels (C) in (A). D and E, Amplification of Akt phosphorylation (D) and GSK3β phosphorylation (E) after insulin stimulation. F, GS immunostaining in liver sections of FADD-D and control mice (n = 3 for each group). Scale bar, 20 μm. G, Immunohistochemistry with anti-Foxo1 antibody (green) and Hoechst (blue) to show the localization of Foxo1 in liver sections of FADD-D and control mice (n = 4 for each group). Scale bar, 20 μm. Data are presented as means ± S.E. Statistical significance was assessed by two-tailed Student's t test, *p < 0.05.
Similar alterations were observed in glycogen synthase kinase (GSK) 3β (Figs. 5A, 5C, and 5E), a key negative regulator in glycogen synthesis downstream of Akt (28, 29). Because of the increased GSK3β phosphorylation under basal conditions, the unphosphorylated GSK3β, which is the active form and can phosphorylate and inactivate GS, was decreased. As a result, the activate form of unphosphorylated glycogen synthase (GS) was highly expressed in FADD-D mice correspondingly, as detected by Western blot and immunohistochemistry analyses of liver sections (Figs. 5A and 5F), leading to enhanced hepatic glycogen synthesis. The increased GS expression in FADD-D liver also confirmed our proteomics data.
In addition, as another downstream target of Akt, forkhead transcription factor (Foxo1) was also phosphorylated and a significant nuclear exclusion of Foxo1 was observed in liver sections of FADD-D mice compared with their littermates (Fig. 5G).
Expression of Genes Involved in Glucose Metabolism in FADD-D Mice
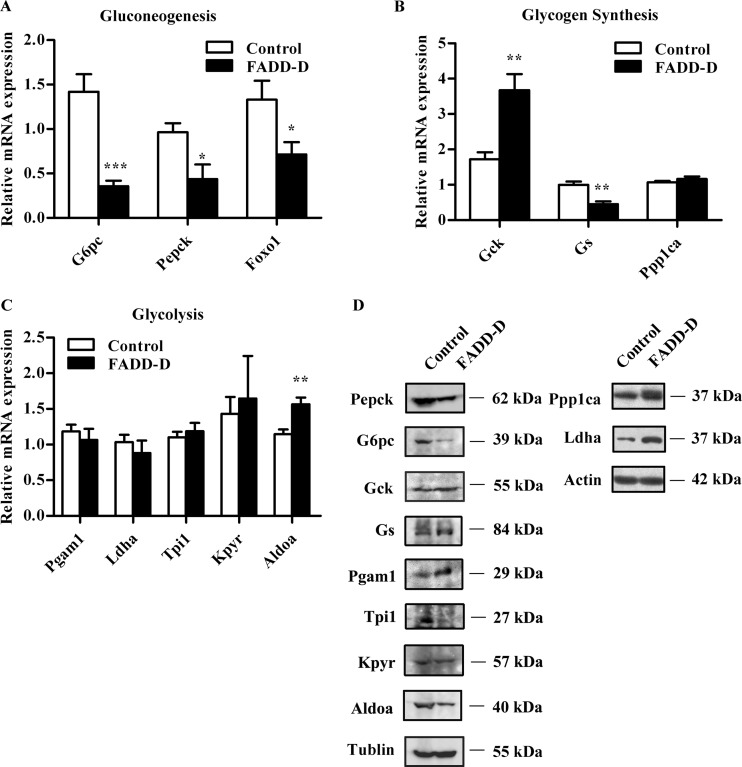
We next preformed qRT-PCR to analyze the expression of genes involved in glucose metabolism. Consistent with changes in insulin signaling and the above proteomics results, the mRNA levels of the two rate-limiting enzymes involved in gluconeogenesis and glycogenolysis glucose-6-phosphatase (G6pc) and phosphoenolpyruvate carboxykinase (Pepck) controlled by Foxo1 (30, 31), were down-regulated in liver tissues from FADD-D mice (Fig. 6A). Furthermore, the decreased hepatic Foxo1 mRNA level in FADD-D mice (Fig. 6A), together with the loss of function of Foxo1 by phosphorylation, led to suppression of G6pc and Pepck expression. Proteomics data indicated that Glucokinase (Gck), which is critical in glycogen synthesis and glycolysis (49), was highly expressed in FADD-D mice liver. The qRT-PCR analysis further confirmed the enhanced expression of Gck (Fig. 6B), accounting for the increased hepatic glycogen and low blood glucose observed in FADD-D mice. We also detected the mRNA expression of other changed enzymes identified in our proteomics analyses (Figs. 6B and 6C). In addition, we performed Western blots to verify the protein expression of these enzymes, which was mostly consistent with our proteomics results (Fig. 6D). In general, immunoblotting, qRT-PCR and physiological analyses confirmed the quantitative results detected by comparative proteomics. Thus, these data demonstrate for the first time that phosphorylation of FADD is associated with glucose homeostasis in vivo.
