Abstract
Iron is a crucial cofactor in numerous redox-active proteins operating in bioenergetic pathways including respiration and photosynthesis. Cellular iron management is essential to sustain sufficient energy production and minimize oxidative stress. To produce energy for cell growth, the green alga Chlamydomonas reinhardtii possesses the metabolic flexibility to use light and/or carbon sources such as acetate. To investigate the interplay between the iron-deficiency response and growth requirements under distinct trophic conditions, we took a quantitative proteomics approach coupled to innovative hierarchical clustering using different “distance-linkage combinations” and random noise injection. Protein co-expression analyses of the combined data sets revealed insights into cellular responses governing acclimation to iron deprivation and regulation associated with photosynthesis dependent growth. Photoautotrophic growth requirements as well as the iron deficiency induced specific metabolic enzymes and stress related proteins, and yet differences in the set of induced enzymes, proteases, and redox-related polypeptides were evident, implying the establishment of distinct response networks under the different conditions. Moreover, our data clearly support the notion that the iron deficiency response includes a hierarchy for iron allocation within organelles in C. reinhardtii. Importantly, deletion of a bifunctional alcohol and acetaldehyde dehydrogenase (ADH1), which is induced under low iron based on the proteomic data, attenuates the remodeling of the photosynthetic machinery in response to iron deficiency, and at the same time stimulates expression of stress-related proteins such as NDA2, LHCSR3, and PGRL1. This finding provides evidence that the coordinated regulation of bioenergetics pathways and iron deficiency response is sensitive to the cellular and chloroplast metabolic and/or redox status, consistent with systems approach data.
The green alga Chlamydomonas reinhardtii has an enormous metabolic versatility (1) and possesses the flexibility to grow in the presence of different carbon sources. It may use carbon dioxide (CO2) for photoautotrophic, acetate for heterotrophic, and both carbon sources for mixotrophic growth. In this alga CO2 is fixed via the Calvin Benson Bassham cycle (2), while acetate can be taken up, converted to acetyl-CoA, and enter the glyoxylate cycle where it may be incorporated into C4 acids (3). In addition to the use of acetate as a source of energy and carbon backbone for biosynthetic processes, acetate can control respiration and photosynthesis in conjunction with the light intensity and CO2 availability (4–6). Moreover, acclimation responses to iron- and copper-deficiencies significantly vary in photoautotrophic versus heterotrophic conditions (7–10), indicating that the metabolic status of the cells influence overall cellular acclimation responses.
Transition metals like copper, manganese, and iron possess the ability to donate and accept electrons, making these metals suitable cofactors in enzymes that catalyze redox reactions. In particular, iron is used as a cofactor in numerous biochemical pathways and is therefore an essential nutrient. Cells require relatively high levels of iron because it is present in heme-, iron-sulfur and other proteins that function in respiratory and photosynthetic energy transducing. Correspondingly, in eukaryotic cells, the mitochondrion is a major iron-utilizing compartment. It is well established that iron is transported into mitochondria for heme synthesis and iron-sulfur cluster assembly. This is required for the formation of a functional respiratory electron transport machinery (11). Therefore, mitochondrial metabolism in mammals, fungi and plants is significantly affected under iron deficiency, as demonstrated by a number of studies (12–14). In plants, the chloroplasts are a primary target of iron deficiency. Changes in chloroplast structure, photosynthetic capacity and the composition of thylakoid membranes have been described for plants deprived of iron (15–21).
Plants have devised various strategies for acquiring iron (22). Generally, iron deficiency leads to the activation of the iron uptake systems in photosynthetic organisms. For example, the accumulation of the ferroxidase, a component of the high affinity iron uptake system in C. reinhardtii, is very rapidly enhanced when iron becomes limiting (23). Inactivation of IRT1, the most prevalent Fe2+ transporter in Arabidopsis thaliana leads to a dramatic iron deficiency that is reflected by chlorosis (24–26). Despite the evolution of elaborate iron-uptake mechanisms in plants, iron deficiency-induced chlorosis remains a major agricultural problem (27, 28).
The global impact of iron deficiency on photosynthetic productivity has been also shown in vast ocean regions, which are severely limited for iron (29, 30). Generally, one can conclude that photosynthesis in the oceans and on land can occur in environment where iron availability is restricted.
Photosystem I (PSI) is a prime target of iron deficiency as it contains 12 atoms of iron per core complex. In algae, the degradation of PSI is also linked to remodeling of PSI-associated light-harvesting antenna (LHCI) (31–33). Cyanobacteria respond to iron deficiency by degradation of light harvesting phycobilisomes (34) and induction of the “iron-stress-induced” gene isiA. The ISIA protein, which has significant sequence similarity with CP43, a chlorophyll a-binding protein of photosystem II (PSII; (35, 36), forms a ring of 18 molecules around a PSI trimeric reaction center, as shown by electron microscopy (37, 38). The overall reorganization of the PSI complex from 900 kDa into 1.7 MDa complex highlights the large adaptive nature of the cellular response to iron deficiency, which helps to optimize the architecture of the photosynthetic apparatus to conditions in which iron is a limiting factor.
The marine diatom Thalassiosira oceanica shows a remarkable retrenchment of cellular metabolism and remodeling of bioenergetic pathways in response to iron availability (39). Low iron triggers a reduction in the level of iron-rich photosynthetic proteins while iron-rich mitochondrial proteins are preserved. Furthermore, iron deprivation causes a remodeling of the photosynthetic machinery resulting in the adjustment of light energy use to an overall decline in the level of photosynthetic electron transport complexes (39). These responses, reported for green algae such as C. reinhardtii (31, 40, 41), are important for minimizing photo-oxidative stress and optimizing photosynthetic function. As observed for T. oceanica, under conditions of low iron availability (in the presence of organic carbon) a hierarchy of iron allocation responses in C. reinhardtii result in the down-regulation of iron-rich photosynthetic complexes while iron-rich mitochondrial complexes remain stable (41). Notably, under photoautotrophic and mixotrophic conditions C. reinhardtii displays distinct iron deprivation responses, suggesting that the cell's response to iron deficiency is also dependent on trophic conditions (7–9). Thus bioenergetics pathways are remodeled in response to iron availability as well as to the type of carbon source available. Moreover, recent data has indicated that the regulation of iron-induced remodeling of the photosynthetic apparatus is linked to energy metabolism. Depletion of Proton Gradient Regulation Like1 protein (PGRL1) in C. reinhardtii has revealed a decreased efficiency of cyclic electron transfer under low iron conditions resulting in higher vulnerability toward iron deprivation (42).
It was our aim to generate a more comprehensive picture of how the proteome of C. reinhardtii varies in response to low iron under distinct trophic conditions and how these changes compare with differences observed for cells grown under photoautotrophic and photoheterotrophic iron replete conditions. Quantitative proteomics in conjunction with a novel hierarchical clustering approach revealed information about the responses of C. reinhardtii to low iron conditions and the iron requirements of photoautotrophic growth. These analyses provide novel insights into the relationships between protein networks required for photosynthesis and iron deprivation-elicited stress responses; these studies are providing the knowledge required for modulating the level of available iron to improve the photosynthetic performance of plants (43, 44).
EXPERIMENTAL PROCEDURES
Strains and Culture Conditions
For all experiments the cell wall-less wild type strain CW15 was used. Cells were grown photoheterotrophically (PH) in Tris-Acetate-Phosphate (TAP) medium (45) and photoautotrophically (PA) in minimal medium (HSM) (45) to a cell density of 1–2 × 106 cells/ml at a light intensity of 60 μE/m2/s. PH- and PA-grown cells were isotopically labeled with 15NH4Cl (Cambridge Isotope Laboratories Inc.) in iron-replete conditions (18 μm iron) and nonlabeled (14NH4Cl) in iron-deficient conditions (0.1 μm iron). The cells were treated as a “steady state” culture and harvested from a liquid culture after 5 days (PH) or 10 days (PA) of iron deficiency. For the comparison of PA to PH (both in 18 μm iron), for swapping experiments PH- or PA- grown cells were isotopically labeled.
For data shown in Fig. 4, the adh1 mutant (46) and its wild type parental strain CC125 (cw+, mt+, nit−) were used. The two strains were grown in TAP medium at a light intensity of 60 μE m−2 s−1 until they reached a cell density of about 2–3 × 106 cells/ml (0 time point). Cells were then pelleted at 4000g for 2 min, washed twice and re-suspended in iron-free TAP medium to a cell density 1 ×106 cells/ml. Samples for protein, cell counting, chlorophyll, and fluorescence emission spectroscopy at 77K were collected at day 1 and 2 after the transfer.
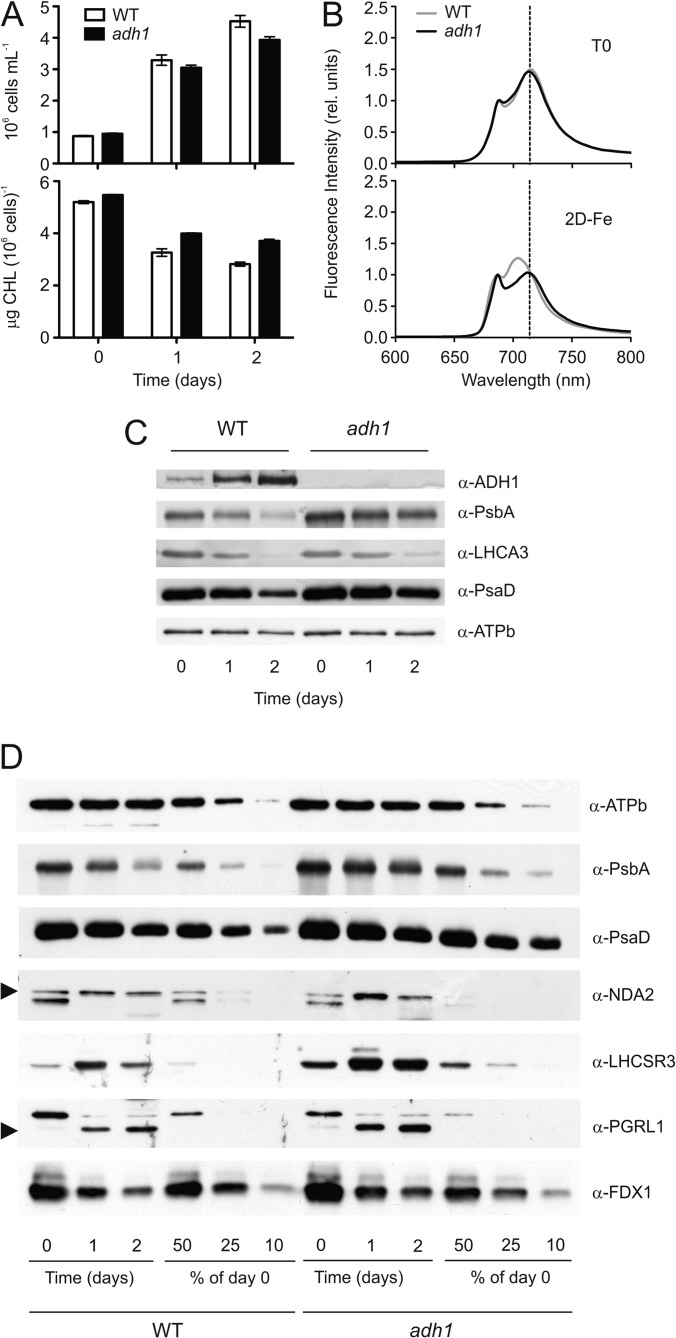
Fig. 4.
Remodeling of bioenergetic pathways under iron-deficiency is affected in the adh1 mutant strain. A, Growth (top) and chlorophyll content (bottom) of adh1 and CC125 (WT) cells transferred from iron-replete (0) to iron-free (1 and 2 days) TAP medium. Data are means of two biological replicates, each with three technical replicates ± S.D. B, Low temperature fluorescence emission spectra of WT and adh1 cells, under iron-replete (top) and iron-free (bottom) conditions. C, D, 25 μg of whole cell protein extracts from WT and mutant cells were fractionated on a 13% SDS-PAGE, and ADH1, PsbA, LHCA3, PsaD, NDA2, LHCSR3, PGRL1, FDX1, and ATPb abundance were analyzed by immunoblot. To highlight differences between WT and adh1 cells in iron starvation-mediated photosystems degradation, dilution series (50–25-10%) relative to the T0 are shown. ATPb was used as a loading control.
Whole Cells and Isolation of Chloroplasts and Mitochondria
Cells were pelleted at 2500 × g (Beckmann Coulter J 20 XP) and chloroplasts and mitochondria were isolated as described in Terashima et al. (47).
Determination of the Protein Amount and Immunodetection
The determination of the protein amount was carried out using the Pierce® Protein assay kit (Thermo Scientific, Perbio Science GmbH, Bonn, Germany). The absorbance of the BCA complex was detected at 562 nm after incubation of the protein with BCA at 55 °C for 30 min. Protein concentrations were calculated based on a BSA calibration curve. Immunodetection was carried out as described in Terashima et al. (47). Antibodies against ADH11 were purchased from Agrisera (ref #AS10 748); anti-NDA2 antibodies were a kind gift of Dr. Pierre Cardol, University of Liège (Belgium).
