Abstract
Yeast Cdc48 (p97/VCP in human cells) is a hexameric AAA ATPase that is thought to use ATP hydrolysis to power the segregation of ubiquitin-conjugated proteins from tightly bound partners. Current models posit that Cdc48 is linked to its substrates through adaptor proteins, including a family of seven proteins (13 in human) that contain a Cdc48-binding UBX domain. However, few substrates for specific UBX proteins are known, and hence the generality of this hypothesis remains untested. Here, we use mass spectrometry to identify ubiquitin conjugates that accumulate in cdc48 and ubx mutants. Different ubx mutants exhibit unique patterns of conjugate accumulation that point to functional specialization of individual Ubx proteins. To validate our findings, we examined in detail the endoplasmic reticulum-bound transcription factor Spt23, which we identified as a putative Ubx2 substrate. Mutant ubx2Δ cells are deficient in both cleaving the ubiquitinated 120 kDa precursor of Spt23 to form active p90 and in localizing p90 to the nucleus, resulting in reduced expression of the target gene OLE1, which encodes fatty acid desaturase. Our findings provide a resource for future investigations on Cdc48, illustrate the utility of proteomics to identify ligands for specific ubiquitin receptor pathways, and uncover Ubx2 as a key player in the regulation of membrane lipid biosynthesis.
Budding yeast Cdc48 is an essential, highly abundant member of the AAA (ATPase associated with various cellular activities) protein family. Cdc48 has been linked to numerous functions throughout the cell but is best known for its critical role in ERAD (endoplasmic reticulum associated protein degradation)1, which occurs via the ubiquitin proteasome system (UPS). It is also involved in cell-cycle progression, homotypic membrane fusion, chromatin remodeling, autophagy, and transcriptional and metabolic regulation (1–4). Human Cdc48, known as p97 or VCP, has been the subject of much attention over the last few years because of its causal links to amyotrophic lateral sclerosis and inclusion body myopathy, Paget's disease of the bone, and frontotemporal dementia as well as its implied role in a variety of diseases including cancer (5–9). As a result, p97 has been the target of multiple drug development efforts (10, 11).
Cdc48/p97 interacts with a large number of putative substrate adaptors and cofactors, including a family of proteins (seven in yeast, 13 in human cells) that contain a UBX domain (12, 13). The UBX domain binds to the N-terminal region of p97 and proteins bearing this domain have been suggested to serve as interchangeable adaptors that target Cdc48/p97 to specific substrates. Although the functions and mechanism of action of Cdc48/p97 remain poorly understood, it is generally presumed that it uses ATP hydrolysis to fuel the extraction of ubiquitinated proteins from multisubunit complexes or membranes as a prelude to their degradation by the proteasome. Cdc48/p97 may also remodel protein–protein and protein–nucleic acid complexes in a manner that is not coupled to either prior ubiquitination (14) or subsequent degradation by the proteasome (15, 16).
Based on the abundance of Cdc48/p97 and the complexity of the network of adaptor proteins for which it serves as the hub, Cdc48/p97 has the potential to exert a profound influence on the UPS. However, the number of known substrates of Cdc48/p97 remains relatively small, and smaller still is the number of substrates that have been linked to a specific UBX domain protein. Indeed, only a handful of specific cases are known, and in half of them proteasomal degradation is not the final outcome (15–22). To understand why there is such a profusion of UBX domain proteins in cells, it will be important to determine what these proteins do to their substrates, which will require knowing what their substrates are. Therefore, a major goal of this work was to enable future investigations into the function and regulation of UBX proteins by assembling a catalogue of candidate substrates/targets.
A key paradigm that guides our current understanding of Cdc48 function emerged from studies on the processing of the transcription factors Spt23 and Mga2 (23–25). These seminal studies revealed the mechanism by which cells control their ratio of saturated to unsaturated fatty acids to maintain an appropriate lipid composition in cellular membranes. Central to this regulation are the transcription factors Spt23 and Mga2, which are released from the endoplasmic reticulum (ER) membrane and translocate to the nucleus to activate expression of OLE1, which encodes the stearoyl-Δ9 desaturase that governs the conversion of saturated fatty acids (SFAs) to unsaturated fatty acids (UFAs) (26, 27). Spt23 and Mga2 are initially produced as 120 kDa precursors (p120) embedded in the ER by C-terminal signal or anchor domains, such that the bulk of each protein projects into the cytosol. The p120 form of each protein is ubiquitinated by Rsp5 and cleaved by the proteasome, which yields a p90 form that lacks the transmembrane signal or anchor (23, 25, 28–30). However, p90 remains tethered to the ER through dimerization to an uncleaved p120, until it is disengaged from its partner by the “segregase” activity of Cdc48, acting in concert with Ufd1-Npl4 (24, 31, 32). The released p90 can then travel to the nucleus, where it activates expression of OLE1 (24). The p90 species are quite unstable, and Cdc48 also promotes their degradation (33). In addition to acting as a transcription factor, Mga2 also influences the stability of OLE1 mRNA (34), although the degree to which this function is carried out by p90 or p120 is not understood.
EXPERIMENTAL PROCEDURES
Yeast Strains and Growth Conditions
Strains, plasmids, and primers used in this study are described in supplemental Tables S3–S5. All yeast strains are derivates of either the wild-type strain RJD 4614 (S288C background from the OpenBiosystems Yeast Knockout Library) or the wild-type strain RJD 360 (W303 background). For Ubx2 domain analysis experiments, mutants were derived from strains gifted by Chao-Wen Wang. Standard genetic techniques were used. Unless otherwise stated, strains were grown at 30 °C and cultured in yeast extract/peptone/dextrose (YPD).
SILAC Labeling of Cells
For SILAC experiments, strains auxotrophic for lysine and arginine were used. Cells expressing His8ubiquitin were grown in complete synthetic medium with 2% dextrose containing 20 mg/L lysine and arginine or in “heavy” medium with 20 mg/L 13C615N2-lysine and 13C6-arginine (Cambridge Isotope Laboratories, Andover, MA). All mutant strains with the exception of cdc48–3 and ubx2Δ expressed His8ubiquitin and endogenous ubiquitin in a 1:1 ratio. Cdc48–3 and ubx2Δ cells expressed lower levels of His8ubiquitin, which we corrected for during quantification. Cells were grown for 10 generations to log-phase (OD600 1.0–2.0) and equal amounts of the light and heavy-labeled cells (as determined by OD600) were mixed, harvested, washed, and flash frozen.
Purification of Ub Conjugates
Purifications were performed similarly to the protocol described previously (35). Two hundred OD600 units of cells were lysed in urea buffer (8 m urea, 300 mm NaCl, 100 mm Na2PO4, 10 mm Tris-HCl pH 8.0, 0.2% Triton X-100, 20 mm imidazole, and 5 mm N-ethylmaleimide (NEM)) by vortexing with glass beads. Ubiquitinated proteins were purified by the addition of nickel-NTA bead slurry (50 μl beads/5-10 mg of lysate) to clarified lysate (13,200 rpm for 15 min in an eppendorf Centrifuge 5417R) and mixed at room temperature for 90 min. Beads were washed three times with 20 bed volumes of buffer. A two-step digestion was performed directly on proteins bound to the beads by first digesting with Lys-C for 4 h, followed by overnight digestion with trypsin (36).
Fractionation and Liquid Chromatography-tandem MS (LC-MS/MS)
The tryptic peptides were desalted on a C18 macrotrap (Michrom Bioresources, Auburn, CA) and concentrated in vacuo. Dried samples were resuspended and subjected to StageTip-based strong anionic exchange (SAX) as previously described (37). Samples were eluted, concentrated, and then acidified before mass spectrometric analysis. Mass spectrometry experiments were performed on an EASY-nLC (Thermo Scientific) connected to a hybrid LTQ-Orbitrap Classic with a nanoelectrospray ion source (Thermo Scientific) using a setup and settings (38) very similar to those previously described (39). Binding and separation of the peptides took place on a 15 cm silica analytical column (75 μm ID) packed in-house with reversed phase ReproSil-Pur C18AQ 3 μm resin (Dr Maisch GmbH, Ammerbuch-Entringen, Germany). Samples were run for 240 min on a 5% to 25% acetonitrile gradient in 0.2% formic acid at a flow rate of 350 nL per minute. The mass spectrometer was programmed to acquire data in a data-dependent mode, automatically switching between full-scan MS and tandem MS acquisition. Survey full scan MS spectra (from m/z 300 to 1700) were acquired in the Orbitrap after the accumulation of 500,000 ions, with a resolution of 60,000 at 400 m/z. The ten most intense ions were sequentially isolated, and after the accumulation of 5000 ions, fragmented in the linear ion trap by collision-induced dissociation (CID) (collisional energy 35% and isolation width 2 Da). Precursor ion charge state screening was enabled and singly charged and unassigned charge states were rejected. The dynamic exclusion list was set for a 90 s maximum retention time, a relative mass window of 10 ppm, and early expiration was enabled.
Mass Spec Analysis and Quantification
Thermo raw data files were analyzed by MaxQuant (v 1.3.0.5) (40) and were searched against the SGD yeast database (5911 sequences) and an in-house contaminant database (259 sequences) including human keratins and proteases. All default options were used except as follows: match between runs was enabled (2 min maximum), GlyGly on Lys (+114.042927), and multiplicity of 2 with heavy labels Arg6 (+6.020129) and Lys8 (+8.014199). Tryptic digest was specified with up to two missed cleavages. Initial precursor mass tolerance was 7 ppm, however MaxQuant calculates tighter individual precursor tolerances after recalibration. Fragment ion tolerance was 0.5 Da. Peptide, protein, and site false discovery rates were fixed at 1% using the target-decoy approach with a reversed database (40). The minimum number of peptides for quantification was 1. Spectra for the small number of proteins identified by a single peptide are included in the supplemental Material section. Further data processing was performed to calculate ratios and standard errors of the ratios for each mutant using in-house scripts described previously (41). Briefly, hierarchical models are constructed for each mutant, in which the overall ratio for each protein is the geometric mean of the biological replicates and the biological replicate ratio is the median of all of the peptide ratios in the replicate. The standard error of the overall protein ratio is calculated by estimating the global peptide ratio standard error using pooled variance (calculated separately for peptide ratios based on requantified isotopic patterns) and using a bootstrapping procedure to resample at each level in the hierarchical model. In cases in which the protein replicate ratios are inconsistent (i.e. up-regulated in one sample and down-regulated in another), the standard error would be very large and the protein would not be considered significantly changed from 1:1.