Fig. 6.
Expression of genes involved in glucose homeostasis in FADD-D mice. A–C, Relative hepatic mRNA expression of genes involved in gluconeogenesis (A), glycogen synthesis (B) and glycolysis (C) (n = 4–6 for each group). D, Western blots of proteins involved in glucose metabolism including Pepck, G6pc, Gck, Gs, Pgam1, Tpi1, Kpyr, Aldoa, Ppp1ca, and Ldha. Data are presented as means ± S.E. Statistical significance was assessed by two-tailed Student's t test, *p < 0.05, **p < 0.01, ***p < 0.005.
DISCUSSION
FADD serves as a critical adaptor in death receptor mediated apoptotic signaling. Further studies have revealed a novel role of “proliferation-apoptosis coupler” of FADD, which is assumed regulated by its phosphorylation (8, 17). Intriguingly, in this study, we uncover an unexpected role of FADD in glucose homeostasis, which may also be regulated by its phosphorylation.
Cellular and mice liver comparative proteomics revealed a special role for FADD in glycolysis and gluconeogenesis. Conventional 2-DE, in combination with advanced mass spectrometric techniques, has facilitated the rapid characterization of thousands of proteins in a single polyacrylamide gel. However, several limitations and problem issues have been recognized: (1) Although silver staining has high sensitivity, it is worth paying special attention that this method has a narrow linear dynamic range, making it less suitable for quantification; (2) Proteins displayed in a single 2D gel represent only a portion of all the proteins that are present in a sample. Thus, in our study, in addition to 2-DE, we further applied HPLC-Chip/Q-TOF technology to perform proteomics screening. The combined use of HPLC-Chip/Q-TOF and 2D-PAGE methodologies afforded a number of advantages over the use of a single proteomic methodology, including the ability to confirm experimental results and accuracy by essentially comparing results from each method. Because each method has its own unique set of advantages, the combined use of both techniques enhanced our confidence in the reported findings. It is important to point out that, though distinct sets of glycolysis and gluconeogenesis proteins were identified with each proteomic platform (HPLC-Chip/Q-TOF and 2D-PAGE), the results observed seemed to point in the same direction. All of the dysregulated proteins identified by proteomics analyses were further validated by qRT-PCR and/or Western blotting assays. For instance, G6pc and Pepck, two enzymes involved in gluconeogenesis, were found to be decreasingly expressed in FADD-D MEFs and livers by proteomics analyses. Later qRT-PCR and Western blots confirmed these results. However, the mRNA levels of a few proteins did not always agree with proteomics results, suggesting that the observed alterations in levels of these proteins were caused by post-transcriptional changes in the stability of mRNA and/or protein. For example, consistent with observations from proteomics, the expression of GS markedly increased at protein levels in FADD-D livers by Western blotting analysis, but the mRNA level was found to be reduced. As we know, although abundant mRNAs usually result in high protein levels (50), the general correlation between mRNA levels and protein abundance is often poor (51), which might be because of the regulatory complexity of mRNA translation and protein stability. Additionally, although the directional changes for all proteins analyzed by Western blotting were consistent with the proteomics data, the intensity differences seen by the immunoblotting analysis did not always agree with the proteomics results. For example, the intensity of pyruvate kinase isozymes R/L (Kpyr) in FADD-D liver was 3.4-fold compared with control liver by proteomics analysis, whereas on the immunoblotting analysis, the intensity of Kpyr in FADD-D liver was only about 1.3-fold of that in control liver. Actually, there might be many technical reasons for differences in the absolute values (e.g. different methods of detection and variations in how software packages define the outlines of bands/spots) as well as biological reasons (protein isoforms differing in charge that are separated by 2-DE but co-migrate in the 1D gels used for immunoblotting). Proteomics analysis is known to suffer from a high rate of false-negative results, thus emphasizing the importance of performing replicate proteomic experiments, and validating the data by orthogonal approaches. In general, qRT-PCR and immunoblotting confirmed most of the quantitative results detected by proteomics screening.
