Abstract
Thermus thermophilus HB27 is an extremely thermophilic eubacteria with a high frequency of natural competence. This organism is therefore often used as a thermophilic model to investigate the molecular basis of type IV pili–mediated functions, such as the uptake of free DNA, adhesion, twitching motility, and biofilm formation, in hot environments. In this study, the phosphoproteome of T. thermophilus HB27 was analyzed via a shotgun approach and high-accuracy mass spectrometry. Ninety-three unique phosphopeptides, including 67 in vivo phosphorylated sites on 53 phosphoproteins, were identified. The distribution of Ser/Thr/Tyr phosphorylation sites was 57%/36%/7%. The phosphoproteins were mostly involved in central metabolic pathways and protein/cell envelope biosynthesis. According to this analysis, the ATPase motor PilF, a type IV pili–related component, was first found to be phosphorylated on Thr-368 and Ser-372. Through the point mutation of PilF, mimic phosphorylated mutants T368D and S372E resulted in nonpiliated and nontwitching phenotypes, whereas nonphosphorylated mutants T368V and S372A displayed piliation and twitching motility. In addition, mimic phosphorylated mutants showed elevated biofilm-forming abilities with a higher initial attachment rate, caused by increasing exopolysaccharide production. In summary, the phosphorylation of PilF might regulate the pili and biofilm formation associated with exopolysaccharide production.
Thermus thermophilus HB27 is a Gram-negative, rod-shaped, and extremely thermophilic eubacterium isolated from a geothermal area (1). This organism grows at temperatures up to 85 °C and has an optimal growth temperature of 70 °C. The thermostable enzymes obtained from members of the genus Thermus are of considerable interest because of their potential in research, biotechnological, and industrial applications (2, 3). In addition, T. thermophilus HB27 is a suitable laboratory model for genetic manipulation, as it is easily cultured under laboratory conditions and has a natural transformation system that is much more efficient than those of other Thermus spp. (4). Intriguingly, thermophiles are also found in biofilms, enclosed within a matrix consisting of extracellular polymeric substances, in various natural and artificial thermal environments (5, 6). Bacteria form biofilms in order to adapt and survive in harsh environments (7, 8). Over the past few decades, biofilm formation has been a major focus of microbial research and, as such, has been studied in relationship to bacterial pathogenesis, immunology, biofouling, microbial technology, and industrial applications (7, 9–12).
Members of the genus Thermus, like many other thermophiles, have evolved two main mechanisms for thermoadaption. One is biofilm formation, which confers protection against environmental stresses such as high temperature and the presence of antibiotics (8). In previous studies, a novel exopolysaccharide, TA-1, was isolated from a T. aquaticus YT-1 biofilm, and both its primary structure and its immunological activity were determined (13). In addition, we showed that the overexpression of uridine diphosphate (UDP)-galactose-4′-epimerase (GalE), which catalyzes the reversible interconversion of UDP-galactose and UDP-glucose, in T. thermophilus HB27 increases biofilm production because of the enzyme's involvement in an important step of exopolysaccharide (EPS)1 biosynthesis (14). The other mechanism that enables Thermus to thrive in extreme habitats is natural transformation (i.e. the ability to take up free DNA). In hot environments, natural transformation allows the horizontal exchange of genetic information between extremophiles, including of genes that promote thermoadaptation (15–17). Recent studies showed that the type IV pili (T4P) on the cell surface of T. thermophilus HB27 not only are required for natural transformation (18, 19), but also mediate adhesion and twitching motility (20). Also, together with the degree of EPS production, the presence of T4P on the bacterial cell surface contributes to the regulation of biofilm formation (21). However, despite extensive research on the physiological, biochemical, and genetic traits of thermophiles, the mechanisms underlying these functions and their role in thermal adaptation have not been fully elucidated (16, 22–24).
Advances in the field of phosphoproteomics have come from high-resolution mass spectrometry and prokaryotic genome sequencing, which have confirmed the phosphorylation of many bacterial proteins on serine/threonine and tyrosine residues (25, 26). In surveys of phosphorylation-related functions, bacterial serine, threonine, and tyrosine phosphoproteins have been shown to regulate many physiological and adaptation processes, such as central carbon catabolism, the heat shock response, osmolarity, starvation, EPS synthesis, virulence, and sporulation (25–27). These observations have been followed by more detailed, species-specific phosphoproteomics investigations, including in Bacillus subtilis (28), Escherichia coli (29), Lactococcus lactis (30), Halobacterium salinarum (31), Klebsiella pneumonia (32), Pseudomonas spp. (33), Rhodopseudomonas palustris (34), and T. thermophilus HB8 (35). In this study, the role played by the global phosphorylation network of the thermophile T. thermophilus HB27 in the physiological processes that mediate the stress responses and thermotolerance of this bacterium was examined. Specifically, we used strong cation exchange (SCX) chromatography and titanium dioxide (TiO2) (28–30) enrichment to characterize the phosphoproteomic map of T. thermophilus HB27. Genetic manipulation of this strain indicated that phosphorylation of the PilF protein, which contains an ATP-binding motif (TTC1622/pilF) and drives T4P formation, is involved in both EPS production and piliation, thereby influencing the biofilm formation during thermophilic adaptation.
MATERIALS AND METHODS
Bacterial Strains and Growth Conditions
Wild-type PilF and its mutant versions were expressed in the host strain T. thermophilus HB27, grown under aerobic conditions at 70 °C in Thermus modified (TM) medium (4). E. coli DH5α and BL21 (DE3) competent cells (Novagen, Madison, WI) were used as hosts for genetic manipulations of plasmids and for the overexpression of proteins, respectively. E. coli strains were grown in Luria-Bertani (LB) medium (36) at 37 °C. When needed, kanamycin (30 μg/ml) and/or ampicillin (100 μg/ml) was added to TM or LB plates for plasmid selection.
Protein Extraction, Digestion, and Phosphopeptide Enrichment
T. thermophilus HB27 was grown aerobically to mid-exponential phase (optical density at 600 nm [OD 600] = 0.8). The cells were harvested, and the resulting pellets were washed twice with PBS and then resuspended in fresh lysis buffer containing 50 mm Tris-HCl (pH 7.5), PhosSTOP phosphatase inhibitor mixture tablets (Roche), 6 m urea, and 2 m thiourea. Cell extracts, obtained using a French press, were subsequently centrifuged at 25,000g for 30 min at 4 °C. The protein concentration was measured using the Bradford protein assay (Bio-Rad). Samples containing 25 mg of protein were disulfide-reduced with 1 mm dithiothreitol for 1 h at 37 °C, alkylated with 5 mm iodoacetamide for 1 h in the dark, and trypsin digested (1:50 w/w) at 37 °C overnight. The tryptic peptides were acidified with 5% trifluoroacetic acid (TFA) to pH 2.7. SCX and TiO2 chromatography were used to fractionate and enrich the phosphopeptides (37, 38). Briefly, the tryptic peptides were loaded onto a 1-ml Resource S column (GE Healthcare) in solvent A (5 mm KH2PO4, 30% acetonitrile (ACN), adjusted to pH 2.7 with TFA) at a flow rate of 1 ml/min and the flow-through was collected. The bound peptides were eluted in a gradient of 0%–30% solvent B (5 mm KH2PO4, 30% ACN, 350 mm KCl, adjusted to pH 2.7 with TFA) over 30 min. For phosphopeptide enrichment, TiO2 beads (GL Sciences, Tokyo, Japan) were pre-incubated with the loading buffer (1 m glycolic acid in 80% ACN and 5% TFA). Each SCX fraction was then mixed and incubated with 2 mg TiO2 beads for 30 min at room temperature. The beads were washed with loading buffer and wash buffer (80% ACN and 5% TFA), and the bound phosphopeptides were eluted with 1% NH4OH in 40% ACN, pH > 10.5. The eluates were desalted and dried for LC-MS/MS analysis.
Liquid Chromatography–Mass Spectrometric Analysis
Nano-HPLC-MS/MS analysis was performed on a nanoAcquity system (Waters, Milford, MA) connected to an LTQ-Orbitrap XL hybrid mass spectrometer (Thermo Electron, Bremen, Germany) equipped with a PicoView nanospray interface (New Objective, Woburn, MA). Peptide mixtures were diluted with 1% formic acid, loaded onto a C18 BEH column (75-μm inner diameter, 25-cm length; Waters, Milford, MA), and separated using a linear gradient of 5%–40% ACN in 0.1% formic acid run over 90 min at a flow rate of 300 nl/min at a column temperature of 35 °C. The mass spectrometer was operated in data-dependent mode. A survey scan of the MS spectra (m/z 300–1800) was acquired in the Orbitrap, with a resolution of 60,000 at m/z 400 after the accumulation of 106 ions. The most intense ions (up to 10) were further sequenced via collision-induced dissociation fragmentation (normalized collision energy 35%) in the LTQ after the accumulation of 7000 ions. An activation q = 0.25 for 30 ms and a multistage activation at 97.98, 48.99, 32.66, and 24.49 Thomson (Th) were applied in the MS2 acquisitions. The dynamic exclusion list was restricted to a maximum of 500 entries with a maximum retention period of 90 s. The Orbitrap measurements were performed with the lock mass option enabled to improve mass accuracy.