To calculate a p value for the number of significantly elevated proteins in at least one experiment, a bootstrapping method was employed. For each bootstrapping iteration, the ratios observed were randomly reassigned to proteins. Next, the pooled variance was calculated based on these reassigned ratios and the hierarchical models were recomputed. Finally, we counted the number of proteins that were significantly elevated using the same criteria as with the real data. A thousand bootstrap iterations were calculated. In most bootstrap iterations, no proteins were found to be significantly elevated in any experiment. We then modeled the distribution of counts of significantly enriched proteins with a Poisson distribution.
All quantified proteins were used to construct Figs. 1B and supplemental Figs. S1A, S1B. Proteins for which the probability that they changed more than 10% in the WT/WT control experiment was >99% or were not significantly enriched (p value ≥ 0.05) in the His8Ub expressing cells compared with the untagged control cells were filtered out because of the high likelihood that they would yield false positive identifications in the mutant/wild type comparisons. Analyses relating to comparisons with the Saccharomyces cerevisiae ubiquitin database (SCUD) and the response of the ubiquitination enzymes in the cdc48–3 mutant used all proteins not filtered out. All proteins not filtered out that were quantified in all mutants were used in the remaining bioinformatic analyses. In Figs. 1C and 1D, we considered proteins to be significantly changed if there was a less than 1% chance that the fold change was less than 10% based on the individual protein standard error. Yeast GO-Slim annotations for Figs. 2A and 2B were downloaded from the SGD. Proteins considered significantly enriched had a more than 10% change from 1:1 (p value < 0.05). Annotations for supplemental Fig. S3A and supplemental Figs. S4A-S4E were downloaded from DAVID (42, 43). Displayed terms were deemed significantly different at a p value < 0.05 (e.g. the 25th percentile of the ratios of the proteins annotated with the term was significantly greater than random) after correcting for multiple hypotheses testing using the Benjamini and Hochberg method(44).
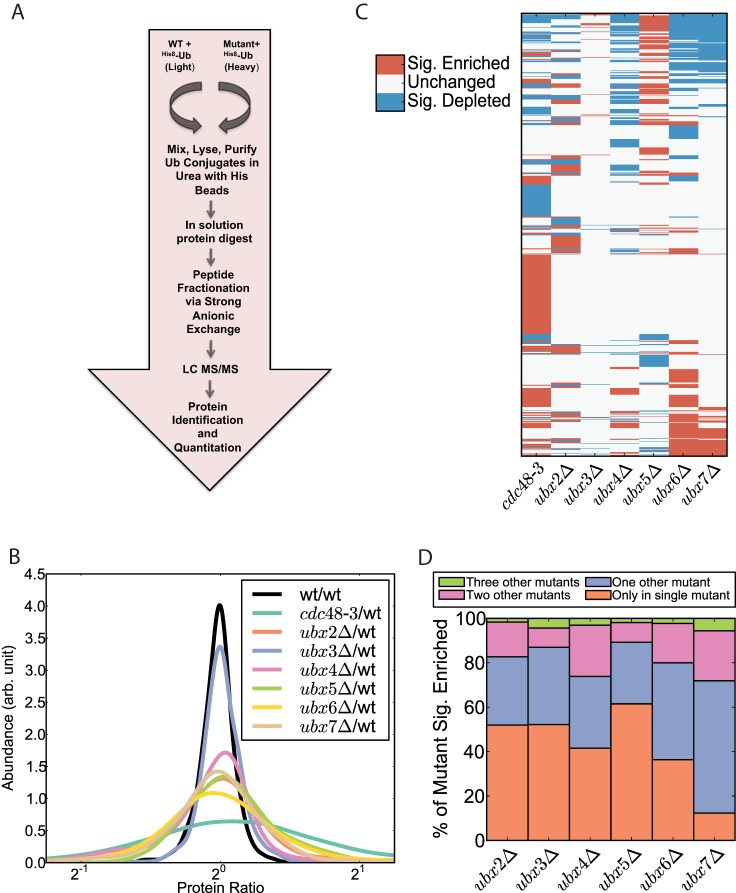
Fig. 1.
Sequencing and quantification of the ubiquitin proteome reveals proteins regulated by Cdc48 and its UBX domain adaptors. A, Experimental workflow for identification and quantification of Ub conjugates. Proteins were quantified using the SILAC method. Light (WT) and heavy (mutant) isotope labeled cells constitutively expressing His8Ub were grown, collected in log-phase, and mixed. The cells were lysed in denaturing urea buffer and ubiquitinated proteins were purified with nickel-NTA magnetic beads. Proteins were digested into peptides, which were fractionated via strong anionic exchange and analyzed by mass spectrometry. B, Smoothed histogram depicting changes in the ubiquitinated proteome in mutants. C, Heatmap of relative changes to the ubiquitin proteome in various mutants. Only proteins that exhibited a significant change from a 1:1 ratio (mutant/wild type) in at least one experiment are colored. Abundance changes were deemed significant if the mutant/wild type ratio deviated from 1:1 with a p value less than .05 corrected for multiple hypothesis testing by the Benjamini and Hochberg method (44). D, Graphical representation of how frequently an ubiquitin conjugate found to accumulate in the query mutant also accumulated in other mutants.
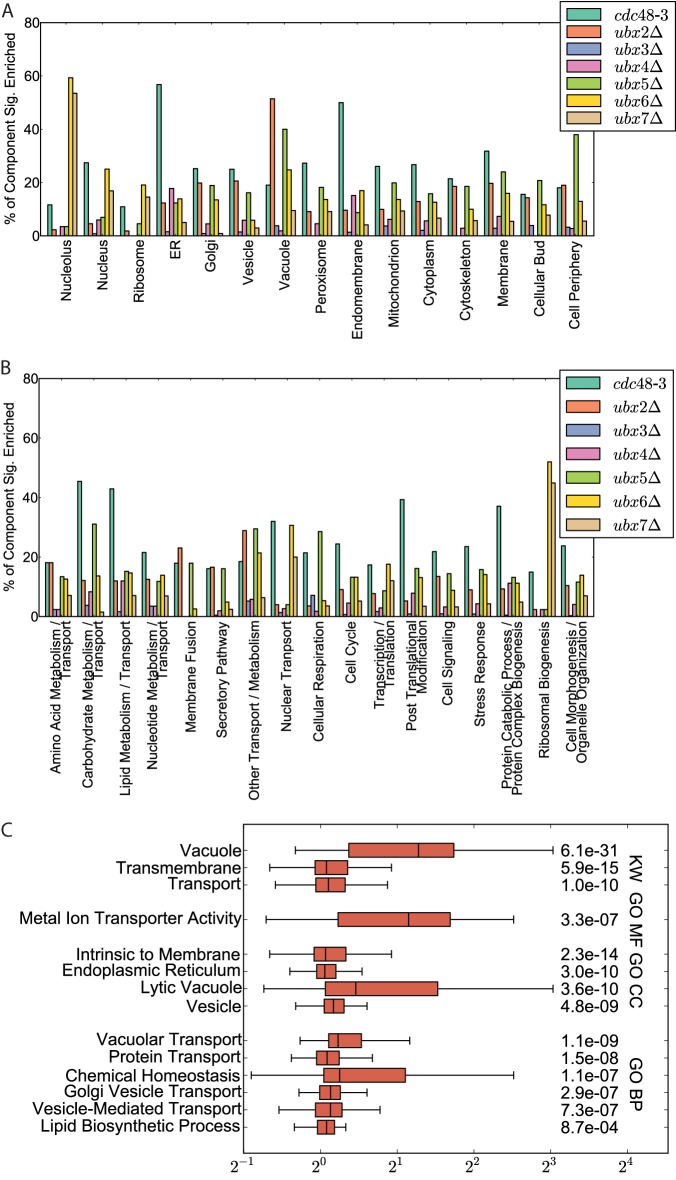
Fig. 2.
Changes to the ubiquitin conjugate proteome in ubxΔ mutants suggest functional specialization of Ubx proteins. A, Overview of how ubiquitin conjugates that accumulate in cdc48-3 and ubxΔ mutants are distributed according to GO-Slim cell components and (B) GO-Slim biological processes. All of the proteins analyzed in (A) and (B) demonstrated enrichment greater than 10% (with ≥95% confidence) in one or more mutants. C, Box and whisker plot of annotation terms demonstrating a significant enrichment in the Ub-Proteome for a majority of their identified members in the ubx2Δ mutant strain. Significance was calculated based on the 25 percentile of the distribution of protein ratios of protein annotated with the term in comparison to all ubx2Δ ratios. Enriched GO Cellular Component (CC), GO Biological Process (BP), GO Molecular Function (MF), and Uniprot Keyword (KW) terms are shown.
Confirmation of Ub Conjugate Accumulation for Spt23
Ubiquitin conjugates were purified from 100 OD600 units of cells expressing tagged ubiquitin as described in the purification of Ub conjugates section. However after washing beads an equal volume of 2× SDS-PAGE buffer was added before boiling the beads for 5 min to elute conjugates. Boiled aliquots were resolved by SDS-PAGE and immunoblotted for endogenously tagged mycSpt23V5.