Protein Analysis and Liquid Chromatography-Tandem MS (LC-MS/MS) Analysis of Proteins
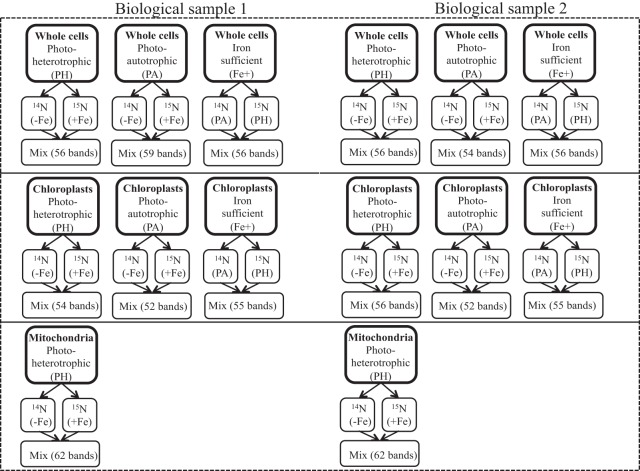
Isolated chloroplasts and whole cell samples were mixed at an equal amount of labeled/unlabeled 1:1 protein. For isolated mitochondria, concentrations of mitochondrial proteins in samples stemming from iron sufficient or iron deficient conditions were enumerated by Western blot analyses (not shown) using the COX2b antibody mentioned above (section above). To account for differences in enrichment of COX2b in the distinct preparations, isolated mitochondria were mixed in the ratio labeled/unlabeled 2.5:1 protein. The mixed samples were fractionated by SDS-PAGE. A different number of bands was excised and digested tryptically as described in Naumann et al. (40). A schematic overview of all mixed preparations and performed measurements is shown in Fig. 1. The LC-MS/MS measurements were carried out as described in (47), except that LTQ Orbitrap XL MS was used in version 2.4 SP2.
Fig. 1.
Schematic overview of the performed measurements for protein identification and quantification via stable 15N isotopic labeling. Whole cells grown in iron deficiency (14N (−Fe)) and iron sufficiency (15N (+Fe)) were harvested from photoheterotrophic conditions (PH) and photoautotrophic conditions (PA). Mixed samples of (15N (+Fe)) and (14N (−Fe)), which were separated via one-dimensional SDS-PAGE, were cut into 56 bands for PH and 54 and 59 bands for PA for two biological replicates, respectively. Isolated chloroplasts of a mix of (15N (+Fe)) and (14N (−Fe)) samples resulted in 54 and 56 bands for PH, 52 bands for PA and for isolated mitochondria in 62 bands for PH for two biological samples, respectively. The mix of samples from 14N PA and 15N PH in iron-sufficient conditions resulted in 56 bands for whole cells and 55 bands for isolated chloroplasts for two biological samples, respectively. For whole cells from iron-sufficient conditions also a label swap experiment was conducted (15N PA and 14N PH). Mitochondria could not be isolated from PA of (15N (+Fe)) and (14N (−Fe)). Therefore no measurements were carried out for isolated mitochondria from PA under these conditions as well as for the comparison of 14N PA to 15N PH.
Acquired Thermo Xcalibur raw files were converted to the open mzML format (48) via msconvert (part of Proteowizard, version 2.0.1905, (49) with 32 bit precision. Standard proteomics data identifications were performed using Proteomatic (50) as a data processing pipeline tool providing a graphical user interface to the later mentioned tools and pymzML (51) for direct access to mzML files. mzML files associated with this manuscript may be downloaded from peptideatlas.org with the database tag ‘Hoehner_2013′. Files in mgf format were used as input for database search algorithms. Mgf files were generated using pymzML (51).
Enhancing Proteomics Data
The detailed procedure of enhancing proteomics data by means of LC-MS/MS alignment and quantifications based on identifications performed in different LC-MS/MS runs within a well-defined elution window will be described in more detail elsewhere (Barth & Fufezan, manuscript in prep.). Briefly, all LC-MS/MS runs were aligned using a combination of the qualities of peptide identifications and peptide quantifications. Peptide identifications were performed by OMSSA (version 2.1.9) (52) and X! Tandem (2010.12.01) (53). Identification parameters were used as described in (47) with the following changes. Precursor mass tolerance was set to 5 ppm and oxidations of methionine and tryptophan (single and double) were set as variable modifications. For OMSSA a linear precursor charge dependence and enzyme specificity was used. For X!Tandem noise suppression was enabled. Protein databases JGI4.3 AUGUSTUS10.2, NCBI BK000554.2, and NCBI NC_001638.1 were used. The resulting database contained 17195 nonredundant protein sequences, with additional 1198 common contaminants. Decoy protein sequences were generated by randomly shuffling tryptic peptides while retaining the redundancy of nonproteotypic peptides. Qvality (version 2.02, (54)) was used for statistical validation of peptide identifications using a posterior error probability (PEP) threshold of less than 0.05. Quantifications were performed using pyQMS (Niehues & Fufezan, manuscript in prep). Preliminary data were quantified using qTrace (47). Ratios obtained by both quantification algorithms were very similar.
Isotopologs, their elution profiles and their elution windows could be assigned to a given peptide. Confident quantifications of the peptides were possible by retaining the strict rule that a peptide was only considered if it could be identified within the aligned retention time window allowing a deviation of ± 30 s. All data analysis were performed using piqDB (Fufezan and coworkers, unpublished), which is very similar to SuperHirn (55) incorporating quality of the identifications and quantifications of a given peptide into the definition of its accurate mass time tag, that is in our case into the definition of the retention time windows.
Protein Ratios
Peptide ratios were calculated using all MS1 scans that contained both sister peptides, i.e. the light (14N) and heavy (15N) peptide. The amounts of light and heavy sister peptides were calculated using pyQMS (Niehues & Fufezan, manuscript in prep). The peptide ratio was calculated by dividing the sum of all light by the sum of all heavy amounts. Amounts are determined for each MS1 spectrum by the sum of all isotope envelope peak intensities of the simulated and scaled envelope, if the m/z and intensity values could be matched by pyQMS with a score higher than 7. Each peptide ratio was weighted by the number of MS1 scans that contributed originally to its ratio, hence abundant peptides with multiple MS1 quantification events contribute more to the protein ratio than single events. Protein ratios were calculated based on the weighted proteotypic peptide ratios. Nonproteotypic peptides were reported combined in protein groups. Protein ratios were calculated in the same way as peptide ratios, i.e. each peptide for a given protein was weighted by the number of MS1 scans that contributed to its ratio, thus peptides with ratios based on few MS1 scans are weighted less. Protein ratios and standard deviations of all proteins in all experiments (log2) are shown in supplemental Table S1.
Hierarchical Clustering of Proteomics Data
Enhanced proteomics data was clustered using the Python (56) and Python modules numpy (57) and scipy (58) combined in the Python module pyGCluster (Jaeger et al., manuscript in prep). Briefly, agglomerative hierarchical clustering for different distance-linkage combinations was performed using a pool of (1) six methods to define the distance among clusters (linkage): “complete,” “average,” “weighted,” “centroid,” “median,” “ward,” and (2) two functions to calculate the distance between clusters (metric): “Euclidean” and “correlation.” Because the ward, centroid and median linkage methods cannot be used with the correlation function, a total of nine distance-linkage combinations were evaluated. Data were clustered independently 250,000 times. During each iteration step, each ratio was altered by randomly sampling a ratio from a Gaussian distribution described by the original ratio mean and its standard deviation. This step injects a high degree of noise into the data set, ensures stringent hierarchical clustering and allows discriminating between core elements of a well-defined high frequency cluster in, e.g. Euclidian space and the adjacent less frequent elements. A cluster is a group of genes or proteins that are similar given the distance definition. For example using Euclidian or correlation distance will group proteins by similarity in ratios over all conditions or by similar trends in ratios comparing the conditions, respectively. Because ratios, i.e. (0, …,∞) are not well suited to estimate distances, the protein ratios were log2 transformed before the clustering.
Obviously, creating a “consensus tree” is neither feasible nor informative using such an approach. The overall frequency in which a certain cluster, i.e. a certain set of proteins, is found is more informative because its position, i.e. clade, can occur on different levels of the tree. In this approach, only clusters with a frequency higher than 0.5% in at least one of the nine hierarchical clustering assays were retained for further analysis. The 0.5% threshold was chosen for the sake of simplicity, i.e. only the top 490 clusters were analyzed further and grouped into communities.
Clusters, their frequency, their relation to each other and the communities that they form are illustrated as a node map using NetworkX (59) and gephi (60). Clusters are shown as nodes and their relations are shown as edges connecting related nodes, i.e. those sharing same proteins. Therefore, the node map shows the high frequency clusters from a combination of multiple hierarchical clustering assays using high number repetitions with a standard deviation weighted degree of variation in each dimension (condition) of each object (protein) during each iteration, and the relation among those high frequency clusters. The communities that are found represent well-defined groups of proteins that show cotranslational regulation. Such node maps are referred to as cNode maps.
Changes in protein levels were plotted for each community as a heat map using the Python scripting language and pyGCluster (Jaeger et al., manuscript in prep). The log2 transformed protein ratios are color-coded and the standard deviation is illustrated by the size of the plotted box in the heat map (see text). The cFrequency is the sum of all cluster frequencies in which the given protein is within the whole community. The cluster frequencies themselves were calculated as the median of the frequencies for all nine assays. The cFrequency illustrates therefore the major elements of each community and the relative frequency among communities. For more detailed information on frequencies for each clustering assay, see supplemental Table S2, and for more details concerning pyGCluster, see Jaeger et al. (2013, manuscript in prep).
Fluorescence Emission Spectroscopy at 77K
The samples were used at a chlorophyll concentration of 5 μg/ml, as described in (42). Fluorescence emission spectra were recorded as an average of four scans within the window of 600–800 nm at a scan speed of 500 nm/min and a slit width of 3 nm. Excitation of the sample was at 435 nm.
Oxygen Exchanges
Oxygen uptake and evolution rates were measured as described in (42). The measurements were performed at 1000 μE/m2/s. After the measurement, 500 μl of the sample was removed from the cuvette for determining cell number to allow for the calculation of pm O2/cell.
RESULTS
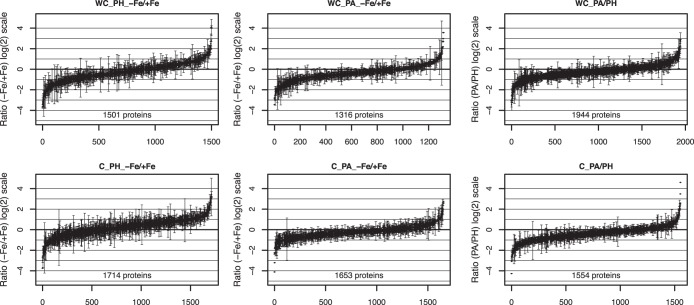
Iron deficiency responses in C. reinhardtii are influenced by the trophic status of the cells (7–9, 41). Yet, although many aspects of the cellular iron deficiency response are driven by low iron availability, other properties appear to be governed by the cell's metabolic status. To dissect these responses and obtain more detailed insights into these regulatory processes we performed an in-depth quantitative proteomics approach using metabolic labeling. To this end iron sufficient and deficient cells were grown under photoheterotrophic (PH) or -autotrophic (PA) growth conditions in 15N or 14N labeled media, respectively. The iron deficiency status was verified using low temperature fluorescence emission spectroscopy and immunoblotting using antibodies directed against marker proteins, diminished in response to iron deprivation (see supplemental Fig. S1). Moreover PH grown iron sufficient cells were labeled with 15N and compared with 14N labeled cells grown under PA conditions. Here also a swapping experiment was performed to detect and avoid isotope effects. Quantitative analysis showed very good correlation among the label swap experiments (see supplemental Fig. S2). 15N labeled iron sufficient cells or isolated chloroplasts were mixed on equal protein basis with the respective complement from 14N labeled iron deficient or iron sufficient material (Fig. 1). In addition mitochondria were isolated from 15N labeled iron sufficient and 14N labeled iron deficient cells grown under PH conditions and also mixed on equal protein basis (Fig. 1). Mixed samples were fractionated by SDS-PAGE and protein bands were excised from the gels. On average about 56 bands were cut per gel. SDS-PAGE bands were in-gel digested with trypsin and peptides were analyzed by LC-MS/MS. We completed and processed two independent biological replicates. For the whole cell and chloroplast sample sets we investigated 337 and 523 excised and tryptically digested protein bands, respectively. Moreover, for isolated mitochondria in total 124 excised and proteolytically digested protein bands were examined by LC-MS/MS. The mass spectrometric analyses resulted in the identification of peptides and proteins and subsequently allowed the quantitation of protein ratios. The results stemming from the two independent biological replicates are summarized in Fig. 2 and are plotted as number of quantified proteins and their respective proteins ratios starting from the lowest to the highest observed ratios. As shown in Fig. 2, from the six independent experimental set-ups at minimum 1316 and at maximum 1944 proteins could be quantified. These high numbers can be attributed to the enhancement procedure using piqDB, (Fufezan and coworkers, unpublished), which is similar to SuperHirn (55). Consequently, such large amount of protein ratios provided a good basis for further exploration. Thus coregulation analyses of the proteomics data was performed using a novel and advanced hierarchical clustering approach, that injects noise into the data set and uses a large number of iterations, using pyGCluster (Jaeger et al., manuscript in prep). Briefly, pyGCluster uses the uncertainty of the data at hand by including the standard deviation (std) into the agglomerative hierarchical clustering approach. For each iteration, a new data set is generated in which each data point is randomly sampled out of the Gauss distribution described by its mean and std. The generated data set is described by the original data yet it will cluster differently. pyGCluster then applies nine different distance-linkage combinations for each of those perturbed data sets to ultimately, after a high number of iterations, isolate the high frequency clusters and merge those into coregulated communities. The advantages lie in less user bias and most importantly in reducing the data set at hand to very few proteins that show coregulations although heavily perturbed based on the uncertainty of their data.