Data Processing and Validation
The raw data were processed using MaxQuant software, version 1.1.1.25 (39), to generate the peak lists. High-resolution profile MS/MS data were deconvoluted before the 10 most abundant peaks per 100 Th were extracted. Using the Andromeda search engine (40), we analyzed the derived peak lists against a composite target-decoy protein database constructed from the NCBI T. thermophilus HB27 protein database of a total of 2210 sequences (accession numbers: AE017221 and AE017222) downloaded from the NCBI Reference Sequence database (www.ncbi.nlm.nih.gov/protein) on March 30, 2009 (15). The database also included commonly observed contaminants of 101 protein sequences (version 1.0 of the common repository of adventitious proteins downloaded from the Global Proteome Machine) and 5880 Saccharomyces cerevisiae protein sequences downloaded from NCBI Reference Sequence database on March 30, 2009, that also serve as possible contaminants during cell culture. A search parameter of cysteine carbamidomethylation was set as the fixed modification; methionine oxidation, protein N-terminal acetylation, and phosphorylation on serine, threonine, and tyrosine residues were set as variable modifications. The digestive enzyme was trypsin, and the maximum number of missed cleavages was two. The minimum peptide length was six amino acids. The Andromeda search results were further processed by MaxQuant. In order to achieve reliable identifications, the maximum false discovery rate of peptides and proteins was set at 0.01, which allowed the posterior error probability of peptide-spectrum matches to be less than 0.05.
Bioinformatics Analysis
The biological function and cellular component of each identified phosphoprotein were grouped using the Blast2GO tool (41). The Blast2GO suite is primarily based on the Gene Ontology annotation to perform functional annotation. Using Fisher's exact test, we obtained the Gene Ontology categories that were statistically overrepresented. All Gene Ontology terms of overrepresented proteins were selected based on p < 0.05. In addition, we used the prediction tool pSORTb (version 3.0) (42) to classify bacterial protein localization. BLASTP, a phylogenomic database (ProtClustDB) (43), protein family and domain databases (UniPort, InterPro, Pfam, and PROSITE), and the pathway database (KEGG) were used in parallel as complementary methods to further classify each identified protein. Information on Enzyme Code, the KEGG pathway, and motifs was also extracted and compiled into a dataset provided in supplemental Table S1.
Construction of PilF Expression Plasmids and Site-directed Mutagenesis
Chromosomal DNA from T. thermophilus HB27 was isolated according to a standard phenol extraction protocol (44). Plasmid DNA was purified using the Bioman plasmid purification kit (Bioman, Taipei, Taiwan). Phusion High-Fidelity DNA Polymerase (Finnzymes, Espoo, Finland) was used for all PCR amplifications. Based on its DNA sequence, the pilF (GeneID: 2774631) gene was PCR-amplified using the primers listed in supplemental Table S2. NdeI and HindIII restriction enzyme sites (MBI Fermentas, Vilnius, Lithuania) were added to the N and C termini of pilF, respectively. The PCR product of pilF was inserted into a linearized pGEM7Z vector (Promega, Madison, WI) for nucleotide sequence determination (Genomics BioSci & Tech, Taipei, Taiwan). Subsequently, pilF was subcloned into the pET28a expression vector (Novagen) at the corresponding NdeI and HindIII restriction sites to generate plasmid pET28a-pilF.
Mutations at the identified phosphorylation sites of PilF were generated using the mutagenic primers in overlap extension PCR (45), in which two rounds of PCR were performed to generate mutations at the desired positions of a gene. pET28a-pilF served as the template in the first round, with two flanking primers and two internal primers (each containing the mutated nucleotides) used to generate the N- and C-terminal PCR products in separate reactions. The primers were designed to create an overlap region in the two resulting PCR fragments. In the second round, the purified PCR products from the first round served as templates. The two flanking primers were used to amplify the full-length mutant PilF gene. Finally, the mutant pilF constructs were ligated with pET28a as described above to create mutant versions of pET28a-pilF.
To aerobically overexpress wild-type and mutant PilFs in T. thermophilus HB27, the inducible nitrate reductase promoter (Pnar) region in plasmid pMKE2 (46) (Biotools B&M, Madrid, Spain) was replaced with the endogenous pilF promoter (PpilF). A 157-bp PpilF promoter containing the flanking region of the XbaI–NcoI sites was PCR-amplified, and the resulting DNA fragment was cloned into the corresponding sites of plasmid pMKE2 to replace the Pnar promoter, yielding pMKE2-PpilF. The expression vector pMEK2, allowing overexpression of His-tagged enzymes in T. thermophilus, was developed based on pET28b(+). To generate pMKE2-Pro-pilF, the wild-type and mutant pilF genes, derived from pET28a-pilF and its mutant versions, were directly subcloned into pMKE2-PpilF containing the same multiple cloning sites (NdeI and HindIII).
Overexpression of Recombinant PilFs in T. thermophilus HB27
Successful recombinant pMKE2-Pro-pilF and its mutant derivatives were transformed into T. thermophilus HB27 using a modified version of the protocol described by Koyama et al. (4). Briefly, T. thermophilus HB27 was grown overnight at 70 °C in 10 ml of TM medium. Overnight cultures were diluted 1:10 in fresh medium and grown for an additional 6 h at 70 °C. One-milliliter aliquots of these cells were transformed with the required amount of plasmids. The transformants were selected 2 h later on 2% agar plates containing 30 μg kanamycin/ml and then incubated for 2 to 4 days at 70 °C. To verify the overproduction of PilF and its mutant forms in T. thermophilus HB27, each transformant colony was inoculated in 5 ml of TM medium containing 30 μg kanamycin/ml and then grown overnight with shaking at 200 rpm for plasmid mini-preparations and anti-His-tag antibody detection.
Expression and Purification of Recombinant PilFs in E. coli
Recombinant wild-type and mutated forms of PilF were obtained using the respective plasmids to transform E. coli BL21 (DE3). E. coli cells containing the distinct plasmids were grown overnight at 37 °C in 100 ml of LB medium containing 30 μg kanamycin/ml. Overnight cultures were transferred into 1000 ml of fresh medium and inoculated until A600 = 0.6 was reached. The cultures were induced with isopropyl-β-d-thiogalactopyranoside (final concentration of 0.5 mm) for 4 h and then harvested via centrifugation at 6000g for 15 min at 4 °C, resuspended in cold lysis buffer (50 mm Tris-HCl pH 7.5, 0.25 m NaCl, 5 mm imidazole), and disrupted by sonication on ice. The E. coli proteins were precipitated by heating the cell lysates at 65 °C for 20 min and then separating the soluble proteins from the debris in a 10-min centrifugation step at 8000g. The clear supernatant was filtered through 0.45-μm pore size filters (Millipore, Bedford, MA) and incubated with resins containing nickel ions (Ni2+). Recombinant PilF and its site-specific mutants carrying the hexahistidine tag were purified via Ni-affinity chromatography (Novagen) and eluted in 250 mm imidazole. Fractions containing the desired protein were identified by means of SDS-PAGE on a 12% gel and concentrated via ultrafiltration through a pore membrane with a 30-kDa cutoff (Amicon Ultra-15, Millipore).
Transmission Electron Microscopy of T4P
Thermus cells were grown on TM agar plates for 2 days and then directly resuspended on a copper grid (300 mesh) containing a drop of PBS buffer. After three washes in drops of PBS buffer, the grids were stained with 2% uranyl acetate for 3 min (the excess stain was drawn off with filter paper) and then dried in vacuo overnight. The cells were then observed using a 120-kV transmission electron microscope (JEOL JEM 1400) equipped with a Gatan UltraScan 4000 CCD camera. Images were taken at a magnification of 25,000× with a defocus of ∼1.5 μm.
Biofilm Formation Assays
Static biofilms of Thermus spp. that developed on microtiter plates or glass slides were characterized as described previously (14). The inoculated broth cultures were grown to stationary phase in 5 ml of TM medium at 70 °C and then used to prepare serial dilutions in fresh medium down to A600 = 0.006. Aliquots of the diluted cells were dispensed into each of the wells of a 96-well plate. A set of eight wells filled only with 300 μl TM medium served as the control in each plate. The plates were incubated at 70 °C for 24 h without shaking, after which 0.1% crystal violet was added to each well for 10 min at room temperature. Unbound crystal violet stain was removed and the wells were washed gently three times with distilled water. The bound crystal violet in each well was solubilized via the addition of 300 μl of 95% ethanol. A microtiter plate reader (MRX Revelation, Dynex Technologies, Denkendorf, Germany) was used to read the optical density at 595 nm. For analyses of biofilm architecture, glass slides were placed into 12-well microtiter plates containing diluted cells at A600 = 0.06. The plates were incubated at 70 °C for 3 h. The slides were then gently taken out, rinsed once in PBS buffer (pH 7.5) to remove nonadherent planktonic cells, air-dried overnight, and processed for scanning electron microscopy (described in the supplementary “Materials and Methods” section).
Quantitative Measurement of EPSs
The sugar content of the biofilms was estimated via the phenol–sulfuric acid method (47). T. thermophilus HB27 strains were grown on TM agar plates at 70 °C for 2 days. The bacterial culture on each agar plate was scraped into 2 ml of PBS buffer, separating the soluble EPS dissolved in the PBS buffer by means of centrifugation at 10,000g for 10 min. The EPS concentration was determined using a phenol–sulfuric acid assay, with dextrans as the standard. The total sugar content was normalized to the bacterial concentration. The EPS preparation was then dialyzed overnight with water for subsequent use in GC-MS analyses.
