Abstract
We report the first proteomic analysis of the SLP76 interactome in resting and activated primary mouse mast cells. This was made possible by a novel genetic approach used for the first time here. It consists in generating knock-in mice that express signaling molecules bearing a C-terminal tag that has a high affinity for a streptavidin analog. Tagged molecules can be used as molecular baits to affinity-purify the molecular complex in which they are engaged, which can then be studied by mass spectrometry. We examined first SLP76 because, although this cytosolic adapter is critical for both T cell and mast cell activation, its role is well known in T cells but not in mast cells. Tagged SLP76 was expressed in physiological amounts and fully functional in mast cells. We unexpectedly found that SLP76 is exquisitely sensitive to mast cell granular proteases, that Zn2+-dependent metalloproteases are especially abundant in mast cells and that they were responsible for SLP76 degradation. Adding a Zn2+ chelator fully protected SLP76 in mast cell lysates, thereby enabling an efficient affinity-purification of this adapter with its partners. Label-free quantitative mass spectrometry analysis of affinity-purified SLP76 interactomes uncovered both partners already described in T cells and novel partners seen in mast cells only. Noticeably, molecules inducibly recruited in both cell types primarily concur to activation signals, whereas molecules recruited in activated mast cells only are mostly associated with inhibition signals. The transmembrane adapter LAT2, and the serine/threonine kinase with an exchange factor activity Bcr were the most recruited molecules. Biochemical and functional validations established the unexpected finding that Bcr is recruited by SLP76 and positively regulates antigen-induced mast cell activation. Knock-in mice expressing tagged molecules with a normal tissue distribution and expression therefore provide potent novel tools to investigate signalosomes and to uncover novel signaling molecules in mast cells.
Mast cells play critical roles in the initiation of IgE-dependent allergic inflammation (1). They express high-affinity receptors for the Fc portion of IgE (FcεRI)1, which are prototypic immunoreceptors (2). Mast cell FcεRI are composed of an IgE-binding subunit, FcεRIα, and of two Immunoreceptor Tyrosine-based Activation Motif (ITAM)-containing subunits, FcRβ and FcRγ (3). FcεRI activate mast cells when receptor-bound IgE antibodies are aggregated by a multivalent specific antigen (4). FcεRI aggregation triggers the constitution of signalosomes in which positive and negative signals are generated, the integration of which determines quantitatively and qualitatively the biological responses of the mast cell. The composition of signalosomes is likely to evolve rapidly, as molecules are sequentially recruited and as enzymes act on their substrates. Determining the composition and describing the dynamics of FcεRI signalosomes is a major challenge for who aims at understanding fundamental mechanisms of allergy and at developing therapeutic tools for controlling allergic reactions.
Mass spectrometry (MS)-based proteomics has emerged as a powerful tool to study signaling networks. Indeed, it enables large-scale analysis of stimulus-induced post-translational modifications (5–7). MS-based proteomics has also been used to identify molecular partners that, at any given time, are associated with a molecular bait of interest (8). This bait carries an affinity-purification tag that markedly enhances the efficiency with which it can be purified from a cell lysate (9). The experimental conditions used may, however, limit the significance of the result. Classically, baits are over-expressed in cells that already express an untagged endogenous version of the bait. Unbalanced expression of corresponding molecules may profoundly alter biological responses. In some cases, baits are expressed in cells that do not normally express the molecule, where they may generate artifactual signalosomes. Finally, baits are often expressed in transformed cells that can be grown in high numbers, so that affinity-purified molecules are obtained in amounts amenable to MS analysis (10, 11). Signaling pathways can be constitutively activated in these cells, because of the expression of transforming oncogenes.
To overcome these problems, we generated a series of knock-in (KI) mice expressing each a key signaling molecule with a C-terminal one-strep-tag (OST) (12). OST-tagged molecules, as well as the molecules with which they interact can be affinity-purified using beads coated with Strep-Tactin. Strep-Tactin is a streptavidin derivative that has a high affinity for OST (12). Affinity-purified molecules can then be identified by MS. As documented here, this novel approach enables one to study the interactome of endogenous OST-tagged molecules that are present in physiological amounts, in nontransformed cells by which they are normally expressed. Combining the interactomes of each tagged molecule analyzed before and at different times after FcεRI engagement should ultimately make it possible to obtain a comprehensive dynamic functional map of the FcεRI signalosome. As a proof-of-concept, we studied the interactome of the Src Homology (SH)2 domain-containing leukocyte protein of 76 kDa (SLP76) (a.k.a. Lymphocyte cytosolic protein 2 or LCP2) in primary mouse mast cells.
SLP76 is a cytosolic adapter that nucleates signaling complexes generated by immunoreceptors (13). It contains an N-terminal leucine Zip motif, a tyrosine-rich domain, a proline-rich domain and a C-terminal SH2 domain. It is constitutively associated with the growth factor receptor-bound protein 2 (Grb2)-related adapter protein 2 (Grap2, a.k.a. GADS) through the interaction of its proline residues with one of the SH3 domains of Grap2 (14). SLP76 was mostly studied in T lymphocytes. Following T-cell receptor (TCR) engagement, Grap2 binds to phosphorylated residues in the raft-associated transmembrane adapter linker of activation of T cells LAT1. SLP76-Grap2 complexes bind to phospholipase C (PLC)γ-1 (15), which stabilizes the recruitment of this enzyme by LAT1. SLP76 tyrosines are phosphorylated by the cytosolic kinase zeta-associated protein of 70 kDa (ZAP70) (16, 17), which provides binding sites for the SH2 domains of Tec kinases such as the IL-2-inducible T-cell kinase (Itk)(18), for the guanine nucleotide exchange factors Vav-1 (17) and for adapters such as the noncatalytic region of tyrosine kinase adapter protein (Nck) (19). Molecules recruited by SLP76 launch the intracellular propagation of signals leading to calcium mobilization, the activation of Mitogen-activated protein (MAP) kinases and, ultimately, full-blown T-cell responses.
SLP76 is also expressed by mast cells, and mast cell activation was markedly impaired in SLP76-deficient mice (20). Surprisingly, signaling molecules that interact with SLP76 in mast cells are not well characterized, and only a few papers have been published on the subject. It was however shown that the function of SLP76 in mast cells may not depend entirely on LAT1, as it does in T cells. Indeed, LAT1 deficiency could be, at least in part, compensated by the presence of LAT2 in mast cells (21). LAT2 (a.k.a. non-T-cell activation linker or NTAL) is a raft-associated tyrosine-rich transmembrane adapter related to LAT1. LAT2 is absent in resting T cells, but present in B cells where it fulfills some of the functions exerted by LAT1 in T cells. In mast cells, LAT1 and LAT2 function as a pair of antagonistic molecules (22). The role of SLP76 in mast cells cannot therefore be deduced from that of SLP76 in T cells, and the mechanism(s) by which it contributes to mast cell activation needs to be established.
We report here the first description of the SLP76 interactome in resting and activated primary cultured mast cells. This was done by combining high-affinity purification and label-free quantitative proteomics in mast cells derived from KI mice expressing SLP-76 with a C-terminal OST (Slp76OST/OST mice). To reach this goal, we first unraveled and solved an unexpected problem because of proteases contained in high amounts in mast cell granules. We found that, compared with the T-cell SLP76 interactome, the mast cell SLP76 interactome is enriched in molecules involved in negative signaling. We also found that the Breakpoint Cluster Region protein Bcr is inducibly recruited by SLP76 and we could validate this unexpected finding by showing that mast cells from Bcr-deficient mice released less granular mediators than mast cells from wt mice. This is the first evidence that Bcr contributes to FcεRI signaling.
EXPERIMENTAL PROCEDURES
Generation of Slp76OST Mice
A 6.2-kb genomic fragment containing exons 19–21 of the Slp76 gene was isolated from a BAC clone (clone # RP23–216O16A; http://www.lifesciences.sourcebioscience) of C57BL/6J origin. A OST-(Stop)2loxP-tACE-CRE-PGK-gb2-neo-loxP cassette (23) was introduced at the 3′ end of the Slp76 coding sequence found in exon 21 using homologous recombination (24). The targeting construct was then abutted to a thymidine kinase expression cassette and linearized with FseI. JM8.F6 C57BL/6N ES cells (25) were electroporated with the Slp76OST targeting vector and selected in G418 and gancyclovir. Colonies were screened for homologous recombination by Southern blot. A neomycin-specific probe was used to ensure that adventitious non-homologous recombination events had not occurred in the selected ES clones. Appropriately targeted ES cells were injected into FVB blastocysts. After germline transmission, screening for the deletion of the neo cassette and the presence of the intended modification was performed by PCR and Southern blot. All experiments were done in accordance with French and European guidelines for animal care.
Cells
Bone marrow-derived mast cells (BMMC): BMMC were generated from femoral bone marrow harvested from Slp76OST/OST and C57BL/6 (Slp76+/+) control mice (Charles River, France), from Bcr−/− and littermate control (Bcr+/+) mice (a gift from Dr. Eunjoon Kim, Korea Advanced Institute of Science and Technology, Daejeon, South Korea), and from Dok-3−/− and littermate control (Dok-3+/+) mice. BMMC were propagated and cultured in OptiMEM (Invitrogen, St Aubin, France) supplemented with 10% FBS (PAA Laboratories, Austria), 50 μm 2-mercaptoethanol (Invitrogen), 100 μg/ml Penicillin-Streptomycin (Invitrogen), and 2 ng/ml recombinant murine IL-3 (Biolegend, San Diego, CA). Homogeneous populations of FcεRI+, Kit+ cells were obtained after 3 weeks of culture. Thymocytes: Thymi were collected from 6-week old C57BL/6 mice. Red cells were lysed in 10 mm Tris-HCL pH 7.5 and 142 mm NH4Cl.
Gene Expression Analysis
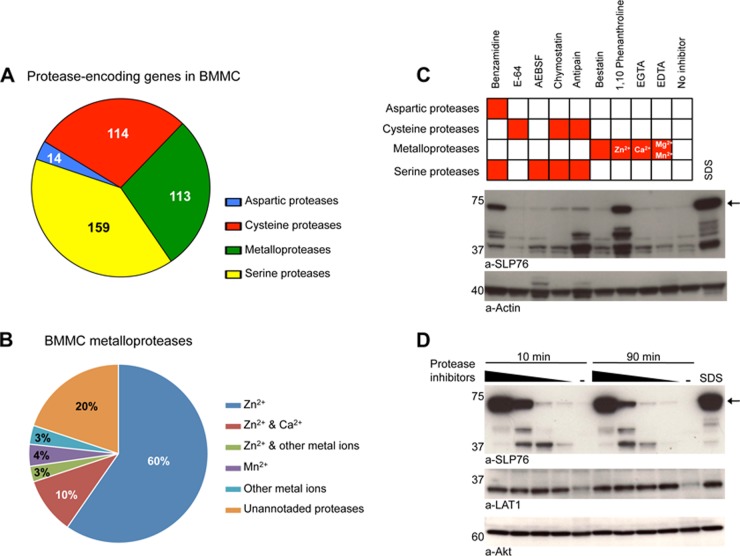
Three independent cultures were seeded from the bone marrow of three individual mice and BMMC were generated as described. Total RNA was extracted from resting BMMC from the three replicate cultures using TRIzol reagent (Invitrogen, Invitrogen), and RNeasy mini kit from Qiagen (Courtaboeuf, France). Purified total RNA was amplified by RT-PCR, and transcripts were analyzed using the Applied Biosystems AB1700 microarrays (Carlsbad, California). We estimated the number of proteases expressed in BMMC, according to the procedure described in Supplementary Materials. A total of 539 out of the 685 annotated protease-coding transcripts were found to be represented in the AB1700 microarrays. Among them, 400 genes were found expressed in BMMC with a confidence level greater than 90% (Fig. 3A). Supplemental Table S2 reports the results for the expression of the 539 protease-coding genes covered by the AB1700 microarrays.