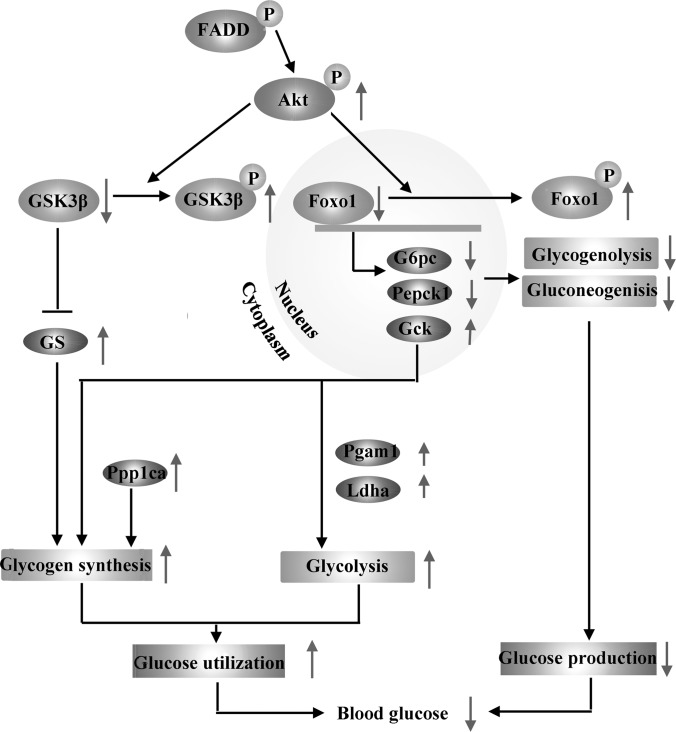
As an important part of metabolism, glucose homeostasis is maintained by the balance of glucose storage as glycogen, glucose utilization by glycolysis and glucose production through gluconeogenesis and glycogenolysis (20). Our analysis of cellular and mice liver comparative proteomics revealed a special role for FADD in glucose metabolism. Glycogen synthase (GS) and glycogen phosphorylase (GP), responsible for glycogen synthesis and breakdown respectively, coordinate to regulate glycogen metabolism (52, 53). The increased expression of protein serine/threonine protein phosphatase PP1-alpha catalytic subunit (Ppp1ca), as indicated in our proteomics and Western bolt assays, will inactivate GP and inhibit glycogen breakdown (52). On the other hand, GS is inactivated by multisite phosphorylation involving several protein kinase, such as GSK3β, which is regulated by Akt (28, 29, 54, 55). In FADD-D mice, the highly phosphorylated Akt increased the active form of GS through elevated GSK3β phosphorylation, leading to enhanced glycogen synthesis as we have detected. Another protein with increased expression identified in the proteomics of MEFs and liver in FADD-D mice was Gck, which catalyzes glucose into glucose-6-phosphate (20), initiating glycogen synthesis. Gck also plays a critical role in glycolysis, together with the changed expression of other enzymes involved in glycolysis (20, 56), such as phosphoglycerate mutase 1 (Pgam1) and l-lactate dehydrogenase A chain (Ldha), enhancing the use of glucose in FADD-D mice. On the contrary, as a result of Akt phosphorylation-induced inactivation of Foxo1, the protein and mRNA expressions of G6pc and Pepck, two key enzymes in gluconeogenesis and glycogenolysis controlled by Foxo1, were suppressed in FADD-D mice liver, leading to decreased glucose production (30). Collectively, the changes in the protein and mRNA levels of these enzymes involved in glucose metabolism contributed to the decreased glucose production, as well as the increased glucose storage and use, finally leading to the hypoglycemia observed in FADD-D mice (Fig. 7).
Fig. 7.
Schematic representation of the altered glucose homeostasis in FADD-D mice. Elevated Akt phosphorylation in FADD-D mice inactivated GSK3β and increased active form of GS, together with the highly expressed Gck and Ppp1ca, leading to enhanced glycogen accumulation. On the other hand, Akt phosphorylated and inactivated Foxo1, which regulates the transcription of G6pc and Pepck, suppressing gluconeogenesis and glycogenolysis. The expression levels of proteins involved in glycolysis were also changed. As a result, there was a low blood glucose level in FADD-D mice. The purple ellipses indicate proteins with changed expression we have found in proteomics and verified by Western blots.