Confirmation of Ub Conjugate Accumulation for Mga2
Cells (100 OD600 units) were disrupted using a FastPrep-24 in lysis buffer containing 50 mm Tris-HCl (pH 7.5), 150 mm NaCl, 1 mm EDTA, 10 mm NEM, 0.5 mm AEBSF, and Protease Inhibitor Mixture Tablet (Roche). Lysates were clarified by centrifugation (14,200 rpm for 20 min in an Eppendorf Centrifuge 5417R) and bound to 30 μl of TUBE2 agarose beads (Boston Biochem, Cambridge, MA)/5–10 mg lysate for 2–3 h with rotation at 4 °C. Beads were washed three times with lysis buffer and an equal volume of 2X SDS-PAGE buffer was added before boiling the beads for 5 min to elute conjugates. Boiled aliquots were resolved by SDS-PAGE and immunoblotted.
Quantitative Reverse Transcription PCR
RNA was isolated from 3 OD600 units of cells using Trizol (Invitrogen) according to the manufacturer's recommendations. cDNA was prepared using the Superscript first strand synthesis kit (Invitrogen) and quantitative PCR was performed using the SYBR GreenER super mix (Invitrogen) with primers described in supplemental Table S5.
Immunoblot Analysis
Cell pellets were first boiled for 3 min and then after the addition of SDS-PAGE buffer supplemented with 5 mm NEM and glass beads, lysed by vortexing in a Fast Prep-24 (MP) for 1 min at a setting of 6.5 and boiled again for 3 min. Boiled lysates were centrifuged at 16,000 × g for 5 min. Aliquots were resolved by SDS-PAGE, transferred to nitrocellulose, and stained with Ponceau S to determine equivalent loading of protein extracts. The nitrocellulose filters were immunoblotted with desired antibody and developed by ECL or SuperSignal (Invitrogen). For detecting mycSpt23V5, mycSpt23HA, and mycMga2, anti-myc (Covance), anti-V5 (Invitrogen), and anti-HA (16B12, Roche) antibodies were used at a dilution of 1:3000. Other immunoblots were performed with antibodies specific for Ubiquitin (Stressgen Biotechnologies, Victoria, BC, Canada) dilution 1:1000, Dpm1 (Invitrogen) dilution 1:3000, tubulin (Santa Cruz, Santa Cruz, CA) dilution 1:30000, TAP tag (Thermo Scientific) dilution 1:3000, Rsp5 (gift from Linda Hicke) dilution 1:3000, Cdc48 (gift from Thomas Sommer and Ernst Jarosch) dilution 1:1000, and Ufd1 (custom polyclonal antibody from Covance) dilution 1:10000.
Growth Assays
For plating assays cells were grown in YPD or SRaffinose-URA and diluted to OD600 of 0.3 in water. Serial fivefold dilutions were prepared in water and spotted onto YPD ± 0.2% oleic acid or minimal medium plates supplemented with various additives as described in the text. Plates were incubated at 30 °C or 37 °C for 2–4 days.
Turnover of Spt23
RJD 6138, 6139, and 6167 cells were grown in YPD to an OD600 ∼1.0, at which point 100 μg/ml cycloheximide was added to initiate a chase. Samples were taken at the time points indicated. To monitor Spt23 following the switch from medium containing oleic acid to medium lacking oleic acid, cells were grown in YPD + 0.2% oleic acid, washed in water, and resuspended in YPD supplemented with cycloheximide. Proteins were extracted as described in the Immunoblot Analysis section with boiling SDS-PAGE buffer supplemented with 5 mm NEM. Lysates were resolved by SDS-PAGE and immunoblotted.
Immunoprecipitation of Ubx2
RJD 3166, 6172, 6173, and 4614 were grown in YP plus 2% galactose (to induce expression of mycSpt23HA from the GAL promoter) to an OD600 ∼1.0, harvested, and flash-frozen in liquid nitrogen. Cells were lysed by grinding in liquid nitrogen. Frozen cell powder was resuspended in lysis buffer containing 50 mm Tris (pH 7.5), 100 mm NaCl, 1 mm EDTA, 5 mm Mg(OAc)2, 0.2% Triton X-100, 5 mm NEM, and Protease Inhibitor Mixture Tablet (Roche), clarified by centrifugation (14,200 rpm for 20 min on an Eppendorf Centrifuge 5417R) and bound to rabbit IgG conjugated to superparamagnetic Dynabeads (Invitrogen) for 3 h with mixing at 4 °C. Beads were washed three times with lysis buffer and bound proteins were eluted by boiling in SDS-PAGE buffer and resolved by SDS-PAGE before immunoblotting.
Immunofluorescence
RJD 6172 and 6182 cells were grown in YP plus 2% galactose (to induce expression of mycSpt23HA from the GAL promoter) to an OD600 ∼0.5–1.0. Cell cultures were adjusted to 4% formaldehyde and incubated for 15 min before cells were collected, washed, and spheroplasted by treatment with 50 units of lyticase (Sigma) for 10 min. Spheroplasts were washed and placed on ConA-coated glass slides. Cells were permeabilized with methanol and acetone washes. Cells were treated with blocking buffer (2% BSA, 0.1% Tween20 in PBS) and incubated with primary antibody overnight at a dilution of 1:1000. FITC-conjugated secondary antibody was added and cells were incubated for another hour followed by DNA staining with DAPI. Mounting medium was placed on cells and images were taken with a Zeiss confocal microscope with a 100× objective.
RESULTS
Identification and Quantification of Changes in the Ubiquitin Proteome
To better understand the breadth of function of Cdc48 and its adaptors that contain an UBX domain, we sought to identify ubiquitin conjugates whose levels increased in yeast cells deficient in these components. The rationale underlying this approach is based on the observation that ubiquitinated Rpb1 substrate accumulates in ubx5Δ and cdc48-3 cells (18). Wild type and mutant yeast cells expressing His8-tagged ubiquitin were labeled with heavy and light lysine and arginine, respectively, and immediately before harvesting and lysis the cultures were mixed. Ubiquitin conjugates were purified on Ni2+-NTA magnetic beads under denaturing conditions (8 m urea) and digested with trypsin. The resulting peptides were subjected to strong anionic exchange (SAX) fractionation before sequencing in a mass spectrometer (Fig. 1A). We performed this analysis in triplicate with cdc48-3 and in duplicate with six of the seven ubxΔ mutant strains (ubx2Δ-ubx7Δ; shp1Δ/ubx1Δ was not analyzed because it grew so poorly).
To evaluate the reproducibility of our methodology we compared biological replicates (supplemental Figs. S1A, S1B), which revealed that our data sets were highly correlated and demonstrated low coefficients of variation. To improve further the quality of our proteomic data set, we performed two separate control experiments, the results of which were used to computationally filter our data before further analysis. In the first experiment both the light and heavy-labeled cultures were wild type. This analysis yielded a narrow distribution of heavy/light ratios centered on 20, confirming the significance of the broader ratio distributions seen in the wild type/mutant comparisons (Fig. 1B, supplemental Fig. S2A). Proteins that exhibited ratios that deviated from 20 by >10% (p value <0.01) in this control experiment were excluded from our overall data set. In the second control, we compared untagged, light-labeled cells and His8Ub-expressing heavy-labeled cells. Proteins that were significantly enriched in the His8Ub-expressing cells (p value<0.05) were deemed to be derived from bona fide ubiquitin conjugates. After computational filtration, our data set contained ratios (ranging between 0.018 and 53.3) for 1916 ubiquitinated proteins identified and quantified at least once and 1733 ubiquitinated proteins identified in at least one replicate of each mutant/wild type comparison (all proteins identified are listed in supplemental Table S1, supplementary File S1 and S2). In the ubx2Δ (1, 677), ubx3Δ (1, 701), ubx4Δ (1, 707), ubx5Δ (1, 581), ubx6Δ (1, 683), and ubx7Δ (1, 625) experiments the indicated number of proteins (out of 1733 possible) were identified in both replicates. In the cdc48-3 experiments, 1588 of the 1733 proteins were identified in triplicate. We additionally identified 67 di-glycine peptide signatures, which are indicative of ubiquitination sites; 25 of the 67 were previously identified (supplemental Table S2, supplementary File S3 and S4). The proteins in this study covered over 50% of the previously identified ubiquitinated proteins in the SCUD (http://scud.kaist.ac.kr/) and included 1226 proteins not previously listed (supplemental Fig. S2B). These metrics suggest that we have sampled a substantial portion of the ubiquitinated proteome.
UBX Proteins Target Different Subsets of Ubiquitin Conjugates
From the initial screen it was evident that cdc48-3 and ubxΔ mutations had wide-ranging and variable effects on the ubiquitin proteome. Comparison of the ratio distributions of the different experiments underscores this conclusion (Fig. 1B). Consistent with Cdc48 being the hub of its eponymous network, cdc48-3 had the greatest effect on the ubiquitinated proteome. Twenty-six percent of ubiquitin conjugates were detected at higher levels in cdc48-3 compared with wild type cells whereas 17% were decreased in amount. We do not know the reason for the latter but it could be because of reduced expression of the protein or increased competition for free ubiquitin, because total conjugated ubiquitin increases substantially in cdc48-3 cells (18). Among the ubxΔ mutants ubx5Δ and ubx6Δ caused the strongest perturbations; by contrast ubx3Δ had the least effect, followed by ubx4Δ. To evaluate the impact of the cdc48-3 and ubxΔ mutations with more granularity, we generated a heatmap that captures the behavior of the changing conjugates quantified in all experiments in our data set (Fig. 1C). Three important points are evident in this graphic. First, ubx6Δ and ubx7Δ displayed remarkable overlap with over 85% of the ubiquitin conjugates accumulating in ubx7Δ also accumulating in ubx6Δ. Very little is known about Ubx6 and Ubx7 but this result is consistent with their close homology and colocalization at the perinuclear membrane (45). Second, cdc48-3 mutants accumulated a set of ubiquitin conjugates (128 proteins) that did not accumulate in any single ubxΔ mutant. This suggests that many substrates may engage Cdc48 without an Ubx protein, and could explain why a relatively low fraction of Cdc48/p97 substrates have been shown to be dependent on an UBX domain cofactor. Third, with the exception of ubx6Δ and ubx7Δ, the different mutations had markedly different effects on the ubiquitin conjugate proteome. This conclusion was also supported by an independent analysis (Fig. 1D), which indicated that for each ubxΔ mutant except ubx6Δ and ubx7Δ, >40% of the accumulating conjugates were elevated only in that mutant and up to ∼90% were elevated in that mutant and at most one other mutant. This supports the hypothesis that individual Ubx proteins target distinct sets of ubiquitin conjugates, which is consistent with their proposed role as substrate adaptors for the Cdc48 engine. Remarkably, of the 1733 proteins in our data set, 62% were elevated significantly in at least one mutant (62% was highly significant in comparison to the null hypothesis in which peptide ratios are randomly assigned to proteins (p value < 1e-100)), suggesting that the Cdc48 network has a profound impact on the UPS that is greater than has been previously appreciated.