Fig. 3.
A new protease inhibitor mixture prevents SLP76 degradation in mast cells. A, Families of proteases detected in BMMC by transcriptomic analysis. B, Families of metal-binding protease encoded by BMMC metalloprotease genes. C, Screening protease inhibitors that can protect SLP76 in BMMC lysates. BMMC were solubilized in LM lysis buffer containing specific protease inhibitors. Equal amounts of lysates were electrophoresed and Western blotted with anti-SLP76 and anti-actin antibodies. D, Total protection of SLP76 degradation. BMMC were solubilized in LM lysis buffer supplemented with a new custom-made inhibitor mixture for 10 min or 90 min. Equal amounts of lysates were electrophoresed and Western blotted with anti-SLP76 and anti-actin antibodies.
Mast Cell Stimulation
BMMC were sensitized overnight with IgE anti-DNP (mAb 2682-I) supernatant in complete culture medium (100 ng/ml IgE, final concentration). Cells were extensively washed with HEPES-containing RPMI (Invitrogen), and challenged for various periods of time at 37 °C with 100 ng/ml DNP-HSA (Sigma-Aldrich, St. Louis, MO) in the same medium.
β-hexosaminidase Release
IgE anti-DNP-sensitized cells were challenged for 20 min at 37 °C with the indicated concentrations of DNP-HSA. β-hexosaminidase released in cell supernatants was measured using an enzymatic assay (26).
Cell lysates
Laurylmaltoside (LM) lysates: BMMC or thymocytes were solubilized on ice for 10 min in lysis buffer consisting of 100 mm Tris-HCL, pH 7.4 or pH 8.0, 150 mm NaCl, 2 mm MgCl2, 5% glycerol, 0.2% LM (Thermo Scientific, Courtaboeuf, France), and 25 U/ml Benzonase (Merck, Darmstadt, Germany), supplemented with specific protease inhibitors (supplemental Table S4) or without. Protease inhibitor cocktails were from Roche (Mannheim, Germany, Cat. #11 697 498 001), and from Sigma-Aldrich (Cat. #P8340. Phosphoramidon (Cat. #R7385), EDTA and EGTA were from Sigma-Aldrich. SDS lysates: BMMC or thymocytes were lysed by boiling at 90 °C for 5 min in 1% SDS and 100 mm Tris-HCL pH 8.0. 25 U/ml Benzonase and 2 mm MgCl2 was added to SDS lysate that was left on ice for 10 min.
Western Blotting Analysis
LM and SDS cell lysates were centrifuged at 14,000 rpm for 15 min at 4 °C and supernatants were electrophoresed on 4–12% Criterion XT precast gel (Bio-Rad, Marnes-la-Coquette, France) using XT MOPS (Bio-Rad) running buffer, and transferred onto nitrocellulose Hybond-P membranes (Amersham Biosciences, Velizy-Villacoublay, France). Membranes were saturated either with 5% BSA (Sigma-Aldrich) or 5% skimmed milk (Régilait, Saint-Martin-Belle-Roche, France) for 1 h at room temperature and blotted overnight at 4 °C with the indicated antibodies. Rabbit anti-SLP76, anti-Akt, anti-Erk1/2, anti-phospho-Erk1/2, anti-phospho-p38 and mouse anti-phospho-Tyrosine (P-Tyr-100) antibodies were from Cell Signaling Technology (St-Quentin-en-Yvelines, France). Rabbit anti-LAT1 and anti-phospho-LAT1, mouse anti-Vav, rabbit anti-Sos antibodies were from Upstate Biotechnology (Lake placid, NY). Goat anti-actin, mouse anti-PLCγ-1, anti-Dok-3 and anti-SHIP1, Rabbit anti-Fyn, anti-Bcr, anti-phopsho-Bcr (pY177), anti-Lyn and anti-phospho-PLCγ-1 antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA). Mouse anti-LAT2 antibodies were from Alexis Biochemical (Lausanne, Switzerland). Rabbit anti-PAG antibodies were from ExBio (Praha, Czech Republic). Mouse anti-Grb2 and anti-phospho SLP76 antibodies were from BD Bioscience (Le-Pont-de-Claix, France). Goat anti-Dok-3 antibody was from Abcam (Paris, France). HRP-conjugated secondary antibodies were from Santa Cruz Biotechnology. Mouse anti-phosphotyrosine antibodies (mAb 4G10) were a gift from Dr. S. Latour (Inserm U.768, Hôpital Necker, Paris, France). HRP was detected using an ECL kit (Thermo Scientific).
Strep-Tactin Affinity-Purification
Four independent affinity purifications were performed on each set of samples (Slp76+/+ and Slp76OST/OST BMMC challenged with antigen or without). One × 108 Slp76+/+ or Slp76OST/OST BMMC were lysed for 10 min at 0 °C with buffer containing 100 mm Tris-HCL, pH 8, 150 mm NaCl, 2 mm MgCl2, 5% glycerol, 0.2% LM, 1 mm sodium orthovanadate, 5 mm sodium floride, 25 U/ml Benzonase and mast cells complete inhibitor mixture (supplemental Table S4). Cell extracts were centrifuged at 14,000 rpm for 15 min at 4 °C. Cleared lysates were incubated with 200 μl prewashed Strep-Tactin Sepharose beads (Iba BioTagnologies, Goettingen, Germany) on a wheel for 1 h at 4 °C. Beads were washed twice in 1 ml LM lysis buffer containing protease and phosphatase inhibitors and three times with 1 ml LM buffer without inhibitors. Beads were eluted 4 times with 150 μl of 2.5 mm d-Biotin (Sigma-Aldrich). Eluates were pooled and precipitated with TCA/Acetone.
Co-Immunoprecipitation
LM lysates of Slp76+/+ BMMC were incubated for 2 h at 4 °C with protein-A Sepharose beads (GE Healthcare BioScience, Sweden) coated with anti-Bcr or anti-Dok-3 antibodies. After washing with lysis buffer, the beads were boiled at 90 °C with loading buffer for 5 min. Eluates were electrophoresed, and Western blotted with anti-SLP76, anti-Bcr, anti-Dok-3, or anti-pY antibodies.
Proteomic Analyses
(1) TCA precipitation. After elution, 125 μl of ice-cold TCA (Sigma-Aldrich) was added (final concentration 20%) to each sample and the mixture was incubated 30 min on ice and centrifuged at 20,000 × g for 20 min at 4 °C. The supernatant was removed and 600 μl of TCA 10% was added to wash the pellet. The samples were centrifuged as above. The supernatant was removed and 1 ml of ice-cold acetone (Sigma-Aldrich) was added to wash the pellet. The samples were centrifuged as above. The acetone-containing supernatant was removed and the pellet was air-dried. The pellet was suspended in 30 μl of sample buffer, consisting of 25 mm Tris-HCl, 4% glycerol, 2% SDS, 5% β-mercaptoethanol, and 0.01% bromphenol blue, and boiled for 5 min. Proteins were stacked by SDS-PAGE in a 6 mm-thick band visualized by staining with Coomassie Brilliant Blue R-250 (Bio-Rad). (2) Protein digestion. The gel band was manually excised and cut in pieces. The following sample incubations were performed automatically using a Freedom EVO150 robot, (Tecan Traging AG, Switzerland). Gel pieces were washed by six successive incubations of 15 min in 25 mm NH4HCO3 and in 25 mm NH4HCO3 (Sigma-Aldrich) containing 50% (v/v) acetonitrile. Gel pieces were then dehydrated with 100% acetonitrile and incubated for 45 min at 53 °C with 10 mm DTT (Sigma-Aldrich) in 25 mm NH4HCO3 and for 35 min in the dark with 55 mm iodoacetamide (Sigma-Aldrich) in 25 mm NH4HCO3. Alkylation was stopped by adding 10 mm DTT in 25 mm NH4HCO3 and mixing for 10 min. Gel pieces were then washed again by incubation in 25 mm NH4HCO3 before dehydration with 100% acetonitrile. For an overnight incubation at 37 °C, 0.2 μg of modified trypsin (Promega, sequencing grade) in 25 mm NH4HCO3 was added to the dehydrated gel pieces. Peptides were then extracted from gel pieces in three 15 min sequential extraction steps in 30 μl of 50% acetonitrile, 30 μl of 5% formic acid (Aristar, VWR International, UK) and finally 30 μl of 100% acetonitrile. The pooled supernatants were then dried under vacuum. (3) Nano-LC-MS/MS analyses. The dried extracted peptides were resuspended in a minimal volume of 5% acetonitrile and 0.1% trifluoroacetic acid (Sigma-Aldrich) and analyzed twice by online nanoLC-MS/MS (Ultimate 3000, Dionex and LTQ-Orbitrap Velos pro, Thermofisher, CA, USA). The nanoLC method consisted in a 115-min gradient ranging from 4% to 45% acetronitrile in 0.1% formic acid at a flow rate of 300 nL/min. Peptides were sampled on a 300 μm × 5 mm PepMap C18 precolumn and separated on a 75 μm × 250 mm C18 PepMap column (Thermofisher). MS and MS/MS data were acquired using Xcalibur (Thermofisher). Spray voltage and heated capillary were respectively set at 1.4 kV and 200 °C. Survey full-scan MS spectra (m/z = 400–1600) were acquired in the Orbitrap with a resolution of 60,000 after accumulation of 106 ions (maximum filling time: 500 ms). The 20 most intense ions from the preview survey scan delivered by the Orbitrap were fragmented by collision induced dissociation (collision energy 35%) in the LTQ after accumulation of 104 ions (maximum filling time: 100 ms). (4) Data analyses. RAW files were processed using MaxQuant (27) version 1.3.0.5. Spectra were searched against the Uniprot database (Mus musculus taxonomy 10090, 78810 sequences, July 2012 version) and the frequently observed contaminants database embedded in MaxQuant. Trypsin was chosen as the enzyme and 2 missed cleavages were allowed. Precursor mass error tolerances were set respectively at 20 ppm and 6 ppm for first and main searches. Fragment mass error tolerance was set to 0.5 Da. Peptide modifications allowed during the search were: carbamidomethylation (C, fixed), acetyl (Protein N-ter, variable), oxidation (M, variable) and deamidation (NQ, variable). Minimum peptide length was set to seven amino acids. Minimum number of peptides, razor + unique peptides and unique peptides were set to 1. Maximum false discovery rates - calculated by employing a reverse database strategy - were set to 0.01 at peptide and protein levels. Raw MS data files, unfiltered protein groups and peptides tables have been uploaded in ProteomeXchange server (www.proteomexchange.org accession PXD000052). Identification and quantification data are also provided for proteins in supplemental Table S7 and for peptides in supplemental Table S8. Proteins identified as “contaminants,” “reverse,” and “only identified by site” were discarded from the list of identified proteins. iBAQ values were calculated using MaxQuant as previously described (28), from MS intensity of unique peptides. Data coming from technical MS replicates were summed automatically. (5) Statistical analyses. Normalization of protein iBAQ values was performed on each sample using median calculated from proteins with values in all 16 samples. Only proteins quantified with a minimum of two unique peptides and for which a quantitative value could be measured for the four affinity purifications replicates in at least one condition were considered for the statistical analysis. The following steps were performed using the Perseus toolbox (version 1.2.0.16) available in the MaxQuant environment. After log2 transformation of iBAQ values and data imputation (replacing missing values by normal distribution as described in (29), protein expression differences were identified using t-testing with a Permutation-based FDR to correct for multiple hypothesis testing, S0 value set to 1 (30) and p value threshold value set to 10−3. Only proteins with a significant t test result and log2 ratio above 2 for enrichment in respectively nonactivated (−) and activated (+) mast cells from Slp76OST/OST versus Slp76+/+ mice were validated in resting and stimulated complex, respectively. Taking into account that contaminant proteins may be not reproducible between nonactivated and activated mast cells from Slp76+/+, we additionally filtered proteins enriched in resting complex (with respect to stimulated complex) if protein iBAQ in activated (+) mast cells from Slp76+/+ mice was above protein iBAQ in nonactivated (+) mast cells from Slp76OST/OST mice (respectively if protein iBAQ in nonactivated (−) mast cells from Slp76+/+ mice was above protein iBAQ in activated (+) mast cells from Slp76OST/OST mice). The enrichment existing between activated (OST+) and nonactivated (OST−) BMMC was also t-tested with S0 value set to 1 (30), p value threshold value set to 10−2. Only proteins from resting or stimulated complex with a significant t test result and log2 ratio above two were further considered.