Western blots of liver tissues showed that FADD-D may regulate glucose homeostasis through increasing the phosphorylation of Akt, which is a key node in insulin signaling (27). Unfortunately, we failed to detect the interaction between exogenously expressed Akt and FADD in vitro. It is likely there might be other proteins integrating the relationship between Akt and FADD, such as type 2A phosphatase (PP2A). Recently, our laboratory has found that FADD interacts with PP2A to facilitate the dephosphorylation of conventional PKC (cPKC), which can be abolished by FADD deficiency or phosphorylation (57). Akt belongs to the AGC kinase family and has extensive homology to protein kinase C (PKC) within kinase domains (58). In addition, Akt has a similar pattern of phosphorylation in PKC (59) and can also be inactivated by PP2A (60, 61). Thus, we suspect that FADD might regulate Akt activity by PP2A in the same way in PKC. Phosphorylation of Akt can also be affected by mammalian tribbles homolog 3 (TRB3) or protein tyrosine phosphatase 1B (PTP1B) (62, 63). In subsequent studies, we will put emphasis on identifying the possible mediators that might serve as the link between FADD-D and enhanced Akt activation.
In conclusion, by comparative proteomics discovery and physiological validation, our study provides evidence for the first time, that FADD, especially its phosphorylation form, has an impact on glucose homeostasis, another nonapoptotic function compared with previous reports. We suggest that FADD-D might affect the expression of enzymes in glucose metabolism, decreasing the blood glucose level. This effect might have a correlation with increased Akt phosphorylation in FADD-D, providing a potential drug target in the treatment of diabetes. Further studies may focus on clarifying the relationships between FADD and Akt, and investigating mechanisms underlying the effect of FADD on glucose metabolism in detail.
Supplementary Material
Acknowledgments
We thank Dr. Luan Shu, Jiangsu Provincial Academy of Chinese Medicine, for critical reading and comments.
Footnotes
* This work was supported by grants from the Chinese National Nature Sciences Foundation (81121062, 31071196, 31200572, 30425009, 30330530, 30270291), the Ministry of Science and Technology of China (2012AA020304, 2008BAI51B01), the Jiangsu Provincial Nature Science Foundation (BK2011228, BZ2011048, BZ2010074, BZ2012050, BK2011573), Bureau of Science and Technology of Changzhou (CZ20100008, CE20115034, CZ20110028, CJ20115006, CM20122003, CZ20120004).
 This article contains supplemental Figs. S1 to S4, Files S1 to S2 and Table S1.
This article contains supplemental Figs. S1 to S4, Files S1 to S2 and Table S1.
1 The abbreviations used are:
- FADD
- Fas-associated death domain-containing protein
- 2-DE
- two-dimensional electrophoresis
- 2-D
- two-dimensional
- LC/MS/MS
- liquid chromatography linked to tandem mass spectrometry
- MEFs
- mouse embryonic fibroblasts
- IPGTT
- intraperitoneal glucose tolerance test
- IPITT
- intraperitoneal insulin tolerance test
- H&E
- hematoxylin and eosin
- PAS
- Periodic Acid-Schiff
- qRT-PCR
- quantitative real-time PCR
- GSK
- glycogen synthase kinase
- GS
- glycogen synthase
- Foxo1
- forkhead transcription factor
- G6pc
- glucose-6-phosphatase
- Pepck
- phosphoenolpyruvate carboxykinase
- Gck
- glucokinase
- Pgam1
- phosphoglycerate mutase 1
- Ldha
- L-lactate dehydrogenase A chain
- Tpi1
- triosephosphate isomerase 1
- Kpyr
- pyruvate kinase isozymes R/L
- Ppp1ca
- serine/threonine protein phosphatase PP1-alpha catalytic subunit
- Aldoa
- fructose-bisphosphate aldolase A.
