Abstract
The majority of mass spectrometry-based protein quantification studies uses peptide-centric analytical methods and thus strongly relies on efficient and unbiased protein digestion protocols for sample preparation. We present a novel objective approach to assess protein digestion efficiency using a combination of qualitative and quantitative liquid chromatography-tandem MS methods and statistical data analysis. In contrast to previous studies we employed both standard qualitative as well as data-independent quantitative workflows to systematically assess trypsin digestion efficiency and bias using mitochondrial protein fractions. We evaluated nine trypsin-based digestion protocols, based on standard in-solution or on spin filter-aided digestion, including new optimized protocols. We investigated various reagents for protein solubilization and denaturation (dodecyl sulfate, deoxycholate, urea), several trypsin digestion conditions (buffer, RapiGest, deoxycholate, urea), and two methods for removal of detergents before analysis of peptides (acid precipitation or phase separation with ethyl acetate). Our data-independent quantitative liquid chromatography-tandem MS workflow quantified over 3700 distinct peptides with 96% completeness between all protocols and replicates, with an average 40% protein sequence coverage and an average of 11 peptides identified per protein. Systematic quantitative and statistical analysis of physicochemical parameters demonstrated that deoxycholate-assisted in-solution digestion combined with phase transfer allows for efficient, unbiased generation and recovery of peptides from all protein classes, including membrane proteins. This deoxycholate-assisted protocol was also optimal for spin filter-aided digestions as compared with existing methods.
MS-based proteomics is an indispensable technology for the characterization of complex biological systems, including relative or absolute protein expression levels and protein post-translational modifications. The most popular method for analyzing medium to high complexity protein samples in large-scale proteomics relies on protein digestion by using the endoprotease trypsin. Analysis and sequencing of tryptic peptides by liquid chromatography-tandem MS (LC-MS/MS)1 then enables identification and determination of protein expression levels based on the peptide ion abundance level or the (fragment) ion intensities of identified peptides. This peptide-centric approach thus strongly relies on efficient, unbiased and reproducible protein digestion protocols. Efficiency is required to maximize the number of detectable peptides per protein (coverage) to distinguish unique proteins within protein families with similar sequences and/or sequence variants, and to detect post-translational modifications. Unbiased generation of peptides is required for the resulting data set to most accurately reflect the relative (stoichiometry) and absolute protein abundance in a sample. A particular protocol should be unbiased with respect to abundance, molecular weight, hydrophobicity and protein class. Membrane proteins for example are often suspected to be underrepresented. For MS-based proteomics approaches several critical steps can be distinguished: (a) disruption and solubilization of cells and protein complexes, (b) protein denaturation and enzymatic proteolysis, (c) MS-compatible peptide recovery, which normally entails removal of reagent leftovers and desalting before MS analysis, (d) adequate peptide separation (achieved by liquid chromatography), and (e) MS peptide analysis and sequencing (MS/MS), including the chosen data acquisition strategy.
Comparative evaluations of digestion protocols generally consist of qualitative studies using standard tandem mass spectrometry. These approaches may reveal efficiency (i.e. more identifications), but are unable to reveal digestion protocol induced bias with respect to peptide and protein abundance, including membrane proteins. In addition, most data-dependent acquisition workflows are intrinsically biased, which is detrimental for making comparisons. The aim of the present study was to systematically assess efficiency and bias of trypsin-based protocols applying both standard qualitative and label-free quantitative MS approaches.
The in-gel digestion protocol for proteomics, established over 15 years ago (1), has been the cornerstone method affording robust protein identifications from many sample types. Although sodium dodecyl sulfate (SDS) interferes with trypsin digestion and hampers LC-MS analysis, this powerful detergent can still be used to achieve complete protein solubilization as gel-separation is an effective way to remove interfering substances. Gel-based approaches are however not optimal for protein samples of increasing complexity and dynamic range (2). Inherent and practical limitations include, for example, concentration-dependent, incomplete peptide recovery and error-prone handling procedures (3–6). This hampers throughput, reproducibility and unbiased protein analysis, which in recent years has prompted a shift toward the application and optimization of in-solution digestion procedures.
Previous comparative studies revealed that for in-solution digestions, the acid labile and MS-compatible detergent RapiGest performed most favorably compared with buffer only, urea, other detergents and organic solvents (7–9). Sodium deoxycholate (SDC), naturally found in mammalian bile (10), has emerged as a cheaper MS-compatible detergent for in-solution digestion (11). Unlike other detergents, SDC was found to enhance trypsin activity almost fivefold at a concentration of 1% (12). Like RapiGest, SDC can also be removed by acidification, but potentially without detrimental peptide loss if a phase separation protocol involving organic solvent is applied (12).
An alternative strategy is to perform protein digestion on spin filter devices, introduced a few years ago by Manza and co-workers (13), and further developed by Wisniewski et al. (14). This approach allows the use of SDS to first achieve complete protein solubilization followed by removal of the detergent through repeated washes with urea (14). This is an effective way to remove interfering chemicals and small molecules after protein solubilization, and before digestion, without substantial sample loss. Although this protocol is touted to be a highly effective and universal method for any type of sample, digestion is performed using urea or buffer only and has so far not been evaluated in combination with detergents such as SDC.
For our comparative study we selected protocols and methods based on spin filter-aided and standard in-solution digestion that were previously reported optimal and we also report novel optimized protocols. We investigated several experimental parameters including reagents for protein solubilization and denaturation (SDS, SDC, urea), spin filter aided removal of SDS before digestion (urea, SDC, buffer), trypsin digestion conditions (buffer, RapiGest, SDC, urea), and methods for removal of detergents before analysis of peptides (acid precipitation or phase separation with ethyl acetate).