Sources of Information and Software Used for Protein Network Analyses
Information on protein interactions was extracted from the STRING (http://string-db.org) and PINA (http://cbg.garvan.unsw.edu.au/pina/) databases. The software Cytoscape was used to construct and visualize the SLP76 network (http://www.cytoscape.org). The LAT2 and Bcr protein–protein interaction networks were obtained from STRING database. We integrated experimentally verified interactions and those described in databases. Information regarding protease-coding genes has been extracted from Expasy web service (http://expasy.org).
RESULTS
SLP76OST is Normally Expressed and Functional in Mast Cells
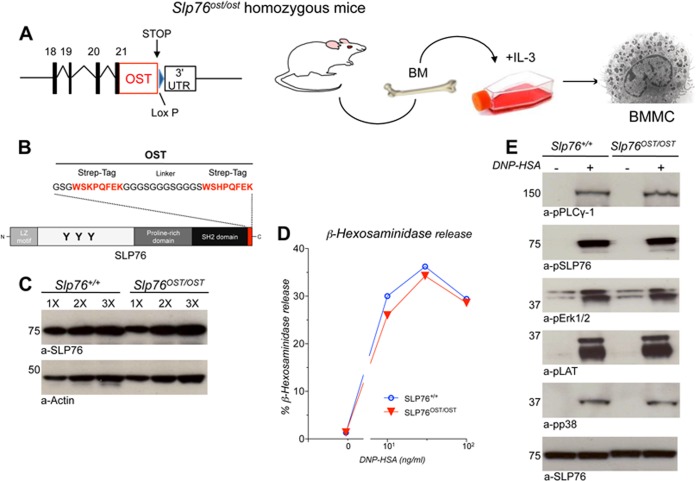
Using gene KI in ES cells, a sequence corresponding to the OST MS tag was introduced at the 3′ end of the Slp76 gene. Following germline transmission of the modified allele (denoted as Slp76OST), KI mice homozygous for the Slp76OST allele were established. Bone Marrow-derived Mast Cells (BMMC) were generated from homozygous Slp76OST/OST KI mice and from wild-type (wt) controls (Slp76+/+ mice) (Fig. 1A and 1B).
Fig. 1.
Expression and function of SLP76OST in BMMC. A, Diagram of the generation of Slp76OST/OST BMMC. B, Schematic representation of SLP76OST molecules. The One-Strep-tag (OST) is composed of two 8-amino acid peptides (in red) separated by a 12-amino acid linker. C, SLP76 expression in Slp76+/+ and Slp76OST/OST BMMC. BMMC were solubilized in SDS lysis buffer. Increasing amounts (1×, 2×, and 3×) of lysates were electrophoresed and Western blotted with anti-SLP76 and anti-actin antibodies. D, β-hexosaminidase release. IgE anti-DNP sensitized Slp76+/+ and Slp76OST/OST BMMC were challenged with the indicated concentrations of DNP-HSA. β-hexosaminidase was measured in supernatant by an enzymatic assay. E, Intracellular signaling. IgE anti-DNP-sensitized Slp76+/+ and Slp76OST/OST BMMC were challenged with or without DNP-HSA for 2 min and lysed in SDS lysis buffer. Equal amounts of proteins were electrophoresed and Western blotted with indicated antibodies.
As judged by Western blotting of SDS lysates, Slp76OST/OST BMMC contained similar amounts of SLP76 molecules as Slp76+/+ BMMC (Fig. 1C), indicating that the presence of the OST did not affect the level of SLP76 expression. When sensitized with mouse IgE anti-DNP and challenged with DNP-HSA, Slp76OST/OST BMMC released similar percentages of the granular mediator β-hexosaminidase as Slp76+/+ BMMC (Fig. 1D). Moreover, SLP76 and key signaling molecules, including LAT1, PLCγ-1, and the MAPK kinases Erk1/2 and p38, which are terminally activated on FcεRI engagement, were similarly phosphorylated in Slp76OST/OST and Slp76+/+ BMMC (Fig. 1E). SLP76OST molecules are thus expressed at the same levels as wt SLP76 molecules in BMMC, and the presence of the OST has no effect on antigen-induced IgE-dependent FcεRI signaling and subsequent mast cell degranulation.
SLP76 is Degraded in BMMC Lysates by Mast Cell-Specific Proteases
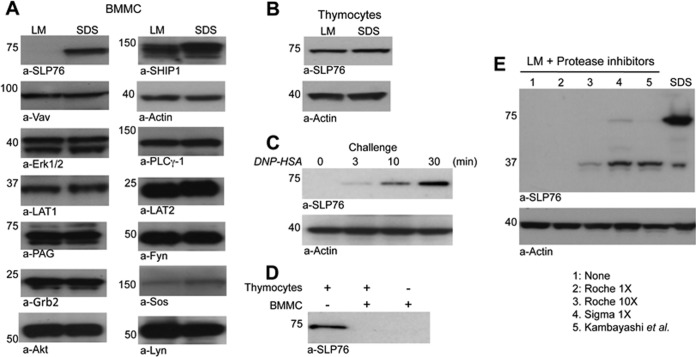
Because we aimed at performing affinity purification of the SLP76OST bait under native conditions, BMMC were solubilized with lysis buffer containing a mild nondenaturing detergent, laurylmaltoside (LM), and a standard protease inhibitor mixture (Roche, Mannheim, Germany). Unexpectedly, wt SLP76, which was readily detected by Western blotting in denaturing SDS lysates, could not be detected in nondenaturing LM lysates. In contrast, all the other signaling molecules examined were found in comparable amounts in SDS and LM lysates (Fig. 2A). SLP76 was similarly undetectable in LM lysates of Slp76OST/OST BMMC (not shown) and in Slp76+/+ BMMC lysates obtained with other mild nondenaturing detergents such as octylglycoside, IGEPAL, Brij35 or Triton X-100 supplemented with standard protease inhibitors (supplemental Fig. S1). Similar amounts of SLP76 were, however, detected in both SDS and LM lysates from thymocytes (Fig. 2B). The inability to detect SLP76 in LM lysates is therefore specific to mast cells.
Fig. 2.
Proteolysis of SLP76 molecules in BMMC. (A, B) BMMC (A) and thymocytes (B) were solubilized in LM lysis buffer or in SDS lysis buffer. Equal amounts of cell lysates were electrophoresed and Western blotted with the indicated antibodies. C, IgE anti-DNP-sensitized BMMC were challenged with DNP-HSA for the indicated times and solubilized in LM lysis buffer. Equal amounts of BMMC lysates were electrophoresed and Western blotted with anti-SLP76 and anti-actin antibodies. D, Equal amounts of proteins from a thymocyte LM lysate, a BMMC LM lysate or a mixture of both were incubated for 10 min on ice, electrophoresed and Western blotted with anti-SLP76 antibodies. E, BMMC were lysed in SDS lysis buffer or in LM lysis buffer containing no inhibitors (1), Roche mixture inhibitors 1× (2), Roche mixture inhibitors 10× (3), Sigma-Adlrich mixture 1× (4) or custom-made mixture inhibitors (21) (5). Lysates were electrophoresed and Western blotted with anti-SLP76 and anti-actin antibodies.
Surprisingly, although SLP76 was undetectable in LM lysates from nonstimulated BMMC sensitized with IgE antibodies, it became increasingly detectable in LM lysates from the same cells challenged with specific antigen for increasing periods of time before lysis (Fig. 2C). As stimulated mast cells degranulate within minutes, we hypothesized that granular proteases might be responsible for SLP76 degradation. Indeed, proteases contained in intact granules in nonstimulated cells, but not in activated, degranulated mast cells, could be freed on lysis and degrade SLP76 in mast cell lysates. To test this hypothesis, we examined SLP76 in a nonactivated BMMC LM lysate, in a thymocyte LM lysate and in a 1 to 1 mixture of both. As expected from the above experiments, SLP76 was observed in thymocyte LM lysate, but not in BMMC LM lysate. Strikingly, SLP76 was also undetectable in the mixture (Fig. 2D). Mast cell-specific proteases present in BMMC could thus degrade thymocyte-derived exogenous SLP76 molecules and, therefore, mast cell-derived endogenous SLP76 molecules following lysis in LM-containing buffer. We attempted to overcome this problem by using other protease inhibitor cocktails and by increasing their concentrations. SLP76 was protected neither by increasing 10 times the concentration of Roche protease inhibitors nor by using other mixtures of inhibitors, whether commercially available (Sigma-Alrdich) or custom-made (21) (Fig. 2E). Altogether, the above set of data indicates that, when mast cells are lysed with mild nondenaturing detergents, SLP76 is degraded by endogenous granular proteases that are resistant to conventional protease inhibitors.
Zinc-Dependent Metalloproteases Account for SLP76 Degradation in Mast Cell Lysates
To investigate the candidate protease(s) responsible for SLP76 degradation, we performed a transcriptomic analysis of BMMC. A total of 400 protease transcripts were identified, belonging to the four main groups of proteases. Serine proteases, cysteine proteases and metalloproteases were the most abundant (Fig. 3A and supplemental Table S1). The metalloprotease genes expressed in BMMC encode 176 metalloproteases (Fig. 3B and supplemental Table S2), 73% of which are zinc-binding proteins (Fig. 3B and supplemental Table S3). We noticed that, although most protease inhibitor cocktails contain inhibitors of proteases that depend on divalent cations such as Ca2+ (EGTA) or Mg2+/Mn2+ (EDTA), they do not contain specific inhibitors of Zn2+-dependent metalloproteases (30). We therefore investigated whether a Zn2+ chelator would protect SLP76 from degradation in mast cell lysates. We found that, indeed, phenanthroline reduced the proteolysis of SLP76 when added to LM lysis buffer. Benzamidine, an inhibitor of aspartic and serine proteases, also protected SLP76, albeit less efficiently (Fig. 3C). The protection of SLP76 by phenanthroline was much more efficient at pH 8.0 (supplemental Fig. S2B) than at pH 7.4 (supplemental Fig. S2A). An increased protection by the Roche mixture was also found at high pH (supplemental Fig. S2C). When used at pH 8.0, phenanthroline protected SLP76 more efficiently than any other inhibitor.