Fig. 2.
The ratio for all quantified proteins of either iron deficient (14N(−Fe)) to iron sufficient conditions (15N (+Fe)) from photoheterotrophic conditions (PH) or photoautotrophic conditions (PA) or the ratio of 14N PA to 15N PH is shown for whole cells (upper panel) and enriched chloroplasts (lower panel). Error bars indicate standard deviations among different combinations of peptide, band, and charge states and between two independent biological samples.
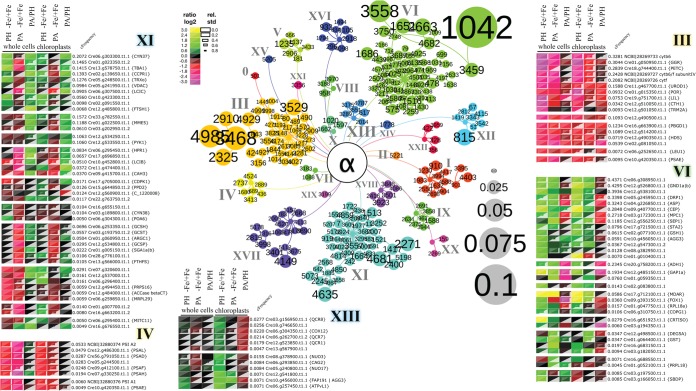
Quantitative mass spectrometric data from whole cell extracts (WC) and isolated chloroplasts (C) have been clustered using pyGCluster (see Material and Methods). It is expected that chloroplast localized proteins harbor similar trends in the six settings compared, which is indeed observed for all chloroplast proteins found in the clusters. Notably among the 109 proteins clustered, 53 were reported to be part of a chloroplast proteome (61) (supplemental Table S3). For nonchloroplast proteins, tendencies seen in whole cells should be representative, whereas changes observed in enriched chloroplast fractions might be biased by the organelle enrichment and purity, yet, jointed clustering might be reached because of these properties. The results are illustrated as cNode maps (Fig. 3) that highlight well-defined high frequency clusters of proteins, their relation to each other and how they are grouped into communities. These communities are colored coded and labeled with roman numerals in the cNode map. A total of 23 communities have been identified. The proteins within each community are illustrated as heat maps, a selection of heat maps is shown in Fig. 3. All heat maps can be found in the supplemental Fig. S3 (communities 0 - XXII). The heat maps also show the cFrequencies of each protein within a community. The cFrequencies are the sum of all cluster frequencies the protein is part of within a community. Because each cluster is found with different frequencies by the 9 hierarchical clustering assays performed here, the median frequency of those was used to represent each cluster. The colors in the heat maps between purple and yellow represent reduced and increased protein levels, respectively, comparing iron deficient (−Fe) and iron sufficient (+Fe) conditions or PA and PH settings. Ratios (i.e. -Fe/+Fe or PA/PH) are log2 transformed and the uncertainty, i.e. the standard deviation of a ratio is reflected in its box size. The higher the standard deviation and therefore the higher the uncertainty of a given ratio, the smaller is the box. This is illustrated in the legend.
Fig. 3.
Results obtained by multiple agglomerative hierarchical clustering approaches using pyGCluster of protein ratios comparing iron deficient (−Fe) with iron sufficient (+Fe) in photoautotrophic growth conditions (PA) or in photoheterotrophic growth conditions (PH) or comparing PA with PH in iron sufficient growth conditions in whole cells or isolated chloroplasts. Shown are the cNode map and five heat maps for selected communities. The cNode map highlights the most frequent clusters with their clusterID and their membership to one of the 23 communities identified. Communities are indicated in different colors and roman numerals. The cluster frequency is illustrated by the node size as shown in the legend (gray nodes). For detailed information on the clusters see Supplemental Table T2. The cFrequency of a protein is equal to the sum of all cluster frequencies the protein is part of, within its community. Heat maps are subdivided to illustrate the separation of subcommunities visible as branches in the cNode map.
Subunits of the cyt b6f Complex and Photosystem I As Well As Proteins Involved in Chlorophyll Biosynthesis Exhibit a Uniform Down-regulation Under Iron Deprivation
Community III (orange, Fig. 3 top right) contains 17 proteins, which are all down-regulated under low iron. This cluster is dominated by four subunits of the cytochrome b6f complex (cyt b6f complex, cytf, Rieske, cytb6 and subunit IV) and five enzymes involved in chlorophyll biosynthesis (the geranyl-geranylreductase (GGR), uroporphyrinogen-III decarboxylase (UROD1), light-dependent protochlorophyllide reductase (POR), copper target homolog 1 (CTH1), and porphobilinogen deaminase (PBGD1)). This cluster supports earlier notions that cyt b6f complex and the chlorophyll biosynthesis pathway are prime targets of iron-deficiency (31, 62–65). A light-harvesting-like protein LIL (Lhl3) shows the same tendency as the chlorophyll biosynthesis proteins. Another photosynthetic protein complex that is preferentially down-regulated under iron-deprivation is photosystem I. In community IV (yellow, Fig. 3 left bottom) seven out of eight proteins belong to the photosystem I complex (PsaA, B, D, E, F, L, H (Cre07.g330250)). The only nonphotosystem I protein in this cluster is a protein of unknown function (Cre05.g244500). All photosystem I complex subunits remain unchanged or slightly increase when iron replete PA/PH conditions are compared.
Core subunits of photosystem II (PSII) and the ATPase and many light-harvesting proteins are not among the clustered proteins. The polypeptides belonging to PSII and ATPase multiprotein complexes as well as to the light-harvesting complement show only moderate down-regulation under low iron availability (supplemental Fig. S4A). Moreover it appears that the impact on these complexes under low iron is generally more pronounced under PA rather than under PH conditions. In contrast, light-harvesting proteins that are associated to PSII, with the exception of Lhcb4 and Lhcb5, and listed in supplemental Fig. S4A are not reduced in amounts by iron deficiency. These data correlate very well with published results (41) and underpin the differential regulation of photosynthetic complex proteins in response to iron deprivation.
Clustering Reveals Response Networks of Acclimation to Iron Deficiency
A complete different picture can be seen in community VI (green, Fig. 3 top right). Here proteins are increased in expression under iron deprivation and mainly decreased under PA/PH iron replete conditions. The protein with the highest cFrequency belongs to the reticulon protein family (Cre06.g308950.t1.1), such proteins are found to be associated with the endoplasmic reticulum (ER) (66). The Chlamydomonas protein possesses a characteristic dilysine motif at the C-terminal and may be associated with ER channel-like membrane complexes as found for the reticulon NSP-A that is connected with an ER Ca2+-ATPase (66). Other members of this community are a 6-phosphogluconate dehydrogenase (GND1a, Cre12.g526800.t1.1), a putative glutathione S-transferase (Cre12.g538100.t1.1) and a dynamin related GTPase (DRP1), followed by two proteases, ASP, an aspartic-type endopeptidase and CEP (a candidate serine carboxypeptidase) and a putative mitochondrial phosphate carrier 1 (MPC1). Lower cFrequency members are a septin (SEP1), a granule bound starch synthase I (STA2), GSH1, a gamma-glutamylcysteine synthetase, a flagellar associated protein (AGG3/AGG4; star behind the name tag in Fig. 3 indicates a merge tag with models Cre10.g456100.t1.1/Cre10.g456050.t1.1 because of non proteotypic peptides matching on both protein sequences), Cre12.g547300.t1.2 (a putative ABC transporter), as well as two proteins of unknown function (Cre08.g382950.t1.1 and Cre01.g010400.t1.1).
Bifunctional alcohol and acetaldehyde dehydrogenase (ADH1), glyceraldehyde-3-phosphate dehydrogenase (plastidic), and monodehydroascorbate reductase (MDAR) are present in correlated subcommunities, having highest cFrequency in their communities. Another related subcommunity consists of four proteins, which differs by the fact that all these proteins are more increased under PA than PH iron-deficiency. These proteins are a DegP protease (DEG5A), a putative glutathione S-transferease (Cre01.g064400.t1.1), a protein of unknown function (Cre16.g683150.t1.1), and a protein possessing an acetyl-CoA-synthetase-like superfamily domain (Cre03.g182050.t1.1).
Overall, members of community VI are involved in bioenergetic remodeling and stress responses as many metabolic enzymes, proteases, and redox related enzymes dominate in this assembly. Importantly the clustering reveals that certain responses are distinct among the different conditions analyzed, a scenario that is also observed in community XI.
Clustering Reveals Responses that are Dependent on the Trophic and Iron Status of the Cells
Proteins that are down-regulated under PA growth conditions (−Fe/+Fe), up or only slightly down-regulated under PH growth (−Fe/+Fe) or up-regulated under PA/PH (+Fe) assembled together and are represented in community XI (light blue, Fig. 3 top left). A candidate Peptidyl-prolyl cis-trans isomerase (CYN37) shows the highest cFrequency in this assembly, followed by a putative metallopeptidase (Cre01.g023350.t1.2), a translation factor for expression of the chloroplast-encoded psbA gene (TBA1) (67), a cytochrome c peroxidase (CCPR1) and a thioredoxin o (TRXo). Proteins having a lower cFrequency are represented by a voltage-dependent anion-selective channel protein (VDAC), a low-CO2-inducible protein (LCIC), a putative peptidase (Cre10.g423300.t1.1), a putative plant-like transcription factor (Cre02.g091550.t1.1), and the AAA-metalloprotease FTSH1.
Three proteins are represented in an adjacent subcommunity. These proteins exhibit a distinctive up-regulation under PA/PH (+Fe) and a diminishment under iron-deficiency. The proteins present in this cluster are a stomatin-like protein (Cre33.g782550.t1.1), a NADP malic enzyme (MME5) as well as a protein that contains a thioredoxin domain and four putative EF-hand motifs (Cre03.g202950.t1.2). According to their regulation, these proteins might be important for phototrophic growth.
An additional related subcommunity shows a characteristic and specific up-regulation under PA/PH (+Fe) and a pronounced down-regulation under PA (−Fe/+Fe) growth conditions. It is represented by five proteins which are a hydroxypuruvate reductase (HPR1), a HVA22-like regulatory protein that was shown to be abscisic acid-induced in barley (Cre17.g696850.t1.1) (68, 69), another low-CO2-inducible protein (LCIB), a protein of unknown function (Cre11.g474400.t1.1), and a carbonic anhydrase 3 (CAH3).
Proteins of another subcommunity show a uniform down-regulation under PA growth conditions (−Fe/+Fe) and exhibit a slight up-regulation in the other two growth settings. Out of seven proteins present in this community, three proteins belong to the glycine cleavage multienzyme complex (GCSH, GCST and GCSP). This complex has glycine decarboxylase activity and is required for efficient photorespiration as demonstrated by analysis of an Arabidopsis thaliana mutant devoid of glycine decarboxylase activity (70). Another photorespiratory enzyme in this cluster is the serine glyoxylate aminotransferase (Cre01.g005150.t1.1) (71). Other members of this assembly are a N-acetyl-gamma-glutamyl-phosphate reductase (ARGC1), the large subunit of carbonyl phosphate synthase (Cre28.g776100.t1.1), and a putative 10-formyltetrahydrofolate synthetase (FTHFS) (Cre13.g566000.t1.1). The latter enzyme is involved in formate dependent synthesis of 10-formyltetrahydrofolate, a precursor for 5,10-methylene tetrahydrofolate that is required for synthesis of serine and thus also linked to the photorespiratory process (72). 5,10 methylene tetrahydrofolate is also produced by glycine decarboxylase activity which is then used as the main single carbon donor for synthesis of serine (73). Yet, the synthesis via the 10-formyltetrahydrofolate synthetase provides independent 5,10 methylene tetrahydrofolate for the synthesis of serine (74). Interestingly using 13C nuclear magnetic resonance, interactions among serine hydroxymethyltransferase, tetrahydrofolate synthase, and glycine decarboxylase complex activities were shown (74). Accordingly our finding of coclustering of the glycine cleavage multienzyme complex, the serine glyoxylate aminotransferase and 10-formyltetrahydrofolate synthetase supports the assumption of coordinated action in serine biosynthesis and reveal that the process of photorespiration is active under PH (−Fe/+Fe) and PA/PH (+Fe), whereas it appears to be not active under PA (−Fe/+Fe).
Respiratory Complexes are More Severely Down-regulated Under Autotrophic Low Iron Conditions
The community XIII (blue, Fig. 3 bottom middle) consists of 12 proteins. Among those, seven proteins are subunits of mitochondrial multiprotein respiratory protein complexes (three subunits of complex I, three subunits of complex III, and one subunit of complex IV). Considering the whole cell data, subunits of the respiratory complexes are slightly induced under PH (−Fe/+Fe) and PA/PH (+Fe), but down-regulated under PA (−Fe/+Fe). Looking at the entire list of mitochondrial respiratory proteins organized in complexes I to IV as well as in ATPase in whole cells or isolated mitochondria (supplemental Fig. S4B), it appears that, except for complex II proteins and subunits of the mitochondrial ATPase, protein expression has a stronger impact under PA (−Fe/+Fe) or PA/PH (+Fe) than under iron deficient PH growth condition. The rather minor down-regulation of respiratory complexes under PH low iron availability has been already described (41) and is confirmed by the current study. However a new aspect is that autotrophic growth under low iron induces a remodeling of the respiratory machinery.