Ubiquitin Conjugates that are Elevated in Different ubxΔ Mutants Map to Diverse Cellular Components and Cellular Processes
To understand the subcellular localization and functions of the ubiquitinated species identified in our analyses, we evaluated Yeast GO-Slim cell components and GO-Slim biological process annotations (Figs. 2A and 2B). Many different trends were apparent from this analysis. Ubiquitin conjugates that were elevated in cdc48-3 cells (referred to hereafter as “cdc48-3 conjugates”) encompassed numerous known targets including Mga2 and Spt23 (24, 28), the HMG-CoA reductases Hmg1 and Hmg2 (46), and the Rpb1 subunit of RNA Polymerase II (18). Consistent with the known roles of Cdc48 in membrane biology, cdc48-3 conjugates were significantly enriched for proteins of the endomembrane system including the ERP and ERV family of proteins involved in ER to Golgi transport, members of the GPI family of proteins, subunits of the Golgi mannosyltransferase complex, and all seven proteins of the PMT (Protein o-mannosyltransferase) family. The cdc48-3 conjugates were also enriched for glycosylated proteins, enzymes of lipid and carbohydrate metabolism, and proteins involved in nuclear pore complex, nuclear organization, stress response, and the ubiquitin-proteasome system (supplemental Fig. S3A). Further investigation of the latter category revealed that cdc48-3 cells contained elevated levels of ubiquitin conjugated species of multiple E3 ubiquitin ligases and F-Box proteins (supplemental Fig. S3B). This is consistent with recent identification of Cdc48 as an SCFMet30 disassembly factor (47) and suggests that Cdc48 may be more widely involved in regulation of SCF complexes.
Analysis of the ubiquitin conjugates that were elevated in ubx2Δ relative to wild type cells (i.e. ubx2Δ conjugates) indicated there was a significant enrichment of vacuolar and metabolic transport proteins, including secretory proteins (Fig. 2A–2C). Many of these proteins are synthesized in the ER, where Ubx2 is localized. Notably, elevated levels of ubiquitinated forms of a number of ER proteins were detected in both ubx2Δ and cdc48-3 cells, including members of the ergosterol biosynthesis pathway and the OLE1 transcription factors Spt23 and Mga2, consistent with a role for Ubx2-Cdc48 in regulating lipid metabolism.
Deletion of UBX3 had little impact on the ubiquitin conjugate proteome with no proteins changing consistently in a significant manner. The pool of ubx4Δ conjugates was likewise small but was enriched for ER proteins involved in lipid metabolism including Spt23, Mga2, Hmg1, and Hmg2 (Figs. 2A, 2B, supplemental Fig. S4A, S4B).
Ubiquitin conjugates that were elevated in ubx5Δ cells were enriched for vacuolar and plasma membrane proteins, including proteins involved in ion and amino acid transport such as the TPO family of proteins involved in polyamine transport (Fig. 2A, 2B, supplemental Fig. S4C). Ubx5 contains an UIM domain that links it to Rub1-conjugated cullin-RING ubiquitin ligases (CRLs) (48, 49), raising the possibility that a CRL regulates trafficking of membrane proteins in yeast.
Lastly, ubiquitin conjugates that were elevated in ubx6Δ and ubx7Δ were highly enriched for nucleolar and ribosomal proteins, including proteins involved in ribosome assembly (Fig. 2A, 2B, supplemental Fig. S4D, S4E). In agreement with a nuclear and nucleolar function, Ubx6 and Ubx7 localize to the nuclear periphery and nucleus (45).
Ubiquitinated Spt23 and Mga2 Accumulate in ubx2Δ
The increase in ubiquitinated Spt23 and Mga2 species in cdc48-3, ubx2Δ and ubx4Δ cells (Fig. 3A) piqued our interest because although it was already known that Cdc48-Ufd1-Npl4 promotes release of Spt23 and Mga2 from the ER membrane and ubiquitinated forms of these transcription factors accumulate in cdc48, npl4, and ufd1 cells (24, 25, 28, 31), Ubx proteins had not previously been implicated in this pathway. To evaluate the ubiquitination status of Spt23 in ubx2Δ and ubx4Δ mutants, we first generated a yeast strain in which endogenous SPT23 was modified to express proteins bearing an N-terminal myc tag and C-terminal V5 tag (mycSpt23V5). We then used Ni2+-NTA magnetic beads to purify ubiquitin conjugates from mycSPT23V5 strains expressing His8-tagged ubiquitin. Immunoblotting for tagged Spt23 revealed robust accumulation of high molecular weight conjugates in both ubx2Δ and ubx4Δ as compared with wild type (Fig. 3B). For reasons we do not understand, overexpression of His8Ub altered accumulation of ubiquitin conjugates on mycSpt23V5; in the presence of His8Ub ubx4Δ accumulated more ubiquitinated mycSpt23V5 than ubx2Δ whereas in the absence of His8Ub the reverse was observed (e.g. Fig. 5A, 5B). To evaluate the ubiquitination status of Mga2 in ubx2Δ and ubx4Δ mutants, we used Tandem Ubiquitin Binding Entity (TUBE2) beads to purify endogenous ubiquitin conjugates from strains transformed with a plasmid that expressed Mga2 with a myc tag appended to its N terminus (mycMga2). Immunoblotting for mycMga2 revealed an accumulation of high molecular weight conjugates in ubx2Δ cells compared with wild type (supplemental Fig. S5). By contrast, few if any high MW ubiquitin conjugates of Mga2 were detected in ubx4Δ cells.
Fig. 3.
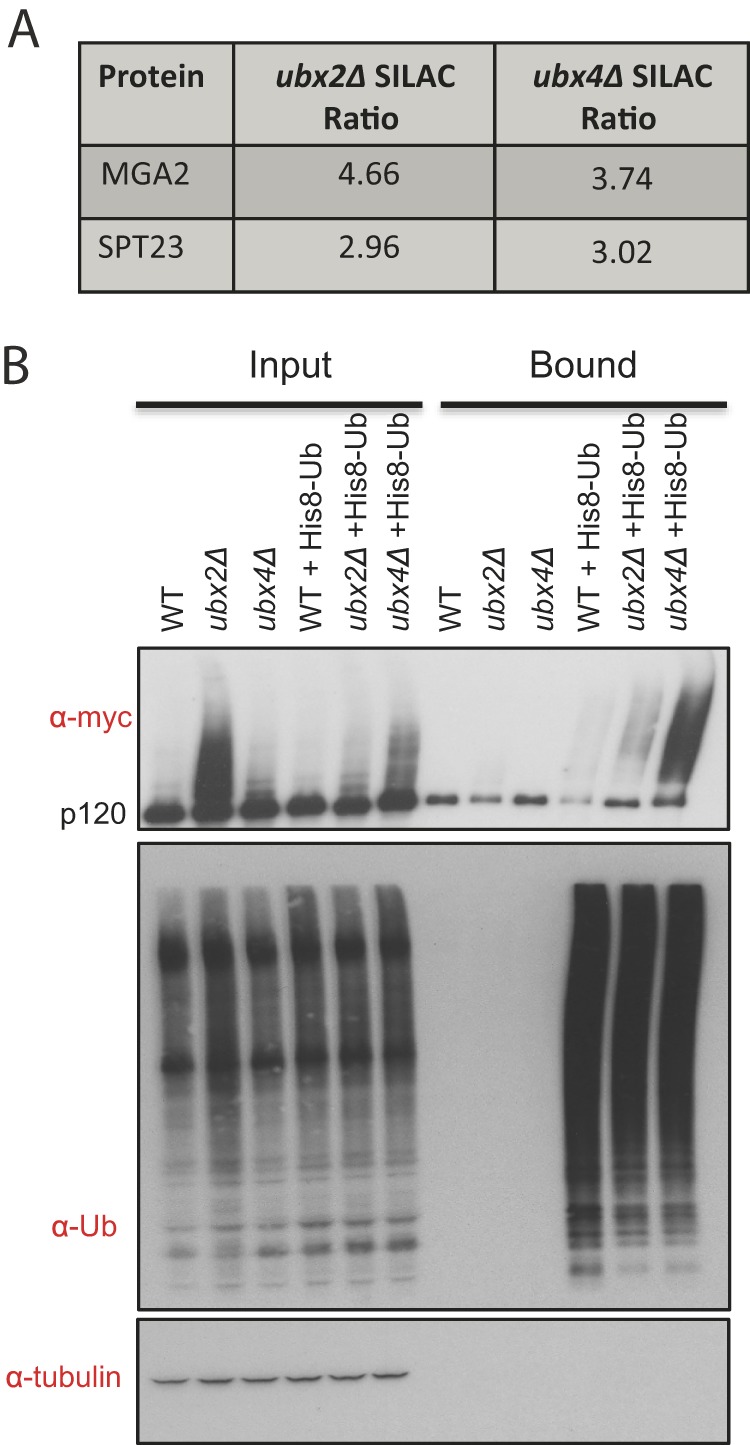
Ubiquitin-conjugated forms of Spt23 and Mga2 accumulate in ubx2Δ cells. A, Table of Spt23 and Mga2 SILAC ratios in ubx2Δ:WT and ubx4Δ:WT comparisons. The reported ratios incorporate many independent measurements (Spt23 ubx2Δ: 7, ubx4Δ: 24; Mga2 ubx2Δ: 21, ubx4Δ: 37). B, Validation of Spt23 ubiquitin conjugate accumulation. Ubiquitin conjugates in WT, ubx2Δ, and ubx4Δ cells expressing mycSpt23V5 from the natural locus and His8Ub from a plasmid were purified with nickel-NTA magnetic beads. The input extract and bound fractions were immunobloted for the myc epitope, Ub, and tubulin (input loading control) as indicated.