On the basis of these results, we devised a mast cell protease inhibitor mixture suitable for the biochemical analysis of signaling molecules in mast cells. This “MC mixture” contained E-64, AEBSF, chymostatin, antipain, bestatin, phenanthroline, and benzamidine (supplemental Table S4). SLP76 was dose-dependently protected when cells were lysed in LM buffer containing increasing concentrations of MC mixture at pH 8.0. Protection was complete when using the highest concentration of MC mixture (supplemental Table S4) because, under these conditions, comparable amounts of SLP76 were recovered in LM lysates and in SDS lysates. Importantly, the level of protection observed at 10 min was fully retained after 90 min at 0 °C (Fig. 3D). The establishment of these optimized lysis conditions was an absolute requirement for an MS-based proteomic analysis of the SLP76 interactome in mast cells, because it is based on the prior affinity purification of tagged SLP76 molecules from LM lysates.
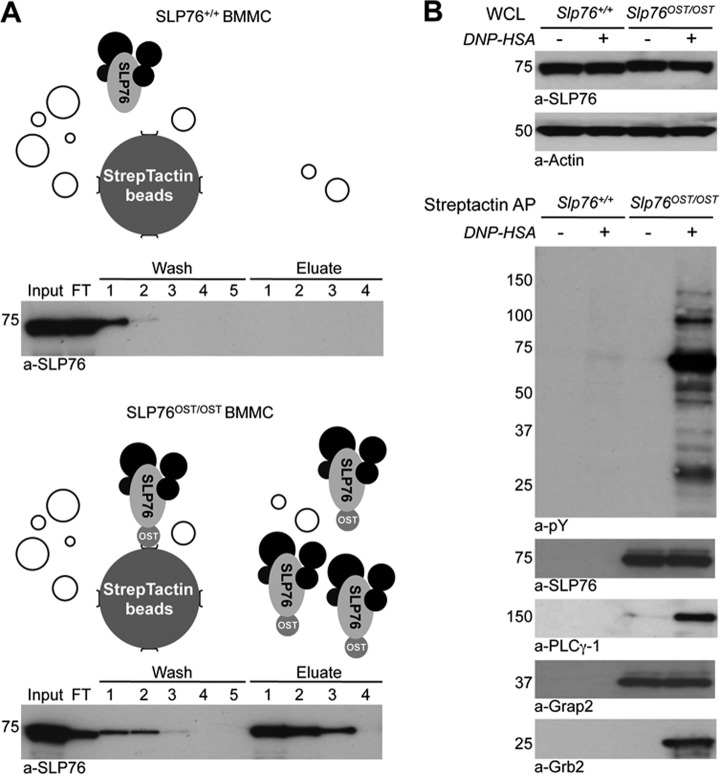
SLP76 and Interacting Proteins Can be Affinity-Purified From Slp76OST/OST BMMC
Slp76OST/OST and Slp76+/+ BMMC were lysed in LM buffer pH 8.0 supplemented with the MC mixture, and SLP76 was affinity-purified from lysates using Strep-Tactin-coated Sepharose beads. Similar amounts of lysates from Slp76OST/OST and Slp76+/+ BMMC were applied to the Strep-Tactin column (Fig. 4A, input). As expected, no SLP76 was recovered from Slp76+/+ BMMC, whereas most of the SLP76 molecules detected in lysate of Slp76OST/OST BMMC were recovered from Strep-Tactin-coated beads using four elutions steps with 2.5 mm d-Biotin, a ligand that binds to Strep-Tactin with a higher affinity than the OST (Fig. 4A). Our protocol therefore enables the recovery of most of the SLP76 species present in mast cell lysates.
Fig. 4.
Strep-Tactin affinity purification of functional SLP76 protein complexes from BMMC lysates. A, StrepTactin affinity-purification in LM lysates of BMMC from Slp76+/+ (upper panel) and Slp76OST/OST (lower panel) mice. Aliquots from each step of the purification procedure were Western blotted with anti-SLP76 antibodies (Input 1%, FT; flow-through 1%, washes 1%; eluates 10%). The upper panels show a schematic of the Strep-Tactin affinity-purification from each cell. B, Copurification of known interacting proteins of SLP76. IgE anti-DNP-sensitized Slp76+/+ and Slp76OST/OST BMMC were challenged with DNP-HSA or without for 2 min and lysed. Lysates were subjected to affinity-purification with Strep-Tactin. Eluates were electrophoresed and Western blotted with anti-phosphotyrosine (pY), anti-SLP76, anti-PLCγ-1, anti-Grap2, and anti-Grb2 antibodies.
To examine which proteins associate with SLP76 on mast cell activation, Slp76OST/OST and Slp76+/+ BMMC were sensitized with IgE antibodies and challenged with antigen or without for 2 min. SLP76 was affinity-purified on Strep-Tactin-coated beads, and eluates were analyzed by Western blotting. Similar amounts of SLP76 were recovered in lysates from challenged and unchallenged Slp76OST/OST BMMC, whereas no SLP76 was recovered from Slp76+/+ BMMC. As expected, the constitutively associated adapter protein Grap2 co-purified with SLP76OST, before and after antigen challenge. SLP76OST was inducibly tyrosyl-phosphorylated on antigen challenge, and two well-known SLP76 partners, Grb2 and PLCγ-1, co-purified with phosphorylated, but not with nonphosphorylated SLP76OST (Fig. 4B). Well-known activation-inducible SLP-76 interactors could therefore be identified by immunoblotting, following Strep-Tactin affinity purification of LM lysates, provided that SLP76 degradation was prevented by the protease inhibitor mixture that we specifically designed. Biochemical conditions having been optimized, the SLP76 interactome was investigated next by label-free quantitative proteomic analysis in Slp76OST/OST BMMC.
The SLP76 Interactome of Resting Mast Cells
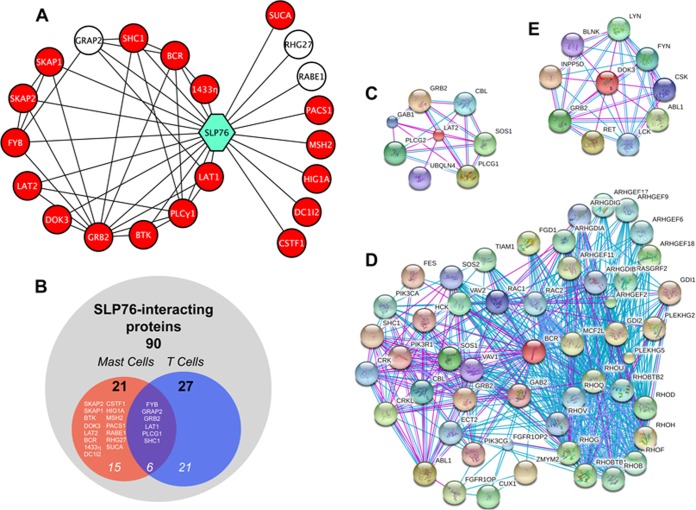
Unlike classical biochemistry approaches, Strep-Tactin-based affinity purification of SLP760ST enabled us to study proteins that interact with SLP76 in resting cells. Indeed, molecules that were co-purified from resting Slp76OST/OST BMMC, but not from resting Slp76+/+ BMMC or in much lower amounts, could be considered as being specifically and constitutively associated with SLP76 in nonstimulated cells. The constitutive SLP76 interactome is largely unknown, especially in mast cells. The statistical treatment of iBAQ values coming from MaxQuant processing of LC-MS/MS data corresponding to four independent affinity purifications involving two MS replicates revealed three proteins significantly enriched in eluates from resting Slp76OST/OST BMMC compared with eluates from resting Slp76+/+ BMMC (Fig. 5A, Table I and supplemental Table S5, p value threshold set to 10−3 with s0 = 1 and log2 ratio OST-/WT- >2). Among them, Grap2, a well-known binding partner of SLP76, that could be seen by Western blotting (Fig. 4B), was 500-fold more abundant in eluates from Slp76OST/OST BMMC than in eluates from Slp76+/+ BMMC. The Rho GTPase-activating proteins RABE1 and RHG27 were enriched to a lesser extent in eluates from Slp76OST/OST BMMC. RABE1 and RHG27 have never been described before as SLP76 interactors.
Fig. 5.
SLP76 protein-protein interaction network in resting and activated mast cells. A, Network view of the SLP76 interactome in mast cells. White nodes: partners identified in resting cells; red nodes: additional partners identified in activated cells. Only proteins significantly associated with SLP76 were included in the network. B, Comparison using Venn diagram of SLP76 partners identified in mast cells in the present study and of previously published SLP76 partners. (C–E) Interactome networks of LAT2 (C), Bcr (D), and Dok-3 (E). Networks were obtained from the STRING database.
Table I. SLP76 interactome in resting BMMC. Proteins were identified in four affinity purifications of SLP76OST complexes in resting cells. Listed molecules were enriched at least fourfold in nonactivated SlpOST/OST compared to Slp+/+ BMMC (OST− vs WT−). The significance (p < 10−3) of enrichment (OST−/WT− ratio) was determined using student's t-test. UniProt Accession numbers, protein symbols and molecular function are also shown.
The SLP76 Interactome is Markedly Enriched in Activated Mast Cells
To determine the activation-induced SLP76 interactome in mast cells, we performed two sets of comparative MS analyses of proteins copurified with SLP76. First, we compared the SLP76 interactome in Slp76OST/OST BMMC and in Slp76+/+ BMMC, sensitized with IgE antibodies and challenged with antigen. The statistical treatment of iBAQ values coming from MaxQuant processing of LC-MS/MS data corresponding to four independent affinity purifications involving two MS replicates revealed 35 proteins significantly enriched in eluates from activated Slp76OST/OST BMMC compared with eluates from activated Slp76+/+ BMMC (p value threshold set to 10−3 with s0 = 1 and log2 ratio OST+/WT+ >2, Table II and supplemental Table S5). This comparison aimed at discriminating molecules that bound specifically to SLP76 from molecules that bound nonspecifically to Strep-Tactin Sepharose beads in activated mast cells. We then compared the SLP76 interactome in Slp76OST/OST BMMC sensitized with IgE antibodies and challenged with antigen or without for 2 min. This comparison aimed at identifying molecules that were recruited specifically by SLP76 on FcεRI engagement (as assessed by the OST+/OST- ratio in Table II and supplemental Table S5).