Deletion of ADH1 Attenuates the Remodeling of the Photosynthetic Machinery in Response to Iron Deficiency and Stimulates Expression of Stress-related Proteins
To substantiate our quantitative proteomic data, we investigated the up-regulation of ADH1 by immunochemical methods and analyzed the iron-deficiency response in an ADH1 deletion mutant that has been recently reported (46). To analyze the induction of ADH1 under low iron conditions and possible acclimation phenotype of the ADH1 deletion mutant in response to the iron-deficiency response we shifted wild type and mutants grown from iron-sufficient acetate-containing medium for 2 days into iron-free culture solution. Such a shift causes a pronounced remodeling of the photosynthetic complexes leading to functional uncoupling of PSI associated LHCI with PSI and diminishment of PSI (31). After shifting to iron-free medium, growth performance of wild type and ADH1-deficient cells was comparable (4.53 ± 0.46 × 106 cells/ml for WT versus 3.93 ± 0.25 × 106 cells/ml for adh1 after 2 days from the shift; Fig. 4A, upper panel). Interestingly, the decrease in chlorophyll per cell content associated with the iron-starvation response appeared to be less pronounced in the mutant devoid of ADH1 (Fig. 4A, lower panel). Low temperature fluorescence spectroscopy of wild type cells after 2 days of iron-deficient growth showed a maximum emission at 705 nm whereas iron sufficient cells peaked at 714 nm (Fig. 4B), revealing that PSI and LHCI are functionally uncoupled under low iron as described (31). Interestingly, this shift in maximal fluorescence emission is not observed in adh1 mutant cells grown in iron-free medium for 2 days. To obtain further insights into the acclimation to iron-deficiency, cells were harvested from the outlined conditions and fractionated by SDS-PAGE, immunoblotted, and analyzed by a set of different antibodies (Figs. 4C, 4D). Analysis of ATPb protein levels by anti-ATPb antibodies was used as a loading control. As expected the immunoblot data revealed for wild type a down-regulation of PSI and PSII as visualized by decreases of PSAD and PSBA in day 1 and 2 of iron deficiency, respectively. On contrary this down-regulation is less pronounced in the ADH1 deletion mutant, particularly, PsbA appears to be two-times more abundant after 2 days of iron-deficiency as compared with the wild type (Fig. 4D). Additionally down-regulation of LHCA3 is less pronounced in adh1 (Fig. 4C), which likely explains the minor blue shift of the low temperature fluorescence emission spectrum in comparison to wild type, as degradation of LHCA3 correlates with changes in low temperature fluorescence emission of PSI-LHCI (40). Interestingly, degradation of FDX1 on the other hand is paralleled in wild type and adh1 mutant. In contrast, the iron-deficiency induced expression of NDA2, LHCSR3 and PGRL1 is significantly stimulated in adh1 (Fig. 4D). Notably, iron-deficiency stimulated expression of the three proteins is also observed in the quantitative proteomics data (supplemental Fig. S4C), thereby confirming the mass spectrometric data.
DISCUSSION
In this work we compared iron deficiency responses under different metabolic conditions with data obtained from iron sufficient cells grown under PA and PH settings. Taking advantage of quantitative mass spectrometric data stemming from these distinct conditions, protein co-expression and the resulting network analyses provided new insights into coordinated remodeling of bioenergetics pathways and iron deficiency responses in C. reinhardtii and revealed interdependencies with the cellular metabolic status. Analyses of an adh1 knockout mutant suggest that the low iron regulatory acclimation networks are particularly sensitive to the chloroplast metabolic and/or redox status.
Iron Deficiency Uniformly Down-regulates Assemblies of Proteins Connected to PSI, the cyt b6f Complex and Chlorophyll Biosynthesis
A hallmark of the iron deficiency response in cyanobacteria, algae and higher plants is the down-regulation of PSI (see above). This is also reflected in community IV shown in Fig. 3, where most of its members are PSI subunits. Moreover, the down-regulation of four subunits belonging to the cyt b6f complex can be observed in community III (Fig. 3). Down-regulation of the cyt b6f complex under iron deprivation is expected (31). Nevertheless, coclustering of subunits belonging to a common multiprotein complex as already seen for PSI subunits underscores the robustness of the quantitative data and the strength of the clustering algorithm, pyGCluster. A protein that clusters with the subunits of the cyt b6f complex is CTH1. CTH1 is a diiron protein that functions as the aerobic cyclase in chlorophyll biosynthesis and is another key chloroplast target under iron deficiency (31, 63, 64, 75, 76). The diminishment of CTH1 under iron deprivation is likely responsible for the accumulation of Mg-protoporphyrin IX and Mg-protoporphyrin IX monomethyl ester under such conditions (62, 63). The regulation of CTH1 goes in line with the down-regulation of GGR, POR, PBGD1, and UROD1 that are also related to the tetrapyrole biosynthesis process. The impact in chlorophyll biosynthesis, however, has a very distinct effect on chlorophyll binding proteins and protein complexes as discussed above. Remarkably, LHCSR3 is up-regulated under PH iron deficiency (Fig. 4D; supplemental Fig. S4C) as well as other LHCII polypeptides (supplemental Fig. S4A) despite a decrease in chlorophyll biosynthesis. Thus the alteration of the abundance of chlorophyll binding proteins is more likely to be a regulatory mechanism than an answer to chlorophyll deficiency under iron deprivation in agreement with previous conclusions (31, 41).
Remodeling of Bioenergetics Pathways Under Iron-deficiency is Modulated by the Cellular Metabolic Status
As found in an earlier study (41) mitochondrial protein complexes are only marginally affected by iron deficiency in C. reinhardtii, although this impact is stronger under PA than under PH growth conditions, which is also reflected in respiratory activity (Table I, supplemental Fig. S4B). Respiratory complex I itself contains at least 26 atoms of iron (77), which is more than the entire photosynthetic machinery possesses, yet this complex as well as the other iron rich respiratory electron transfer complexes remain unaffected by iron deficiency (supplemental Fig. S4B). Down-regulation of the photosynthetic electron transfer complexes PSI as well as cyt b6f is therefore a result of a coordinated regulation. Notably, a bifunctional alcohol and acetaldehyde dehydrogenase (ADH1) is induced under iron deprivation (Fig. 3, Fig. 4C). Interestingly expression of ADH1 follows a day/night cycle and was mainly insensitive to oxygen availability ((78); Barth and Fufezan, unpublished results). A proteomic study found the chloroplast localized ADH1 to be 1.5 fold up-regulated under anaerobic conditions (47). Moreover, ADH1 is also induced under copper and zinc deficiency (79). As discussed for anoxic growth in C. reinhardtii (46), ADH1 could oxidize two molecules of NAD(P)H per acetyl-CoA under low iron availability and thereby modulate the redox poise of the chloroplast by replenishing it with NAD(P)+. The induction of cyclic photosynthetic electron flow (CEF) (42) would likewise decrease the flow of electrons into the NADPH pool, acidify the thylakoid lumen and activate energy dependent nonphotochemical quenching, going along with the described induction of LHCSR3 (80), particularly under PH settings of low iron availability (8, 41); Fig. 4D). Most interestingly deletion of ADH1 leads to an increased expression of LHCSR3, PGRL1 and NDA2 after the onset of iron deprivation as compared with wild type (Fig. 4D), indicating that the redox poise of the chloroplast in adh1 knockout strain is more reducing than in the wild type. Beside increase of PGRL1 also induction of NDA2 can be seen as a response to diminish the chloroplast redox poise, as NDA2 can lower the stromal redox poise by oxidizing NADPH and reducing plastoquinone (81). The attenuated degradation of PSI and PSII and the absence of the low iron induced low temperature fluorescence emission shift of PSI-LHCI in the ADH1 deletion mutant could be explained by the fact that stress protection in form of LHCSR3, PGRL1 and NDA2 is faster induced and therefore more protective. Alternatively, iron deficiency induced proteases could be less active and/or less induced in the adh1 mutant. On the other hand, iron deficiency induced degradation of FDX1 is unaltered when wild type and adh1 are compared (Fig. 4D), indicating that the iron deficiency response is active in the mutant. Summarizing these considerations, we can at least conclude that the deletion of ADH1 has a profound impact on the remodeling of the photosynthetic apparatus as well as in the iron dependent regulation of LHCSR3, PGRL1 and NDA2 expression. Similarly, the activation of a metabolic by-pass resulting in glycerol synthesis had been shown for the adh1 mutant placed under dark anoxic conditions. This new metabolic pathway has been suggested to compensate for the inability of adh1 to synthesize ethanol and concomitantly reoxidize NAD(P)H (46).
Table I. Photosynthetic and respiratory rates of PH and PA grown cells in two different iron concentrations. The photosynthetic rate was measured at 1000 μE/m2/s2. Standard deviation is based on biological triplicates.
| Fe [μm] | PH |
PA |
||
|---|---|---|---|---|
| Photosynthetic rate [pm O2/cell] | Respiration rate [pm O2/cell] | Photosynthetic rate [pm O2/cell] | Respiration rate [pm O2/cell] | |
| 0.1 | 0.180 ± 0.060 | −0.191 ± 0.063 | 0.119 ± 0.036 | −0.048 ± 0.007 |
| 18 | 0.601 ± 0.076 | −0.305 ± 0.037 | 1.000 ± 0.260 | −0.207 ± 0.077 |
ADH1 clustered with other metabolic enzymes namely 6-phosphogluconate dehydrogenase (GND1a) and glyceraldehyde-3-phosphate dehydrogenase (GAP1, plastidic) and shared up-regulation under low iron and down-regulation by PA/PH under iron sufficiency (Fig. 3). GND1a catalyzes the second step of the oxidative Pentose-Phosphate-Pathway (OPPP). The glycolytic enzyme GAP1 catalyzes the first step of the second phase of glycolysis and was found in vascular plants to be induced in expression under anaerobiosis and heat stress (82) as well as under iron-deficiency (83). Impaired iron availability clearly decreases photosynthetic activity, so that plants as well as algae probably enter into an anaerobic growth mode. The increased expression of GAP1 probably mirrors increased request to synthesize ATP under conditions where ATP synthesis is diminished because of decreased photosynthetic and respiratory activities. In line, it was found that specific activities of GAP and GND1 were greater in leaf tissues unable to generate reducing equivalents and ATP by photosynthesis (84). Thus induction of GAP1 and GND1 in C. reinhardtii under low iron likely reflects the necessity to generate NADH, NADPH, and ATP from the gluconeogenesis and successive oxidation of glucose. Under such circumstances ADH1 is required to oxidize NAD(P)H to allow advancement of glucose oxidizing by replenishing NAD(P)+. Its deletion, under conditions when GAP1 and GND1 are induced, on the other hand will therefore certainly increase the cellular redox poise, supporting the conclusions made above. As ADH1, GAP1, and GND1a are chloroplast localized (47), it is likely that the redox poise of the chloroplast is involved in communication with the nucleus to modulate gene expression. Interestingly, transcripts of genes encoding ADH1, GAP1, and GND1a are down-regulated by iron-deficiency (9), whereas the protein amounts are up-regulated (herein; (9)), thus strengthening our interpretation that these proteins are in the same regulatory circuit. Moreover, the divergence of transcript and protein expression may implicate post-transcriptional regulation for expression control. It is tempting to speculate that this may occur via the cytosolic translational machinery and the redox poise of the chloroplast. Besides remodeling of bioenergetics pathways, the low iron response also induced proteases, redox- and oxidative stress related enzymes.
The Iron Deficiency Response Induces Proteases, Redox- and Oxidative Stress Related Enzymes - Nonetheless Differences Between PH and PA Conditions are Evident
In the iron deficiency response of C. reinhardtii several proteases are up-regulated. This up-regulation is especially pronounced under PH conditions (Fig. 3), as seen for a putative metallopeptidase (Cre01.g023350.t1.2) and a serine-type protease (Cre10.g423300.t1.1) as well as a peptidase M (Cre20.g758550.t1.2, supplemental Fig. S3 community XVIII). Two other proteases, a cysteine protease, CEP, and an aspartic acid protease, ASP, clustered together and are also induced under PA iron-deficiency but down-regulated under PA/PH iron sufficient conditions. Notably, CEP and ASP proteases are chloroplast-localized (47). Generally plant proteases are involved in many cellular functions, including chloroplast related properties (85). However, the precise functions of the four proteases found in our protein assemblies are unknown. For strain C. reinhardtii W80, a cysteine protease was isolated that was also induced under photo-oxidative stress conditions (86). Photo-oxidative stress in the chloroplast of iron-deficient photoheterotrophic cells can be conceived, as PSI and the cytochrome b6f complex are down-regulated, whereas the PSII related light-harvesting protein complement remains stable or either increases or decreases in amount as in the case of LHCBM1 and LHCBM6/3/7 or LHCBM4, respectively (supplemental Fig. S3 community XVII). It was reported that under PH growth and iron deprivation the light-harvesting antenna was still functionally coupled to PSII, which caused significant photoinhibition (41). Thus induction of the proteases could be related to the light-induced photo-oxidative stress that is particular for iron deficient PH grown cells and potentially involved in the degradation of Fe-containing proteins (e.g. FDX1, FAB2, cytf etc.) At the same time respiratory activity of these cells, as measured by oxygen uptake activity in the dark, is only 1.5 times lower as compared with iron sufficient PH grown cells (Table I). In contrast, respiratory activity of iron deficient PA grown cells is four times lower in regard to iron sufficient cells. Importantly the oxygen evolution activity of iron-sufficient PA grown cells is increased almost twofold compared with the activity of PH grown cells, whereas iron deficient PA grown cells produce less oxygen as compared with iron deficient PH cells (Table I). Thus iron deficient PH grown cells possess a higher photosynthetic activity as well as a higher respiratory rate than iron deficient PA grown cells. Apparently, PH grown cells suffer a higher degree of photo-oxidative stress for their pronounced bioenergetic capacity. Notably, we found proteins with a flavodoxin domain (AGG3 (Cre10.g456000.t1.1) and particularly AGG4 (Cre10.g456050.t1.1)) that are highly up-regulated under PH low iron conditions (Fig. 3, supplemental Fig. S3 community XVIII). AGG3 and AGG4 were found to be enriched in the flagella proteome and the membrane-matrix fraction of C. reinhardtii (87). AGG3 was shown to be involved in mediating the orientation of the alga toward a directional light source (88). It was concluded that down-regulation of AGG3 blocks positive phototaxis (88). It is unclear what impact overexpression of AGG3 would have, also the concrete function of AGG4, a close paralog of AGG3, is currently unknown but it is tempting to speculate that increase of AGG3 and AGG4 under iron deficiency impacts the phototaxis of the alga, thereby contributing to the overall strategy to minimize photooxidative stress and optimize photosynthetic function. Interestingly, under iron deficiency, flavodoxin substitutes for ferredoxin as electron acceptor of PSI in diatoms and cyanobacteria (89–92). However, because of their localization and low sequence similarity to photosynthetic flavodoxins, AGG3 and AGG4 are rather involved in regulation of phototaxis than in photosynthesis.