RESULTS
Phosphoproteome of Exponentially Growing T. thermophilus HB27
To identify phosphorylated proteins in extremely thermophilic eubacteria, a phosphoproteomic analysis of T. thermophilus HB27 was carried out using the gel-free shotgun LC-MS/MS approach (28–31). Extracts of trypsin-digested proteins from exponentially growing T. thermophilus HB27 were fractionated via SCX. The resulting 15 SCX peptide fractions, including the flow-through, were subjected to phosphopeptide enrichment using TiO2 and then analyzed five times on a high-accuracy LTQ-Orbitrap MS. Ninety-three unique phosphopeptides were identified from 53 proteins, with an estimated false-positive rate of less than 1% (Table I). Among the identified phosphopeptides, the locations of 67 nonredundant phosphorylation sites were determined with the highest level of confidence (class I sites: 99% peptide identification confidence (48)): 38 (56.7%) on serine, 24 (35.8%) on threonine, and 5 (7.4%) on tyrosine. The relative Ser/Thr/Tyr phosphorylation ratio was similar to that of other bacterial phosphoproteomes (28, 29, 31, 33, 35). Annotated MS/MS spectra of all identified phosphopeptides and a detailed list of the identified phosphoproteins are provided in supplemental Fig. S1 and supplemental Table S1, respectively.
Table I. List of identified phosphoproteins and phosphopeptides from T. thermophilus HB27.
| Locus name/gene name | Protein description | Phosphopeptide | Phosphosites |
|---|---|---|---|
| TTC1279/metK | S-adenosylmethionine synthetase | TVVVSAQHpSPEVEQEQLREDLIR | pS238 |
| p[TVVVSAQHS]PEVEQEQLR | p[T230, S234, S238] | ||
| TTC0088/aroA | 3-phosphoshikimate 1-carboxyvinyltransferase | LRVPGDKpSVTHR | pS23 |
| FKEPEDVLDCGNAGpTLMR | pT94 | ||
| VPGDKp[SVT]HR | p[S23, T25] | ||
| TTC0089/cmk | Cytidylate kinase | GIVTIDGPpSASGK | pS11 |
| GIVTIDGPp[SASGKSS]VAR | p[S11, S13, S16, S17] | ||
| TTC0117 | Threonine synthase | LTEAFPVALVNpSVNPHRLEGQK | pS155 |
| TTC0159 | Riboflavin kinase/FMN adenylyltransferase | GVANVGpTRPTLGGGER | pT230 |
| TTC0166/ask | Aspartate kinase | YGGpT[S]VGDLER | pT11, p[S12] |
| TTC0169/sucD | Succinyl-CoA synthetase alpha chain | KPVVGFIGGRpSAPK | pS237 |
| TTC0291 | Phosphoglucomutase/phosphomannomutase | AAGGAMLTApSHNPPQYLGVK | pS102 |
| HLKAAGGAMLpTAp[S]HNPPQYLGVK | pT100, p[S102] | ||
| TTC0368 | Hypothetical protein TTC0368 | EEPTPpSRPR | pS219 |
| RLPVEVEAAEAp[SPPASPGPPREEPT]PpSRPR | pS219, p[S204, S208, T217] | ||
| RLPVEVEAAEASPPApSPGPPREEPp[TPS]RPR | pS208, p[T217, S219] | ||
| RLPVEVEAAEASPPASPGPPREEPpTPp[S]RPR | pT217, p[S219] | ||
| TTC0408/oah1 | O-acetyl-l-(homo)serine sulfhydrylase | FETLQLHAGYEPEPp[TTLS]R | p[T17, T18, S20] |
| TTC0553 | Ornithine aminotransferase | Fp[TGLDRVESHAEGPY]VWDTQGKR | p[T24, S31, Y37] |
| TTC0691/glmM | Phosphoglucosamine mutase/phosphoacetylglucosamine | ATAGAVIpSAp[S]HNPYQDNGIK | pS99, p[S101] |
| Mutase/phosphomannomutase | ATAGAVISApSHNPYQDNGIK | pS101 | |
| Ap[TAGAVISASHNPY]QDNGIKFFGPTGEKLPDEAEEEIER | p[T93, S99, S101, Y105] | ||
| Ap[TAGAVISASHNPY]QDNGIK | p[T93, S99, S101, Y105] | ||
| TTC0731/thyX | FAD-dependent thymidylate synthase | TASVNEIpSGR | pS92 |
| TTC0777 | Two-component response regulator | IREEAGPPVLAVpSAp[SVS]R | pS78, p[S80, S82] |
| EEAGPPVLAVSApSVSR | pS80 | ||
| IREEAGPPVLAVSAp[S]VpSR | pS82, p[S80] | ||
| TTC0800/trpC | Indole-3-glycerol phosphate synthase | GFGGVLVAEpSGYAR | pS215 |
| KQp[S]PpSAGVIR | pS58, p[S56] | ||
| TTC0805/valS | Valyl-tRNA synthetase | pSKGNVIDPLEMVER | pS532 |
| TTC0927 | Putative ribosomal associated protein | RHpSp[Y]QGPPPPEVR | pS105, p[Y106] |
| RHp[SY]QGPPPPEVR | p[S105, Y106] | ||
| TTC0992 | Hypothetical protein TTC0992 | FpTKDLDLWVDAEEAPRVLAAIR | pT36 |
| TTC1012/hicd | Homoisocitrate dehydrogenase | ILSCHATLFGAAp[T]pSPp[T]R | pS72, p[T71, T74] |
| TTC1029 | Hypothetical protein TTC1029 | pTKp[T]LEEFGHLAALLDGSR | pT180, p[T182] |
| KTKpTLEEFGHLAALLDGSR | pT182 | ||
| TTC1063 | Phosphoglucomutase/phosphomannomutase | EAAGLYLTGpSR | pS99 |
| AKEAAGLp[YLTGS]R | p[Y95, T97, S99] | ||
| EAAGLp[YLTGS]RRPFAFQGVK | p[Y95, T97, S99] | ||
| TTC1102/pyrG | CTP synthetase | RGEp[YLSQT]VQVIPHITDEIKER | p[Y117, S119, T121] |
| RGEp[YLSQTVQVIPHIT]DEIKER | p[Y117, S119, T121, T129] | ||
| TTC1136 | Phosphoenolpyruvate synthase | GGRp[TS]HAAIVAR | p[T419, S420] |
| GGRpTp[S]HAAIVAR | pT419, p[S420] | ||
| TTC1149 | Putative adenylosuccinate lyase protein | TEVLEAQEPFHEGQp[TGSSS]MPHKK | p[T268, S270, S271, S272] |
| TEVLEAQEPFHEGQp[TGSSS]MPHKKNPVGLENLTGVAR | p[T268, S270, S271, S272] | ||
| p[TEVLEAQEPFHEGQTGSSS]MPHKK | p[T254, T268, S270, S271, S272] | ||
| TEVLEAQEPFHEGQp[TGSSSMPHKKNPVGLENLT]GVAR | p[T268, S270, S271, S272, T286] | ||
| TTC1330/tuf | Translation elongation factor tu | GVpSREEVER | pS295 |
| TTC1351 | Hypothetical protein TTC1351 | HLEGNpSHAHLK | pS81 |
| TTC1381/pyrF | Orotidine 5′-phosphate decarboxylase | RGDIGpSp[T]AEAYAR | pS92, p[T93] |
| TTC1414 | Fructose-bisphosphate aldolase | INTDpTDLR | pT254 |
| TTC1420 | Metal dependent phosphohydrolase | NKVHpYLK | pY162 |
| TTC1488 | ABC transporter ATP-binding protein | pSELEIR | pS2 |
| TTC1541 | Acetylglutamate kinase | LLLVHGGpSAEp[T]NKVAEALGHPPR | pS38, p[T41] |
| TTC1600 | Inorganic pyrophosphatase | LDHIQDIGDVPEGVKQEIQHFFEp[TY]KALEAK | p[T139, Y140] |
| LDHIQDIGDVPEGVKQEIQHFFEp[T]pYKALEAK | pT139, p[Y140] | ||
| TTC1601 | Hypothetical protein TTC1601 | APVWEEEPPAKpTPp[SVELAPEEPPPAPT]PTPPPPAQGTR | pT127, p[S129, T142] |
| KApTAAPRPEPPKPPAQEAK | pT99 | ||
| TTC1611 | Pyruvate kinase | LNFpSHGAPEDHRR | pS39 |
| AIVVFp[TATGGS]AR | p[T380, T382, S385] | ||
| AIVVFp[T]ApTGGSAR | pT382, p[T380] | ||
| TTC1619 | Phosphoribosylaminoimidazole synthetase | EGDALLALPSSGPHp[TNGYS]LIR | p[T187, Y190, S191] |
| TTC1622/pilF | ATP-binding motif-containing protein PilF | RGGGRLEDTLVQpSGK | pS372 |
| RGGGRLEDpTLVQSGK | pT368 | ||
| TTC1630 | Phosphoglucomutase | ADGVLLTPpSHNPPEDGGFK | pS130 |
| ADGVLLpTPp[S]HNPPEDGGFK | pT128, p[S130] | ||
| TTC1659 | Phosphopentomutase | MREVNPGKDpTp[T]TGHWEFVGIHLEKPFR | pT80, p[T81] |
| EVNPGKDp[TT]pTGHWEFVGIHLEKPFR | pT82, p[T80, T81] | ||
| TTC1709/pckA | Phosphoenolpyruvate carboxylase | EGDVAVFFGLpSGp[T]GK | pS234, p[T236] |
| TTC1714/groEL | Chaperonin GroEL | ESTPApSAGAGDMDF | pS535 |
| TTC1772 | Kinase | EAVAPFGDpSLLVVGAEGYVK | pS249 |
| TTC1776/rplL | 50S ribosomal protein L7/L12 | IKEELSQApTVLELK | pT16 |
| TTC1798/ndk | Nucleoside diphosphate kinase | IpSQELAERHp[Y]AEHR | pS41, p[Y49] |
| ISQELAERHpYAEHR | pY49 | ||
| TTC1827 | Hypothetical protein TTC 1827 | KGELKApTAVPQGPLK | pT106 |
| TTC1852 | Peptide chain release factor 2 | LpYELERKK | pY297 |
| TTC1855 | Hypothetical protein TTC1855 | LVpSKSEVVR | pS36 |
| LVSKpSEVVR | pS38 | ||
| TTC1856 | Serine/threonine protein kinase | VMDFGLAYLLQEpSR | pS159 |
| HLpTRp[TGYT]LGTPTYMAPEQAK | pT163, p[T165, Y167, T168] | ||
| pTGYTLGTPTYMAPEQAK | pT165 | ||
| HLp[TRTGYTLGT]PTYMAPEQAK | p[T163, T165, YI67, T168, T171] | ||
| HLp[TRTGYTLGTPTY]MAPEQAK | p[T163, T165, Y167, T168, T171, T173, Y174] | ||
| HLpTRp[T]GpYTLGTPTYMAPEQAK | p[T163, T165, Y167, T168] | ||
| VMDFGLAp[YLLQES]R | p[Y154, S159] | ||
| TTC1888/apgM | Cofactor-independent phosphoglycerate mutase | LAEESALGLLTPVYPGLAPGpSGPGHLALFGYDPFR | pS70 |
| LTPAAIASYPMpYK | pY256 | ||
| TTC1904 | UDP-N-acetylmuramoylalanyl-d-glutamate-2,6-diaminopimelate | LGEEARPPVGHFpTTPEAPEVYAFLR | pT155, pT156 |
| TTC1957/bmrU | Protein bmrU | VLGVVPIGpSGNDFAR | pS86 |
| TTC1960 | Aspartate aminotransferase | AVRPp[SGAFYVLMDTS]PIAPDEVR | p[S318, Y322, T327, S328] |
| AVRPp[S]GAFpYVLMDTSPIAPDEVR | pY322, p[S318] | ||
| TT P0022 | Histidinol-phosphate aminotransferase | RALAEAHGVSPEQVAVGpTGp[T]SELIHR | pT78, p[T80] |
| TT P0024/phoA | Alkaline phosphatase | YPNGLINTYSLTSYVTEp[SS]AAGNAFSCGVK | p[S95, pS96] |
Sites of phosphorylation with 99% peptide identification confidence assigned by MaxQuant software are indicated in bold: pS, phosphoserine; pT, phosphothreonine; pY, phosphotyrosine. Ambiguous sites are indicated in bold with brackets as p[S], p[T], and p[Y]. Detailed information on the detected phosphopeptides can be found in supplemental Table S1.