Table II. SLP76 interactome in activated BMMC. The proteins listed were recurrently identified in four affinity purifications of SLP76OST complexes in activated cells. Listed molecules were enriched at least fourfold in activated SlpOST/OST (OST+) compared to Slp+/+ BMMC (WT+). The enrichment was measured by the OST+/WT+ ratio and its significance (p < 10−3) determined using student's t-test. The enrichment existing between activated (OST+) and nonactivated (OST−) BMMC is also shown and its significance was determined using student's t-test (at least fourfold enrichment: **p < 10−2, ***p < 10−3). UniProt Accession numbers, protein symbols, and molecular function are also shown.
| Protein symbol Activated BMMC | Protein name | UniProt Acc | Function | Ratio |
|
|---|---|---|---|---|---|
| OST+/WT+ | OST+/OST− | ||||
| LAT2 | Linker for activation of T-cells family member 2 | Q9JHL0 | Adapter Protein | 7146 | 4820*** |
| BCR | Breakpoint cluster region protein | Q6PAJ1 | Kinase/GTPasa activation | 168 | 148** |
| HIG1A | HIG1 domain family member 1A | Q9JLR9 | Unknown | 144 | 104*** |
| CDC23 | Cell division cycle protein 23 homolog | Q8BGZ4 | Cell division | 245 | 55 |
| DC1I2 | Cytoplasmic dynein 1 intermediate chain 2 | O88487 | Cytoskeleton regulation | 15 | 38** |
| LAT1 | Linker for activation of T-cells family member 1 | O54957 | Adapter Protein | 36 | 37** |
| PLCγ1 | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase gamma-1 | Q62077 | Phospholipase | 36 | 36*** |
| SHC1 | SHC-transforming protein 1 | P98083 | Adapter Protein | 22 | 30*** |
| DOK3 | Docking protein 3 | Q9QZK7 | Adapter Protein | 8,6 | 29*** |
| SKAP2 | Src kinase-associated phosphoprotein 2 | Q3UND0 | Src Kinase regulator | 34 | 22*** |
| SUCA | Succinyl-CoA ligase [ADP/GDP-forming] subunit alpha, mitochondrial | Q9WUM5 | Ligase | 20 | 22** |
| GRB2 | Growth factor receptor-bound protein 2 | Q60631 | Adapter Protein | 18 | 16*** |
| SKAP1 | Src kinase-associated phosphoprotein 1 | Q3UUV5 | Src Kinase regulator | 22 | 14*** |
| HECD3 | E3 ubiquitin-protein ligase HECTD3 | Q3U487 | Ligase | 47 | 12 |
| MSH2 | DNA mismatch repair protein Msh2 | P43247 | DNA-binding | 12 | 11** |
| CSTF1 | Cleavage stimulation factor subunit 1 | Q99LC2 | mRNA processing | 10 | 11** |
| FYB | FYN-binding protein | O35601 | Adapter Protein | 17 | 9.5** |
| BTK | Tyrosine-protein kinase BTK | P35991 | Kinase | 6,1 | 6.7** |
| PACS1 | Phosphofurin acidic cluster sorting protein 1 | Q8K212 | Coat protein | 13 | 5.4** |
| 1433η | 14–3-3 protein eta | P68510 | Adapter Protein | 26 | 4** |
| 1433ϴ | 14–3-3 protein theta | P68254 | Adapter Protein | 13 | 3,8 |
| Q99M59 | Interferon gamma inducible protein 47 | Q99M59 | Hydrolase activity | 6,9 | 3,7 |
| CLNK | Cytokine-dependent hematopoietic cell linker | Q9QZE2 | Adapter Protein | 40 | 3,7 |
| VAV | Proto-oncogene vav | P27870 | Guanine-nucleotide releasing factor | 10 | 3,3 |
| MA2C1 | Alpha-mannosidase 2C1 | Q91W89 | Mannosidase | 13 | 3,1 |
| 1433γ | 14–3-3 protein gamma | P61982 | Adapter Protein | 45 | 3,0 |
| 1433ϵ | 14–3-3 protein epsilon | P62259 | Adapter Protein | 20 | 2,8 |
| E9Q4N7 | Protein Arid1b | E9Q4N7 | Unknown | 28 | 2,4 |
| 1433β | 14–3-3 protein beta/alpha | Q9CQV8 | Adapter Protein | 19 | 2,3 |
| 1433ζ | 14–3-3 protein zeta/delta | P63101 | Adapter Protein | 15 | 2,0 |
| RHG27 | Rho GTPase-activating protein 27 | A2AB59 | GTPase activation | 123 | 1,8 |
| TP53B | Tumor suppressor p53-binding protein 1 | Q99LC2 | mRNA processing | 10 | 1,3 |
| FA49B | Protein FAM49B | Q921M7 | Unknown | 29 | 1,2 |
| GRAP2 | GRB2-related adaptor protein 2 | O89100 | Adapter Protein | 1086 | 0,9 |
| RABE1 | Rab GTPase-binding effector protein 1 | O35551 | GTPase activation | 20 | 0,4 |
Eighteen proteins were significantly (p value threshold set to 10−2 with s0 = 1 and log2 ratio OST+/OST- >2) and specifically associated with SLP76 in activated Slp76OST/OST BMMC (compared with nonactivated Slp76OST/OST BMMC) (red nodes in Fig. 5A). In addition, the three SLP76 partners identified in resting BMMC (white nodes in Fig. 5A) also co-purified with SLP76 in activated BMMC. Grap2 remained associated in comparable amounts, RHG27 was enriched twofold, and RABE1 decreased more than half. Among the 21 proteins that constitute the SLP76 interactome of activated BMMC (Fig. 5B), nine molecules were inducibly recruited with a high statistical significance (p < 10−3). Surprisingly, the transmembrane adapter LAT2 (NTAL), which was not detected in nonactivated Slp76OST/OST BMMC, was dramatically enriched in activated Slp76OST/OST BMMC, much more than any other molecule. In comparison, LAT1, which was also not detected in nonactivated Slp76OST/OST BMMC, was detected in activated Slp76OST/OST BMMC, but it was much less enriched (p < 10−2) than LAT2. The molecular basis of this dramatic difference remains to be determined. A limited number of proteins are known to interact with LAT2 (Fig. 5C). Two unexpected molecules, the Breakpoint Cluster Region protein (Bcr) and HIG1A, were also highly enriched. Bcr is a protein with compound functions which interacts with numerous molecules (Fig. 5D). HIG1A is a HIG1-domain family member whose function is not known. Other partners recruited with a high statistical significance were also enriched but to a lower extent. They included molecules known to be recruited by SLP76 in activated T cells such as PLC-γ1, Shc1 and Grb2, but also molecules that were not previously identified as SLP76 partners, such as the Src kinase-associated phopshoproteins SKAP1/2 and the cytosolic adapter Dok-3. Other molecules that were statistically significantly recruited (p < 10−2) included LAT1, as already mentioned, a member of the 14–3-3 family, 14–3-3η, the Fyn-binding protein Fyb, and the Tec kinase BTK.
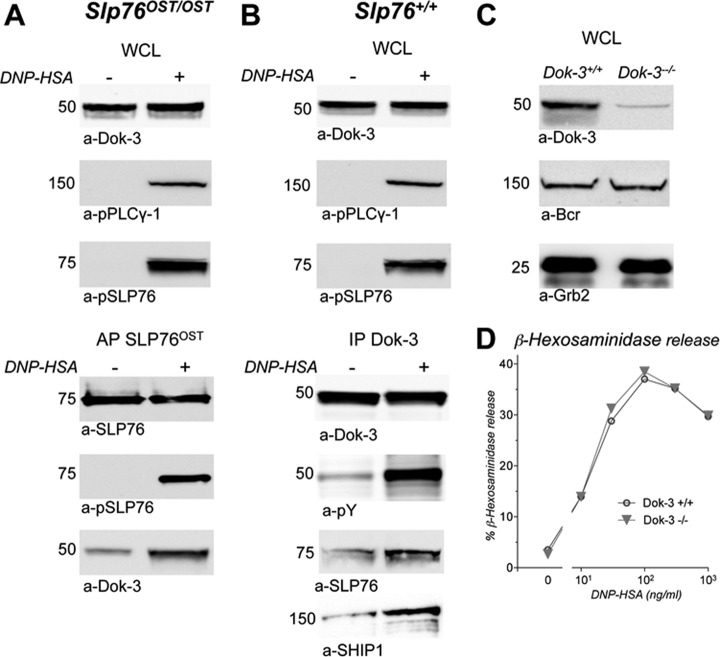
The cytosolic adapter Dok-3 interacts with several signaling molecules among which the SH2 domain-containing inositol 5-phosphatase SHIP1 (INPP5D) (Fig. 5D). It belongs to the same family as Dok-1, which is a major player in SHIP1-dependent negative regulation of both B cell (31) and mast cell (32) activation. Dok-3 was found in lysates of nonactivated and activated BMMC, from Slp76OST/OST (Fig. 6A, upper panel) and from Slp76+/+ (Fig. 6B, upper panel) mice. Confirming our AP-MS analysis, Dok-3 copurified with SLP76 in activated cells in higher amount than in nonactivated cells (Fig. 6A, lower panel). Conversely, SLP76 co-precipitated with Dok-3 in activated cells more than in resting cells, and Dok-3 was markedly phosphorylated on antigen challenge. Noticeably, SHIP1 also co-precipitated with phosphorylated Dok-3 in activated cells (Fig. 6B, lower panel). To further investigate the involvement of Dok-3 in FcεRI signaling, we compared β-hexosaminidase released by BMMC from Dok-3−/− mice and by BMMC from Dok-3+/+ mice (Fig. 6C). Antigen-induced mediator release was of the same magnitudes in Bcr−/− and Bcr+/+ BMMC sensitized with IgE (Fig. 6D). The deletion of Dok-3 therefore did not detectably affect mast cell activation.
Fig. 6.
Validation of the SLP76-Dok-3 interaction in activated mast cells. A, Co-purification of Bcr with SLP76OST. Slp76OST/OST BMMC sensitized with IgE anti-DNP were challenged with DNP-HSA (+) or without (−) for 2 min at 37 °C and lysed. Samples of whole cell lysates (WCL) were Western blotted with anti-Dok-3 antibodies (upper panel). Remaining lysates were subjected to affinity-purification with Strep-Tactin. Eluates were electrophoresed and Western blotted with anti-SLP76, anti-phosphotyrosine (pY), and anti-Dok-3 antibodies (lower panel). B, Co-immunoprecipitation of SLP76 and SHIP1 with phosphorylated Dok-3. Slp76+/+ BMMC sensitized with IgE anti-DNP were challenged with DNP-HSA (+) or without (−) for 2 min at 37 °C and lysed. Samples of whole cell lysates (WCL) were Western blotted with anti-Dok-3 or anti-phospho-PLCγ-1 antibodies (upper panel). Remaining lysates were immunoprecipitated with anti-Dok-3 antibody. Eluates were electrophoresed and Western blotted with anti-Dok-3, anti-phosphotyrosine (pY), anti-SLP76 or anti-SHIP1 antibodies (lower panel). (C, D) Lack of genetic evidence that Dok-3 is involved in FcεRI signaling. Aliquots of BMMC from Dok-3−/− and from Dok-3+/+ mice were lysed in SDS. whole cell lysates (WCL) were Western blotted with anti-Dok-3 antibodies and, as positive controls, with anti-Bcr or anti-Grb2 antibodies (C). Aliquots of the same cells were sensitized with IgE anti-DNP, and challenged with the indicated concentrations of DNP-HSA. β-hexosaminidase was measured in supernatant 10 min later (D).