REFERENCES
- 1. Boldin M. P., Varfolomeev E. E., Pancer Z., Mett I. L., Camonis J. H., Wallach D. (1995) A novel protein that interacts with the death domain of Fas/APO1 contains a sequence motif related to the death domain. J. Biol. Chem. 270, 7795–7798 [DOI] [PubMed] [Google Scholar]
- 2. Chinnaiyan A. M., Orourke K., Tewari M., Dixit V. M. (1995) FADD, A Novel Death Domain-Containing Protein, Interacts With the Death Domain of Fas and Initiates Apoptosis. Cell. 81, 505–512 [DOI] [PubMed] [Google Scholar]
- 3. Zhang J., Winoto A. (1996) A mouse Fas-associated protein with homology to the human Mort1/FADD protein is essential for Fas-induced apoptosis. Mol. Cell. Biol. 16, 2756–2763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kim P. K., Dutra A. S., Chandrasekharappa S. C., Puck J. M. (1996) Genomic structure and mapping of human FADD, an intracellular mediator of lymphocyte apoptosis. J. Immunol. 157, 5461–5466 [PubMed] [Google Scholar]
- 5. Schneider P., Thome M., Burns K., Bodmer J. L., Hofmann K., Kataoka T., Holler N., Tschopp J. (1997) TRAIL receptors 1 (DR4) and 2 (DR5) signal FADD-dependent apoptosis and activate NF-kappaB. Immunity 7, 831–836 [DOI] [PubMed] [Google Scholar]
- 6. Kuang A. A., Diehl G. E., Zhang J., Winoto A. (2000) FADD is required for DR4- and DR5-mediated apoptosis: lack of trail-induced apoptosis in FADD-deficient mouse embryonic fibroblasts. J. Biol. Chem. 275, 25065–25068 [DOI] [PubMed] [Google Scholar]
- 7. Zhang J., Cado D., Chen A., Kabra N. H., Winoto A. (1998) Fas-mediated apoptosis and activation-induced T-cell proliferation are defective in mice lacking FADD/Mort1. Nature 392, 296–300 [DOI] [PubMed] [Google Scholar]
- 8. Hua Z. C., Sohn S. J., Kang C., Cado D., Winoto A. (2003) A function of Fas-associated death domain protein in cell cycle progression localized to a single amino acid at its C-terminal region. Immunity 18, 513–521 [DOI] [PubMed] [Google Scholar]
- 9. Matsuyoshi S., Shimada K., Nakamura M., Ishida E., Konishi N. (2006) FADD phosphorylation is critical for cell cycle regulation in breast cancer cells. Br. J. Cancer 94, 532–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Osborn S. L., Sohn S. J., Winoto A. (2007) Constitutive phosphorylation mutation in Fas-associated death domain (FADD) results in early cell cycle defects. J. Biol. Chem. 282, 22786–22792 [DOI] [PubMed] [Google Scholar]
- 11. Yeh W. C., Pompa J. L., McCurrach M. E., Shu H. B., Elia A. J., Shahinian A., Ng M., Wakeham A., Khoo W., Mitchell K., El-Deiry W. S., Lowe S. W., Goeddel D. V., Mak T. W. (1998) FADD: essential for embryo development and signaling from some, but not all, inducers of apoptosis. Science 279, 1954–1958 [DOI] [PubMed] [Google Scholar]
- 12. Zhang H., Zhou X., McQuade T., Li J., Chan F. K. M., Zhang J. (2011) Functional complementation between FADD and RIP1 in embryos and lymphocytes. Nature 471, 373–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shimada K., Matsuyoshi S., Nakamura M., Ishida E., Konishi N. (2005) Phosphorylation status of Fas-associated death domain-containing protein (FADD) is associated with prostate cancer progression. J. Pathol. 206, 423–432 [DOI] [PubMed] [Google Scholar]
- 14. Welz P. S., Wullaert A., Vlantis K., Kondylis V., Fernández-Majada V., Ermolaeva M., Kirsch P., Sterner-Kock A., van Loo G., Pasparakis M. (2011) FADD prevents RIP3-mediated epithelial cell necrosis and chronic intestinal inflammation. Nature 477, 330–334 [DOI] [PubMed] [Google Scholar]