Mitochondria are organelles carrying out key metabolic processes fundamental for cellular function (15). The mitochondrial proteome is predicted to contain up to a thousand proteins (16) and is very heterogeneous with a wide range of protein pI, molecular weight and hydrophobicity values (17). We selected mitochondrial preparations to serve as model sample of medium complexity, containing a favorable combination of peptide and protein classes, including soluble and insoluble membrane-anchored or integral proteins.
Using standard qualitative as well as data-independent quantitative LC-MS/MS workflows we demonstrate that SDC-based protocols combined with phase separation are the most optimal for both in-solution and filter-aided tryptic digestion, yielding the highest efficiency and lowest bias. This workflow enabled quantitative and objective assessment of various protein digestion conditions, identifying optimal protocols for efficient and unbiased protein analysis.
EXPERIMENTAL PROCEDURES
Materials
RapiGest acid-labile surfactant was purchased from Waters Corporation (Milford, MA). Trypsin (modified, sequencing grade) was obtained from Promega (Madison, WI). Poros®20 R2 reverse phase material was purchased from Applied Biosystems (Invitrogen, Applied Biosystems). Sodium deoxycholate (SDC), sodium dodecyl sulfate (SDS) and other chemicals were purchased from Sigma. Solvents used were minimally HPLC grade.
Rat Liver Mitochondria-Enriched Fractions
Crude mitochondrial samples from 25 animals were generated in the group of Professor Rolf Kristian Berge, University of Bergen, Norway as recently described (18). The model sample was generated by pooling equal amounts of the 25 preparations. Samples were stored at −80 °C and all manipulations were performed in a cold room to avoid degradation during sample preparation. Protein quantitation was carried out by Qubit™ Fluorometric Quantitation (Invitrogen, Invitrogen).
Sample Preparation Protocols
Hundred microgram aliquots of pooled mitochondrial sample were used for each individual experiment and all procedures were performed in triplicate. In order to keep the description of all the various protocols brief, only general buffer names are mentioned. All protocol-specific buffer compositions are provided in Table I.
Table I. Overview of the sample preparation and digestion conditions for the spin filter-aided (SF) and standard in-solution digestion (ISD) protocols. All listed values are end-concentrations; the marked values (*) represent an effective 10-fold dilution over initial conditions. If present during digestion, detergent removal was achieved by acid precipitation (AP) or phase transfer (PT).

In-solution Digestion (ISD)
Five μl aliquots, equivalent to 100 μg of protein, were mixed with 10 μl of “denaturation & solubilization” buffer and incubated for 10 min at 80 °C (except for the ISD:Urea protocol in which the temperature was controlled to not exceed 30 °C). Subsequently, 5 μl of 45 mm dithiotreitol solution (in H2O) was added followed by incubation for 20 min at 60 °C. Reduced cysteine residues were alkylated by adding 5 μl of 100 mm iodoacetamide solution (in H2O) and incubation proceeded for 30 min at room temperature, in the dark. The sample was diluted with water and the protease trypsin was added in a 1:100 (enzyme/protein) ratio to a final volume of 100 μl. This is an effective 10-fold dilution over initial conditions and the end-concentrations are indicated in Table I. The digestions took place for 5–7 h at 37 °C. Trypsin activity was inhibited by acidification with 5 μl 10% TFA, which also induced precipitation of the surfactant, if added. RapiGest was removed according to the protocol supplied by the manufacturer (incubation for 30 min at 37 °C, followed by centrifugation). Standard and phase transfer assisted removal of SDC was performed as described (12).
Spin Filter Aided In-Solution Digestion (SF-ISD)
Five μl aliquots, equivalent to 100 μg of protein, were mixed with 50 μl of denaturation and solubilization buffer and incubated for 30 min at 60 °C. After protein denaturation, the sample was transferred to a Microcon spin filter device (YM-30, Millipore) and mixed with 200 μl of “remove SDS” buffer (Table I). The device was centrifuged at 10,000 × g for 15 min. This step was repeated once. All subsequent centrifugation steps were performed under the same conditions, allowing maximum concentration. Subsequently, 100 μl of iodoacetamide solution (0.05 m) was added to the concentrated protein mixture followed by 1 min shaking and 20 min incubation without shaking, at room temperature in the dark. All devices were centrifuged to remove excess iodoacetamide solution. Two additional wash steps were performed by adding 100 μl of buffer (Table I) followed by centrifugation. The concentrated protein mixture was subjected to tryptic digestion by adding 50 μl of 0.02 μg/μl trypsin solution (enzyme to protein ratio 1:100, in buffer; see Table I) and mixed at 600 rpm in a thermomixer for 1 min. Digestion was performed by incubation in a wet chamber at 37 °C for 5–7 h. Afterward, peptides were collected in a low-binding tube using centrifugation, and the filter device was rinsed with 50 μl of buffer (Table I). When applicable, standard and phase transfer assisted removal of SDC was performed as described (12).
Protein Database
Peptide and protein identifications for both approaches were obtained using the same UniProt database (release 2011_01, 16432 entries) with rat Swiss-Prot and TrEMBL entries that were modified to include known N-terminal processing and maturation of proteins (19, 20). Common contaminants were appended, as well as several protein standards that serve as internal standard for the label-free absolute quantitative approach enabling determination of protein concentrations and to address technical variation (21).