Having acetate as a fueling source, the iron deprived PH grown cells produce energy via respiration and apparently invest in a costly stress response to maintain photosynthetic activity. In line with this argumentation, a Peptidyl-prolyl cis-trans isomerases (CYN37), a translation factor for expression of the chloroplast-encoded psbA gene (TBA1) (67), a cytochrome c peroxidase (CCPR1) and a thioredoxin o (TRXo) were induced solely under PH iron-deficiency and clustered together (Fig. 3, XI). Notably these proteins are also induced under PA iron plus conditions. CYN37 shares highest similarity with Arabidopsis TLP38, a luminal chloroplast protein (93). PPIases catalyze the cis-trans isomerization of the peptidyl-prolyl bond during protein folding, a rate-limiting step in this process (94). Interestingly, Vener and colleagues (95) found redox-dependent activity of chloroplast PPIase implying a link to the redox poise of the stroma and suggesting that protein folding catalysis under conditions of active photosynthesis is important. The finding of a constant up-regulation of CYN37 under iron deficiency stress or PA conditions underpins this notion. TBA1 encodes a putative oxidoreductase required for translation of the chloroplast psbA mRNA (67). Up-regulation of chloroplast localized TBA1 may reflect high turnover of the D1 protein, thereby requiring increased translation of psbA RNA. The oxidoreductase domain may connect TBA1 activity to the redox poise of the stroma and implicate a regulation similar to CYN37. The mitochondrial cytochrome c peroxidase (CCPR1) is located at the intermembrane space, reduces hydrogen peroxide to water using the electrons provided by cytochrome c and is thereby involved in scavenging of reactive oxygen species in the mitochondria (96). The mitochondrial thioredoxin o (TRXo) that is found in the same cluster is another player in this scavenging system. It acts as an electron donor to mitochondrial type II peroxiredoxin F, which has been shown to be essential for redox homeostasis and root growth of A. thaliana under stress (97). Correspondingly, the mitochondrial manganese containing superoxide dismutases (MSD1 and MSD2) are strongly induced under PH low iron, likely combating superoxide formed by the respiratory electron transport (supplemental Fig. S3 community XVIII, supplemental Fig. S4D). Beside these responses, Fig. 3 reveals other redox and oxidative stress related proteins that are up-regulated under PH and PA iron-deficiency, though up-regulation of these proteins is stronger under PH low iron. These proteins are represented by a putative glutathione S-transferase (Cre12.g526800.t1.1) and GSH1, the gamma-glutamylcysteine synthetase. This enzyme catalyzes the second step of glutathione (GSH) biosynthesis by adding glycine to the C-terminal site of γ-glutamylcysteine to form GSH (98). GSH operates as a redox buffer but contributes also to other cellular functions; in particular it participates in heavy metal detoxification and in ROS scavenging (99). GSH is involved in the generation of reduced ascorbate, another cellular redox buffer (100) and antioxidant (101), which is required e.g. for the detoxification of ROS produced by PSI (102, 103). Likewise monodehydroascorbate reductase (MDAR), which also participates in regeneration of ascorbate (104) by reducing monodehydroascorbate using NADPH, is up-regulated under PH low iron growth condition (Fig. 3). The putative glutathione S-transferase (Cre12.g526800.t1.1) might be functionally related to the glutathione S-transferase, GST1. GST1 expression is induced as an acclimation response to singlet oxygen treatment and its overexpression was sufficient to enforce resistance against singlet oxygen stress (105). Ledford and colleagues (105) discuss that GST1 could function as lipid peroxidase, thereby increasing lipid peroxidase activity of the cells, which in turn would protect against reactive oxygen species (ROS) and/or participate in signaling activity.
Interestingly another glutathione S-transferase (Cre01.g064400.t1.1) and a DegP protease (DEG5A) are induced under PA iron-deprivation (Fig. 3), indicating functional relation between these two but on the contrary distinct tasks for the one induced under PH iron deficiency.
Carbon-concentrating Mechanism, Photorespiration, and Arginine Biosynthesis are Linked Under Photoautotrophic Growth Conditions
A common motif of regulation for growth under photoautotrophic condition is the induction of carbon-concentrating mechanism (CCM) related proteins. In C. reinhardtii and other aquatic unicellular organism, CCM is induced under low CO2 availability to improve photosynthesis by increasing the concentration of intracellular inorganic carbon to elevate the CO2 concentration at the site of ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco; for reviews see (106, 107)). Rubisco catalyzes the first step of photosynthetic CO2 fixation and beside the carboxylation of its substrate ribulose-1,5-bisphosphate (RuBP), it may also oxygenate it. Oxygenation produces the toxic component 2-phosphoglycolate (2-PG), which is recycled to 3-phosphoglycerate (3-PGA) during the process of photorespiration. Consequently an increase in the CO2 to O2 ratio will decrease photorespiration. Among the proteins that are found to be up-regulated under PA conditions compared with PH (communities VI and XI, Fig. 3) and found in a shared cluster are LCIC and LCIB. Studies of mRNA expression in response to low CO2 revealed that transcripts of lcib and lcic are among the most induced mRNA molecules recorded (108, 109). Genetic analyses indicated that LCIB is involved in the accumulation of inorganic carbon in the chloroplast (108) and that correspondingly to their co-expression, LCIB and LCIC form a 350 kDa hexameric complex in the chloroplast (110). Induction of CCM proteins is also strongly supported by the fact that CAH3 (for review see (106, 107) is significantly increased under iron sufficient PA settings. In accordance, LCIB/LCIC and CAH3 were found in CCM clusters when transcriptome-wide changes in relation to CO2 and the CO2-CCM regulators CIA5/CCM1 were analyzed (111). Despite the fact that C. reinhardtii possesses a carbon-concentrating mechanism, photorespiratory products have been detected after shifting cells to low CO2 by using gas chromatography-coupled time of flight mass spectrometry (112). Accordingly, transcript levels for the photorespiratory enzymes, including HRP1, were increased in low CO2 grown cells (109, 111). HPR1 is involved in photorespiration as explained above, and is found in a coregulated community with LCIB and LCIC (Fig. 3, community XI). Thus a close connection between photorespiration and the CCM can be interfered as already suggested (109, 111). Coclustering of the glycine cleavage multienzyme complex, the serine glyoxylate aminotransferase and 10-formyltetrahydrofolate synthetase, as suggested above, indicates coordinated action in serine biosynthesis and supports that photorespiration is more active under PH low iron and PA/PH as compared with PH low iron. Other members of this assembly are a N-acetyl-gamma-glutamyl-phosphate reductase (ARGC1) and the large subunit of carbonyl phosphate synthase (Cre28.g776100.t1.1) (CPS), two enzymes that are involved in arginine biosynthesis. It is of note that these enzyme are all found in the chloroplast proteome of C. reinhardtii (47) and Arabidopsis thaliana (113, 114). Knock-out of the small or large subunit of CPS resulted in a reticulate leaf phenotype that was correlated with a defect in mesophyll development (115), implicating that CPS function and arginine may play a role in these developmental processes. CPS is involved in the conversion of glutamine and bicarbonate into carbamoyl phosphate (CP) and glutamate (116). CP formation is the first committed step in the biosynthesis of arginine and in de novo biosynthesis of pyrimidines (117). Because CPS, some CCM proteins, HPR1, three subunits of the glycine cleavage multienzyme complex, the serine glyoxylate aminotransferase and 10-formyltetrahydrofolate synthetase are coregulated, the question arises about the connection between these biological processes and the arginine and pyrimidine biosynthesis. During photorespiration, the formation of glycine from glyoxylate via the glutamate:glyoxylate aminotransferase (GGT) requires glutamate as a cosubstrate (71). Thus the creation of CP as a committed step in the arginine biosynthetic pathway could participate in glutamate production as substrate for the photorespiratory process, linking thereby arginine biosynthesis and photorespiration. This goes hand in hand with the observed increase in 2-oxogluterate, which is produced via GGT action induced after shifting C. reinhardtii cells into low CO2 conditions (112), whereas glutamate is decreased because it is steadily consumed. Notably arginine accumulates under stress in A. thaliana (118, 119) and is considered an important intermediate for nitrogen storage (120). In line our data suggest that arginine biosynthesis, as well as photorespiration, is characteristic for cells actively performing photosynthesis.
Photoautotrophic Growth Regulation May Require Calcium and Redox Signaling
Beside proteins involved in CCM, photorespiration or arginine biosynthesis as presented in community XI, other proteins were particularly up-regulated under PA iron sufficient conditions (Fig. 3). These proteins are a stomatin-like protein (Cre33.g782550.t1.1), a protein that contains a thioredoxin domain and four putative EF-hand motifs (Cre03.g202950.t1.2) as well as a NADP malic enzyme (MME5). MME5 is a chloroplast localized NADP-malic enzyme (61) that either operates in decarboxylating malate to pyruvate or in catalyzing the carboxylation of pyruvate to malate thereby generating NADPH or oxidizing it to NADP+. As the direction is dependent on the chloroplast redox poise, the enzyme could thereby be important for the fine-tuning of the chloroplast redox status.
In mammalians, stomatin and its paralogs are described, as homo-oligomeric, lipid raft-associated, integral membrane proteins that may interact with and modulate various ion channels and transporters (121). More particularly, stomatin was found to interact with a Ca2+ ATPase (122). However, whether a link between C. reinhardtii stomatin and calcium exists remains to be shown.
Four EF hands domains were found in Cre03.g202950.t1.2. EF hands present a common motif for proteins involved in the binding of Ca2+. The EF is a helix-loop-helix motif, which mostly occurs in pairs (not identical; up to 6 pairs in one protein), allowing high affinity binding to Ca2+. The pairing is important for the Ca2+-binding mechanism and Ca2+-induced conformational change, translating the Ca2+concentration into an output response (123). EF-hand containing proteins operate in all aspects of cell function and more than 3000 EF-hand related entries can be found in the NCBI RefSeq database. Likewise thioredoxin domains are contained in many proteins and enzymatically thioredoxin acts as a protein disulphideoxidoreductase that catalyzes the reversible oxidation of two cysteine thiol groups to a disulfide, thereby transferring two electrons and two protons (124). Thus combining EF-hand motifs with a thioredoxin domain would link calcium and redox signaling. From network analyses it distinctly appears that thioredoxins are central to chloroplast redox signaling (125). Recent data revealed that chloroplast calcium signaling may also contribute to retrograde signaling between chloroplast and the nucleus. In C. reinhardtii, the expression of the nuclear encoded LHCSR3 is regulated via the thylakoid membrane protein CAS (Ca2+-sensing receptor) and Ca2+ (126). In A. thaliana, CAS is involved in chloroplast mediated activation of plant immune signaling (127) and in stomatal regulation in response to elevations of external Ca2+ through the modulation of cytoplasmic Ca2+ dynamics (128, 129), thereby regulating the availability of carbon dioxide and thus indirectly also the redox poise of the chloroplast stroma. Importantly, CAS has been also implicated in regulation of cyclic electron transfer (130). Here consistently a link between redox poise and calcium can be anticipated.
Notably the induction of a Peptidyl-prolyl cis-trans isomerase (CYN37) and a redox-related translation factor for expression of the chloroplast-encoded psbA gene (TBA1) (67), under PA/PH conditions, as already discussed for PH iron deficiency, support the conclusion that photoautotrophic growth regulation is related to the chloroplast redox poise.
Supplementary Material
Acknowledgments
We thank Dr. Y. Takahashi and Dr. Eugen Urzica for fruitful comments and lively discussions.
Footnotes
* M.H. and C.F. acknowledge support from the Deutsche Forschungsgemeinschaft. M.H. also acknowledges support from the FP7-funded Sunbiopath Project (GA245070). L.M. acknowledges support from the Alexander von Humboldt Stiftung/Foundation during his stay at the Institute of Plant Biology and Biotechnology (University of Münster, Germany). A.R.G. acknowledges support from the Department of Energy, Grant No. DE-FG02-12ER16338.