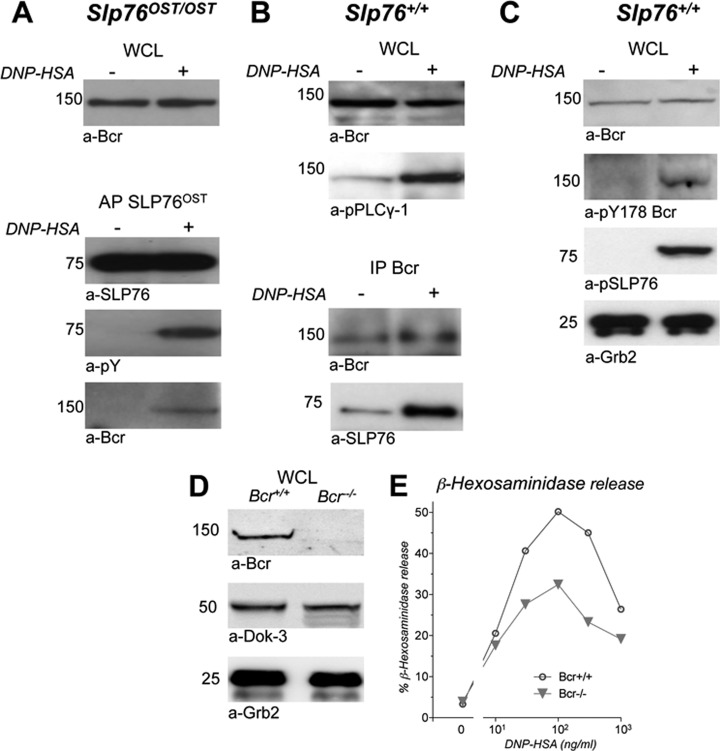
Because Bcr was an unexpected partner of SLP76, because it was inducibly recruited in high amounts on FcεRI engagement, because it was not previously linked to FcεRI signaling, because it interacts with numerous molecules and because it can exert multiple functions that may have a major impact in FcεRI signaling, we validated biochemically the interaction between SLP76 and Bcr. We indeed found that Bcr was similarly observed in lysates of nonactivated and activated BMMC, whether from Slp76OST/OST (Fig. 7A, upper panel) or from Slp76+/+ (Fig. 7B, upper panel and Fig. 7C) mice. Bcr was co-purified with phospshorylated SLP76OST in activated, but not in nonactivated Slp76OST/OST BMMC (Fig. 7A, lower panel). Conversely, SLP76 was co-immunoprecipitated with Bcr in Slp76+/+ BMMC, and in higher amount in activated cells than in nonactivated cells (Fig. 7B, lower panel). Also, Bcr was inducibly phosphorylayed at Y178 on antigen challenge (Fig. 7C). To further investigate the involvement of Bcr in FcεRI signaling, we compared β-hexosaminidase released by BMMC from Bcr−/− mice and by BMMC from Bcr+/+ mice (Fig. 7D). Antigen-induced mediator release was decreased in Bcr−/− BMMC, compared with Bcr+/+ BMMC, sensitized with IgE (Fig. 7E). The deletion of Bcr therefore impaired mast cell activation.
Fig. 7.
Validation of the SLP76-Bcr interaction in activated mast cells. A, Co-purification of Bcr with SLP76OST. Slp76OST/OST BMMC sensitized with IgE anti-DNP were challenged with DNP-HSA (+) or without (−) for 2 min at 37 °C and lysed. Samples of whole cell lysates (WCL) were Western blotted with anti-Bcr antibodies. Remaining lysates were subjected to affinity-purification with Strep-Tactin. Eluates were electrophoresed and Western blotted with anti-SLP76, anti-phosphotyrosine (pY), and anti-Bcr antibodies. B, Co-immunoprecipitation of SLP76 with Bcr. Slp76+/+ BMMC sensitized with IgE anti-DNP were challenged with DNP-HSA (+) or without (−) for 2 min at 37 °C and lysed. Samples of whole cell lysates (WCL) were Western blotted with anti-Bcr or anti-phospho-PLCγ-1 antibodies. Remaining lysates were immunoprecipitated with anti-Bcr antibody. Eluates were electrophoresed and Western blotted with anti-Bcr, or anti-SLP76 antibodies. C, Inducible Bcr phosphorylation in activated mast cells. BMMC sensitized with IgE anti-DNP were challenged with DNP-HSA (+) or without (−) for 2 min at 37 °C and lysed. Samples of whole cell lysates (WCL) were Western blotted with anti-Bcr, anti-pY178 Bcr, anti-phospho-SLP76 or anti-Grb2 antibodies. (D, E) Genetic evidence that Bcr is involved in FcεRI signaling. Aliquots of BMMC from Bcr−/− and from Bcr+/+ littermate control mice were lysed in SDS. whole cell lysates (WCL) were Western blotted with anti-Bcr antibodies and, as positive controls, with anti-Dok-3 or anti-Grb2 antibodies (D). Aliquots of the same cells were sensitized with IgE anti-DNP, and challenged with the indicated concentrations of DNP-HSA. β-hexosaminidase was measured in supernatant 10 min later (E).
DISCUSSION
We report here the first proteomic analysis of FcεRI signaling in primary mouse mast cells. Specifically, we describe the SLP76 interactome in resting and in activated cultured mast cells, using affinity purification coupled to label-free quantitative proteomics. Data presented in this paper are the first results of a large-scale program aiming at assembling a comprehensive dynamic map of signaling events triggered by FcεRI engagement in mast cells.
SLP76 has been extensively investigated following TCR engagement in T cells, but much less following FcεRI engagement in mast cells. SLP76 deletion was however found to profoundly affect IgE-induced mast cell activation (20). As uncovered in the present study, one reason of this poor knowledge on an adapter molecule that is essential for FcεRI signaling may be linked to its unexpected sensitivity to proteolysis by mast cell granular proteases. We indeed found that SLP76 was undetectable in LM lysates of nonactivated mast cells, whereas it was readily detected in SDS lysates of the same cells. No other intracellular molecule among the 14 that we tested behaved like SLP76. This reminded us that, under similar lysis conditions, SHIP1 was degraded by unknown proteases in peritoneal cell-derived mast cells, a model of mature serosal-type mast cells that are especially rich in granular proteases, but not in BMMC (33). We noticed in the present study that similar amounts of SLP76 were detected in LM and SDS lysates of thymocytes and in LM lysates of activated but not of nonactivated mast cells. To explain these differences, we hypothesized that proteases contained in the granules might digest SLP76 in lysates of nonactivated mast cells, if granular membranes are disrupted by detergents. This hypothesis was confirmed by experiments showing that thymocyte-derived SLP76 was also degraded in a 1:1 mixture of LM lysates of thymocytes and of nonactivated BMMC. Even if restricted to SLP76, this proteolysis was a major obstacle for our project. The SLP76 molecule is indeed functioning as a scaffold protein, and its degradation is likely to dramatically alter the signaling complex to be investigated. To circumvent this problem, we undertook an extensive analysis of proteases in BMMC.
Proteases were reported to account for up to 25% of total proteins in mast cells (34). They are stored in granules under a catalytically active form. They are released in the extracellular medium under physiological conditions during antigen-induced degranulation and they can have both pro-inflammatory effects in IgE-induced allergic inflammation, and anti-inflammatory effects in infection-associated inflammation (35). They can also be released under nonphysiological conditions, when granular membranes are solubilized by mild detergents during cell lysis. Intracellular molecules are thus suddenly exposed to large amounts of biologically active proteases that they normally never meet.
According to our transcriptomic study, more than a quarter (28%) of proteases expressed in BMMC are metalloproteases. Among these, almost three quarters (73%) are Zn2+-dependent. Commercially available protease inhibitor cocktails, however, do not or poorly inhibit Zn2+-dependent metalloproteases. We found that adding a specific Zn2+ chelator to other inhibitors fully protected SLP76, even for prolonged periods of times (90 min at 0 °C) that are required for affinity purification.
The lysis conditions defined in the present study enabled us to analyze the SLP76 interactome, not only in activated mast cells, but also in nonactivated mast cells. The unique tagged SLP76 molecules that we developed via a KI approach and used here enabled us to compare mast cells from Slp76+/+ and from Slp76OST/OST mice. Under these conditions, molecules eluted from Strep-Tactin-coated beads in Slp76+/+ BMMC were considered as binding nonspecifically, i.e. as background, whereas molecules eluted from Strep-Tactin-coated beads in Slp76OST/OST BMMC, but not in Slp76+/+ BMMC, could be considered as binding specifically to SLP76 in resting mast cells. Three molecules were affinity-purified along with SLP76OST from nonstimulated Slp76OST/OST BMMC but not, or in much lower amounts, from nonstimulated Slp76+/+ BMMC. These molecules corresponded to the adapter protein Grap2 and to the two GTPase-activating proteins, RABE1 and RHG27.
Among the molecules that displayed a statistically significant enrichment (p < 10−2) on FcεRI engagement, one can distinguish molecules that have been already described as SLP76 partners in T cells and novel SLP76 partners that are identified for the first time here in mast cells.
Six molecules interacted with SLP76 in both activated T cells and activated mast cells. These were mostly adapter molecules (LAT1, Grap2, Grb2, Shc1, Fyb) and one enzyme (PLCγ-1). One notices that all these molecules concur to intracellular pathways that lead to cell activation. LAT1 is an essential scaffold protein that organizes the signalosomes generated on TCR or FcεRI engagement. The cytosolic adapters Shc, Grb2, and Grap2 are widely used by activating receptors and are involved in numerous signaling pathways. Phospholipase Cγ-1 (PLCγ-1) is a widely expressed major signaling molecule, which, when activated on tyrosyl-phosphorylation, generates inositol tris-phosphate (IP3) and diacyl-glycerol (DAG). IP3 triggers the efflux of Ca2+ from endothelial reticulum, leading to the influx of extracellular Ca2+, whereas DAG activates Protein Kinase C. By phosphorylating PLCγ-1, the Bruton's Tyrosine Kinase of the Tec family BTK plays a critical role in the activation of this phospholipase. SLP76 also associates with Clnk, a cytosolic adapter of the SLP76 family which promotes TCR-dependent activation of T cells (36, 37) and FcεRI-dependent responses of mast cells (38). Clnk deficiency, however, impaired neither TCR signaling in T cells nor FcεRI signaling in mast cells (39). Clnk interacts with two phosphoproteins expressed in mast cell lines, Fyb, a.k.a. SLP76-associated protein of 130 kDa (SLAP-130) or adhesion- and degranulation-promoting adapter protein (ADAP), and SKAP1. These adapters can interact with the SH2 domain of Src kinases to form a Clnk-Fyb-SKAP1 complex in mast cells (40). Fyb and SKAP1 were among the molecules that were recruited by SLP76 in mast cells, following FcεRI engagement (p < 10−3 and p < 10−2, respectively). Some proteins previously reported to associate with SLP76 in T cells were not found with a high enough statistical significance in our MS-based proteomic analysis of the SLP76 interactome in mast cells. Lyn and Nck-1 could however be detected by Western blot analysis among molecules that were affinity-purified with SLP0ST (not shown). The reason for the discrepancy lies in the severe filters used for establishing statistical significance in our proteomic analysis (MS data and statistical analysis of these results are shown in supplemental Table S6).
Fifteen molecules that were inducibly recruited by SLP76 on FcεRI engagement in mast cells have not been reported to be recruited by SLP76 on TCR engagement in T cells. These comprised adapter molecules (LAT2, 14–3-3F, CSTF1 and Dok-3), a Tec family protein tyrosine kinase (Btk), Src kinase regulators (SKAP1 and SKAP2), GTPase-activating molecules (RABE1 and RHG37), a serine/threonine kinase with a guanine-nucleotide exchange factor activity for Rho GTPases (Bcr) (41), and molecules with poorly defined or unknown functions (HIG1A, SUCA, MSH2, PACS1 and DC1I2). One notices that most molecules with a known function that were inducibly recruited in high amounts by SLP76 in mast cells, but not in T cells, are associated with negative regulation.
The transmembrane adapter LAT2 was inducibly recruited by SLP76 exceedingly more than any other molecules in antigen-stimulated Slp76OST/OST BMMC. LAT2 is a well-known antagonist of LAT1 in mouse mast cells, and LAT2-deficient mice exhibited enhanced secretory responses on FcεRI engagement (42). The exact mechanisms by which LAT2 inhibits mast cell activation is, however, largely unknown (22), and it has been reported to interact with six proteins only.