Standard nanoLC-MS/MS Analysis
Peptide digest mixtures (corresponding to about 250 ng) were desalted using Poros®20 R2 reversed phase microcolumns as previously described (22) and SpeedVac lyophilized before LC-MS. Peptides were dissolved in mobile phase A (0.1% formic acid in water) and applied onto an in-house made 17 cm fused silica capillary column (100 μm ID) packed with 3 μm Reprosil-C18 reverse phase material (Dr. Maisch GmbH, Ammerbuch-Entringen, Germany) and fitted to an Easy-nLC (Thermo Scientific/Proxeon, Odense, Denmark). Peptides were separated using a 100 min gradient from 0% to 34% of mobile phase B (90% acetonitrile, 0.1% formic acid) at 250 nl/min. Eluting peptides were analyzed using automated data-dependent acquisition on a LTQ-Orbitrap XL mass spectrometer (Thermo Scientific, Bremen, Germany). Each MS scan (400–1800 m/z) was acquired at a resolution of 60000 FWHM and was followed by 5 MS/MS scans triggered above an intensity of 20000 using CID (normalized collision energy 35). The maximum ion injection time was 500 ms for MS and 300 ms for MS/MS scans. The automatic gain control (AGC) target value was 1000000 for MS scans in the Orbitrap and 10000 for MS/MS scans in the LTQ.
Standard LC-MS/MS Data Processing and Protein Identification
Raw data from the LTQ-Orbitrap-MS were processed in Proteome Discoverer v1.3.0.339 (Thermo Scientific) using default parameters. The rat N-matured UniProt database was searched using an in-house Mascot server, version 2.2.03 (Matrix Science, London, U.K.) with 10 ppm peptide and 0.6 Da fragment ion tolerances. Trypsin with the possibility of two missed cleavages was selected as enzyme. Carbamidomethyl cysteine was specified as fixed modification. The following variable modifications were allowed: oxidation (M), deamidation (N/Q) and N-terminal protein acetylation. The Percolator tool was used for peptide validation based on the PEP score. A cutoff value of peptide rank = 1 and high confidence was chosen, corresponding to a 1% false discovery rate (FDR) on peptide-level.
Quantitative nanoLC-MSE Analysis
Prior to analysis, 0.5 μl of each tryptic peptide solution (corresponding to about 500 ng) was diluted with an aqueous 0.1% trifluoroacetic acid solution to which protein digest internal standards were added, serving both as reference and internal standard. Included predigested standard proteins were 100 fmol bovine albumin and 50 fmol rabbit glycogen phosphorylase B (MassPrep standards, Waters, Milford, MA). Each sample was analyzed in triplicate. Online desalting and nanoscale LC separation of tryptic peptides was performed with a NanoAcquity ultraperformance liquid chromatography (UPLC) system (Waters) equipped with a Symmetry C18 trapping column (180 μm x 20 mm, 5 μm particle size; Waters) and a Bridged Ethyl Hybrid (BEH) C18 analytical reversed phase column (75 μm x 250 mm, 1.7 μm particle size; Waters). Mobile phase A was water with 0.1% formic acid and mobile phase B was 0.1% formic acid in acetonitrile. The peptides were separated with a gradient of 3–40% mobile phase B over 90 min at a flow rate of 300 nl/min. The auxiliary pump of the NanoAcquity system provided [Glu1]fibrinopeptide B as standard to the reference sprayer, which was sampled during the acquisition at 60 s intervals for postdata acquisition lock mass correction.
Eluting peptides were analyzed using a Q-ToF Synapt HDMS mass spectrometer (Waters Corporation, Manchester, UK). Data were acquired in data independent acquisition (DIA) mode, also referred to as MSE, which is an unbiased, alternating mode of acquisition in which the mass spectrometer does not select specific precursors, but alternates between low and elevated collision energy states (23, 24). Low and elevated energy MS spectra were both acquired from m/z 50 to 1990 for 0.7 s each with a 0.02-s interscan delay. Low energy MS scans were collected at constant collision energy of 4 eV whereas the collision energy during elevated energy MS scans was ramped from 15 to 40 eV.
LC-MSE Data Processing and Protein Identification
DIA LC-MS raw data files were processed using ProteinLynx GlobalServer (PLGS) version 2.4 (Waters) and subsequent database searching was performed using the Ion Accounting algorithm (25), embedded in PLGS, searching the rat N-matured UniProt database. The search tolerances were set to automatic (typically 10 ppm for precursor and 20 ppm for product ions), with trypsin as enzyme (allowing up to two missed cleavages), fixed carbamidomethyl modification for cysteine residues, and N-terminal acetylation, oxidation of methionine, and deamidation of asparagine and glutamine as variable modifications. Other settings included, number of product ion matches per peptide ≥ 3, number of product ion matches per protein ≥ 6, number of missed tryptic cleavage sites ≤ 2, and protein false positive rate (FPR) ≤ 5%. The protein-level FPR is calculated during the search depletion loops based on appearance of random matches observed during the search of the concatenated forward and corresponding randomized database (25). Identifications were filtered to only accept proteins that were detected in at least two out of three replicate injections. As a result, the final false positive identification rate for the complete data set was well below 1%.
Label Free (Absolute) Quantification
PLGS was configured to only report and quantify homologous proteins when unique, discriminating peptides were also detected for each protein. PLGS was also configured to output csv-files for further analysis of absolute quantitative levels in Excel (Microsoft). For increased accuracy, label-free quantitation and subsequent statistical analysis, the raw DIA LC-MS data files were loaded into Progenesis LC-MS (Nonlinear Dynamics, UK) to align all detected features among all runs, and determine their abundance, followed by outlier insensitive median normalization. After import of the PLGS search results into Progenesis LC-MS, the complete data set was filtered to use only unique (proteotypic) peptides for protein quantitation and was exported as a csv-file.