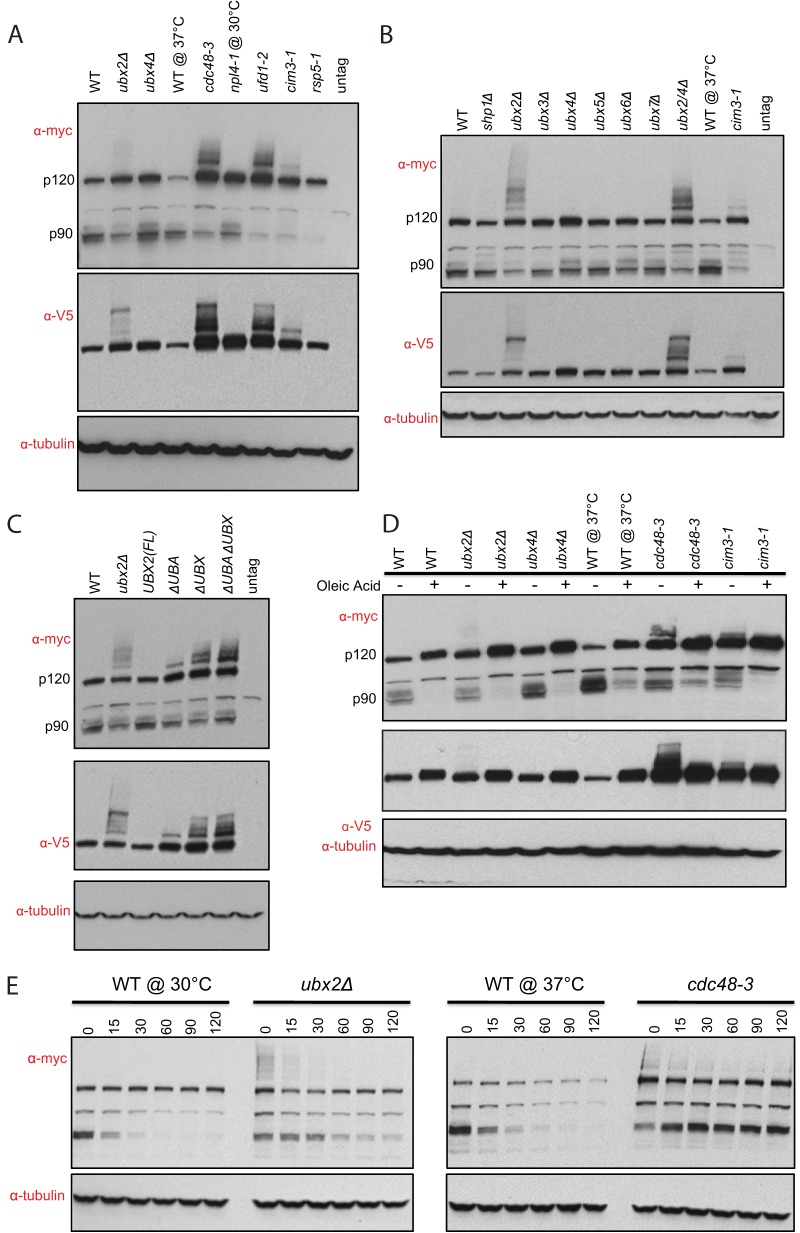
Fig. 5.
Ubx2 regulates processing and stability of the transcription factor Spt23. A, Steady-state level of Spt23 in OLE1 pathway mutants. The indicated mutant strains expressing mycSpt23V5 from the endogenous locus were grown in YPD at 30 °C (ubx2Δ, ubx4Δ, and npl4-1) and 37 °C (cdc48-3, ufd1-2, cim-3, and rsp5-1). Cell lysates were fractionated by SDS-PAGE and immunoblotted with anti-myc, anti-V5, and anti-tubulin antibodies. B, Steady-state levels of Spt23 in ubxΔ mutants. The indicated strains were grown in YPD at 30 °C (except for cim3–1 at 37 °C) and analyzed as in (A). ubx2/4Δ indicates an ubx2Δ ubx4Δ double mutant. C, Influence of Ubx2's UBA and UBX domains on the steady-state level of Spt23. Same as (A) except that strains in lanes 2–6 were ubx2Δ bearing a copy of full-length (FL) UBX2 or a version lacking the indicated domain integrated at LEU2. D, Effect of UFAs on modification and processing of Spt23 in various mutants. Same as (A) except that strains were grown in normal medium or medium supplemented with 0.2% oleic acid as indicated. Cells were grown at 30 °C (ubx2Δ) and 37 °C (cdc48-3). E, Role of Ubx2 and Cdc48 in p90 degradation. Wild type, ubx2Δ, and cdc48-3 cells expressing mycSpt23V5 from the endogenous locus were grown in YPD at 30 °C (left panel) or at 25 °C followed by a shift to 37 °C for 2 h (right panel) before addition of cycloheximide to initiate a chase. At the indicated times (minutes) cells were harvested and processed for SDS-PAGE followed by immunoblotting with anti-myc, or anti-tubulin antibodies.
Ubx2 is a Component of the OLE1 Pathway
In rapidly growing cells that are producing lipids for membrane assembly, the transmembrane p120 forms of Spt23 and Mga2 are ubiquitinated by the ubiquitin ligase Rsp5 and cleaved by the proteasome (23). Subsequently, Cdc48-Ufd1-Npl4 promotes release of cleaved p90 forms of Spt23 and Mga2 from the ER membrane, allowing these proteins to translocate to the nucleus where they activate transcription of OLE1, which encodes the essential enzyme that generates UFAs (26). The accumulation of ubiquitin-conjugated Spt23 and Mga2 in ubx2Δ raised the question of whether Ubx2 also functions in this pathway. To address this question we first tested for genetic interactions between rsp5-1 and ubxΔ mutants. Similar to the previously reported synthetic lethality of cdc48 and ufd1 with rsp5 (24), ubx2Δ rsp5-1 cells failed to grow on YPD at 30 °C but were rescued by addition of the UFA oleic acid (Fig. 4A). By contrast, ubx4Δ rsp5-1 did not exhibit synthetic lethality. To test more directly the hypothesis that Ubx2 promotes expression of OLE1, we measured OLE1 mRNA levels in mutants grown in YPD. During normal log-phase growth in YPD, ubx2Δ cells showed a fivefold reduction in OLE1 mRNA (Fig. 4B). Moreover, as was previously reported for rsp5 mutants (23), both ubx2Δ and rsp5Δ cells failed to accumulate OLE1 mRNA on being shifted from a medium containing oleic acid to a medium lacking UFAs (Fig. 4C). By contrast, ubx4Δ was not defective in OLE1 regulation and did not exacerbate the effect of ubx2Δ (Figs. 4B, 4C).
Fig. 4.
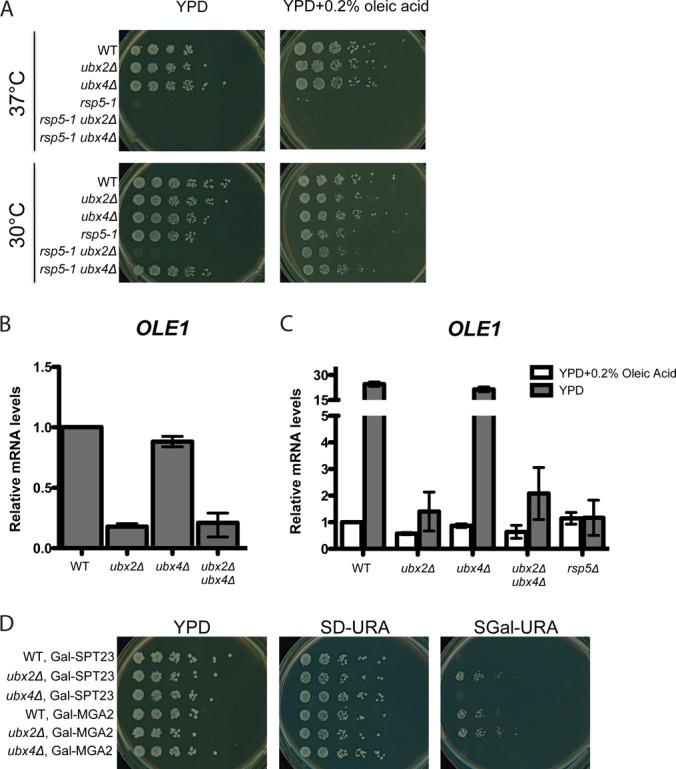
Ubx2 promotes inducible expression of OLE1. A, Synthetic lethality of rsp5-1 and ubx2Δ mutations. Fivefold serial dilutions of WT, rsp5-1, ubx2Δ, ubx4Δ, rsp5-1 ubx2Δ, and rsp5-1 ubx4Δ cells were plated on YPD and YPD+0.2% oleic acid and incubated at 30 °C or 37 °C for 2 days. B, C, Expression of OLE1 mRNA in ubx2Δ cells grown in YPD (B) or grown first in YPD +0.2% oleic acid and shifted to YPD for two hours (C). RNA from cells grown to log-phase in YPD medium was used for RT-qPCR to assess OLE1 mRNA levels. Values were normalized to ACT1. Error bars denote S.D.; n = 2. D, Toxicity because of Spt23 and Mga2 overexpression is suppressed by ubx2Δ. WT, ubx2Δ, and ubx4Δ strains transformed with a 2 μm plasmid that contains either GAL-SPT23 or GAL-MGA2 were plated onto YPD and synthetic dextrose- or galactose- containing media and grown for 4 days at 30 °C.