Classification of the Identified Phosphoproteins
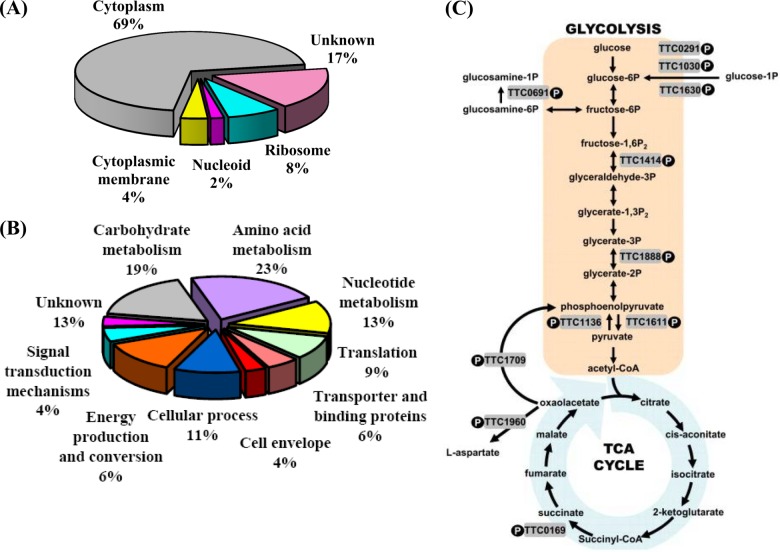
The identified phosphoproteins in T. thermophilus HB27 were categorized according to their cellular localization and biological function (Fig. 1), as determined using the databases noted in “Materials and Methods.” Accordingly, as shown in Fig. 1A, 37 phosphoproteins could be assigned to the cytoplasm, 4 to the ribosome, 2 to the cytoplasmic membrane, and 1 to the nucleoid. Of the 53 phosphoproteins identified in this study, 44 (86.8%) could be functionally annotated, and 7 remained hypothetical proteins and were therefore placed within the “Unknown” category. From the biological process perspective, the 46 phosphoproteins were mostly involved in metabolic pathways, including amino acid (22.6%), carbohydrate (18.9%), and nucleotide (13.2%) metabolism (Fig. 1B). These results are consistent with those of other studies, and the functional distribution was similar to that determined for other bacteria, in which proteins involved in housekeeping pathways and central metabolisms are significantly overrepresented (28–30, 32, 33, 49).
Fig. 1.
Classification of the identified phosphoproteins in T. thermophilus HB27. The phosphoproteins were grouped by (A) cellular location and (B) biological function based on Gene Ontology terms as assigned by Blast2GO. Hypothetical proteins were grouped into the “Unknown” category of the pie chart. C, this simplified diagram shows the central metabolic pathways that link glycolysis and the tricarboxylic acid cycle for glucose. Other carbohydrates can also be metabolized, via glucose-1-phosphate or glucosamine-1-phosphate. The shaded boxes show the phosphorylated enzymes identified in this study.
Key glycolytic, tricarboxylic acid cycle, and gluconeogenesis enzymes that were determined to be phosphorylated are shown in Fig. 1C. These phosphoproteins and phosphosites in the central metabolic pathways of T. thermophilus HB27 were found to be highly conserved with respect to their counterparts in other bacteria (supplementary Table S3) (28–31, 35). Moreover, several phosphoproteins essential to nucleotide metabolism, protein biosynthesis, and cell envelope biosynthesis were also annotated (supplemental Table S4), including the phosphoproteins Ndk (TTC1798), Cmk (TTC0089), and phosphopentomutase (TTC1659), involved in the homeostasis of nucleoside triphosphates and their deoxy derivatives (13, 50–52); EF-Tu (TTC1330), RF2 (TTC1852), and chaperonin GroEL (TTC1714), which facilitate translation and protein synthesis (53–55); and DAG kinase (TTC1957), MurE (TTC1904), and PilF (TTC1622), which are involved in phospholipid, peptidoglycan, and T4P biogenesis, respectively (56–59). This comprehensive analysis provided the first insights into the importance of phosphoproteins in regulating the biological functions of thermophiles that allow their efficient adaptation to extreme and nutrient-limiting environments.
Phosphorylation of Thr368 and Ser372 Residues of the ATP-binding Motif-containing Protein PilF
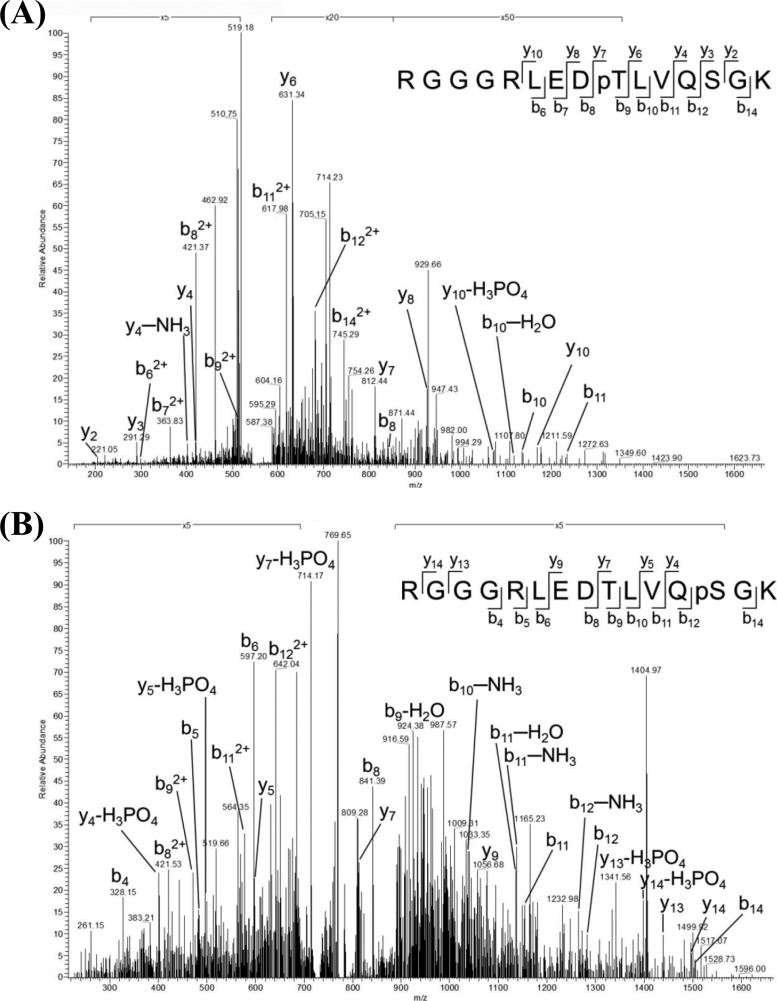
In many species of bacteria, biofilm formation and natural competence have been shown to depend on the retractable protein appendages of T4P, located on the cell surface (19, 24, 60, 61). One of the phosphoproteins identified in this analysis was PilF, which contains an ATP-binding motif (TTC1622/pilF). PilF is essential for correct T4P synthesis and, through its ATPase activity, drives the natural transformation system, as shown in several species of bacteria (59). This protein is phosphorylated at two sites of phosphorylation, Thr-368 and Ser-372, located on the two individual phosphopeptides RGGGRLEDpTLVQSGK and RGGGRLEDTLVQpSGK (Table I). The MS/MS spectra of these peptides, indicating the exact sites of their phosphorylation, are shown in Fig. 2. In a previously described sequence alignment of ATPase families within type II and type IV secretion systems (59), the double phosphorylation sites of PilF were localized to the third conserved GSPII (general secretion pathway) motif at the N terminus of the protein. This domain is crucial for normal protein functioning in type II and IV secretion systems (62, 63).