- 15. Imtiyaz H. Z., Rosenberg S., Zhang Y., Rahman Z. S., Hou Y. J., Manser T., Zhang J. (2006) The Fas-associated death domain protein is required in apoptosis and TLR-induced proliferative responses in B cells. J. Immunol. 176, 6852–6861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhande R., Dauphinee S. M., Thomas J. A., Yamamoto M., Akira S., Karsan A. (2007) FADD negatively regulates lipopolysaccharide signaling by impairing interleukin-1 receptor-associated kinase 1-MyD88 interaction. Mol. Cell. Biol. 27, 7394–7404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Alappat E. C., Feig C., Boyerinas B., Volkland J., Samuels M., Murmann A. E., Thorburn A., Kidd V. J., Slaughter C. A., Osborn S. L., Winoto A., Tang W. J., Peter M. E. (2005) Phosphorylation of FADD at serine 194 by CKIalpha regulates its nonapoptotic activities. Mol. Cell 19, 321–332 [DOI] [PubMed] [Google Scholar]
- 18. Zhang J., Zhang D., Hua Z. (2004) FADD and its phosphorylation. IUBMB Life. 56, 395–401 [DOI] [PubMed] [Google Scholar]
- 19. Scaffidi C., Volkland J., Blomberg I., Hoffmann I., Krammer P. H., Peter M. E. (2000) Phosphorylation of FADD/ MORT1 at serine 194 and association with a 70-kDa cell cycle-regulated protein kinase. J. Immunol. 164, 1236–1242 [DOI] [PubMed] [Google Scholar]
- 20. Nordlie R. C., Foster J. D., Lange A. J. (1999) Regulation of glucose production by the liver. Annu. Rev. Nutr. 19, 379–406 [DOI] [PubMed] [Google Scholar]
- 21. Kasuga M., Ogawa W., Ohara T. (2003) Tissue glycogen content and glucose intolerance. J. Clin. Invest. 111, 1282–1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hers H. G., Hue L. (1983) Gluconeogenesis and related aspects of glycolysis. Annu. Rev. Biochem. 52, 617–653 [DOI] [PubMed] [Google Scholar]
- 23. Barthel A., Schmoll D. (2003) Novel concepts in insulin regulation of hepatic gluconeogenesis. Am. J. Physiol. Endocrinol. Metab. 285, E685–692 [DOI] [PubMed] [Google Scholar]
- 24. Hutton J. C., O'Brien R. M. (2009) Glucose-6-phosphatase catalytic subunit gene family. J. Biol. Chem. 284, 29241–29245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yabaluri N., Bashyam M. D. (2010) Hormonal regulation of gluconeogenic gene transcription in the liver. J. Biosci. 35, 473–484 [DOI] [PubMed] [Google Scholar]
- 26. Saltiel A. R., Kahn C. R. (2001) Insulin signalling and the regulation of glucose and lipid metabolism. Nature 414, 799–806 [DOI] [PubMed] [Google Scholar]
- 27. Taniguchi C. M., Emanuelli B., Kahn C. R. (2006) Critical nodes in signalling pathways: insights into insulin action. Nat. Rev. Mol. Cell Biol. 7, 85–96 [DOI] [PubMed] [Google Scholar]
- 28. Cohen P., Frame S. (2001) The renaissance of GSK3. Nat. Rev. Mol. Cell Biol. 2, 769–776 [DOI] [PubMed] [Google Scholar]
- 29. Frame S., Cohen P. (2001) GSK3 takes centre stage more than 20 years after its discovery. Biochem. J. 359, 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nakae J., Kitamura T., Silver D. L., Accili D. (2001) The forkhead transcription factor Foxo1 (Fkhr) confers insulin sensitivity onto glucose-6-phosphatase expression. J. Clin. Invest. 108, 1359–1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gross D. N., van den Heuvel A. P., Birnbaum M. J. (2008) The role of FoxO in the regulation of metabolism. Oncogene 27, 2320–2336 [DOI] [PubMed] [Google Scholar]
- 32. Brusic V., Marina O., Wu C. J., Reinherz E. L. (2007) Proteome informatics for cancer research: from molecules to clinic. Proteomics 7, 976–991 [DOI] [PubMed] [Google Scholar]
- 33. Gorg A., Weiss W., Dunn M. J. (2004) Current two-dimensional electrophoresis technology for proteomics. Proteomics 4, 3665–3685 [DOI] [PubMed] [Google Scholar]
- 34. Aebersold R., Mann M. (2003) Mass spectrometry-based proteomics. Nature 422, 198–207 [DOI] [PubMed] [Google Scholar]