The results above suggested that Ubx2 is involved in activation of Spt23 and Mga2. As further confirmation, we examined the effect of Spt23 and Mga2 overexpression in mutant cells (Fig. 4D). Although UFAs are required for survival, the excess production of oleic acid that occurs on hyperactivation of OLE1 expression is toxic (50). Whereas overexpression of Spt23 or Mga2 from a galactose-inducible promoter inhibited growth of wild type cells, ubx2Δ cells, like rsp5-1 cells (51), were more tolerant (Fig. 4D), even though they equally overexpressed these OLE1 activators (data not shown).
Ubx2 Regulates Processing of Spt23
We next wanted to determine if Ubx2 is involved in processing of the OLE1 activators from their p120 precursor forms to their p90 active forms. For these experiments we focused on Spt23 because we found it difficult to consistently detect tagged Mga2 expressed from its endogenous locus. We examined steady state levels of mycSpt23V5 in YPD-grown cells deficient in various components of the Cdc48 and proteasome pathways (Fig. 5A). As anticipated from prior results (24), the cim3-1 and rsp5-1 mutants exhibited a severe defect in p90 accumulation (Fig. 5A, 5B; cim3-1 is a temperature-sensitive allele of the gene that encodes proteasome subunit Rpt6). Notably, p90 accumulation was also significantly impaired in cdc48-3 and ufd1-2 strains. In ubx2Δ (Fig. 5A, 5B) but not in other ubxΔ mutants (Fig. 5B), ubiquitinated high MW forms of mycSpt23V5 accumulated and the p90:p120 ratio was significantly reduced. However, the degree to which processing to p90 was defective varied in different experiments. In the prevailing model, Cdc48-Ufd1-Npl4 acts primarily after the proteasome to promote release of p90 from the ER membrane (24, 31, 52). However, it has also been reported that Cdc48-Ufd1-Npl4 is required for processing of p120 to p90 (25). Our data are consistent with those of Hitchcock et al. and suggest that Cdc48-Ufd1 and Ubx2 make important contributions to efficient formation of p90.
After establishing a role for Ubx2 in Spt23 processing, we next investigated the contribution of its UBA and UBX domains to this reaction. We immunoblotted for mycSpt23V5 in ubx2Δ strains in which full-length and various deletion mutants of UBX2 were integrated at the LEU2 locus under control of the UBX2 promoter (53) (Fig. 5C). This experiment confirmed that the defect in Spt23 processing in ubx2Δ cells was because of lack of Ubx2, and suggested that the UBX domain played a more important role in processing than the UBA domain. RT-PCR analysis of these same strains grown in oleic acid and switched to oleic acid-free medium revealed a large decrease in OLE1 mRNA induction in the UBAΔ UBXΔ mutant but not the single mutants (data not shown).
Ubx2 is orthologous to human Ubxd8, which may directly sense UFAs (54, 55). We therefore tested whether Ubx2 was required to sense UFAs in yeast. To address this question we grew wild type and mutant cells with and without oleic acid and evaluated processing of mycSpt23V5. We reasoned if Ubx2 is required to sense UFAs, that the reduced level of p90 formed in ubx2Δ would be insensitive to their presence. Processing of mycSpt23V5 in wild type cells was strongly repressed by addition of oleic acid (Fig. 5D). Likewise, the residual formation of p90 observed in cim3-1, cdc48-3, and ubx2Δ cells remained sensitive to inhibition by oleic acid. Consistent with this, our analysis of OLE1 mRNA revealed that although its levels were severely reduced in ubx2Δ and ubx2Δ ubx4Δ, they were diminished further on addition of oleic acid (Fig. 4C). Taken together, these data indicate that there must be at least one UFA sensor that remained in ubx2Δ cells. Careful inspection of the modification state of mycSpt23V5 in cdc48-3 and ubx2Δ cells grown with or without oleic acid suggested that sensing of UFAs occurred at or before Rsp5-dependent ubiquitination (Fig. 5D).
To investigate in greater depth the role of Ubx2 in Spt23 processing, we performed cycloheximide chase experiments (Fig. 5E). The p90 form of mycSpt23V5 was rapidly degraded in wild-type but was modestly stabilized in ubx2Δ and strongly stabilized in cdc48-3 cells. By contrast, p120 was more stable than p90 in all conditions whereas the high MW ubiquitin conjugates that accumulated in ubx2Δ and cdc48-3 were rapidly metabolized. Given that: (1) p90 was unstable in wild type, (2) more than half of mycSpt23V5 detected at zero-time in wild type was processed to p90, and (3) conversion of p120 to p90 was very slow and inefficient, we suggest that in wild type cells grown at 30 °C p120 became conjugated with ubiquitin and rapidly processed to p90 either during or very shortly after completion of synthesis, such that newly synthesized molecules that escaped modification were largely refractory to subsequent processing.
Ubx2 Associates with Spt23 and affects its Subcellular Localization
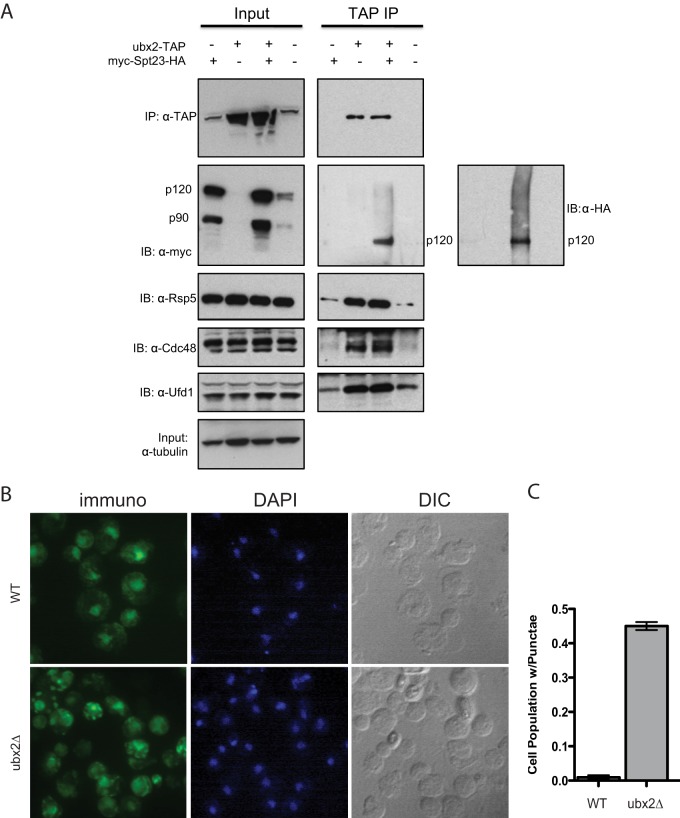
To test whether the effects of ubx2Δ on Spt23 were likely to be direct, TAP-tagged Ubx2 was immunoprecipitated from cells and immunoblotted for associated proteins (Fig. 6A). This experiment revealed a direct interaction between Ubx2 and unmodified p120 as well as higher MW ubiquitinated mycSpt23HA. Additionally, as expected (33), Cdc48 and Ufd1 were detected at levels above background in the Ubx2TAP immunoprecipitate, as was Rsp5, which had not previously been shown to interact with Ubx2.
Fig. 6.
Ubx2 interacts with Spt23 and influences its subcellular localization. A, Ubx2 directly interacts with Spt23 and Rsp5. RJD 6173 cells expressing Ubx2TAP from the natural locus were grown at 30 °C in YP plus 2% galactose to induce expression of Spt23. Native cell lysates were subjected to anti-TAP immunoprecipitation (IP) and the immunoprecipitates and input lysates were immunoblotted (IB) for TAP, mycSpt23HA, Cdc48, Ufd1, and Rsp5. Input extracts were also blotted for tubulin. B, Immunofluorescence localization of Spt23 in WT and ubx2Δ cells. WT (RJD 6172) and ubx2Δ (RJD 6182) cells expressing Spt23 from the GAL promoter were grown at 30 °C for 6 h in YP plus 2% galactose. All forms of Spt23 containing an intact N terminus were detected with anti-myc antibody and the nucleus was marked by staining DNA with DAPI. Nomarski optics (DIC) were used to identify the outline of each cell. C, Quantification of fraction of cell population with from (B) that contain Spt23 punctae. Seven fields of ∼300 cells were counted per strain. Error bars denote S.D.
To address the effect of ubx2Δ on subcellular localization of Spt23, we performed immunofluorescence on cells that expressed a pulse of mycSpt23 from the GAL1 promoter (attempts to detect mycSpt23V5 expressed from the natural locus were unsuccessful). Consistent with our observation that Ubx2 was required for OLE1 expression, mycSpt23 demonstrated a decrease in localization to the nucleus in most ubx2Δ cells (Fig. 6B). Additionally, in ∼45% of ubx2Δ cells, Spt23 was observed to accumulate in cytosolic punctae (Fig. 6C). Although we do not understand their exact molecular nature, the existence of these punctae in ubx2Δ together with poor nuclear accumulation point to a role for Ubx2 in proper localization of Spt23.
DISCUSSION
The Cdc48 Network Plays a Broad Role in Shaping the Ubiquitin Conjugate Proteome
To address globally the impact of Cdc48 and its Ubx adaptors on the UPS, we evaluated changes to the ubiquitin conjugate proteome in yeast cells deleted for individual UBX genes or carrying a temperature-sensitive cdc48-3 allele. Our findings support a broad role for the Cdc48 network in the UPS. Overall, we identified 1,916 putative ubiquitin conjugates and 1,733 were quantified in all experiments. Of the 1733, over 62% were found at statistically significant elevated levels relative to wild type in at least one of the mutant strains. Mutation of different UBX genes led to markedly distinct effects on the ubiquitin conjugate proteome. For example, our findings suggested novel and unexpected roles for Ubx2, Ubx5, and Ubx6/7 in regulation of vacuolar, plasma membrane, and nucleolar/ribosomal proteins, respectively. However, only a fraction of ubiquitin conjugates annotated with a given term were up-regulated in a particular ubxΔ mutant (e.g. not all ubiquitin-conjugated vacuolar proteins were found at increased levels in ubx2Δ). Thus, localization alone does not render a ubiquitin conjugate subject to regulation by a particular Ubx cofactor.