Dok-3, a cytosolic adapter of the Dok family is also associated with negative regulation and was inducibly recruited in the SLP76 interactome. This interaction of Dok-3 with SLP76 could be confirmed biochemically as Dok-3 co-purified with SLP760ST. Conversely, SLP76 co-precipitated with Dok-3 in wild-type cells. Noticeably, Dok-3 was dramatically phosphorylated in activated mast cells. Dok-1 is well known to become phopshorylated on immunoreceptor engagement in both B cells (31) and mast cells (32), and to mediate the recruitment of RasGAP which, by increasing the autocatalytic activity of Ras-GTP, inhibits the Ras pathway. Dok-2 plays a similar role as Dok-1 in T cells (43), but Dok-1 and Dok-3 were found to have nonredundant roles in B cells (44). Dok-3 was found among the proteins that are phosphorylated on FcεRI engagement in mouse mast cells (45). When tyrosyl-phosphorylated in B cells and macrophages, Dok-3 mediated the membrane recruitment of SHIP1 and Csk, two molecules that have inhibitory properties. Dok-3 over-expression reduced B Cell Receptor-dependent B cell activation whereas the expression of Dok-3 the four C-terminal tyrosines of which were mutated, i.e. rendered unable to bind to the SH2 domain of SHIP1, enhanced B cell activation (46). We show here that Dok-3 can also inducibly recruit SHIP1 on FcεRI engagement in mast cells as SHIP1 co-precipitated with phosphorylated Dok-3 in activated mast cells. SHIP1 is a major negative regulator of immunoreceptor signaling, and especially of FcεRI signaling (reviewed in (47)). The deletion of Dok-3, however, had no detectable effect on mast cell degranulation, probably because Dok-1, and possibly Dok-2, were sufficient to recruit SHIP1.
The second most inducibly recruited molecule was the Breakpoint Cluster Region protein Bcr. It was enriched almost 150 fold in eluates from antigen-stimulated Slp76OST/OST BMMC, as compared with eluates from resting Slp76OST/OST BMMC. Importantly, we could confirm the inducible interaction of Bcr with SLP76 with biochemical techniques. Bcr was indeed co-affinity-purified with SLP76OST in lysates of activated Slp76OST/OST BMMC, and SLP76 was co-immunoprecipitated with Bcr in lysates of activated Slp76+/+ BMMC. Supporting a functional role of Bcr in FcεRI signaling, we found that it was inducibly phopshorylated at Y178 in activated mast cells. Brc was similarly found to be phosphorylated at Y177 on TCR engagement in human Jurkat cells (48). Finally, we obtained a compelling evidence that Bcr positively regulates IgE-dependent mast cell activation as mediator release was reduced in BMMC from Bcr-deficient mice. To our knowledge, this is the first demonstration that Bcr is involved in and contributes to FcεRI signaling. Bcr binds to multiple adapter molecules, including Grb2, Hck, Shc1, and Cbl as well as Dok-1, Dok-2, and therefore, possibly Dok-3. One notices several other interesting molecular partners in the Bcr interactome that can be constructed from published databases (Fig. 5D). These include Kit, which critically determines the proliferation and differentiation of mast cells, molecules that control cytokine receptor signaling such as Jak2 and STAT5A/B, the exchange factors Sos and Vav, and, strikingly, a whole cluster of small G proteins of the Rho and Rac families that exert major cytoskeletal effects by controlling the dynamics of actin. Bcr has itself a serine-threonine kinase activity. Bcr is also a GTPase-activating protein for Rac1 and cdc42 and an exchange factor for Rho GTPases. Normally, the Bcr exchange factor activity is auto-inhibited by flanking sequences (49). It is constitutively activated in the Bcr-Abl fusion protein (41) that accounts for the transformation of myeloid cells in chronic myeloid leukemia. The GTPase activity of Bcr is positively regulated by a Protein Tyrosine Phosphatase Receptor in neurons. Bcr is indeed highly expressed in the central nervous system where it decreases dendrite formation by negatively regulating actin polymerization in neurons (50). Along this line, the genetic ablation of Bcr and Abr, another Rac/cdc42-specific GTPase-activating exchange factor, increased cytoskeleton-dependent motility and phagocytosis in primary macrophages (51).
In conclusion, the present work first describes the SLP76 interactome in primary mast cells and unravels that it markedly differs from the SLP76 interactome previously described in T cells. Our work also provides the first evidence that Bcr is involved in FcεRI-dependent mast cell activation. It also validates OST-KI mice as powerful tools for the proteomic analysis of cell signaling. As SLP76OST was expressed with a normal tissue distribution, it could be studied in nontransformed mast cells. SLP76OST was expressed in mast cells from Slp76OST/OST mice in similar amounts as wt SLP76 in mast cells from Slp76+/+ mice. A unique advantage of the present approach is that, unlike in cells transfected with cDNA encoding a tagged molecule, no competition and no dilution with endogenous nontagged molecules occurs in cells from OST-KI mice as all molecules of interest are tagged. Importantly, OST-SLP76 was also functionally similar as wt-SLP76 because β-hexosaminidase release and intracellular phosphorylations of a comparable magnitude were induced on antigen challenge in Slp76OST/OST and Slp76+/+ mast cells sensitized wit IgE antibodies. On the basis of these results, one can apply the same approach to study the interactomes of other key signaling molecules in mouse mast cells. One expects not only to order known molecules in space and time, and to untangle the complex network of their interactions, but also to discover molecules not previously known as being involved in mast cell activation and, ultimately, to build up a comprehensive molecular map of the FcεRI signalosome.
Supplementary Material
Acknowledgments
We thank Dr. Eleanora Heisterkamp (Childrens Hospital of Los Angeles, Los Angeles CA) for having generated Bcr-deficient mice; Dr. Eunjoon Kim, (Center for Synaptic Brain Dysfunctions, Institute for Basic Science, and Department of Biological Sciences, Korea Advanced Institute of Science and Technology, Daejeon 305-601, Korea) for Bcr-deficient bone marrow; Dr. Koon-Guan Lee, (Immunology Group, Bioprocessing Technology Institute, A*STAR, Singapore) for Dok-3-deficient bone marrow; Dr. Frédéric Fiore (Centre d'Immunophénomique, Marseille) for supervising the construction of Slp76OST/OST mice, Dr. Sylvain Latour (Inserm U768, Hôpital Necker, Paris) for 4G10 antibodies; Dr. Vincenzo di Bartolo (Unité de Biologie Cellulaire des Lymphocytes, Département d'Immunologie, Institut Pasteur, Paris) for helpful advice and for mouse anti-SLP76 antibodies; Dr. Taku Kambayashi, (Abramson Family Cancer Research Institute, University of Pennsylvania, Philadelphia, PA) for sharing experience on SLP76 biochemistry; Dr. Christophe Bruley (iRTSV/BGE, CEA Grenoble) for helpful discussions. We are grateful to Dr. Matthew Albert (Département d'Immunologie, Institut Pasteur, Paris) for hosting YB at the Centre d'Immunologie Humaine, Inserm UMS20.
Footnotes
* This work was supported by CNRS, Inserm and ANR (Projet iSa). The Slp76OST/OST mice have been developed in the frame of the Centre d'Immunophénomique (INSERM US012, CNRS UMS3367, Université d'Aix Marseille).
 This article contains supplemental Figs. S1 and S2 and Tables S1 to S8.
This article contains supplemental Figs. S1 and S2 and Tables S1 to S8.
1 The abbreviations used are:
- FcεRI
- high-affinity receptors for the Fc portion of IgE
- ADAP
- adhesion- and degranulation-promoting adapter protein
- Bcr
- breakpoint cluster region protein
- BMMC
- Bone marrow-derived mast cells
- DAG
- diacyl-glycerol
- DNP
- Dinitrophenyl
- ES
- Embryonic stem
- FDR
- false discovery rate
- Fyb
- Fyn-binding protein
- Grap2
- Grb2-related adapter protein 2
- Grb2
- growth factor receptor-bound protein 2
- HSA
- human serum albumin
- iBAQ
- intensity-based absolute quantification
- IP3
- inositol tris-phosphate
- ITAM
- immunoreceptor tyrosine-based activation motif
- Itk
- IL-2-inducible T-cell kinase
- KI
- knock-in
- LAT
- linker of activation of T cells
- LCP2
- lymphocyte cytosolic protein 2
- LM
- laurylmaltoside
- mAb
- monoclonal antibody
- MAP
- mitogen-activated protein
- MS/MS
- tandem mass spectrometry
- nanoLC
- nano liquid chromatography
- Nck
- noncatalytic region of tyrosine kinase adapter protein
- NTAL
- non-T cell activation linker
- OST
- one-strep-tag
- PAG
- phosphoprotein associated with glycosphingolipid-enriched microdomains
- PLC
- phospholipase C
- SH
- Src homology
- SHIP
- SH2 domain-containing inositol 5′-phosphatase
- SKAP
- Src kinase-associated phopshoproteins
- SLP76
- SH2 domain-containing leukocyte protein of 76 kDa
- SDS
- sodium dodecyl sulfate
- TCA
- trichloroacetic acid
- TCR
- T cell receptor
- WT
- wild-type
- ZAP70
- zeta-associated protein of 70 kDa.