Additional Data Processing and Biostatistics
ProteinCenter (Professional Edition, Proxeon, Odense, Denmark) was used to calculate the peptide grand average of hydropathicity (GRAVY) values and to determine the number of protein transmembrane domains for both the qualitative and quantitative data set. Significant up/down-regulations among experimental stages were determined by means of q-values (p values corrected for multiple testing) obtained from a t test among results from a digestion protocol compared with the mean over all protocols. Hierarchical clustering (standard function hclust in R, http://www.R-project.org) was performed on normalized peptide and protein abundance data sets and presented as heat maps including reordering of the peptides/proteins. Fuzzy c-means clustering analysis was carried out on standardized data to allow identification of common trends in data sets after correct parameter estimation. Parameters were set according to a previous publication (26) where the number of clusters was estimated by inspection of the minimum centroid distance and the Xie-Beni index (27). Colors correspond to the degree a peptide/protein belongs to a cluster, represented by the so-called membership value.
RESULTS
Study Design
We devised a comparative study to identify the most efficient and unbiased protein digestion protocol, by applying both standard qualitative and data-independent label-free quantitative MS approaches. Using mitochondria-enriched fractions as sample model, we optimized and assessed both existing and novel combinations of conditions for trypsin-based digestion methods, using both spin filter-aided (SF) and standard in-solution digestion (ISD). A flowchart illustrating the combination of different steps and digestion conditions compared in this study is depicted in Fig. 1. These combinations were chosen to evaluate several critical parameters of a digestion protocol including protein solubilization and denaturation, conditions during trypsin digestion as well as removal of detergents, if any, before digestion and/or before MS analysis. The protocols were designed and selected based on the use of the best, but principally MS-incompatible detergent for protein solubilization (SDS) versus MS-compatible surfactants (SDC, RapiGest) and chaotropic reagents (urea) considered optimal for protein digestion. Digestion of 100 μg of mitochondrial protein sample was performed in triplicate for each of the nine investigated protocols as detailed in Table I.
Fig. 1.
Study design. Flowchart illustrating the combination of different steps and digestion conditions compared in this study.
Standard Qualitative Evaluation of Digestion Protocol Efficiency
We first focused on the qualitative comparison of digestion protocols including only standard removal of MS-compatible surfactants (RapiGest and SDC) by acid precipitation as well as urea-based protocols. In addition we investigated the effectiveness of removing SDS before trypsin digestion in SF-based protocols by washing with urea, SDC or buffer only. A total of 21 protein digests corresponding to seven different protocols were analyzed by LC-MS/MS on a LTQ-Orbitrap XL using standard data dependent acquisition (DDA). Under-sampling and missing values are common problems related to the stochastic nature of standard DDA approaches and therefore combining 3–5 replicates is at least recommended to characterize a sample (28). The unique peptide and protein identifications for each single replicate as well as the combined result of triplicate experiments are provided in Table II. A qualitative summary of technical and protocol triplicates is supplied in the supplemental Material (supplemental Figs. S1 and S2). The number of summed identifications over three replicates for each of the seven different protocols ranged from 121 to 484 proteins (Fig. 2A) and 208 to 3489 peptides (Fig. 2C). When considering all the presented parameters including the number of identified proteins, peptides and overall coverage, the SF-ISD protocol with SDC is the most efficient protocol in this comparison. With this result we also evaluated for the first time the application of SDC as surfactant in SF-ISD protocols. This combination outperformed the standard SF-ISD:SDS-Urea/- protocol (14), commonly regarded as a highly effective and universal sample preparation method. The difference is most notable when considering the average and distribution of protein sequence coverage (Fig. 2B), and the number of identified peptides (Fig. 2C). Although the number of protein identifications ranks as the second best result when using the SF-ISD:SDS-Urea/- protocol (Fig. 2A), the number of peptides per protein is among the lowest when using that protocol (Table II).
Table II. Comparison of trypsin-based digestion protocols after analysis by standard qualitative nanoLC-MS/MS.
| Digestion protocol | Peptide identifications |
Protein identifications |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Rep 1 | Rep 2 | Rep 3 | Suma | Average pI | Average GRAVY | Rep 1 | Rep 2 | Rep 3 | Suma | Average MW (kDa)a | Average coverageb | Peptides/protein | |
| ISD:RG | 2225 | 2241 | 2148 | 2988 | 6.31 | −0.28 | 404 | 414 | 367 | 455 | 41.6 | 18.7 | 6.6 |
| ISD:Urea | 2334 | 2319 | 2387 | 3153 | 6.35 | −0.23 | 372 | 366 | 361 | 421 | 43.6 | 21.6 | 7.5 |
| ISD:SDC | 1958 | 2515 | 2762 | 3359 | 6.30 | −0.21 | 345 | 387 | 411 | 444 | 42.3 | 21.3 | 7.6 |
| SF-ISD:SDC | 2682 | 2536 | 2750 | 3489 | 6.33 | −0.22 | 420 | 407 | 430 | 484 | 43.4 | 20.8 | 7.2 |
| SF-ISD:SDS-Urea/- | 1589 | 1642 | 1952 | 2348 | 6.13 | −0.35 | 397 | 426 | 381 | 468 | 44.0 | 13.5 | 5.0 |
| SF-ISD:SDS/SDC | 944 | 1124 | 770 | 1248 | 6.11 | −0.29 | 252 | 270 | 206 | 290 | 43.2 | 12.7 | 4.3 |
| SF-ISD:SDS/- | 154 | 149 | 48 | 208 | 5.57 | −0.45 | 106 | 98 | 42 | 121 | 50.4 | 4.5 | 1.7 |
a Identified proteins and reported molecular weights represent mature protein forms as non-mature sequences such as signal peptides and mitochondrial transit sequences have been removed, as described in Material and Methods.
b Values represent the sum of unique peptide sequences and protein identifications from triplicate experiments.