 This article contains supplemental Figs. S1 to S4 and Tables S1 to S3.
This article contains supplemental Figs. S1 to S4 and Tables S1 to S3.
Authors declare no competing commercial interests.
1 The abbreviations used are:
- ADH1
- bifunctional alcohol/acetaldehyde dehydrogenase
- AGG3/AGG4
- flagellar associated protein
- ARGC1
- N-acetyl-gamma-glutamyl-phosphate reductase
- ASP
- aspartic-type endopeptidase
- C
- chloroplast
- CAH3
- carbonic anhydrase 3
- CAS
- Ca2+-sensing receptor
- CCPR1
- cytochrome c peroxidase
- CCM
- carbon-concentrating mechanism
- CCM1/ CIA5
- carbon-concentrating mechanism protein 1
- CEF
- cyclic photosynthetic electron flow
- CEP
- serine carboxypeptidase
- CP
- carbamoyl phosphate
- CPS
- carbonyl phosphate synthase
- CTH1
- copper target homolog 1
- CYN37
- candidate peptidyl-prolyl cis-trans isomerase
- cyt b6f
- cytochrome b6f complex
- DEG5A
- DegP protease
- DRP1
- dynamin related GTPase
- FAB2
- plastid acyl-ACP desaturase
- FDX1
- ferredoxin 1
- FTHFS
- putative 10-formyltetrahydrofolate synthetase
- FTSH1
- AAA-metalloprotease
- GAP1
- glyceraldehyde-3-phosphate dehydrogenase
- GCSH
- GCST, GCSP, glycine cleavage multienzyme complex proteins
- GGR
- geranyl-geranylreductase
- GGT
- glutamate:glyoxylate aminotransferase
- GND1a
- 6-phosphogluconate dehydrogenase
- GSH
- glutathione
- GSH1
- gamma-glutamylcysteine synthetase
- HPR1
- hydroxypuruvate reductase
- HSM
- high salt medium
- IRT1
- iron regulatory protein 1
- ISIA
- iron-stress-induced protein A
- Lhcb
- light-harvesting protein b
- LHCBM
- major light-harvesting protein b
- LHCI
- light-harvesting antenna I
- LHCII
- light-harvesting antenna II
- LCIB
- low-CO2-inducible protein B
- LCIC
- low-CO2-inducible protein C
- LHCSR3
- light harvesting complex stress related 3
- Lhl3
- light-harvesting-like protein LIL
- M
- mitochondrium
- MDAR
- monodehydroascorbate reductase
- MME5
- NADP malic enzyme
- MPC1
- mitochondrial phosphate carrier 1
- MSD1
- MSD2, mitochondrial manganese containing superoxide dismutases
- NCBI
- National Center for Biotechnology Information
- NDA2
- platoquinon-reducing type II NAD(P)H dehydrogenase
- OPPP
- oxidative Pentose-Phosphate-Pathway
- PA
- photoautotrophic
- PBGD1
- porphobilinogen deaminase
- PGRL1
- Proton Gradient Regulation Like1 protein
- PH
- photohetrotrophic
- POR
- protochlorophyllide reductase
- PPIase
- peptidylprolyl isomerase
- PsaA
- B, D, E, F, L, H, photosystem I complex subunits
- PsbA
- photosystem II complex subunit A
- PSI
- photosystem I
- PSII
- photosystem II
- ROS
- reactive oxygen species; Rubisco ribulose-1,5-bisphosphate carboxylase/oxygenase
- SEP1
- septin, STA2, granule bound starch synthase I
- std
- standard deviation
- TAP
- Tris-Acetate-Phosphate
- TBA1
- translation factor for expression of the chloroplast-encoded psbA gene
- TLP38
- Arabidopsis lumenal chloroplast protein
- TRXo
- thioredoxin o
- UROD1
- uroporphyrinogen-III decarboxylase
- VDAC
- voltage-dependent anion-selective channel
- WC
- whole cells.
REFERENCES
- 1. Merchant S. S., Prochnik S. E., Vallon O., Harris E. H., Karpowicz S. J., Witman G. B., Terry A., Salamov A., Fritz-Laylin L. K., Marechal-Drouard L., Marshall W. F., Qu L. H., Nelson D. R., Sanderfoot A. A., Spalding M. H., Kapitonov V. V., Ren Q., Ferris P., Lindquist E., Shapiro H., Lucas S. M., Grimwood J., Schmutz J., Cardol P., Cerutti H., Chanfreau G., Chen C. L., Cognat V., Croft M. T., Dent R., Dutcher S., Fernandez E., Fukuzawa H., Gonzalez-Ballester D., Gonzalez-Halphen D., Hallmann A., Hanikenne M., Hippler M., Inwood W., Jabbari K., Kalanon M., Kuras R., Lefebvre P. A., Lemaire S. D., Lobanov A. V., Lohr M., Manuell A., Meier I., Mets L., Mittag M., Mittelmeier T., Moroney J. V., Moseley J., Napoli C., Nedelcu A. M., Niyogi K., Novoselov S. V., Paulsen I. T., Pazour G., Purton S., Ral J. P., Riano-Pachon D. M., Riekhof W., Rymarquis L., Schroda M., Stern D., Umen J., Willows R., Wilson N., Zimmer S. L., Allmer J., Balk J., Bisova K., Chen C. J., Elias M., Gendler K., Hauser C., Lamb M. R., Ledford H., Long J. C., Minagawa J., Page M. D., Pan J., Pootakham W., Roje S., Rose A., Stahlberg E., Terauchi A. M., Yang P., Ball S., Bowler C., Dieckmann C. L., Gladyshev V. N., Green P., Jorgensen R., Mayfield S., Mueller-Roeber B., Rajamani S., Sayre R. T., Brokstein P., Dubchak I., Goodstein D., Hornick L., Huang Y. W., Jhaveri J., Luo Y., Martinez D., Ngau W. C., Otillar B., Poliakov A., Porter A., Szajkowski L., Werner G., Zhou K., Grigoriev I. V., Rokhsar D. S., Grossman A. R. (2007) The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318, 245–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bassham J. A., Benson A. A., Calvin M. (1950) The path of carbon in photosynthesis. J. Biol. Chem. 185, 781–787 [PubMed] [Google Scholar]
- 3. Kornberg H. L., Krebs H. A. (1957) Synthesis of cell constituents from C2-units by a modified tricarboxylic acid cycle. Nature 179, 988–991 [DOI] [PubMed] [Google Scholar]
- 4. Heifetz P. B., Forster B., Osmond C. B., Giles L. J., Boynton J. E. (2000) Effects of acetate on facultative autotrophy in Chlamydomonas reinhardtii assessed by photosynthetic measurements and stable isotope analyses. Plant Physiol. 122, 1439–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Matsuo M., Hachisu R., Tabata S., Fukuzawa H., Obokata J. (2011) Transcriptome analysis of respiration-responsive genes in Chlamydomonas reinhardtii: mitochondrial retrograde signaling coordinates the genes for cell proliferation with energy-producing metabolism. Plant Cell Physiol. 52, 333–343 [DOI] [PubMed] [Google Scholar]
- 6. Nogales J., Guijo M. I., Quesada A., Merchan F. (2004) Functional analysis and regulation of the malate synthase from Chlamydomonas reinhardtii. Planta 219, 325–331 [DOI] [PubMed] [Google Scholar]
- 7. Busch A., Rimbauld B., Naumann B., Rensch S., Hippler M. (2008) Ferritin is required for rapid remodeling of the photosynthetic apparatus and minimizes photo-oxidative stress in response to iron availability in Chlamydomonas reinhardtii. Plant J. 55, 201–211 [DOI] [PubMed] [Google Scholar]
- 8. Terauchi A. M., Peers G., Kobayashi M. C., Niyogi K. K., Merchant S. S. (2010) Trophic status of Chlamydomonas reinhardtii influences the impact of iron deficiency on photosynthesis. Photosynth. Res. 105, 39–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Urzica E. I., Casero D., Yamasaki H., Hsieh S. I., Adler L. N., Karpowicz S. J., Blaby-Haas C. E., Clarke S. G., Loo J. A., Pellegrini M., Merchant S. S. (2012) Systems and trans-system level analysis identifies conserved iron deficiency responses in the plant lineage. Plant Cell 24, 3921–3948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Castruita M., Casero D., Karpowicz S. J., Kropat J., Vieler A., Hsieh S. I., Yan W., Cokus S., Loo J. A., Benning C., Pellegrini M., Merchant S. S. (2011) Systems biology approach in Chlamydomonas reveals connections between copper nutrition and multiple metabolic steps. Plant Cell 23, 1273–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Napier I., Ponka P., Richardson D. R. (2005) Iron trafficking in the mitochondrion: novel pathways revealed by disease. Blood 105, 1867–1874 [DOI] [PubMed] [Google Scholar]
- 12. Ackrell B. A., Maguire J. J., Dallman P. R., Kearney E. B. (1984) Effect of iron deficiency on succinate- and NADH-ubiquinone oxidoreductases in skeletal muscle mitochondria. J. Biol. Chem. 259, 10053–10059 [PubMed] [Google Scholar]
- 13. Pascal N., Douce R. (1993) Effect of Iron Deficiency on the Respiration of Sycamore (Acer pseudoplatanus L.) Cells. Plant Physiol. 103, 1329–1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Onishi T., Asakura T., Yonetani T., Chance B. (1971) Electron paramagnetic resonance studies at temperatures below 77 degrees K on iron-sulfur proteins of yeast and bovine heart submitochondrial particles. J. Biol. Chem. 246, 5960–5964 [PubMed] [Google Scholar]
- 15. Winder T. L., Nishio J. N. (1995) Early iron deficiency stress response in leaves of sugar beet. Plant Physiol. 108, 1487–1494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Machold O. (1971) Lamellar proteins of green and chlorotic chloroplasts as affected by iron deficiency and antibiotics. Biochim. Biophys. Acta 238, 324–331 [DOI] [PubMed] [Google Scholar]
- 17. Nishio J. N., Taylor S. E., Terry N. (1985) Changes in Thylakoid Galactolipids and Proteins during Iron Nutrition-Mediated Chloroplast Development. Plant Physiol. 77, 705–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Taylor S. E., Terry N., Huston R. P. (1982) Limiting Factors in Photosynthesis. III Effects of Iron Nutrition on the Activities of Three Regulatory Enzymes of Photosynthetic Carbon Metabolism. Plant Physiol. 70, 1541–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Terry N. (1983) Limiting factors in photosynthesis. IV. Iron stress-mediated changes on light-harvesting and electron transport capacity and its effects on photosynthesis in vivo. Plant Physiol. 71, 855–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Terry N. (1980) Limiting Factors in Photosynthesis. I. Use of Iron Stress to Control Photosynthetic Capacity In Vivo. Plant Physiol. 65, 114–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nenova V., Stoyanov I. (1993) Physiological and Biochemical Changes in Young Maize Plants Under Iron Deficiency. I. Growth and Photosynthesis. J. Plant Nutr. 16, 835–849 [Google Scholar]
- 22. Palmer C. M., Guerinot M. L. (2009) Facing the challenges of Cu, Fe and Zn homeostasis in plants. Nat. Chem. Biol. 5, 333–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. La Fontaine S., Quinn J. M., Nakamoto S. S., Page M. D., Gohre V., Moseley J. L., Kropat J., Merchant S. (2002) Copper-dependent iron assimilation pathway in the model photosynthetic eukaryote Chlamydomonas reinhardtii. Eukaryot Cell 1, 736–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Henriques R., Jasik J., Klein M., Martinoia E., Feller U., Schell J., Pais M. S., Koncz C. (2002) Knock-out of Arabidopsis metal transporter gene IRT1 results in iron deficiency accompanied by cell differentiation defects. Plant Mol. Biol. 50, 587–597 [DOI] [PubMed] [Google Scholar]
- 25. Varotto C., Maiwald D., Pesaresi P., Jahns P., Salamini F., Leister D. (2002) The metal ion transporter IRT1 is necessary for iron homeostasis and efficient photosynthesis in Arabidopsis thaliana. Plant J. 31, 589–599 [DOI] [PubMed] [Google Scholar]
- 26. Vert G., Grotz N., Dedaldechamp F., Gaymard F., Guerinot M. L., Briat J. F., Curie C. (2002) IRT1, an Arabidopsis Transporter Essential for Iron Uptake from the Soil and for Plant Growth. Plant Cell 14, 1223–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Guerinot M. L., Salt D. E. (2001) Fortified Foods and Phytoremediation. Two Sides of the Same Coin. Plant Physiol. 125, 164–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Murray-Kolb L. E., Takaiwa F., Goto F., Yoshihara T., Theil E. C., Beard J. L. (2002) Transgenic rice is a source of iron for iron-depleted rats. J. Nutr. 132, 957–960 [DOI] [PubMed] [Google Scholar]
- 29. Martin J. H., Fitzwater S. E. (1988) Iron deficiency limits phytoplankton growth in the north-east Pacific subarctic. Nature 331, 341–343 [Google Scholar]
- 30. Behrenfeld M. J., Kolber Z. S. (1999) Widespread iron limitation of phytoplankton in the south pacific ocean. Science 283, 840–843 [DOI] [PubMed] [Google Scholar]
- 31. Moseley J. L., Allinger T., Herzog S., Hoerth P., Wehinger E., Merchant S., Hippler M. (2002) Adaptation to Fe-deficiency requires remodeling of the photosynthetic apparatus. EMBO J.. 21, 6709–6720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Desquilbet T. E., Duval J. C., Robert B., Houmard J., Thomas J. C. (2003) In the unicellular red alga Rhodella violacea iron deficiency induces an accumulation of uncoupled LHC. Plant Cell Physiol. 44, 1141–1151 [DOI] [PubMed] [Google Scholar]