Fig. 2.
MS/MS spectra of the threonine and serine phosphorylated peptides belonging to ATP-binding-motif-containing protein PilF. Rich backbone fragmentation (RGGGRLEDpTLVQSGK and RGGGRLEDTLVQpSGK) was shown in the MS/MS spectra, in which (A) Thr-368 and (B) Ser-372 were identified as the sites of PilF phosphorylation.
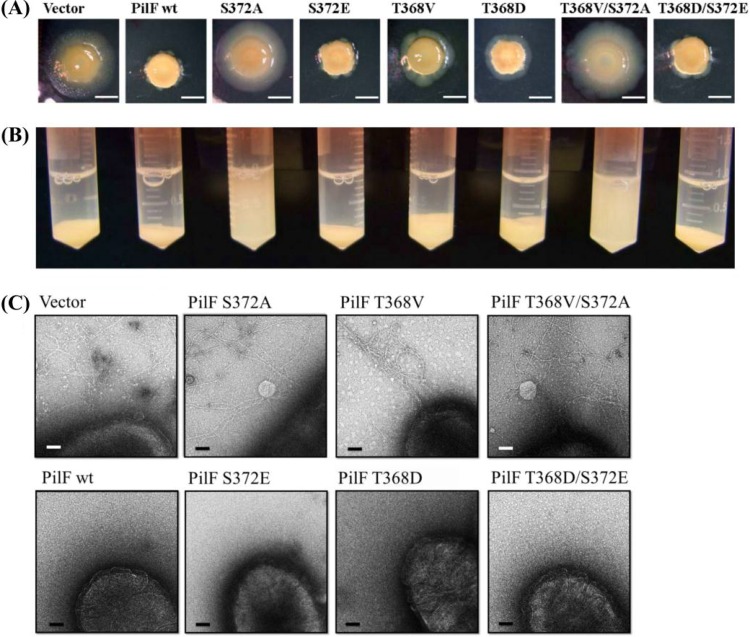
Effects of Site-directed Mutagenesis of PilFs on Colony Morphology and Piliation
In order to elucidate the role of Thr-368 and Ser-372 phosphorylation in PilF, plasmids expressing wild-type or site-specific mutagenized PilFs (T368V, T368D, S372A, S372E, and the double mutants T368V/S372A and T368D/S372E) under the control of the constitutively active pilF promoter were generated and individually transformed into T. thermophilus HB27. A transformant containing the unmodified vector (pMEK2) was used as the control strain. All of the transformants were examined for twitching motility, discernible in T. thermophilus HB27 by the formation of large, flat, spreading colonies on agar surfaces (20). Interestingly, the results showed that the colonies formed by the control strain and by mutants expressing a nonphosphorylatable form of PilF (i.e. T368V, S372A, and T368V/S372A) were large, flat, and spreading, whereas the colonies of mutants with mimic phosphorylated forms of PilF (T368D, S372E, and T368D/S372E) and the strain overexpressing wild-type PilF (PilF wt) were smaller (Fig. 3A). Moreover, both PilF wt and the mutant strains T368D, S372E, and T368D/S372E, carrying phosphorylation mimic PilFs, lost their twitching motility and exhibited pili-formation-deficient phenotypes (Fig. 3C). When the strains were grown in liquid medium, the nonphosphorylated mutant strains (S372A and T368V/S372A) had a lower gravity sedimentation rate, indicating that the mutants form pili and have higher twitching motility (Fig. 3B).
Fig. 3.
Colony morphology and piliation of T. thermophilus HB27 strains. A, the effects of the overexpression of various PilF proteins in T. thermophilus HB27. Strains containing a plasmid expressing the indicated PilF allele were incubated on 1.6% Thermus medium plates for 3 days at 70 °C under humid conditions. Scale bar represents 5 mm. B, precipitation of the overexpressed PilF proteins in T. thermophilus HB27. After a 6-h incubation at 70 °C and an initial A600 of 0.6, the cells were transferred to 2-ml tubes containing PBS buffer and subjected to gravity sedimentation at 4 °C for 1 day. C, images of T. thermophilus HB27 strains overexpressing the various PilF proteins. Cells from 2-day cultures on agar plates of the indicated strains were directly transferred to a grid and stained with 2% uranyl acetate for transmission electron microscopy. Approximately 20 cells of each strain were analyzed for piliation. Bar, 0.1 μm.
To further examine the stoichiometry of phosphorylation in the PilF wt strain, the relative quantities of the dephosphorylated phosphopeptides (RGGGRLEDTLVQSGK and GGGRLEDTLVQSGK) in phosphorylated and phosphatase-treated PilF were measured via mass spectrometry, and the areas of the respective peaks were compared. The results showed 63% phosphorylation site occupancy in cells grown to mid-exponential phase (supplemental Table S5).
PilF ATPase activity powers the mechanical force necessary for T4P pilus extension; conversely, following a reduction in ATPase activity, T4P function is interrupted (64). We found that both the PilF wt strain and the serial mutant PilF strains retained weak ATPase activity, as measured by γ-phosphate release from ATP hydrolysis (supplemental Fig. S2). Also, the secondary and quaternary structures of these strains were similar with respect to their circular dichroism spectra and the results of analytical ultracentrifugation analyses, respectively (supplemental Figs. S3 and S4 and supplemental Table S6). Thus, all of the recombinant proteins formed hexameric complexes, and there was no disruption of their ATPase activity. These results implied that seryl/threonyl phosphorylation of PilF is essential to pili formation and pili-mediated twitching motility but does not determine or alter the conformation, quaternary assembly, and ATPase activity of the protein.
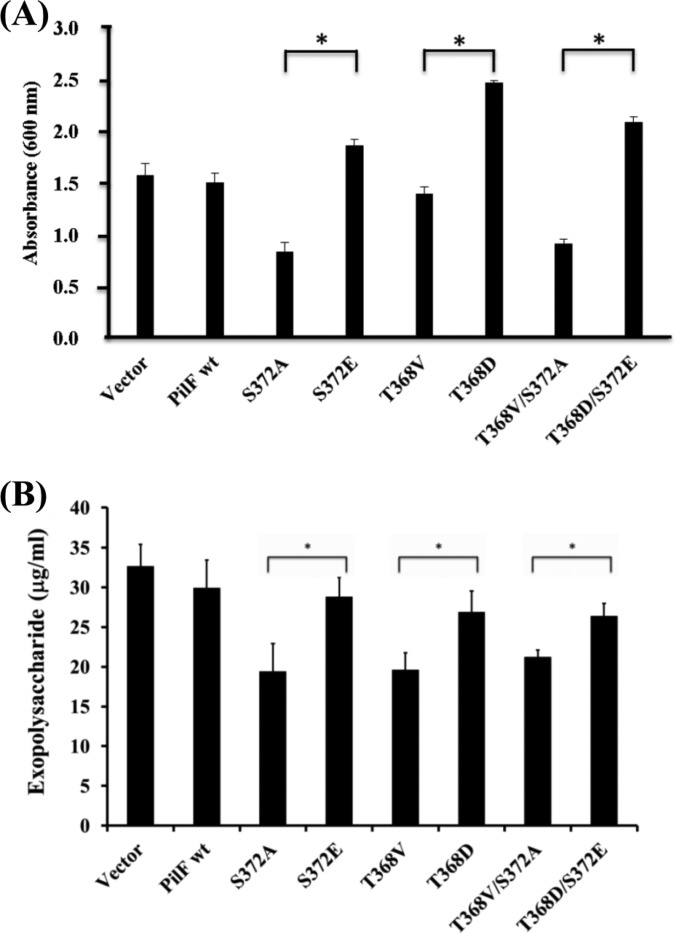
Effects of Site-directed Mutagenesis of PilFs on Biofilm Formation and EPS Production
Apart from twitching motility, T4P is known to involve in the formation of complex colonial structures in biofilms (60). In a previous study, we demonstrated the ability of T. thermophilus HB27 to form biofilms (14). Thus, in the present work we further investigated the effect of PilF phosphorylation on biofilm-formation ability, using crystal violet staining and scanning electron microscopy. The results showed significant reductions (p < 0.05) in the biofilm-formation ability of strains expressing nonphosphorylatable forms of PilF (T368V, S372A, and T368V/S372A) relative to the PilF wt strain or the strains with phosphorylation simulated PilFs (T368D, S372E, and T368D/S372E), in which biofilm formation was normal (Fig. 4A). Additionally, strains T368V, S372A, and T368V/S372A had a lower initial attachment rate during biofilm formation (supplemental Fig. S5), and their cell surfaces had a granular appearance (supplemental Fig. S6), unlike the thick and smooth matrix that normally coats the bacterium and that was also seen in the transformants with normal biofilm production. These differences in biofilm formation and the nature of the cell surface led us to further explore EPS production and the maintenance of biofilm structure (7, 8). In an EPS quantitative assay, the PilF mutant strains T368V, S372A, and T368V/S372A produced less EPS than strains S372E, T368D, and S372E/T368D with the mimic phosphorylated PilF, strain PilF wt, or vector control (Fig. 4B). The relative quantities of EPS measured via GC-MS were similar to the amounts determined using the phenol–sulfuric acid method (supplemental Table S7). In terms of sugar composition, the glycosyl residues of the EPS produced by all of the examined strains consisted of 75% galactose, 15% mannose, and 7% glucose. These results indicated a relationship between PilF phosphorylation and EPS production in regulating biofilm formation in thermophilic bacteria.