Our approach assumes that ubiquitin conjugates that are elevated in a particular mutant relative to wild type represent intermediates that accumulate on attenuation of Cdc48 activity or deletion of the adaptor that normally links them to Cdc48. However, increased levels of a conjugate may arise from increased expression of the protein. Of interest, numerous ubiquitin conjugates were selectively depleted in specific ubxΔ strains. This was not investigated further, but during the course of this work it was reported that the human UBX protein SAKS1 protects ERAD substrates from deubiquitination (56) and overproduced UBXD7 binds to and increases the fraction of Nedd8-conjugated Cul2 in human cells (48, 49). Hence, the large number of conjugate depletion events that we observed in ubxΔ cells may represent species whose ubiquitin-conjugated forms are stabilized on binding of Ubx proteins. Our extensive dataset provides a rich resource for future investigations into this and other questions related to the functions and regulation of the Cdc48 network.
Ubx2 and Cdc48 Promote Proteasome-Dependent Processing of Spt23
Prior work from the Jentsch and Haines laboratories emphasized a role for Cdc48-Ufd1-Npl4 in release of the processed forms of Spt23 (24) and Mga2 (31) from the ER membrane. By contrast, data from the Silver laboratory emphasized an upstream role for Cdc48-Ufd1-Npl4 in processing of both proteins (25). Our data support a role for Cdc48-Ufd1 and Ubx2 in both steps. We observe diminished levels of p90 in mutant cells. However, the reduced p90 accumulation in ubx2Δ is variable and is less striking than the defect in OLE1 induction. We propose that Cdc48-Ufd1-Npl4-Ubx2 acts sequentially to promote both the processing of Spt23 p120 to p90, and localization of p90 to the nucleus. A prominent role for Cdc48-Ufd1 and Ubx2 in proteasome-dependent cleavage of p120 resonates with data on other Cdc48-proteasome substrates, which consistently place Cdc48/p97 function upstream of (or contemporaneous with) the proteasome. Sequential roles for Ubx2 in the formation and activation of p90 helps explain why ubx2Δ exhibits such a severe defect in OLE1 regulation.
Consistent with the prior data on rsp5 mutants (24), ubx2Δ cells are severely deficient in induction of OLE1 expression and double mutants deficient in Ubx2 and Rsp5 activity fail to grow on normal medium at 30 °C but are rescued by the addition of oleic acid, pointing to a crucial role for Ubx2 in maintaining sufficient Ole1 activity to meet the cellular requirement for UFAs. An important and interesting question that remains unresolved is, how are levels of fatty acids sensed so that the cell maintains an appropriate ratio of SFAs:UFAs? Addition of exogenous UFAs suppresses processing of p120 to p90 by the Rsp5-Ubx2-Cdc48 pathway, which reduces expression of the desaturase enzyme encoded by OLE1. It was suggested in prior work that the human ortholog of Ubx2, UBXD8, directly senses UFAs (54, 55, 57). However, addition of UFAs suppresses accumulation of ubiquitinated Spt23 and further reduces the residual expression of OLE1 in ubx2Δ, indicating that these cells retain the ability to sense UFAs. Thus, if Ubx2 is a fatty acid sensor, it cannot be the only sensor in the OLE1 pathway. Very recently, it was reported that Ire1 serves as a sensor for membrane lipid saturation in mouse cells (58). Our data suggest that either Rsp5 itself or a step upstream of Rsp5-dependent ubiquitination must be capable of sensing UFAs.
Supplementary Material
Acknowledgments
We thank S. Jentsch, L. Hicke, T. Sommer, E. Jarosch, and C.-W. Wang for gifts of strains, plasmids, antibodies, and W. den Besten for help with strain construction and valuable discussions. We also thank R. Ernst for communicating results before publication and M. Mann for hosting N.K. for a visit.
Footnotes
* This work was supported by the Howard Hughes Medical Institute, of which R.J.D. is an Investigator. The Proteome Exploration Laboratory is supported by the Gordon and Betty Moore Foundation through Grant GBMF775 and the Beckman Institute.
 This article contains supplemental Figs. S1 to S4 and Tables S1 to S5.
This article contains supplemental Figs. S1 to S4 and Tables S1 to S5.
1 The abbreviations used are:
- ERAD
- endoplasmic reticulum associated protein degradation
- UPS
- ubiquitin protease system
- ER
- endoplasmic reticulum
- SFA
- saturated fatty acids
- UFA
- unsaturated fatty acids
- YPD
- yeast extract/peptone/dextrose.
REFERENCES
- 1. Stolz A., Hilt W., Buchberger A., Wolf D. H. (2011) Cdc48: a power machine in protein degradation. Trends Biochem. Sci. 36, 515–523 [DOI] [PubMed] [Google Scholar]
- 2. Meusser B., Hirsch C., Jarosch E., Sommer T. (2005) ERAD: the long road to destruction. Nat. Cell Biol. 7, 766–772 [DOI] [PubMed] [Google Scholar]
- 3. Meyer H., Bug M., Bremer S. (2012) Emerging functions of the VCP/p97 AAA-ATPase in the ubiquitin system. Nat. Cell Biol. 14, 117–123 [DOI] [PubMed] [Google Scholar]
- 4. Ye Y. (2006) Diverse functions with a common regulator: ubiquitin takes command of an AAA ATPase. J. Struct. Biol. 156, 29–40 [DOI] [PubMed] [Google Scholar]
- 5. Johnson J. O., Mandrioli J., Benatar M., Abramzon Y., Van Deerlin V. M., Trojanowski J. Q., Gibbs J. R., Brunetti M., Gronka S., Wuu J., Ding J., McCluskey L., Martinez-Lage M., Falcone D., Hernandez D. G., Arepalli S., Chong S., Schymick J. C., Rothstein J., Landi F., Wang Y. D., Calvo A., Mora G., Sabatelli M., Monsurrò M. R., Battistini S., Salvi F., Spataro R., Sola P., Borghero G., Galassi G., Scholz S. W., Taylor J. P., Restagno G., Chiò A., Traynor B. J. (2010) Exome sequencing reveals VCP mutations as a cause of familial ALS. Neuron 68, 857–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Neumann M., Kwong L. K., Truax A. C., Vanmassenhove B., Kretzschmar H. A., Van Deerlin V. M., Clark C. M., Grossman M., Miller B. L., Trojanowski J. Q., Lee V. M. (2007) TDP-43-positive white matter pathology in frontotemporal lobar degeneration with ubiquitin-positive inclusions. J. Neuropathol. Exp. Neurol. 66, 177–183 [DOI] [PubMed] [Google Scholar]
- 7. Chapman E., Fry A. N., Kang M. (2011) The complexities of p97 function in health and disease. Mol. Biosyst. 7, 700–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Haines D. S. (2010) p97-containing complexes in proliferation control and cancer: emerging culprits or guilt by association? Genes Cancer 1, 753–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Watts G. D., Thomasova D., Ramdeen S. K., Fulchiero E. C., Mehta S. G., Drachman D. A., Weihl C. C., Jamrozik Z., Kwiecinski H., Kaminska A., Kimonis V. E. (2007) Novel VCP mutations in inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia. Clin. Genet. 72, 420–426 [DOI] [PubMed] [Google Scholar]
- 10. Chou T. F., Deshaies R. J. (2011) Development of p97 AAA ATPase inhibitors. Autophagy 7, 1091–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bursavich M. G., Parker D. P., Willardsen J. A., Gao Z. H., Davis T., Ostanin K., Robinson R., Peterson A., Cimbora D. M., Zhu J. F., Richards B. (2010) 2-Anilino-4-aryl-1,3-thiazole inhibitors of valosin-containing protein (VCP or p97). Bioorg. Med. Chem. Lett. 20, 1677–1679 [DOI] [PubMed] [Google Scholar]
- 12. Kloppsteck P., Ewens C. A., Förster A., Zhang X., Freemont P. S. (2012) Regulation of p97 in the ubiquitin-proteasome system by the UBX protein-family. Biochim. Biophys. Acta 1823, 125–129 [DOI] [PubMed] [Google Scholar]
- 13. Schuberth C., Buchberger A. (2008) UBX domain proteins: major regulators of the AAA ATPase Cdc48/p97. Cell Mol. Life Sci. 65, 2360–2371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wilcox A. J., Laney J. D. (2009) A ubiquitin-selective AAA-ATPase mediates transcriptional switching by remodelling a repressor-promoter DNA complex. Nat. Cell Biol. 11, 1481–1486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Böhm S., Buchberger A. (2013) The budding yeast Cdc48(Shp1) complex promotes cell cycle progression by positive regulation of protein phosphatase 1 (Glc7). PLoS One 8, e56486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kirchner P., Bug M., Meyer H. (2013) Ubiquitination of the N-terminal Region of Caveolin-1 Regulates Endosomal Sorting by the VCP/p97 AAA-ATPase. J. Biol. Chem. 288, 7363–7372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Alexandru G., Graumann J., Smith G. T., Kolawa N. J., Fang R., Deshaies R. J. (2008) UBXD7 binds multiple ubiquitin ligases and implicates p97 in HIF1alpha turnover. Cell 134, 804–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Verma R., Oania R., Fang R., Smith G. T., Deshaies R. J. (2011) Cdc48/p97 mediates UV-dependent turnover of RNA Pol II. Mol. Cell 41, 82–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang L., Zhou F., Li Y., Drabsch Y., Zhang J., van Dam H., ten Dijke P. (2012) Fas-associated factor 1 is a scaffold protein that promotes beta-transducin repeat-containing protein (beta-TrCP)-mediated beta-catenin ubiquitination and degradation. J. Biol. Chem. 287, 30701–30710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Barbin L., Eisele F., Santt O., Wolf D. H. (2010) The Cdc48-Ufd1-Npl4 complex is central in ubiquitin-proteasome triggered catabolite degradation of fructose-1,6-bisphosphatase. Biochem. Biophys. Res. Commun. 394, 335–341 [DOI] [PubMed] [Google Scholar]