- 33. Doan J. M., Schoefs B., Ruban A. V., Etienne A. L. (2003) Changes in the LHCI aggregation state during iron repletion in the unicellular red alga Rhodella violacea. FEBS Lett. 533, 59–62 [DOI] [PubMed] [Google Scholar]
- 34. Guikema J. A. a. S., Sherman L. A. (1983) Influence of iron deprivation on the membrane composition of Anacystis nidulans. Plant Physiol. 74, 90–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Laudenbach D. E., Straus N. A. (1988) Characterization of a cyanobacterial iron stress-induced gene similar to psbC. J. Bacteriol 170, 5018–5026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Burnap R. L., Troyan T., Sherman L. A. (1993) The highly abundant chlorophyll-protein complex of iron-deficient Synechococcus sp. PCC7942 (CP43′) is encoded by the isiA gene. Plant Physiol. 103, 893–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Boekema E. J., Hifney A., Yakushevska A. E., Piotrowski M., Keegstra W., Berry S., Michel K. P., Pistorius E. K., Kruip J. (2001) A giant chlorophyll-protein complex induced by iron deficiency in cyanobacteria. Nature 412, 745–748 [DOI] [PubMed] [Google Scholar]
- 38. Bibby T. S., Nield J., Barber J. (2001) Iron deficiency induces the formation of an antenna ring around trimeric photosystem I in cyanobacteria. Nature 412, 743–745 [DOI] [PubMed] [Google Scholar]
- 39. Lommer M., Specht M., Roy A. S., Kraemer L., Andreson R., Gutowska M. A., Wolf J., Bergner S. V., Schilhabel M. B., Klostermeier U. C., Beiko R. G., Rosenstiel P., Hippler M., Laroche J. (2012) Genome and low-iron response of an oceanic diatom adapted to chronic iron limitation. Genome Biol. 13, R66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Naumann B., Stauber E. J., Busch A., Sommer F., Hippler M. (2005) N-terminal processing of Lhca3 Is a key step in remodeling of the photosystem I-light-harvesting complex under iron deficiency in Chlamydomonas reinhardtii. J. Biol. Chem. 280, 20431–20441 [DOI] [PubMed] [Google Scholar]
- 41. Naumann B., Busch A., Allmer J., Ostendorf E., Zeller M., Kirchhoff H., Hippler M. (2007) Comparative quantitative proteomics to investigate the remodeling of bioenergetic pathways under iron deficiency in Chlamydomonas reinhardtii. Proteomics 7, 3964–3979 [DOI] [PubMed] [Google Scholar]
- 42. Petroutsos D., Terauchi A. M., Busch A., Hirschmann I., Merchant S. S., Finazzi G., Hippler M. (2009) PGRL1 participates in iron-induced remodeling of the photosynthetic apparatus and in energy metabolism in Chlamydomonas reinhardtii. J. Biol. Chem. 284, 32770–32781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Murchie E. H., Pinto M., Horton P. (2009) Agriculture and the new challenges for photosynthesis research. New Phytol. 181, 532–552 [DOI] [PubMed] [Google Scholar]
- 44. Murchie E. H., Niyogi K. K. (2011) Manipulation of photoprotection to improve plant photosynthesis. Plant Physiol. 155, 86–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Harris E. H. (1989) The Chlamydomonas sourcebook. A comprehensive guide to biology and laboratory use., Academic Press, San Diego: [DOI] [PubMed] [Google Scholar]
- 46. Magneschi L., Catalanotti C., Subramanian V., Dubini A., Yang W., Mus F., Posewitz M. C., Seibert M., Perata P., Grossman A. R. (2012) A mutant in the ADH1 gene of Chlamydomonas reinhardtii elicits metabolic restructuring during anaerobiosis. Plant Physiol. 158, 1293–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Terashima M., Specht M., Naumann B., Hippler M. (2010) Characterizing the anaerobic response of Chlamydomonas reinhardtii by quantitative proteomics. Mol. Cell. Proteomics 9, 1514–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Martens L., Chambers M., Sturm M., Kessner D., Levander F., Shofstahl J., Tang W. H., Rompp A., Neumann S., Pizarro A. D., Montecchi-Palazzi L., Tasman N., Coleman M., Reisinger F., Souda P., Hermjakob H., Binz P. A., Deutsch E. W. (2011) mzML–a community standard for mass spectrometry data. Mol. Cell. Proteomics 10, R110 000133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chambers M. C., Maclean B., Burke R., Amodei D., Ruderman D. L., Neumann S., Gatto L., Fischer B., Pratt B., Egertson J., Hoff K., Kessner D., Tasman N., Shulman N., Frewen B., Baker T. A., Brusniak M. Y., Paulse C., Creasy D., Flashner L., Kani K., Moulding C., Seymour S. L., Nuwaysir L. M., Lefebvre B., Kuhlmann F., Roark J., Rainer P., Detlev S., Hemenway T., Huhmer A., Langridge J., Connolly B., Chadick T., Holly K., Eckels J., Deutsch E. W., Moritz R. L., Katz J. E., Agus D. B., MacCoss M., Tabb D. L., Mallick P. (2012) A cross-platform toolkit for mass spectrometry and proteomics. Nature Biotechnol. 30, 918–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Specht M., Kuhlgert S., Fufezan C., Hippler M. (2011) Proteomics to go: Proteomatic enables the user-friendly creation of versatile MS/MS data evaluation workflows. Bioinformatics 27, 1183–1184 [DOI] [PubMed] [Google Scholar]
- 51. Bald T., Barth J., Niehues A., Specht M., Hippler M., Fufezan C. (2012) pymzML–Python module for high-throughput bioinformatics on mass spectrometry data. Bioinformatics 28, 1052–1053 [DOI] [PubMed] [Google Scholar]
- 52. Geer L. Y., Markey S. P., Kowalak J. A., Wagner L., Xu M., Maynard D. M., Yang X., Shi W., Bryant S. H. (2004) Open mass spectrometry search algorithm. J. Proteome Res. 3, 958–964 [DOI] [PubMed] [Google Scholar]
- 53. Craig R., Beavis R. C. (2004) TANDEM: matching proteins with tandem mass spectra. Bioinformatics 20, 1466–1467 [DOI] [PubMed] [Google Scholar]
- 54. Kall L., Storey J. D., Noble W. S. (2009) QVALITY: non-parametric estimation of q-values and posterior error probabilities. Bioinformatics 25, 964–966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Mueller L. N., Rinner O., Schmidt A., Letarte S., Bodenmiller B., Brusniak M. Y., Vitek O., Aebersold R., Muller M. (2007) SuperHirn - a novel tool for high resolution LC-MS-based peptide/protein profiling. Proteomics 7, 3470–3480 [DOI] [PubMed] [Google Scholar]
- 56. van Rossum G., Python tutorial, Technical Report CS-R9526, Centrum voor Wiskunde en Informatica (CWI), Amsterdam, May 1995 [Google Scholar]
- 57. Dubois Paul F., Konrad Hinsen, James Hugunin. (1996) Numerical Python, Computers in Physics 10 [Google Scholar]
- 58. Jones E., Oliphant T., Peterson P. (2001) SciPy - Open source scientific tools for Python. [Google Scholar]
- 59. Hagberg A. A., Schult D. A., Swart P. J. (2008) Exploring network structure, dynamics, and function using NetworkX. In: Gäel Varoquaux T. V., Jarrod Millman. (Eds), ed. Proceedings of the 7th Python in Science Conference (SciPy2008), Pasadena, CA U.S.A [Google Scholar]
- 60. Bastian M., Heymann S., Jacomy M. (2009) Gephi: an open source software for exploring and manipulating networks. International AAAI Conference on Weblogs and Social Media May 17–20, San Jose, California, USA [Google Scholar]
- 61. Terashima M., Specht M., Hippler M. (2011) The chloroplast proteome: a survey from the Chlamydomonas reinhardtii perspective with a focus on distinctive features. Current Genetics 57, 151–168 [DOI] [PubMed] [Google Scholar]
- 62. Spiller S. C., Castelfranco A. M., Castelfranco P. A. (1982) Effects of Iron and Oxygen on Chlorophyll Biosynthesis : I. In Vivo observations on iron and oxygen-deficient plants. Plant Physiol. 69, 107–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Chereskin B. M., Castelfranco P. A. (1982) Effects of Iron and Oxygen on Chlorophyll Biosynthesis : II. Observations on the biosynthetic pathway in isolated etiochloroplasts. Plant Physiol. 69, 112–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Pinta V., Picaud M., Reiss-Husson F., Astier C. (2002) Rubrivivax gelatinosus acsF (previously orf358) codes for a conserved, putative binuclear-iron-cluster-containing protein involved in aerobic oxidative cyclization of Mg-protoporphyrin IX monomethylester. J. Bacteriol. 184, 746–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Moseley J., Quinn J., Eriksson M., Merchant S. (2000) The Crd1 gene encodes a putative di-iron enzyme required for photosystem I accumulation in copper deficiency and hypoxia in Chlamydomonas reinhardtii. EMBO J. 19, 2139–2151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. van de Velde H. J., Roebroek A. J., Senden N. H., Ramaekers F. C., Van de Ven W. J. (1994) NSP-encoded reticulons, neuroendocrine proteins of a novel gene family associated with membranes of the endoplasmic reticulum. J. Cell Sci. 107 (Pt 9), 2403–2416 [DOI] [PubMed] [Google Scholar]
- 67. Somanchi A., Barnes D., Mayfield S. P. (2005) A nuclear gene of Chlamydomonas reinhardtii, Tba1, encodes a putative oxidoreductase required for translation of the chloroplast psbA mRNA. Plant J. 42, 341–352 [DOI] [PubMed] [Google Scholar]
- 68. Shen Q., Uknes S. J., Ho T. H. (1993) Hormone response complex in a novel abscisic acid and cycloheximide-inducible barley gene. J. Biol. Chem. 268, 23652–23660 [PubMed] [Google Scholar]
- 69. Shen Q., Chen C. N., Brands A., Pan S. M., Ho T. H. (2001) The stress- and abscisic acid-induced barley gene HVA22: developmental regulation and homologues in diverse organisms. Plant Mol. Biol. 45, 327–340 [DOI] [PubMed] [Google Scholar]
- 70. Somerville C. R., Ogren W. L. (1982) Mutants of the cruciferous plant Arabidopsis thaliana lacking glycine decarboxylase activity. Biochem. J. 202, 373–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Bauwe H., Hagemann M., Fernie A. R. (2010) Photorespiration: players, partners and origin. Trends Plant Sci. 15, 330–336 [DOI] [PubMed] [Google Scholar]
- 72. Shingles R., Woodrow L., Grodzinski B. (1984) Effects of Glycolate Pathway Intermediates on Glycine Decarboxylation and Serine Synthesis in Pea (Pisum sativum L.). Plant Physiol. 74, 705–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Rebeille F., Neuburger M., Douce R. (1994) Interaction between glycine decarboxylase, serine hydroxymethyltransferase and tetrahydrofolate polyglutamates in pea leaf mitochondria. Biochem. J. 302 (Pt 1), 223–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Prabhu V., Chatson K. B., Abrams G. D., King J. (1996) 13C nuclear magnetic resonance detection of interactions of serine hydroxymethyltransferase with C1-tetrahydrofolate synthase and glycine decarboxylase complex activities in Arabidopsis. Plant Physiol. 112, 207–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Tottey S., Block M. A., Allen M., Westergren T., Albrieux C., Scheller H. V., Merchant S., Jensen P. E. (2003) Arabidopsis CHL27, located in both envelope and thylakoid membranes, is required for the synthesis of protochlorophyllide. Proc. Natl. Acad. Sci. U.S.A. 100, 16119–16124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Moseley J. L., Page M. D., Pergam N., Eriksson M., Quinn J., Soto J., Theg S., Hippler M., Merchant S. (2002) Reciprocal expression of two di-iron enzymes affecting photosystem I and light-harvesting complex accumulation. Plant Cell 14, 673–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Brandt U. (2006) Energy converting NADH:quinone oxidoreductase (complex I). Annu. Rev. Biochem. 75, 69–92 [DOI] [PubMed] [Google Scholar]
- 78. Whitney L. A., Loreti E., Alpi A., Perata P. (2011) Alcohol dehydrogenase and hydrogenase transcript fluctuations during a day-night cycle in Chlamydomonas reinhardtii: the role of anoxia. New Phytol. 190, 488–498 [DOI] [PubMed] [Google Scholar]
- 79. Hsieh S. I., Castruita M., Malasarn D., Urzica E., Erde J., Page M. D., Yamasaki H., Casero D., Pellegrini M., Merchant S. S., Loo J. A. (2013) The proteome of copper, iron, zinc, and manganese micronutrient deficiency in Chlamydomonas reinhardtii. Mol. Cell. Proteomics 12, 65–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Peers G., Truong T. B., Ostendorf E., Busch A., Elrad D., Grossman A. R., Hippler M., Niyogi K. K. (2009) An ancient light-harvesting protein is critical for the regulation of algal photosynthesis. Nature 462, 518–521 [DOI] [PubMed] [Google Scholar]