REFERENCES
- 1. Kinet J. P. (1999) The high-affinity IgE receptor (Fc epsilon RI): from physiology to pathology. Annu. Rev. Immunol. 17, 931–972 [DOI] [PubMed] [Google Scholar]
- 2. Reth M. (1989) Antigen receptor tail clue. Nature 338, 383–384 [PubMed] [Google Scholar]
- 3. Blank U., Ra C., Miller L., White K., Metzger H., Kinet J. P. (1989) Complete structure and expression in transfected cells of high affinity IgE receptor. Nature 337, 187–189 [DOI] [PubMed] [Google Scholar]
- 4. Turner H., Kinet J. P. (1999) Signalling through the high-affinity IgE receptor Fc epsilonRI. Nature 402, B24–B30 [DOI] [PubMed] [Google Scholar]
- 5. Sardiu M. E., Washburn M. P. (2011) Building protein-protein interaction networks with proteomics and informatics tools. J. Biol. Chem. 286, 23645–23651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bensimon A., Heck A. J., Aebersold R. (2012) Mass spectrometry-based proteomics and network biology. Ann. Rev. Biochem. 81, 379–405 [DOI] [PubMed] [Google Scholar]
- 7. Choudhary C., Mann M. (2010) Decoding signalling networks by mass spectrometry-based proteomics. Nat. Rev. 11, 427–439 [DOI] [PubMed] [Google Scholar]
- 8. Collins M. O., Choudhary J. S. (2008) Mapping multiprotein complexes by affinity purification and mass spectrometry. Curr. Opin. Biotechnol. 19, 324–330 [DOI] [PubMed] [Google Scholar]
- 9. Rigaut G., Shevchenko A., Rutz B., Wilm M., Mann M., Seraphin B. (1999) A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17, 1030–1032 [DOI] [PubMed] [Google Scholar]
- 10. Daulat A. M., Maurice P., Froment C., Guillaume J. L., Broussard C., Monsarrat B., Delagrange P., Jockers R. (2007) Purification and identification of G protein-coupled receptor protein complexes under native conditions. Mol. Cell. Proteomics 6, 835–844 [DOI] [PubMed] [Google Scholar]
- 11. Oellerich T., Bremes V., Neumann K., Bohnenberger H., Dittmann K., Hsiao H. H., Engelke M., Schnyder T., Batista F. D., Urlaub H., Wienands J. (2011) The B-cell antigen receptor signals through a preformed transducer module of SLP65 and CIN85. EMBO J. 30, 3620–3634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schmidt T. G., Skerra A. (2007) The Strep-tag system for one-step purification and high-affinity detection or capturing of proteins. Nat Protoc 2, 1528–1535 [DOI] [PubMed] [Google Scholar]
- 13. Jackman J. K., Motto D. G., Sun Q., Tanemoto M., Turck C. W., Peltz G. A., Koretzky G. A., Findell P. R. (1995) Molecular cloning of SLP-76, a 76-kDa tyrosine phosphoprotein associated with Grb2 in T cells. J. Biol. Chem. 270, 7029–7032 [DOI] [PubMed] [Google Scholar]
- 14. Liu S. K., Fang N., Koretzky G. A., McGlade C. J. (1999) The hematopoietic-specific adaptor protein gads functions in T-cell signaling via interactions with the SLP-76 and LAT adaptors. Curr. Biol. 9, 67–75 [DOI] [PubMed] [Google Scholar]
- 15. Yablonski D., Kadlecek T., Weiss A. (2001) Identification of a phospholipase C-gamma1 (PLC-gamma1) SH3 domain-binding site in SLP-76 required for T-cell receptor-mediated activation of PLC-gamma1 and NFAT. Mol. Cell. Biol. 21, 4208–4218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bubeck Wardenburg J., Fu C., Jackman J. K., Flotow H., Wilkinson S. E., Williams D. H., Johnson R., Kong G., Chan A. C., Findell P. R. (1996) Phosphorylation of SLP-76 by the ZAP-70 protein-tyrosine kinase is required for T-cell receptor function. J. Biol. Chem. 271, 19641–19644 [DOI] [PubMed] [Google Scholar]
- 17. Raab M., da Silva A. J., Findell P. R., Rudd C. E. (1997) Regulation of Vav-SLP-76 binding by ZAP-70 and its relevance to TCR zeta/CD3 induction of interleukin-2. Immunity 6, 155–164 [DOI] [PubMed] [Google Scholar]
- 18. Su Y. W., Zhang Y., Schweikert J., Koretzky G. A., Reth M., Wienands J. (1999) Interaction of SLP adaptors with the SH2 domain of Tec family kinases. Eur. J. Immunol. 29, 3702–3711 [DOI] [PubMed] [Google Scholar]
- 19. Wunderlich L., Farago A., Downward J., Buday L. (1999) Association of Nck with tyrosine-phosphorylated SLP-76 in activated T lymphocytes. Eur. J. Immunol. 29, 1068–1075 [DOI] [PubMed] [Google Scholar]
- 20. Pivniouk V. I., Martin T. R., Lu-Kuo J. M., Katz H. R., Oettgen H. C., Geha R. S. (1999) SLP-76 deficiency impairs signaling via the high-affinity IgE receptor in mast cells. J. Clin. Invest. 103, 1737–1743 [PMC free article] [PubMed] [Google Scholar]
- 21. Kambayashi T., Okumura M., Baker R. G., Hsu C. J., Baumgart T., Zhang W., Koretzky G. A. (2010) Independent and cooperative roles of adaptor molecules in proximal signaling during FcepsilonRI-mediated mast cell activation. Mol Cell. Biol. 30, 4188–4196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Roget K., Malissen M., Malbec O., Malissen B., Daeron M. (2008) Non-T cell activation linker promotes mast cell survival by dampening the recruitment of SHIP1 by linker for activation of T cells. J. Immunol. 180, 3689–3698 [DOI] [PubMed] [Google Scholar]
- 23. Mingueneau M., Roncagalli R., Gregoire C., Kissenpfennig A., Miazek A., Archambaud C., Wang Y., Perrin P., Bertosio E., Sansoni A., Richelme S., Locksley R. M., Aguado E., Malissen M., Malissen B. (2009) Loss of the LAT adaptor converts antigen-responsive T cells into pathogenic effectors that function independently of the T cell receptor. Immunity 31, 197–208 [DOI] [PubMed] [Google Scholar]
- 24. Zhang Y., Muyrers J. P., Testa G., Stewart A. F. (2000) DNA cloning by homologous recombination in Escherichia coli. Nat. Biotechnol. 18, 1314–1317 [DOI] [PubMed] [Google Scholar]
- 25. Pettitt S. J., Liang Q., Rairdan X. Y., Moran J. L., Prosser H. M., Beier D. R., Lloyd K. C., Bradley A., Skarnes W. C. (2009) Agouti C57BL/6N embryonic stem cells for mouse genetic resources. Nat. Methods 6, 493–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Malbec O., Malissen M., Isnardi I., Lesourne R., Mura A. M., Fridman W. H., Malissen B., Daeron M. (2004) Linker for activation of T cells integrates positive and negative signaling in mast cells. J. Immunol. 173, 5086–5094 [DOI] [PubMed] [Google Scholar]
- 27. Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 28. Schwanhausser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., Chen W., Selbach M. (2011) Global quantification of mammalian gene expression control. Nature 473, 337–342 [DOI] [PubMed] [Google Scholar]
- 29. Deeb S. J., D'Souza R. C., Cox J., Schmidt-Supprian M., Mann M. (2012) Super-SILAC allows classification of diffuse large B-cell lymphoma subtypes by their protein expression profiles. Mol. Cell. Proteomics 11, 77–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Felber J. P., Coombs T. L., Vallee B. L. (1962) The mechanism of inhibition of carboxypeptidase A by 1,10-phenanthroline. Biochemistry 1, 231–238 [DOI] [PubMed] [Google Scholar]
- 31. Tamir I., Stolpa J. C., Helgason C. D., Nakamura K., Bruhns P., Daeron M., Cambier J. C. (2000) The RasGAP-binding protein p62dok is a mediator of inhibitory FcgammaRIIB signals in B cells. Immunity 12, 347–358 [DOI] [PubMed] [Google Scholar]
- 32. Ott V. L., Tamir I., Niki M., Pandolfi P. P., Cambier J. C. (2002) Downstream of kinase, p62(dok), is a mediator of Fc gamma IIB inhibition of Fc epsilon RI signaling. J. Immunol. 168, 4430–4439 [DOI] [PubMed] [Google Scholar]
- 33. Malbec O., Roget K., Schiffer C., Iannascoli B., Dumas A. R., Arock M., Daeron M. (2007) Peritoneal cell-derived mast cells: an in vitro model of mature serosal-type mouse mast cells. J. Immunol. 178, 6465–6475 [DOI] [PubMed] [Google Scholar]
- 34. Pejler G., Ronnberg E., Waern I., Wernersson S. (2010) Mast cell proteases: multifaceted regulators of inflammatory disease. Blood 115, 4981–4990 [DOI] [PubMed] [Google Scholar]
- 35. Caughey G. H. (2011) Mast cell proteases as protective and inflammatory mediators. Adv. Exp. Med. Biol. 716, 212–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cao M. Y., Davidson D., Yu J., Latour S., Veillette A. (1999) Clnk, a novel SLP-76-related adaptor molecule expressed in cytokine-stimulated hemopoietic cells. J. Exp. Med. 190, 1527–1534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yu J., Riou C., Davidson D., Minhas R., Robson J. D., Julius M., Arnold R., Kiefer F., Veillette A. (2001) Synergistic regulation of immunoreceptor signaling by SLP-76-related adaptor Clnk and serine/threonine protein kinase HPK-1. Mol. Cell. Biol. 21, 6102–6112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Goitsuka R., Kanazashi H., Sasanuma H., Fujimura Y., Hidaka Y., Tatsuno A., Ra C., Hayashi K., Kitamura D. (2000) A BASH/SLP-76-related adaptor protein MIST/Clnk involved in IgE receptor-mediated mast cell degranulation. Int. Immunol. 12, 573–580 [DOI] [PubMed] [Google Scholar]
- 39. Utting O., Sedgmen B. J., Watts T. H., Shi X., Rottapel R., Iulianella A., Lohnes D., Veillette A. (2004) Immune functions in mice lacking Clnk, an SLP-76-related adaptor expressed in a subset of immune cells. Mol. Cell. Biol. 24, 6067–6075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Fujii Y., Wakahara S., Nakao T., Hara T., Ohtake H., Komurasaki T., Kitamura K., Tatsuno A., Fujiwara N., Hozumi N., Ra C., Kitamura D., Goitsuka R. (2003) Targeting of MIST to Src-family kinases via SKAP55-SLAP-130 adaptor complex in mast cells. FEBS Lett. 540, 111–116 [DOI] [PubMed] [Google Scholar]
- 41. Sahay S., Pannucci N. L., Mahon G. M., Rodriguez P. L., Megjugorac N. J., Kostenko E. V., Ozer H. L., Whitehead I. P. (2008) The RhoGEF domain of p210 Bcr-Abl activates RhoA and is required for transformation. Oncogene 27, 2064–2071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Volna P., Lebduska P., Draberova L., Simova S., Heneberg P., Boubelik M., Bugajev V., Malissen B., Wilson B. S., Horejsi V., Malissen M., Draber P. (2004) Negative regulation of mast cell signaling and function by the adaptor LAB/NTAL. J. Exp. Med. 200, 1001–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dong S., Corre B., Foulon E., Dufour E., Veillette A., Acuto O., Michel F. (2006) T cell receptor for antigen induces linker for activation of T cell-dependent activation of a negative signaling complex involving Dok-2, SHIP-1, and Grb-2. J. Exp. Med. 203, 2509–2518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ng C. H., Xu S., Lam K. P. (2007) Dok-3 plays a nonredundant role in negative regulation of B-cell activation. Blood 110, 259–266 [DOI] [PubMed] [Google Scholar]
- 45. Cao L., Yu K., Banh C., Nguyen V., Ritz A., Raphael B. J., Kawakami Y., Kawakami T., Salomon A. R. (2007) Quantitative time-resolved phosphoproteomic analysis of mast cell signaling. J. Immunol. 179, 5864–5876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lemay S., Davidson D., Latour S., Veillette A. (2000) Dok-3, a novel adapter molecule involved in the negative regulation of immunoreceptor signaling. Mol. Cell. Biol. 20, 2743–2754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Daeron M., Lesourne R. (2006) Negative signaling in Fc receptor complexes. Adv. Immunol. 89, 39–86 [DOI] [PubMed] [Google Scholar]
- 48. Mayya V., Lundgren D. H., Hwang S. I., Rezaul K., Wu L., Eng J. K., Rodionov V., Han D. K. (2009) Quantitative phosphoproteomic analysis of T cell receptor signaling reveals system-wide modulation of protein-protein interactions. Sci. Signal. 2, ra46. [DOI] [PubMed] [Google Scholar]
- 49. Korus M., Mahon G. M., Cheng L., Whitehead I. P. (2002) p38 MAPK-mediated activation of NF-kappaB by the RhoGEF domain of Bcr. Oncogene 21, 4601–4612 [DOI] [PubMed] [Google Scholar]
- 50. Park A. R., Oh D., Lim S. H., Choi J., Moon J., Yu D. Y., Park S. G., Heisterkamp N., Kim E., Myung P. K., Lee J. R. (2012) Regulation of dendritic arborization by BCR Rac1 GTPase-activating protein, a new substrate of protein tyrosine phosphatase receptor T. J. Cell Sci. 125, 4518–4531 [DOI] [PubMed] [Google Scholar]
- 51. Cho Y. J., Cunnick J. M., Yi S. J., Kaartinen V., Groffen J., Heisterkamp N. (2007) Abr and Bcr, two homologous Rac GTPase-activating proteins, control multiple cellular functions of murine macrophages. Mol. Cell. Biol. 27, 899–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.