- 21. Lee J. J., Park J. K., Jeong J., Jeon H., Yoon J. B., Kim E. E., Lee K. J. (2013) Complex of Fas-associated Factor 1 (FAF1) with Valosin-containing Protein (VCP)-Npl4-Ufd1 and Polyubiquitinated Proteins Promotes Endoplasmic Reticulum-associated Degradation (ERAD). J. Biol. Chem. 288, 6998–7011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lee J. J., Kim Y. M., Jeong J., Bae D. S., Lee K. J. (2012) Ubiquitin-associated (UBA) domain in human Fas associated factor 1 inhibits tumor formation by promoting Hsp70 degradation. PLoS One 7, e40361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hoppe T., Matuschewski K., Rape M., Schlenker S., Ulrich H. D., Jentsch S. (2000) Activation of a membrane-bound transcription factor by regulated ubiquitin/proteasome-dependent processing. Cell 102, 577–586 [DOI] [PubMed] [Google Scholar]
- 24. Rape M., Hoppe T., Gorr I., Kalocay M., Richly H., Jentsch S. (2001) Mobilization of processed, membrane-tethered SPT23 transcription factor by CDC48(UFD1/NPL4), a ubiquitin-selective chaperone. Cell 107, 667–677 [DOI] [PubMed] [Google Scholar]
- 25. Hitchcock A. L., Krebber H., Frietze S., Lin A., Latterich M., Silver P. A. (2001) The conserved npl4 protein complex mediates proteasome-dependent membrane-bound transcription factor activation. Mol. Biol. Cell 12, 3226–3241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Martin C. E., Oh C. S., Jiang Y. (2007) Regulation of long chain unsaturated fatty acid synthesis in yeast. Biochim. Biophys. Acta 1771, 271–285 [DOI] [PubMed] [Google Scholar]
- 27. Zhang S., Skalsky Y., Garfinkel D. J. (1999) MGA2 or SPT23 is required for transcription of the delta9 fatty acid desaturase gene, OLE1, and nuclear membrane integrity in Saccharomyces cerevisiae. Genetics 151, 473–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shcherbik N., Zoladek T., Nickels J. T., Haines D. S. (2003) Rsp5p is required for ER bound Mga2p120 polyubiquitination and release of the processed/tethered transactivator Mga2p90. Curr. Biol. 13, 1227–1233 [DOI] [PubMed] [Google Scholar]
- 29. Piwko W., Jentsch S. (2006) Proteasome-mediated protein processing by bidirectional degradation initiated from an internal site. Nat. Struct. Mol. Biol. 13, 691–697 [DOI] [PubMed] [Google Scholar]
- 30. Shcherbik N., Kee Y., Lyon N., Huibregtse J. M., Haines D. S. (2004) A single PXY motif located within the carboxyl terminus of Spt23p and Mga2p mediates a physical and functional interaction with ubiquitin ligase Rsp5p. J. Biol. Chem. 279, 53892–53898 [DOI] [PubMed] [Google Scholar]
- 31. Shcherbik N., Haines D. S. (2007) Cdc48p(Npl4p/Ufd1p) binds and segregates membrane-anchored/tethered complexes via a polyubiquitin signal present on the anchors. Mol. Cell 25, 385–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Braun S., Matuschewski K., Rape M., Thoms S., Jentsch S. (2002) Role of the ubiquitin-selective CDC48(UFD1/NPL4)chaperone (segregase) in ERAD of OLE1 and other substrates. EMBO J. 21, 615–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rumpf S., Jentsch S. (2006) Functional division of substrate processing cofactors of the ubiquitin-selective Cdc48 chaperone. Mol. Cell 21, 261–269 [DOI] [PubMed] [Google Scholar]
- 34. Kandasamy P., Vemula M., Oh C. S., Chellappa R., Martin C. E. (2004) Regulation of unsaturated fatty acid biosynthesis in Saccharomyces: the -endoplasmic reticulum membrane protein, Mga2p, a transcription activator of the OLE1 gene, regulates the stability of the OLE1 mRNA through exosome-mediated mechanisms. J. Biol. Chem. 279, 36586–36592 [DOI] [PubMed] [Google Scholar]
- 35. Mayor T., Deshaies R. J. (2005) Two-step affinity purification of multiubiquitylated proteins from Saccharomyces cerevisiae. Methods Enzymol. 399, 385–392 [DOI] [PubMed] [Google Scholar]
- 36. Gomez T. A., Kolawa N., Gee M., Sweredoski M. J., Deshaies R. J. (2011) Identification of a functional docking site in the Rpn1 LRR domain for the UBA-UBL domain protein Ddi1. BMC Biol. 9, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wiśniewski J. R., Zougman A., Mann M. (2009) Combination of FASP and StageTip-based fractionation allows in-depth analysis of the hippocampal membrane proteome. J. Proteome Res. 8, 5674–5678 [DOI] [PubMed] [Google Scholar]
- 38. Lee J. E., Sweredoski M. J., Graham R. L., Kolawa N. J., Smith G. T., Hess S., Deshaies R. J. (2011) The steady-state repertoire of human SCF ubiquitin ligase complexes does not require ongoing Nedd8 conjugation. Mol. Cell. Proteomics 10, M110.006460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kalli A., Hess S. (2012) Effect of mass spectrometric parameters on peptide and protein identification rates for shotgun proteomic experiments on an LTQ-orbitrap mass analyzer. Proteomics 12, 21–31 [DOI] [PubMed] [Google Scholar]
- 40. Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 41. Pierce N. W., Lee J. E., Liu X., Sweredoski M. J., Graham R. L., Larimore E. A., Rome M., Zheng N., Clurman B. E., Hess S., Shan S. O., Deshaies R. J. (2013) Cand1 Promotes Assembly of New SCF Complexes through Dynamic Exchange of F Box Proteins. Cell 153, 206–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Huang da W., Sherman B. T., Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 43. Huang da W., Sherman B. T., Lempicki R. A. (2009) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Benjamini Y., Drai D., Elmer G., Kafkafi N., Golani I. (2001) Controlling the false discovery rate in behavior genetics research. Behav. Brain Res. 125, 279–284 [DOI] [PubMed] [Google Scholar]
- 45. Decottignies A., Evain A., Ghislain M. (2004) Binding of Cdc48p to a ubiquitin-related UBX domain from novel yeast proteins involved in intracellular proteolysis and sporulation. Yeast 21, 127–139 [DOI] [PubMed] [Google Scholar]
- 46. DeBose-Boyd R. A. (2008) Feedback regulation of cholesterol synthesis: sterol-accelerated ubiquitination and degradation of HMG CoA reductase. Cell Res. 18, 609–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yen J. L., Flick K., Papagiannis C. V., Mathur R., Tyrrell A., Ouni I., Kaake R. M., Huang L., Kaiser P. (2012) Signal-induced disassembly of the SCF ubiquitin ligase complex by Cdc48/p97. Mol .Cell 48, 288–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. den Besten W., Verma R., Kleiger G., Oania R. S., Deshaies R. J. (2012) NEDD8 links cullin-RING ubiquitin ligase function to the p97 pathway. Nat. Struct. Mol. Biol. 19, 511–516, S511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bandau S., Knebel A., Gage Z. O., Wood N. T., Alexandru G. (2012) UBXN7 docks on neddylated cullin complexes using its UIM motif and causes HIF1alpha accumulation. BMC Biol. 10, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Stukey J. E., McDonough V. M., Martin C. E. (1989) Isolation and characterization of OLE1, a gene affecting fatty acid desaturation from Saccharomyces cerevisiae. J. Biol. Chem. 264, 16537–16544 [PubMed] [Google Scholar]
- 51. Kee Y., Lyon N., Huibregtse J. M. (2005) The Rsp5 ubiquitin ligase is coupled to and antagonized by the Ubp2 deubiquitinating enzyme. EMBO J. 24, 2414–2424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Jentsch S., Rumpf S. (2007) Cdc48 (p97): a “molecular gearbox” in the ubiquitin pathway? Trends Biochem. Sci. 32, 6–11 [DOI] [PubMed] [Google Scholar]
- 53. Wang C. W., Lee S. C. (2012) The ubiquitin-like (UBX)-domain-containing protein Ubx2/Ubxd8 regulates lipid droplet homeostasis. J. Cell Sci. 125, 2930–2939 [DOI] [PubMed] [Google Scholar]
- 54. Lee J. N., Zhang X., Feramisco J. D., Gong Y., Ye J. (2008) Unsaturated fatty acids inhibit proteasomal degradation of Insig-1 at a postubiquitination step. J. Biol. Chem. 283, 33772–33783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lee J. N., Kim H., Yao H., Chen Y., Weng K., Ye J. (2010) Identification of Ubxd8 protein as a sensor for unsaturated fatty acids and regulator of triglyceride synthesis. Proc. Natl. Acad. Sci. U. S. A. 107, 21424–21429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. LaLonde D. P., Bretscher A. (2011) The UBX protein SAKS1 negatively regulates endoplasmic reticulum-associated degradation and p97-dependent degradation. J. Biol. Chem. 286, 4892–4901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Olzmann J. A., Richter C. M., Kopito R. R. (2013) Spatial regulation of UBXD8 and p97/VCP controls ATGL-mediated lipid droplet turnover. Proc. Natl. Acad. Sci. U. S. A. 110, 1345–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Volmer R., van der Ploeg K., Ron D. (2013) Membrane lipid saturation activates endoplasmic reticulum unfolded protein response transducers through their transmembrane domains. Proc. Natl. Acad. Sci. U. S. A. 110, 4628–4633 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.