Fig. 2.

Qualitative comparison of seven digestion protocols analyzed with standard nanoLC-MS/MS. Protein (A) and peptide (C) identifications are reported as the merged result of three replicates per protocol. Depicted are the distributions of summed protein sequence coverage (B) and the percentage of missed cleavages among the evaluated protocols (D).
The improved performance of our new SF-ISD:SDC protocol can most likely be attributed to the use of additives during digestion, as in this comparison the SF-ISD:Urea/- protocol is generally outperformed by any of the other protocols involving RapiGest, SDC, or urea. When comparing detergent performance, the use of SDC seemingly outperforms the RapiGest-based protocol. Although the number of protein identifications is similar, the number of identified peptides (Fig. 2C) and overall protein coverage (Fig. 2B) are in favor of the SDC-based protocols. The protein coverage for the ISD:Urea protocol is the second best, but also displays less protein identifications and an obvious larger number of missed cleavages (Fig. 2D). No obvious differences were noted for the percentage of modified peptides (supplemental Fig. S3D), average peptide GRAVY score, average peptide pI and average molecular weight (Table II) for which graphical distributions are also provided (supplemental Fig. S3). Combined we thus concluded that our SF-ISD:SDC protocol is preferable to the other methods tested.
Evaluation of the SDS Removing Efficiency before Digestion with SF-ISD Protocols
SDS is a powerful solubilization agent used at the start of SF-ISD protocols. Any leftovers of SDS will however affect trypsin digestion and can hamper chromatographic separation of peptides and subsequent MS analysis. Urea was shown to be effective in the quantitative removal of SDS (14), but we attempted to streamline the SF-ISD protocol for use with SDC by avoiding urea. This would simplify and shorten the procedure by reducing the number of steps required. Three protocols were tested for this purpose. A total of 468 proteins were identified after using the published standard SF-ISD protocol. Far fewer proteins were identified for the SF-ISD protocols, which use washes with SDC or buffer only for removing SDS before trypsin digestion (290 and 121 proteins, respectively). The chromatographic runs for these two samples were clearly disrupted (data not shown), indicative of interfering residual SDS present in the digests. The original SF-ISD protocol with urea as wash solvent clearly indicates that only urea is effective enough to completely remove SDS and cannot be replaced by washing in the presence of SDC.
In summary, the standard qualitative results indicate a general advantage of detergent-assisted (SDC) digestion and that only urea is sufficient enough to remove the SDS used in spin filter-aided protocols. Any particular differences between for example the best performing filter-aided and standard in-solution digestion protocol cannot be discerned. The variation among those protocols lies within the general systematic variation caused by the experimental procedures and data-dependent MS/MS acquisition (supplemental Figs. S1 and S2).
Qualitative Evaluation of Digestion Protocols Following Data-Independent Acquisition
The main limitations of standard MS/MS approaches in shotgun proteomics are under-sampling and missing values as the precursor selection process is favorable to the more abundant components present in a sample and different pools of peptides are targeted in each (replicate) experiment (28). We therefore employed MSE, a data-independent mode of acquisition, which allows for the detection and multiplexed fragmentation of all ions without selection of precursors (23, 24). This mode of acquisition also enables accurate label-free relative and absolute quantitation (21). We proceeded with this qualitative and quantitative approach to identify the most efficient protocol, which at the same time shows the least bias to a particular peptide or protein class (e.g. highly abundant, hydrophobic or membrane associated). We decided to proceed with the four most promising and efficient protocols (ISD:RG, ISD:Urea, ISD:SDC, SF-ISD:SDC) and added two additional conditions to investigate any bias of removing SDC by either acid precipitation (AP) or phase transfer (PT). A previous qualitative study reported that during acidic precipitation of SDC and hydrolysis of RapiGest, potential bias is introduced because of coprecipitation events (12). The authors introduced a phase transfer protocol for SDC using ethyl acetate to prevent detrimental peptide loss, which was suggested to particularly affect hydrophobic peptides (12).
A total of six different protocols were analyzed in triplicate by LC-MSE on a QTOF tandem mass spectrometer as described in the materials and methods. A detailed overview of results is provided in Fig. 3 and Table III. We applied stringent filtering to the qualitative and quantitative results obtained from the PLGS software. Only unique peptide and protein identifications replicating in at least two out of three replicate runs were reported and used for further analysis. The number of identifications replicating in at least two out of three runs for each of the six different protocols ranged from 204 to 272 proteins (Fig. 3A) and from 1729 to 2706 peptides (Fig. 3C). These qualitative results follow a similar trend as compared with the initial analysis (see previous sections). All SDC-based protocols outperformed the other protocols (ISD:RG and ISD:Urea). The SF-ISD protocols with SDC are the most efficient when considering the number of identified peptides, proteins, and overall coverage (Table III). Interestingly, for both the ISD:SDC and SF-ISD:SDC protocols, removal of SDC after digestion using the phase transfer protocol (PT) is advantageous over standard removal by acid precipitation (AP). Although the protein identifications are similar, the number of identified peptides clearly differs, which is reflected in the protein coverage box plots (Fig. 3B). The protein coverage for the ISD:Urea protocol again displays a relatively high protein coverage, but also the lowest number of identified peptides and proteins (Table III), in addition to a comparably large number of missed cleavages (Fig. 3D). No differences were observed on the level of peptide modifications or protein molecular weight (supplemental Fig. S4).
Fig. 3.