- 35. Pandey A., Mann M. (2000) Proteomics to study genes and genomes. Nature 405, 837–846 [DOI] [PubMed] [Google Scholar]
- 36. Liao L., McClatchy D. B., Yates J. R. (2009) Shotgun proteomics in neuroscience. Neuron 63, 12–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lu P., Vogel C., Wang R., Yao X., Marcotte E. M. (2007) Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation. Nat. Biotechnol. 25, 117–124 [DOI] [PubMed] [Google Scholar]
- 38. Old W. M., Meyer-Arendt K., Aveline-Wolf L., Pierce K. G., Mendoza A., Sevinsky J. R., Resing K. A., Ahn N. G. (2005) Comparison of label-free methods for quantifying human proteins by shotgun proteomics. Mol. Cell. Proteomics 4, 1487–1502 [DOI] [PubMed] [Google Scholar]
- 39. Meng F., Wiener M. C., Sachs J. R., Burns C., Verma P., Paweletz C. P., Mazur M. T., Deyanova E. G., Yates N. A., Hendrickson R. C. (2007) Quantitative analysis of complex peptide mixtures using FTMS and differential mass spectrometry. J. Am. Soc. Mass Spectrom. 18, 226–233 [DOI] [PubMed] [Google Scholar]
- 40. Vasudevan K. M., Garraway L. A. (2010) AKT signaling in physiology and disease. Curr. Top. Microbiol. Immunol. 347, 105–133 [DOI] [PubMed] [Google Scholar]
- 41. Li C., Tan Y. X., Zhou H., Ding S. J., Li S. J., Ma D. J., Man X. B., Hong Y., Zhang L., Li L., Xia Q. C., Wu J. R., Wang H. Y., Zeng R. (2005) Proteomic analysis of hepatitis B virus-associated hepatocellular carcinoma: Identification of potential tumor markers. Proteomics 5, 1125–1139 [DOI] [PubMed] [Google Scholar]
- 42. Yu L. R., Zeng R., Shao X. X., Wang N., Xu Y. H., Xia Q. C. (2000) Identification of differentially expressed proteins between human hepatoma and normal liver cell lines by two-dimensional electrophoresis and liquid chromatography-ion trap mass spectrometry. Electrophoresis 21, 3058–3068 [DOI] [PubMed] [Google Scholar]
- 43. Yang W., Liu P., Liu Y., Wang Q., Tong Y., Ji J. (2006) Proteomic analysis of rat pheochromocytoma PC12 cells. Proteomics 6, 2982–2990 [DOI] [PubMed] [Google Scholar]
- 44. Gobom J., Schuerenberg M., Mueller M., Theiss D., Lehrach H., Nordhoff E. (2001) Alpha-cyano- 4-hydroxycinnamic acid affinity sample preparation. A protocol for MALDI-MS peptide analysis in proteomics. Anal. Chem. 73, 434–438 [DOI] [PubMed] [Google Scholar]
- 45. Bogenhagen D., Clayton D. A. (1974) The number of mitochondrial deoxyribonucleic acid genomes in mouse L and human HeLa cells. Quantitative isolation of mitochondrial deoxyribonucleic acid. J. Biol. Chem. 249, 7991–7995 [PubMed] [Google Scholar]
- 46. Liu Q., Peng Y. B., Qi L. W., Cheng X. L., Xu X. J., Liu L. L., Liu E. H., Li P. (2012) The Cytotoxicity Mechanism of 6-Shogaol-Treated HeLa Human Cervical Cancer Cells Revealed by Label-Free Shotgun Proteomics and Bioinformatics Analysis. Evid. Based Complement. Alternat. Med. 2012, 278652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Okamoto H., Latres E., Liu R., Thabet K., Murphy A., Valenzeula D., Yancopoulos G. D., Stitt T. N., Glass D. J., Sleeman M. W. (2007) Genetic deletion of Trb3, the mammalian Drosophila tribbles homolog, displays normal hepatic insulin signaling and glucose homeostasis. Diabetes 56, 1350–1356 [DOI] [PubMed] [Google Scholar]