- 81. Jans F., Mignolet E., Houyoux P. A., Cardol P., Ghysels B., Cuine S., Cournac L., Peltier G., Remacle C., Franck F. (2008) A type II NAD(P)H dehydrogenase mediates light-independent plastoquinone reduction in the chloroplast of Chlamydomonas. Proc. Natl. Acad. Sci. U.S.A. 105, 20546–20551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Russell D. A., Sachs M. M. (1989) Differential Expression and Sequence-Analysis of the Maize Glyceraldehyde-3-Phosphate Dehydrogenase Gene Family. Plant Cell 1, 793–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Herbik A., Giritch A., Horstmann C., Becker R., Balzer H. J., Baumlein H., Stephan U. W. (1996) Iron and copper nutrition-dependent changes in protein expression in a tomato wild type and the nicotianamine-free mutant chloronerva. Plant Physiol. 111, 533–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Wurtele E. S., Nikolau B. J. (1986) Enzymes of Glucose Oxidation in Leaf Tissues : The Distribution of the Enzymes of Glycolysis and the Oxidative Pentose Phosphate Pathway between Epidermal and Mesophyll Tissues of C(3)-Plants and Epidermal, Mesophyll, and Bundle Sheath Tissues of C(4)-Plants. Plant Physiol. 82, 503–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. van der Hoorn R. A. (2008) Plant proteases: from phenotypes to molecular mechanisms. Ann. Rev. Plant Biol. 59, 191–223 [DOI] [PubMed] [Google Scholar]
- 86. Usui M., Tanaka S., Miyasaka H., Suzuki Y., Shioi Y. (2007) Characterization of cysteine protease induced by oxidative stress in cells of Chlamydomonas sp. strain W80. Physiol. Plant. 131, 519–526 [DOI] [PubMed] [Google Scholar]
- 87. Pazour G. J., Agrin N., Leszyk J., Witman G. B. (2005) Proteomic analysis of a eukaryotic cilium. J. Cell Biol. 170, 103–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Iomini C., Li L., Mo W., Dutcher S. K., Piperno G. (2006) Two flagellar genes, AGG2 and AGG3, mediate orientation to light in Chlamydomonas. Current Biol. 16, 1147–1153 [DOI] [PubMed] [Google Scholar]
- 89. LaRoche J., Boyd P. W., McKay R. M. L., Geider R. J. (1996) Flavodoxin as an in situ marker for iron stress in phytoplankton. Nature 382, 802–805 [Google Scholar]
- 90. Leonhardt K., Straus N. A. (1992) An iron stress operon involved in photosynthetic electron transport in the marine cyanobacterium Synechococcus sp. PCC 7002. J. Gen. Microbiol. 8, 1613–1621 [DOI] [PubMed] [Google Scholar]
- 91. Straus N. A. (1994) Iron deprivation: physiology and gene regulation, Kluwer Academic Publisher, Dordrecht, The Netherlands [Google Scholar]
- 92. Ghassemian M., Straus N. A. (1996) Fur regulates the expression of iron-stress genes in the cyanobacterium Synechococcus sp. strain PCC 7942. Microbiology 142, 1469–1476 [DOI] [PubMed] [Google Scholar]
- 93. Schubert M., Petersson U. A., Haas B. J., Funk C., Schroder W. P., Kieselbach T. (2001) Proteome map of the chloroplast lumen of Arabidopsis thaliana. J. Biol. Chem. 21, 21. [DOI] [PubMed] [Google Scholar]
- 94. Gothel S. F., Marahiel M. A. (1999) Peptidyl-prolyl cis-trans isomerases, a superfamily of ubiquitous folding catalysts. Cell. Mol. Life Sci. 55, 423–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Shapiguzov A., Edvardsson A., Vener A. V. (2006) Profound redox sensitivity of peptidyl-prolyl isomerase activity in Arabidopsis thylakoid lumen. FEBS Lett. 580, 3671–3676 [DOI] [PubMed] [Google Scholar]
- 96. Volkov A. N., Nicholls P., Worrall J. A. (2011) The complex of cytochrome c and cytochrome c peroxidase: the end of the road? Biochim. Biophys. Acta 1807, 1482–1503 [DOI] [PubMed] [Google Scholar]
- 97. Finkemeier I., Goodman M., Lamkemeyer P., Kandlbinder A., Sweetlove L. J., Dietz K. J. (2005) The mitochondrial type II peroxiredoxin F is essential for redox homeostasis and root growth of Arabidopsis thaliana under stress. J. Biol. Chem. 280, 12168–12180 [DOI] [PubMed] [Google Scholar]
- 98. May M. J., Leaver C. J. (1994) Arabidopsis thaliana gamma-glutamylcysteine synthetase is structurally unrelated to mammalian, yeast, and Escherichia coli homologs. Proc. Natl. Acad. Sci. U.S.A. 91, 10059–10063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Rouhier N., Lemaire S. D., Jacquot J. P. (2008) The role of glutathione in photosynthetic organisms: emerging functions for glutaredoxins and glutathionylation. Ann. Rev. Plant Biol. 59, 143–166 [DOI] [PubMed] [Google Scholar]
- 100. Pignocchi C., Foyer C. H. (2003) Apoplastic ascorbate metabolism and its role in the regulation of cell signalling. Curr. Opinion Plant Biol. 6, 379–389 [DOI] [PubMed] [Google Scholar]
- 101. Gutteridge J. M., Halliwell B. (2000) Free radicals and antioxidants in the year 2000. A historical look to the future. Ann. N.Y. Acad. Sci. 899, 136–147 [DOI] [PubMed] [Google Scholar]
- 102. Ort D. R., Baker N. R. (2002) A photoprotective role for O(2) as an alternative electron sink in photosynthesis? Curr. Opin. Plant Biol. 5, 193–198 [DOI] [PubMed] [Google Scholar]
- 103. Asada K. (2000) The water-water cycle as alternative photon and electron sinks. Philos. Trans. R. Soc. Lond. 355, 1419–1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Hossain M. A., Nakano Y., Asada K. (1984) Monodehydroascorbate reductase in spinach-chloroplasts and its participation in regeneration of ascorbate for scavenging hydrogen-peroxide. Plant Cell Physiol. 25, 385–395 [Google Scholar]
- 105. Ledford H. K., Chin B. L., Niyogi K. K. (2007) Acclimation to singlet oxygen stress in Chlamydomonas reinhardtii. Eukaryotic Cell 6, 919–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Wang Y., Duanmu D., Spalding M. H. (2011) Carbon dioxide concentrating mechanism in Chlamydomonas reinhardtii: inorganic carbon transport and CO2 recapture. Photosynthesis Res. 109, 115–122 [DOI] [PubMed] [Google Scholar]
- 107. Yamano T., Fukuzawa H. (2009) Carbon-concentrating mechanism in a green alga, Chlamydomonas reinhardtii, revealed by transcriptome analyses. J. Basic Microbiol. 49, 42–51 [DOI] [PubMed] [Google Scholar]
- 108. Wang Y., Spalding M. H. (2006) An inorganic carbon transport system responsible for acclimation specific to air levels of CO2 in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. U.S.A. 103, 10110–10115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Yamano T., Miura K., Fukuzawa H. (2008) Expression analysis of genes associated with the induction of the carbon-concentrating mechanism in Chlamydomonas reinhardtii. Plant Physiol. 147, 340–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Yamano T., Tsujikawa T., Hatano K., Ozawa S., Takahashi Y., Fukuzawa H. (2010) Light and low-CO2-dependent LCIB-LCIC complex localization in the chloroplast supports the carbon-concentrating mechanism in Chlamydomonas reinhardtii. Plant Cell Physiol. 51, 1453–1468 [DOI] [PubMed] [Google Scholar]
- 111. Fang W., Si Y. Q., Douglass S., Casero D., Merchant S. S., Pellegrini M., Ladunga I., Liu P., Spalding M. H. (2012) Transcriptome-wide changes in Chlamydomonas reinhardtii gene expression regulated by carbon dioxide and the CO2-concentrating mechanism regulator CIA5/CCM1. Plant Cell 24, 1876–1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Renberg L., Johansson A. I., Shutova T., Stenlund H., Aksmann A., Raven J. A., Gardestrom P., Moritz T., Samuelsson G. (2010) A metabolomic approach to study major metabolite changes during acclimation to limiting CO2 in Chlamydomonas reinhardtii. Plant Physiol. 154, 187–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Kleffmann T., Russenberger D., von Zychlinski A., Christopher W., Sjolander K., Gruissem W., Baginsky S. (2004) The Arabidopsis thaliana chloroplast proteome reveals pathway abundance and novel protein functions. Curr. Biol. 14, 354–362 [DOI] [PubMed] [Google Scholar]
- 114. Peltier J. B., Cai Y., Sun Q., Zabrouskov V., Giacomelli L., Rudella A., Ytterberg A. J., Rutschow H., van Wijk K. J. (2006) The oligomeric stromal proteome of Arabidopsis thaliana chloroplasts. Mol. Cell. Proteomics 5, 114–133 [DOI] [PubMed] [Google Scholar]
- 115. Molla-Morales A., Sarmiento-Manus R., Robles P., Quesada V., Perez-Perez J. M., Gonzalez-Bayon R., Hannah M. A., Willmitzer L., Ponce M. R., Micol J. L. (2011) Analysis of ven3 and ven6 reticulate mutants reveals the importance of arginine biosynthesis in Arabidopsis leaf development. Plant J. 65, 335–345 [DOI] [PubMed] [Google Scholar]
- 116. Holden H. M., Thoden J. B., Raushel F. M. (1999) Carbamoyl phosphate synthetase: an amazing biochemical odyssey from substrate to product. Cell. Mol. Life Sci. 56, 507–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Zrenner R., Stitt M., Sonnewald U., Boldt R. (2006) Pyrimidine and purine biosynthesis and degradation in plants. Ann. Rev. Plant Biol. 57, 805–836 [DOI] [PubMed] [Google Scholar]
- 118. Kalamaki M. S., Alexandrou D., Lazari D., Merkouropoulos G., Fotopoulos V., Pateraki I., Aggelis A., Carrillo-Lopez A., Rubio-Cabetas M. J., Kanellis A. K. (2009) Over-expression of a tomato N-acetyl-L-glutamate synthase gene (SlNAGS1) in Arabidopsis thaliana results in high ornithine levels and increased tolerance in salt and drought stresses. J. Exp. Bot. 60, 1859–1871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Kalamaki M. S., Merkouropoulos G., Kanellis A. K. (2009) Can ornithine accumulation modulate abiotic stress tolerance in Arabidopsis? Plant Signaling Behavior 4, 1099–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Llacer J. L., Fita I., Rubio V. (2008) Arginine and nitrogen storage. Current Opinion Structural Biol. 18, 673–681 [DOI] [PubMed] [Google Scholar]
- 121. Lapatsina L., Brand J., Poole K., Daumke O., Lewin G. R. (2012) Stomatin-domain proteins. Eur. J. Cell. Biol. 91, 240–245 [DOI] [PubMed] [Google Scholar]
- 122. Rungaldier S., Oberwagner W., Salzer U., Csaszar E., Prohaska R. (2013) Stomatin interacts with GLUT1/SLC2A1, band 3/SLC4A1, and aquaporin-1 in human erythrocyte membrane domains. Biochim. Biophys. Acta 1828, 956–966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Kretsinger R. H. (1976) Calcium-binding proteins. Ann. Rev. Biochem. 45, 239–266 [DOI] [PubMed] [Google Scholar]
- 124. Holmgren A. (1985) Thioredoxin. Ann. Rev. Biochem. 54, 237–271 [DOI] [PubMed] [Google Scholar]
- 125. Dietz K. J., Jacquot J. P., Harris G. (2010) Hubs and bottlenecks in plant molecular signalling networks. New Phytol. 188, 919–938 [DOI] [PubMed] [Google Scholar]
- 126. Petroutsos D., Busch A., Janssen I., Trompelt K., Bergner S. V., Weinl S., Holtkamp M., Karst U., Kudla J., Hippler M. (2011) The Chloroplast Calcium Sensor CAS Is Required for Photoacclimation in Chlamydomonas reinhardtii. Plant Cell 23, 2950–2963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Nomura H., Komori T., Uemura S., Kanda Y., Shimotani K., Nakai K., Furuichi T., Takebayashi K., Sugimoto T., Sano S., Suwastika I. N., Fukusaki E., Yoshioka H., Nakahira Y., Shiina T. (2012) Chloroplast-mediated activation of plant immune signalling in Arabidopsis. Nat. Commun. 3, 926. [DOI] [PubMed] [Google Scholar]
- 128. Weinl S., Held K., Schlucking K., Steinhorst L., Kuhlgert S., Hippler M., Kudla J. (2008) A plastid protein crucial for Ca2+-regulated stomatal responses. New Phytol. 179, 675–686 [DOI] [PubMed] [Google Scholar]
- 129. Nomura H., Komori T., Kobori M., Nakahira Y., Shiina T. (2008) Evidence for chloroplast control of external Ca2+-induced cytosolic Ca2+ transients and stomatal closure. Plant J. 53, 988–998 [DOI] [PubMed] [Google Scholar]
- 130. Terashima M., Petroutsos D., Hudig M., Tolstygina I., Trompelt K., Gabelein P., Fufezan C., Kudla J., Weinl S., Finazzi G., Hippler M. (2012) Calcium-dependent regulation of cyclic photosynthetic electron transfer by a CAS, ANR1, and PGRL1 complex. Proc. Natl. Acad. Sci. U.S.A. 109, 17717–17722 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.