Qualitative evaluation of six digestion protocols analyzed with data independent LC-MSE. Average number of unique protein (A) and peptide (C) identifications from technical triplicates (white bars), number of quantified proteins and peptides present in at least two out of three replicates (gray bars), and average number of unique, quantified protein and peptide identifications after alignment of all data-independent runs using Progenesis LC-MS software (black bars). The distributions of summed protein sequence coverage are depicted (B) as well as the percentage of missed cleavages among the evaluated protocols (D).
Table III. Comparison of trypsin-based digestion protocols after analysis by label-free quantitative nanoLC-MSE.
| Digestion protocol | Peptide identifications |
Protein identifications |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Averagea (sd) | ID1 (≥2)b | ID2 (≥2)c | Averaged pI (≥2) | Averaged GRAVY (≥2) | Averagea (sd) | Quantified1 (≥2)b | Quantified2 (≥2)c | Averaged MW (kDa) | Averaged coverage | Peptides/Protein 1b–2c | |
| ISD:RG (AP) | 2584 (56) | 1869 | 3452 | 6.42 | −0.19 | 246 (9) | 226 | 327 | 45.7 | 30.4 | 8.3–10.6 |
| ISD:Urea | 2502 (95) | 1729 | 3536 | 6.55 | −0.22 | 222 (9) | 204 | 331 | 46.7 | 33.9 | 8.5–10.7 |
| ISD:SDC (AP) | 2782 (129) | 2087 | 3559 | 6.44 | −0.15 | 261 (9) | 245 | 331 | 44.5 | 32.2 | 8.5–10.8 |
| ISD:SDC (PT) | 3144 (295) | 2394 | 3696 | 6.50 | −0.18 | 262 (15) | 239 | 336 | 46.7 | 36.9 | 10.0–11.0 |
| SF-ISD:SDC (AP) | 3020 (119) | 2293 | 3574 | 6.45 | −0.13 | 257 (13) | 244 | 328 | 44.5 | 35.0 | 9.4–10.9 |
| SF-ISD:SDC (PT) | 3453 (63) | 2706 | 3626 | 6.35 | −0.16 | 287 (5) | 272 | 334 | 45.6 | 38.2 | 9.9–10.9 |
a Average number of unique peptide sequences or protein identifications from three replicates.
b Number of unique peptide sequences or proteins identified by PLGS in at least two out of three replicates.
c Number of unique peptide sequences or proteins identified in at least two out of three replicates after run-alignment (Progenesis LC-MS).
d Averaged values are calculated from peptides and proteins present in at least two out of three replicates.
These results indicate a general advantage of SDC-assisted digestion, and the SF-ISD:SDC protocol in particular, with a clear preference for removing SDC using phase transfer. The MSE approach generally resulted in higher average protein coverage with more peptides identified per protein and among runs as compared to DDA (Table III). This improved qualitative information alone can however not reveal the details of any protocol bias. We therefore continued with the quantitative information of this data set to investigate protocol dependent abundance changes of peptide and protein classes.
Quantitative Evaluation of Digestion Protocols
To enable label-free relative and absolute quantification, unlabeled digested protein standards were added to each sample before the LC-MSE analyses (29). A complete overview of the quantitative results is provided in supplemental Table SI and supplemental Fig. S4, including coefficients of variation (CV) and average total protein and peptide abundances. For example, the total amount loaded on column was estimated to be around 500 ng as determined by a protein assay before digestion. The measured amounts determined by label-free absolute quantification range from 409 to 479 ng (with an average CV of 15%), indicating recoveries (digestion efficiency) between 82 and 96% (supplemental Table S1).
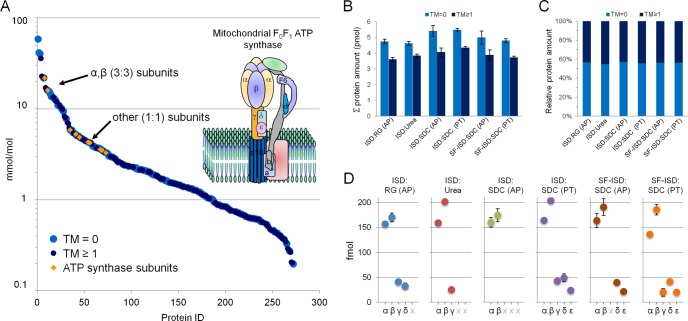
Fig. 4A depicts the dynamic range distribution and stoichiometry of absolute quantified mitochondrial proteins in mmol/mol for one of the best performing protocols, ISD:SDC (PT), based on the qualitative assessment. The individual quantitative protein values, in molar amount on-column as reported by the PLGS search engine, were divided by the sum. Membrane proteins, with one or more transmembrane domains, appear equally distributed among all detected proteins over the measured dynamic range. The total detected molar amount in each protocol for proteins with (TM ≥ 1) or without (TM = 0) transmembrane domains is shown in Fig. 4B. Although the standard ISD protocols display the highest yield for both protein classes, the relative contribution is very similar among the protocols (Fig. 4C). The number of membrane proteins (TM ≥ 1) constitute less than a third of all identifications (supplemental Fig. S4E), but represent almost half of the total molar protein amount (Fig. 4C). Expression in molar amount allows the estimation of stoichiometry for which an example is provided in Fig. 4. The alpha and beta subunits of the mitochondrial ATPsynthase complex are present in three copies compared with the majority of other subunits present in only one copy. This 3:1 stoichiometry is indeed reflected by the experimentally determined absolute protein values. We further extended the example to all protocols by investigating the presence and absolute amount of the catalytic core F1 complex subunits α, β,γ,δ,and ε with an expected stoichiometry of 3:3:1:1:1 (Fig. 4D). Although the identified F0 complex subunits were uniformly detected among the protocols (supplemental Protein Data Table), this is not the case for the F1 complex subunits (indicated by gray crosses in Fig. 4D). The missing subunits did not meet the criteria for absolute quantitation of 3 or more peptides per protein and presence in two out of three replicates. Although the detected subunits generally conform to the expected stoichiometry, all five F1 subunits are only detected in the two SDC-based ISD and SF-ISD protocols combining SDC with phase transfer. After this selective analysis we set out to perform a complete and in-depth differential comparison.