- 48. Cabrera O., Jacques-Silva M. C., Speier S., Yang S. N., Köhler M., Fachado A., Vieira E., Zierath J. R., Kibbey R., Berman D. M., Kenyon N. S., Ricordi C., Caicedo A., Berggren P. O. (2008) Glutamate is a positive autocrine signal for glucagon release. Cell Metab. 7, 545–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhang W., Patil S., Chauhan B., Guo S., Powell D. R., Le J., Klotsas A., Matika R., Xiao X., Franks R., Heidenreich K. A., Sajan M. P., Farese R. V., Stolz D. B., Tso P., Koo S. H., Montminy M., Unterman T. G. (2006) FoxO1 regulates multiple metabolic pathways in the liver: effects on gluconeogenic, glycolytic, and lipogenic gene expression. J. Biol. Chem. 281, 10105–10117 [DOI] [PubMed] [Google Scholar]
- 50. Lundberg E., Fagerberg L., Klevebring D., Matic I., Geiger T., Cox J., Algenäs C., Lundeberg J., Mann M., Uhlen M. (2010) Defining the transcriptome and proteome in three functionally different human cell lines. Mol. Syst. Biol. 6, 450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gygi S. P., Rochon Y., Franza B. R., Aebersold R. (1999) Correlation between protein and mRNA abundance in yeast. Mol. Cell. Biol. 19, 1720–1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Agius L. (2008) Glucokinase and molecular aspects of liver glycogen metabolism. Biochem. J. 414, 1–18 [DOI] [PubMed] [Google Scholar]
- 53. Suzuki Y., Lanner C., Kim J. H., Vilardo P. G., Zhang H., Yang J., Cooper L. D., Steele M., Kennedy A., Bock C. B., Scrimgeour A., Lawrence J. C., Jr., DePaoli-Roach A. A. (2001) Insulin control of glycogen metabolism in knockout mice lacking the muscle-specific protein phosphatase PP1G/RGL. Mol. Cell. Biol. 21, 2683–2694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Pugazhenthi S., Khandelwal R. L. (1995) Regulation of glycogen synthase activation in isolated hepatocytes. Mol. Cell. Biochem. 149–150, 95–101 [DOI] [PubMed] [Google Scholar]
- 55. Roach R. J., Larner J. (1977) Covalent phosphorylation in the regulation glycogen synthase activity. Mol. Cell. Biochem. 15, 179–200 [DOI] [PubMed] [Google Scholar]
- 56. Kurland I. J., Pilkis S. J. (1989) Indirect versus direct routes of hepatic glycogen synthesis. FASEB J. 3, 2277–2281 [DOI] [PubMed] [Google Scholar]
- 57. Cheng W., Wang L., Zhang R., Du P., Yang B., Zhuang H., Tang B., Yao C., Yu M., Wang Y., Zhang J., Yin W., Li J., Zheng W., Lu M., Hua Z. (2012) Regulation of protein kinase C inactivation by Fas-associated protein with death domain. J. Biol. Chem. 287, 26126–26135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Manning B. D., Cantley L. C. (2007) AKT/PKB signaling: navigating downstream. Cell 129, 1261–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Alessi D. R., Cohen P. (1998) Mechanism of activation and function of protein kinase B. Curr. Opin. Genet. Dev. 8, 55–62 [DOI] [PubMed] [Google Scholar]
- 60. Meier R., Thelen M., Hemmings B. A. (1998) Inactivation and dephosphorylation of protein kinase Balpha (PKBalpha) promoted by hyperosmotic stress. EMBO J. 17, 7294–7303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Brazil D. P., Yang Z. Z., Hemmings B. A. (2004) Advances in protein kinase B signalling: AKTion on multiple fronts. Trends Biochem. Sci. 29, 233–242 [DOI] [PubMed] [Google Scholar]
- 62. Du K., Herzig S., Kulkarni R. N., Montminy M. (2003) TRB3: a tribbles homolog that inhibits Akt/PKB activation by insulin in liver. Science 300, 1574–1577 [DOI] [PubMed] [Google Scholar]
- 63. Zinker B. A., Rondinone C. M., Trevillyan J. M., Gum R. J., Clampit J. E., Waring J. F., Xie N., Wilcox D., Jacobson P., Frost L., Kroeger P. E., Reilly R. M., Koterski S., Opgenorth T. J., Ulrich R. G., Crosby S., Butler M., Murray S. F., McKay R. A., Bhanot S., Monia B. P., Jirousek M. R. (2002) PTP1B antisense oligonucleotide lowers PTP1B protein, normalizes blood glucose, and improves insulin sensitivity in diabetic mice. Proc. Natl. Acad. Sci. U.S.A. 99, 11357–11362 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.