Fig. 4.
Biofilm formation and EPS production by T. thermophilus HB27 strains. A, biofilm formation by strains of T. thermophilus HB27 overexpressing various PilF proteins. Biofilms formed on 96-well microtiter plates after 24 h were stained with crystal violet. Surface-attached cells were quantified by solubilizing the dye in ethanol and determining the absorbance at 600 nm. Data are the means of three independent experiments, with each sample tested in triplicate. The asterisks (*) indicate statistical significance (p < 0.05). B, the total EPS content extracted from T. thermophilus HB27 strains expressing wild-type or mutant PilF proteins normalized to each of their bacterial concentrations, as determined in a phenol–sulfuric acid assay. Error bar represents the standard deviations of three independent measurements. Asterisks indicate statistical significance (p < 0.05).
DISCUSSION
To date, global phosphoproteome analyses from archaea (31) to eubacteria (28–30, 32, 33) have identified quite a few phosphoproteins. Despite the presence of the same phosphosites, there is very little overlap between these phosphoproteins and thus between the respective phosphoproteomes (49). In a comparison of phosphoproteomes within the same species (i.e. T. thermophilus strains HB27 (this study) and HB8 (35)), ∼45% of the phosphoproteins were found to be identical (supplemental Table S8), and, with the exception of valyl-tRNA synthetase (TTC0805) and chaperonin GroEL (TTC1714), the phosphorylation sites of these proteins were the same. The remaining 55% of the phosphoproteins of these two closely related thermophiles were distinct, perhaps accounting for strain-specific differences in regulatory mechanisms and biological functions.
Protein Phosphorylation Regulates T4P-mediated Functions in T. thermophilus HB27
In this study, the phosphoprotein PilF (TTC1622) was shown to have two phosphorylation sites, Thr-368 and Ser-372, located on distinct monophosphopeptides (Table I and Fig. 2). In many species of bacteria, PilF is essential to T4P-mediated natural transformation, which in T. thermophilus HB27 is required for DNA uptake from thermal environments and has allowed thermoadaption (17–19). Furthermore, PilF forms hexameric complexes at the bacterial inner membrane and is the only ATPase driving T4P-mediated functions (59). This study is the first to describe PilF phosphorylation and thus to suggest a mechanism for its regulation with respect to T4P.
Phosphorylated PilF Interferes with Pilus Formation and Retards Motility in T. thermophilus HB27
In a previous study, it was found that long pilus structures are still formed but competence is defective in T. thermophilus with a deleted pilF gene (65). We found that overexpression of wild-type PilF resulted in a nonpiliated phenotype (Fig. 3C, PilF wt). This inhibition of pilus formation was likely due to the higher levels of phosphorylated PilF, as the phosphorylation stoichiometry of strain PilF wt was 63% higher than that of the control (vector) strain (supplemental Table S5). Interestingly, natural transformation ability was nonetheless maintained in strain PilF wt, as evidenced by transformation frequencies of 10−4 to 10−5 transformants per viable count. This result is consistent with that of the previous report (20).
In terms of the role of PilF phosphorylation, an interesting point is that the phosphorylation-simulated mutants including single or double acidic residue substituted PilFs (T368D, S372E, and T368D/S372E) all displayed nonpiliated and nontwitching phenotypes (Fig. 3). Conversely, in these nonphosphorylated mutants (T368V, S372A, and T368V/S372A), pili were formed and, at the same time, twitching motilities were retained (Fig. 3). In other words, the nonphosphorylation of PilF might possess a functional pilus apparatus because of its twitching motility. Likewise, in our results, natural competence was independent of piliation and preserved in all of the transformants at frequencies similar to those of the wild-type control (∼10−4 to 10−5 transformants per viable count) (20). Additionally, neither ATPase activity nor hexameric complex formation was lost in any of the mutated strains, including PilF wt (supplemental Figs. S2–S4). Taken together, these findings suggest that PilF functions regulated by serine/threonine phosphorylation are required for T4P formation and T4P-driven motility but not for natural transformation and ATPase activity. Accordingly, the two independent phosphorylation sites, Thr-368 and Ser-372, in PilF may individually have the same regulatory ability in the switch from the assembly to the retraction of T4P.
Phosphorylated PilF Increases EPS Production and Thus Enhances Biofilm Formation
T4P is a multifunctional protein, involved not only in natural competence but also in adherence, movement, biofilm formation, and virulence in a wide variety of bacterial species, including Myxococcus xanthus, P. aeruginosa, Neisseria spp., E. coli, and Vibrio cholerae (60, 66, 67). In T. thermophilus HB27, T4P was shown to be important in natural transformation (17–19), adhesion, and twitching motility (20). In this study, we examined not only the regulation of these T4P-mediated functions by PilF phosphorylation but also the role of T4P-mediated motility in promoting the initiation of biofilm formation. Our results suggested that the retarded motility exhibited by mutants carrying the mimic phosphorylated PilF proteins (T368D, S372E, and T368D/S372E) enhanced biofilm formation (Fig. 4A). This observation is in contrast to reports on P. aeruginosa (68) and M. xanthus (69) in which bacterial movement by twitching motility and social motility, respectively, was shown to facilitate surface attachment, microcolony formation, and therefore biofilm maturation. In our mutants, it is tempting to consider that phosphorylated PilFs enhance biofilm formation by interfering with pili formation, which limits motility and thereby increases initial adhesion (supplemental Fig. S5).
In M. xanthus, the membrane accumulation of pilin subunits (PilA) was shown to alter EPS production and social motility (70), whereas in our T. thermophilus HB27 mutants the expression of mimic phosphorylated or phosphorylatable PilFs influenced the degree of EPS production and thus of biolfim formation (Fig. 4B). According to Cava et al. (71), the composition of polysaccharides detected in T. thermophilus cell wall is 74% reducing sugars. In addition, previous studies indicated that in T. thermophilus HB27, the overexpression of GalE, which participates in the Leloir pathway for galactose metabolism, leads to an increase in biofilm formation (14). In this study, these mutant strains also incorporated a higher proportion (∼75%) of galactose into the EPS on their cell wall (supplemental Table S7). These findings are consistent with the results of our study, in which galactose was the major component extracted from EPS and its relative amount was positively related to effects on the ability to form biofilm. In terms of the relationship between bacterial phosphorylation and EPS biosynthesis, current evidence suggests that either deletion of specific bacterial tyrosine kinases or mutation of the phosphorylation sites on EPS biosynthetic proteins results in the loss of EPS production, especially in pathogens (32, 72, 73). Consequently, in T. thermophilus HB27, phosphorylated PilF might influence sugar incorporation into the cell wall and thereby biofilm production capacity.
T4P-driven motility is typically powered by the two-protein systems ATPase PilB/PilF (pilin polymerase) and PilT/PilU (pilin depolymerase), controlling pilus fiber extension and retraction, respectively (64, 67, 74). This is the case in mesophilic bacteria such as P. aeruginosa (64), Neisseria spp. (75), and M. xanthus (74), whereas in T. thermophilus HB27 there is only one T4P-related ATPase, PilF (17, 59). Although we did not carry out structure-function analyses of T. thermophilus PilF, this protein is unusual because although its primary sequence is closely related to PilB/PilF, it is functionally closer to PilT/PilU (59). How Thermus cells coordinate pilus fiber retraction and extension using only one ATPase and the mechanism by which protein phosphorylation coordinates these functions in thermophilic bacteria are challenging questions to be addressed in future studies.
The phosphoproteins identified in our global phosphoproteome analysis of T. thermophilus HB27 were shown to be involved in a diverse range of cellular processes. One such phosphoprotein, PilF, powers the multiple functions of T4P in this bacterium, including the acquisition of DNA for transformation, in addition to playing an essential role in thermoadaptation. PilF phosphorylation was shown to be related to the sugar composition of the EPSs, which are secreted into the cell wall to assist in cell attachment preceding biofilm formation. Our results provide insights into the mechanisms underlying the well-regulated balance in thermophiles between phosphorylation-regulated T4P and EPS production.
Supplementary Material
Acknowledgments
LTQ-Orbitrap data were acquired at the Academia Sinica Common Mass Spectrometry Facilities located at the Institute of Biological Chemistry. Additional technical assistance was provided by the NRPGM Core Facilities for Proteomics and Glycomics, located at the Institute of Biological Chemistry, Academia Sinica. We thank the Cryo-EM Common Facility of the Scientific Instrument Center, Academia Sinica, for transmission electron microscopy services and Tzu-Chi University (Hualien, Taiwan) for carrying out scanning electron microscopy.
Footnotes
* Portions of this work were supported by a National Science Council grant (NSC 98-3112-B-001-023 and NSC 100-2113-M-001-022-M3).
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- ACN
- acetonitrile
- EPS
- exopolysaccharide
- HB27
- Thermus thermophilus strain HB27
- LB
- Luria-Bertani (medium)
- PilF
- ATP-binding motif-containing protein
- SCX
- strong cation exchange
- T4P
- type IV pili
- TFA
- trifluoroacetic acid
- TiO2
- titanium dioxide
- TM
- Thermus modified.