Fig. 4.
Dynamic range distribution and stoichiometry of quantified mitochondrial proteins. The plotted abundance distribution (A) depicts results from the ISD:SDC (PT) protocol. Light blue circles represent proteins without transmembrane domains (TM = 0), whereas dark blue circles represent proteins containing one or more transmembrane domains (TM ≥ 1). Yellow diamonds represent quantified ATP synthase subunits, which agree with their known stoichiometry. The ATPsynthase complex is schematically depicted in the top right corner (A). The bar charts represent the summed absolute (B) and relative (C) protein amounts obtained for each protocol categorized by proteins with (TM ≥ 1) or without (TM = 0) transmembrane domains. (D) The detection and absolute amount of the catalytic core F1 complex subunits α3 β3 γ1 δ1 and ε1 for each of the investigated protocols. The error bars indicate standard deviation of triplicate measurements.
To maximize the quality and completeness of the data set for quantitative and statistical analyses, we first applied further data processing including run alignment, normalization, and filtering. The previously acquired data files were imported into the Progenesis LC-MS software package (Nonlinear Dynamics) to align and match all detectable features among all runs. This was followed by outlier insensitive median normalization and import of the PLGS search results. The complete data set was then filtered to use only unique (proteotypic) peptides for protein quantitation, and only proteins and peptides present in at least two out of three replicate runs were reported (Table III and supplemental Table S1). Despite these stringent filters, the resulting data set has very few missing values (∼1–4%), which demonstrates the strength of combining run alignment with data-independent MSE data sets. The very comparable numbers of quantified identifications from the six different protocols ranged from 327 to 336 proteins (Fig. 3A) and 3452 to 3696 peptides (Fig. 3C). The highly complete discovery-based DIA data set is reminiscent of targeted approaches and includes 3729 distinct peptides quantified with 96% completeness between all protocols and replicates (64,224 out of 67,122). An average of 11 peptides were identified per protein, providing a very high average coverage of 40% for the 336 proteins, observed with 99% completeness (5967 out of 6048). An identification-based analysis would obviously not reveal any differences, but this high quality data set is well suited for a quantitative and statistical evaluation to uncover relevant abundance differences in and among protocols.
We first evaluated general protocol bias aiming to identify the most reproducible and unbiased method. We therefore investigated the variation of generated peptide and protein abundance levels in each protocol. For a measure of abundance bias, histograms were created depicting the distribution of ratios, calculated as the change in abundance for each peptide or protein in each protocol relative to the mean of all digestion protocols (Fig. 5). Next, an interval was defined consisting of one standard deviation, calculated over the complete quantitative data set. This represents no significant change in abundance and the relative amount (in percentage) of peptides (Fig. 5A) and proteins (Fig. 5B) within that interval is indicated. Statistical approaches are generally not designed to demonstrate non-significance, but the volcano plots generated using log ratios versus log q-values (p values corrected for multiple testing) are provided in the supplemental Material for comparison (supplemental Fig. S5). The highest percentage indicates the least bias, which is the case for the ISD:SDC protocols on both protein and peptide level (83–86% and 90–92%, respectively). The ISD:Urea protocol has the lowest percentages (69 and 76% for peptides and proteins, respectively) and thus relatively the highest bias. Interestingly, the phase transfer removal of SDC in all cases results in higher percentages compared with standard acid precipitation. For peptides this percentage increased from 82.6 to 86.1% for ISD:SDC and from 75.2 to 77.3% for SF-ISD:SDC protocols. For proteins the increase is from 90.4 to 91.7% and from 85.4 to 85.5% for ISD:SDC and SF-ISD:SDC, respectively (Fig. 5).
Fig. 5.
Quantitative evaluation of the relative protein and peptide abundance bias for each digestion protocol. Histograms represent the peptide (A) and protein (B) distribution among the generated bins of the log2(ratio). This ratio was calculated as the change in abundance for each peptide or protein in each protocol relative to the average of all digestion protocols. The interval among the dashed lines represents no significant change in peptide or protein abundances, which is defined as ± one standard deviation, calculated over the complete quantitative data set. The percentages above each histogram represent the peptides or proteins that are included in this interval. Box plot distribution of percentage ‘deviation from average’ for peptides (C) and proteins (D). The deviation from average is defined as the relative deviation of each peptide or protein in a particular protocol from the mean abundance of all protocols.
To investigate this further we assessed the distribution of the observed variation for each protocol on both peptide and protein level. The box plots shown in Fig. 5 depict the percentage of ‘deviation from average’ distribution for peptides (Fig. 5C) and proteins (Fig. 5D). The ‘deviation from average’ is defined as the deviation of each peptide or protein in a particular protocol relative to the mean abundance of all protocols. For comparison, the average technical variation (CV) of the peptide and protein abundances over three replicate measurements is as low as 4 and 9% on protein and peptide level, respectively (supplemental Table S1). Again, the ISD:SDC protocols display the most favorable results with the lowest average variation and smallest distribution, whereas ISD:Urea has the highest variation and largest distribution. Phase transfer removal of SDC clearly aids in reducing some of the variation, although this is not as pronounced in case of the SF-ISD:SDC protocol. Together these results suggest that the standard ISD method with SDC and phase transfer is the most reproducible and least biased protocol.