REFERENCES
- 1. Oshima T. (1974) Comparative studies on biochemical properties of an extreme thermophile, Thermus thermophilus HB 8. Seikagaku 46, 887–907 [PubMed] [Google Scholar]
- 2. Pantazaki A. A., Pritsa A. A., Kyriakidis D. A. (2002) Biotechnologically relevant enzymes from Thermus thermophilus. Appl. Microbiol. Biotechnol. 58, 1–12 [DOI] [PubMed] [Google Scholar]
- 3. Cava F., Hidalgo A., Berenguer J. (2009) Thermus thermophilus as biological model. Extremophiles 13, 213–231 [DOI] [PubMed] [Google Scholar]
- 4. Koyama Y., Hoshino T., Tomizuka N., Furukawa K. (1986) Genetic transformation of the extreme thermophile Thermus thermophilus and of other Thermus spp. J. Bacteriol. 166, 338–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Degryse E., Glansdorff N., Pierard A. (1978) Comparative analysis of extreme thermophilic bacteria belonging to genus Thermus. Archiv. Microbiol. 117, 189–196 [DOI] [PubMed] [Google Scholar]
- 6. Brock T. D., Freeze H. (1969) Thermus aquaticus gen. n. and sp. n., a nonsporulating extreme thermophile. J. Bacteriol. 98, 289–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vu B., Chen M., Crawford R. J., Ivanova E. P. (2009) Bacterial extracellular polysaccharides involved in biofilm formation. Molecules 14, 2535–2554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Flemming H. C., Wingender J. (2010) The biofilm matrix. Nat. Rev. Microbiol. 8, 623–633 [DOI] [PubMed] [Google Scholar]
- 9. Costerton J. W., Lewandowski Z., Caldwell D. E., Korber D. R., Lappin-Scott H. M. (1995) Microbial biofilms. Annu. Rev. Microbiol. 49, 711–745 [DOI] [PubMed] [Google Scholar]
- 10. Costerton J. W. (1995) Overview of microbial biofilms. J. Ind. Microbiol. 15, 137–140 [DOI] [PubMed] [Google Scholar]
- 11. Hall-Stoodley L., Costerton J. W., Stoodley P. (2004) Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2, 95–108 [DOI] [PubMed] [Google Scholar]
- 12. Mattilasandholm T., Wirtanen G. (1992) Biofilm formation in the industry—a review. Food Rev. Int. 8, 573–603 [Google Scholar]
- 13. Lin M. H., Yang Y. L., Chen Y. P., Hua K. F., Lu C. P., Sheu F., Lin G. H., Tsay S. S., Liang S. M., Wu S. H. (2011) A novel exopolysaccharide from the biofilm of Thermus aquaticus YT-1 induces the immune response through Toll-like receptor 2. J. Biol. Chem. 286, 17736–17745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Niou Y. K., Wu W. L., Lin L. C., Yu M. S., Shu H. Y., Yang H. H., Lin G. H. (2009) Role of galE on biofilm formation by Thermus spp. Biochem. Biophys. Res. Commun. 390, 313–318 [DOI] [PubMed] [Google Scholar]
- 15. Henne A., Bruggemann H., Raasch C., Wiezer A., Hartsch T., Liesegang H., Johann A., Lienard T., Gohl O., Martinez-Arias R., Jacobi C., Starkuviene V., Schlenczeck S., Dencker S., Huber R., Klenk H. P., Kramer W., Merkl R., Gottschalk G., Fritz H. J. (2004) The genome sequence of the extreme thermophile Thermus thermophilus. Nat. Biotechnol. 22, 547–553 [DOI] [PubMed] [Google Scholar]
- 16. Omelchenko M. V., Wolf Y. I., Gaidamakova E. K., Matrosova V. Y., Vasilenko A., Zhai M., Daly M. J., Koonin E. V., Makarova K. S. (2005) Comparative genomics of Thermus thermophilus and Deinococcus radiodurans: divergent routes of adaptation to thermophily and radiation resistance. BMC Evol. Biol. 5, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Averhoff B. (2009) Shuffling genes around in hot environments: the unique DNA transporter of Thermus thermophilus. FEMS Microbiol. Rev. 33, 611–626 [DOI] [PubMed] [Google Scholar]
- 18. Averhoff B., Friedrich A. (2003) Type IV pili-related natural transformation systems: DNA transport in mesophilic and thermophilic bacteria. Arch. Microbiol. 180, 385–393 [DOI] [PubMed] [Google Scholar]
- 19. Averhoff B. (2004) DNA transport and natural transformation in mesophilic and thermophilic bacteria. J. Bioenerg. Biomembr. 36, 25–33 [DOI] [PubMed] [Google Scholar]
- 20. Burkhardt J., Vonck J., Langer J. D., Salzer R., Averhoff B. (2012) Unusual N-terminal ααβαββα fold of PilQ from Thermus thermophilus mediates ring formation and is essential for piliation. J. Biol. Chem. 287, 8484–8494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Donlan R. M. (2002) Biofilms: microbial life on surfaces. Emerg. Infect. Dis. 8, 881–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. McDonald J. H. (2001) Patterns of temperature adaptation in proteins from the bacteria Deinococcus radiodurans and Thermus thermophilus. Mol. Biol. Evol. 18, 741–749 [DOI] [PubMed] [Google Scholar]
- 23. Basak S., Mukhopadhyay P., Gupta S. K., Ghosh T. C. (2010) Genomic adaptation of prokaryotic organisms at high temperature. Bioinformation 4, 352–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schwarzenlander C., Haase W., Averhoff B. (2009) The role of single subunits of the DNA transport machinery of Thermus thermophilus HB27 in DNA binding and transport. Environ. Microbiol. 11, 801–808 [DOI] [PubMed] [Google Scholar]
- 25. Soufi B., Jers C., Hansen M. E., Petranovic D., Mijakovic I. (2008) Insights from site-specific phosphoproteomics in bacteria. Biochim. Biophys. Acta 1784, 186–192 [DOI] [PubMed] [Google Scholar]
- 26. Macek B., Mijakovic I. (2011) Site-specific analysis of bacterial phosphoproteomes. Proteomics 11, 3002–3011 [DOI] [PubMed] [Google Scholar]
- 27. Jers C., Soufi B., Grangeasse C., Deutscher J., Mijakovic I. (2008) Phosphoproteomics in bacteria: towards a systemic understanding of bacterial phosphorylation networks. Expert. Rev. Proteomics 5, 619–627 [DOI] [PubMed] [Google Scholar]
- 28. Macek B., Mijakovic I., Olsen J. V., Gnad F., Kumar C., Jensen P. R., Mann M. (2007) The serine/threonine/tyrosine phosphoproteome of the model bacterium Bacillus subtilis. Mol. Cell. Proteomics 6, 697–707 [DOI] [PubMed] [Google Scholar]
- 29. Macek B., Gnad F., Soufi B., Kumar C., Olsen J. V., Mijakovic I., Mann M. (2008) Phosphoproteome analysis of E. coli reveals evolutionary conservation of bacterial Ser/Thr/Tyr phosphorylation. Mol. Cell. Proteomics 7, 299–307 [DOI] [PubMed] [Google Scholar]
- 30. Soufi B., Gnad F., Jensen P. R., Petranovic D., Mann M., Mijakovic I., Macek B. (2008) The Ser/Thr/Tyr phosphoproteome of Lactococcus lactis IL1403 reveals multiply phosphorylated proteins. Proteomics 8, 3486–3493 [DOI] [PubMed] [Google Scholar]
- 31. Aivaliotis M., Macek B., Gnad F., Reichelt P., Mann M., Oesterhelt D. (2009) Ser/Thr/Tyr protein phosphorylation in the archaeon Halobacterium salinarum—a representative of the third domain of life. PLoS One 4, e4777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lin M. H., Hsu T. L., Lin S. Y., Pan Y. J., Jan J. T., Wang J. T., Khoo K. H., Wu S. H. (2009) Phosphoproteomics of Klebsiella pneumoniae NTUH-K2044 reveals a tight link between tyrosine phosphorylation and virulence. Mol. Cell. Proteomics 8, 2613–2623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ravichandran A., Sugiyama N., Tomita M., Swarup S., Ishihama Y. (2009) Ser/Thr/Tyr phosphoproteome analysis of pathogenic and non-pathogenic Pseudomonas species. Proteomics 9, 2764–2775 [DOI] [PubMed] [Google Scholar]
- 34. Hu C. W., Lin M. H., Huang H. C., Ku W. C., Yi T. H., Tsai C. F., Chen Y. J., Sugiyama N., Ishihama Y., Juan H. F., Wu S. H. (2012) Phosphoproteomic analysis of Rhodopseudomonas palustris reveals the role of pyruvate phosphate dikinase phosphorylation in lipid production. J. Proteome Res. 11, 5362–5375 [DOI] [PubMed] [Google Scholar]
- 35. Takahata Y., Inoue M., Kim K., Iio Y., Miyamoto M., Masui R., Ishihama Y., Kuramitsu S. (2012) Close proximity of phosphorylation sites to ligand in the phosphoproteome of the extreme thermophile Thermus thermophilus HB8. Proteomics 12, 1414–1430 [DOI] [PubMed] [Google Scholar]
- 36. Lennox E. S. (1955) Transduction of linked genetic characters of the host by bacteriophage P1. Virology 1, 190–206 [DOI] [PubMed] [Google Scholar]
- 37. Thingholm T. E., Larsen M. R. (2009) The use of titanium dioxide micro-columns to selectively isolate phosphopeptides from proteolytic digests. Methods Mol. Biol. 527, 57–66, xi [DOI] [PubMed] [Google Scholar]
- 38. Olsen J. V., Macek B. (2009) High accuracy mass spectrometry in large-scale analysis of protein phosphorylation. Methods Mol. Biol. 492, 131–142 [DOI] [PubMed] [Google Scholar]
- 39. Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 40. Cox J., Neuhauser N., Michalski A., Scheltema R. A., Olsen J. V., Mann M. (2011) Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10, 1794–1805 [DOI] [PubMed] [Google Scholar]
- 41. Conesa A., Gotz S., Garcia-Gomez J. M., Terol J., Talon M., Robles M. (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676 [DOI] [PubMed] [Google Scholar]
- 42. Yu N. Y., Wagner J. R., Laird M. R., Melli G., Rey S., Lo R., Dao P., Sahinalp S. C., Ester M., Foster L. J., Brinkman F. S. (2010) PSORTb 3.0: improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 26, 1608–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Klimke W., Agarwala R., Badretdin A., Chetvernin S., Ciufo S., Fedorov B., Kiryutin B., O'Neill K., Resch W., Resenchuk S., Schafer S., Tolstoy I., Tatusova T. (2009) The national center for biotechnology information's protein clusters database. Nucleic Acids Res. 37, D216–D223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chan J. W., Goodwin P. H. (1995) Extraction of genomic DNA from extracellular polysaccharide-synthesizing gram-negative bacteria. BioTechniques 18, 418–422 [PubMed] [Google Scholar]
- 45. Aiyar A., Xiang Y., Leis J. (1996) Site-directed mutagenesis using overlap extension PCR. Methods Mol. Biol. 57, 177–191 [DOI] [PubMed] [Google Scholar]
- 46. Moreno R., Haro A., Castellanos A., Berenguer J. (2005) High-level overproduction of His-tagged Tth DNA polymerase in Thermus thermophilus. Appl. Environ. Microbiol. 71, 591–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. M. Dubois K. A. G., Hamilton J. K., Rebers P. A., Smith F. (1956) Colorimetric method for determination of sugars and related substances. Anal. Chem. 28, 350–356 [Google Scholar]
- 48. Olsen J. V., Blagoev B., Gnad F., Macek B., Kumar C., Mortensen P., Mann M. (2006) Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127, 635–648 [DOI] [PubMed] [Google Scholar]
- 49. Gnad F., Forner F., Zielinska D. F., Birney E., Gunawardena J., Mann M. (2010) Evolutionary constraints of phosphorylation in eukaryotes, prokaryotes, and mitochondria. Mol. Cell. Proteomics 9, 2642–2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Munoz-Dorado J., Almaula N., Inouye S., Inouye M. (1993) Autophosphorylation of nucleoside diphosphate kinase from Myxococcus xanthus. J. Bacteriol. 175, 1176–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Otero A. S. (2000) NM23/nucleoside diphosphate kinase and signal transduction. J. Bioenerg. Biomembr. 32, 269–275 [DOI] [PubMed] [Google Scholar]
- 52. Panosian T. D., Nannemann D. P., Watkins G. R., Phelan V. V., McDonald W. H., Wadzinski B. E., Bachmann B. O., Iverson T. M. (2011) Bacillus cereus phosphopentomutase is an alkaline phosphatase family member that exhibits an altered entry point into the catalytic cycle. J. Biol. Chem. 286, 8043–8054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lippmann C., Lindschau C., Vijgenboom E., Schroder W., Bosch L., Erdmann V. A. (1993) Prokaryotic elongation factor Tu is phosphorylated in vivo. J. Biol. Chem. 268, 601–607 [PubMed] [Google Scholar]
- 54. Wang Y., Jiang Y., Meyering-Voss M., Sprinzl M., Sigler P. B. (1997) Crystal structure of the EF-Tu.EF-Ts complex from Thermus thermophilus. Nat. Struct. Biol. 4, 650–656 [DOI] [PubMed] [Google Scholar]
- 55. Zoldak G., Redecke L., Svergun D. I., Konarev P. V., Voertler C. S., Dobbek H., Sedlak E., Sprinzl M. (2007) Release factors 2 from Escherichia coli and Thermus thermophilus: structural, spectroscopic and microcalorimetric studies. Nucleic Acids Res. 35, 1343–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Miller D. J., Jerga A., Rock C. O., White S. W. (2008) Analysis of the Staphylococcus aureus DgkB structure reveals a common catalytic mechanism for the soluble diacylglycerol kinases. Structure 16, 1036–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Jerga A., Lu Y. J., Schujman G. E., de Mendoza D., Rock C. O. (2007) Identification of a soluble diacylglycerol kinase required for lipoteichoic acid production in Bacillus subtilis. J. Biol. Chem. 282, 21738–21745 [DOI] [PubMed] [Google Scholar]
- 58. Gordon E., Flouret B., Chantalat L., van Heijenoort J., Mengin-Lecreulx D., Dideberg O. (2001) Crystal structure of UDP-N-acetylmuramoyl-L-alanyl-D-glutamate: meso-diaminopimelate ligase from Escherichia coli. J. Biol. Chem. 276, 10999–11006 [DOI] [PubMed] [Google Scholar]
- 59. Rose I., Biukovic G., Aderhold P., Muller V., Gruber G., Averhoff B. (2011) Identification and characterization of a unique, zinc-containing transport ATPase essential for natural transformation in Thermus thermophilus HB27. Extremophiles 15, 191–202 [DOI] [PubMed] [Google Scholar]
- 60. Mattick J. S. (2002) Type IV pili and twitching motility. Annu. Rev. Microbiol. 56, 289–314 [DOI] [PubMed] [Google Scholar]
- 61. Wall D., Kaiser D. (1999) Type IV pili and cell motility. Mol. Microbiol. 32, 1–10 [DOI] [PubMed] [Google Scholar]
- 62. Chen Y., Shiue S. J., Huang C. W., Chang J. L., Chien Y. L., Hu N. T., Chan N. L. (2005) Structure and function of the XpsE N-terminal domain, an essential component of the Xanthomonas campestris type II secretion system. J. Biol. Chem. 280, 42356–42363 [DOI] [PubMed] [Google Scholar]
- 63. Abendroth J., Murphy P., Sandkvist M., Bagdasarian M., Hol W. G. (2005) The X-ray structure of the type II secretion system complex formed by the N-terminal domain of EpsE and the cytoplasmic domain of EpsL of Vibrio cholerae. J. Mol. Biol. 348, 845–855 [DOI] [PubMed] [Google Scholar]
- 64. Chiang P., Sampaleanu L. M., Ayers M., Pahuta M., Howell P. L., Burrows L. L. (2008) Functional role of conserved residues in the characteristic secretion NTPase motifs of the Pseudomonas aeruginosa type IV pilus motor proteins PilB, PilT and PilU. Microbiology 154, 114–126 [DOI] [PubMed] [Google Scholar]
- 65. Friedrich A., Prust C., Hartsch T., Henne A., Averhoff B. (2002) Molecular analyses of the natural transformation machinery and identification of pilus structures in the extremely thermophilic bacterium Thermus thermophilus strain HB27. Appl. Environ. Microbiol. 68, 745–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Pelicic V. (2008) Type IV pili: e pluribus unum? Mol. Microbiol. 68, 827–837 [DOI] [PubMed] [Google Scholar]
- 67. Burrows L. L. (2012) Pseudomonas aeruginosa twitching motility: type IV pili in action. Annu. Rev. Microbiol. 66, 493–520 [DOI] [PubMed] [Google Scholar]
- 68. Klausen M., Heydorn A., Ragas P., Lambertsen L., Aaes-Jorgensen A., Molin S., Tolker-Nielsen T. (2003) Biofilm formation by Pseudomonas aeruginosa wild type, flagella and type IV pili mutants. Mol. Microbiol. 48, 1511–1524 [DOI] [PubMed] [Google Scholar]
- 69. Li Y., Sun H., Ma X., Lu A., Lux R., Zusman D., Shi W. (2003) Extracellular polysaccharides mediate pilus retraction during social motility of Myxococcus xanthus. Proc. Natl. Acad. Sci. USA 100, 5443–5448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Yang Z., Lux R., Hu W., Hu C., Shi W. (2010) PilA localization affects extracellular polysaccharide production and fruiting body formation in Myxococcus xanthus. Mol. Microbiol. 76, 1500–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Cava F., de Pedro M. A., Schwarz H., Henne A., Berenguer J. (2004) Binding to pyruvylated compounds as an ancestral mechanism to anchor the outer envelope in primitive bacteria. Mol. Microbiol. 52, 677–690 [DOI] [PubMed] [Google Scholar]
- 72. Cozzone A. J. (2005) Role of protein phosphorylation on serine/threonine and tyrosine in the virulence of bacterial pathogens. J. Mol. Microbiol. Biotechnol. 9, 198–213 [DOI] [PubMed] [Google Scholar]
- 73. Vincent C., Duclos B., Grangeasse C., Vaganay E., Riberty M., Cozzone A. J., Doublet P. (2000) Relationship between exopolysaccharide production and protein-tyrosine phosphorylation in gram-negative bacteria. J. Mol. Biol. 304, 311–321 [DOI] [PubMed] [Google Scholar]
- 74. Wu S. S., Wu J., Kaiser D. (1997) The Myxococcus xanthus pilT locus is required for social gliding motility although pili are still produced. Mol. Microbiol. 23, 109–121 [DOI] [PubMed] [Google Scholar]
- 75. Aas F. E., Lovold C., Koomey M. (2002) An inhibitor of DNA binding and uptake events dictates the proficiency of genetic transformation in Neisseria gonorrhoeae: mechanism of action and links to Type IV pilus expression. Mol. Microbiol. 46, 1441–1450 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.