Next, we performed an in-depth quantitative analysis to investigate significant differences among protocols. Initial principal component analysis (supplemental Fig. S6) revealed that the various protocol replicates are very similar whereas the different protocols can easily be distinguished. Hierarchical and fuzzy c-means clustering was subsequently applied to visualize and identify relative changes in specific groups of proteins and peptides for each digestion protocol (Fig. 6). When inspecting the hierarchical peptide and protein clusters (Fig. 6A and 6D, respectively), several groups of abundance changes can be discerned. It is also noticeable that both the ISD:SDC (PT) and SF-ISD:SDC (PT) protocols display the least variation as was also concluded from Fig. 5. Fuzzy c-means clustering was subsequently applied to define and visualize the significant groups of proteins and peptides that display similar changes in abundance. In total, four peptide clusters could be defined (Fig. 6B) and three protein clusters (Fig. 6E) and for each the number of members is indicated. To assess the properties of clustered peptides and proteins, we investigated several physicochemical parameters including peptide sequence length, pI, hydrophobicity (GRAVY), protein molecular weight, abundance and number of transmembrane domains. Several interesting features could be noted and the overall results are schematically summarized in Fig. 6C and 6F, whereas the supporting evidence is provided in supplemental Fig. S7. We observed that peptide cluster 3 and protein cluster 2 mainly consists of features underrepresented using the ISD:Urea protocol. This includes relatively small and on average higher abundant proteins with few transmembrane domains (Fig. 6F), and peptides with the longest average sequence length (Fig. 6C and supplemental Fig. S7). Some proteins and peptides in the ISD:Urea protocol are overrepresented (both cluster 1), which represent relatively large and lower abundant proteins with a high number of TM domains (membrane proteins) but with smaller, more hydrophilic peptides. This cluster also indicates that, unless phase transfer is applied, the abundance of these particular proteins and peptides is diminished when SDC or RapiGest is removed by standard acid precipitation. Peptide cluster 2 and protein cluster 3 contain features that are generally better represented in SDC-based protocols but not in any of the others. These clusters contain proteins with an intermediate amount of TM domains (Fig. 6F), but on average the most hydrophobic peptides with longest average sequence length (Fig. 6C and supplemental Fig. S7). This is the only peptide cluster that revealed a significant peptide sequence profile after scanning with motif-x (30). Methionine residues were overrepresented in peptide cluster 2 (supplemental Fig. S8), probably reflecting that methionine residues are abundant in transmembrane segments which generate, on average, longer hydrophobic peptides. This concurs with the properties uncovered for peptide cluster 2 (Fig. 6C). Finally, peptide cluster 4 contains features that are negatively influenced by the phase transfer protocols. As in this quantitative approach all unique peptides were used for protein quantitation, certain peptide and protein clusters display similar trends. No corresponding protein cluster was however found for peptide cluster 4. This indicates that the peptides in cluster 4 do not significantly contribute to the protein quantitation, most likely because of their relative lower intensities.
Fig. 6.
In-depth quantitative analysis of significant differences observed among protocols. A, D, Hierarchical clustering of peptide (A) and protein (D) normalized log ratios among each digestion protocol, measured in triplicate. The heatmap color limits are set to ± two standard deviations, calculated over the complete quantitative data set. B, E, Fuzzy c-means clustering of changes in peptide (B) and protein (E) standardized abundance among all protocols, presented in the same order as listed for the heatmaps. C, F, Summary of differences observed among each cluster for several physicochemical peptide (C) and protein (F) parameters, including peptide sequence length, pI, hydrophobicity (GRAVY), protein molecular weight (MW) and number of transmembrane (TM) domains. G, Bar graphs representing the summed log ratio of significantly changed protein and peptide abundances in each protocol plotted against several binned physicochemical parameters. Significance level was defined as q-value <0.05 (p value corrected for multiple testing) and ± one standard deviation calculated over the average of all protocols. This corresponds to a 1.5-fold and 2.2-fold change on peptide and protein level, respectively.
Lastly, we aimed to further investigate the most prominent differences between each protocol and their individual characteristics in more detail. For this purpose, we plotted only the significantly changed proteins and peptides against binned physicochemical parameters, summing the ratios to obtain a weighted representation (Fig. 6G). The significance level thresholds were defined as q-value <0.05 and >1.5-fold and >2.2-fold change on peptide and protein level, respectively, which corresponds to ± one standard deviation calculated over the average of all protocols. Identical parameters were applied to create the volcano plots provided in supplemental Fig. S5, where the numbers of significantly changed proteins and peptides are indicated for each protocol. As a result, several prominent protocol characteristics can be gleaned from Fig. 6G. The urea-based ISD protocol results in significantly more missed cleavages compared with any other protocol and displays a bias toward proteins with multiple transmembrane domains. An overall low yield of non-membrane proteins is particularly apparent for protocols based on Rapigest or urea. The SDC-based protocols appear generally superior, but an obvious observation is that phase transfer removal of SDC is quite crucial to obtain the efficiently high and unbiased recovery of peptides. This effect appears most prominent for peptides of medium hydrophobicity with neutral to acidic isoelectric points. When comparing the performance of spin filters to the standard in-solution protocol, it seems that the use of filters may result in the loss of certain larger, hydrophobic proteins. Whereas a lower recovery of very small proteins could potentially be expected with the use of 30 kDa cutoff filters, this is not observed.
In summary, our extensive quantitative comparative analysis indicates that ISD:SDC (PT) is the most efficient and accurate standard in-solution protocol with the least bias, closely followed by the spin filter-aided variant of the protocol, SF-ISD:SDC (PT). In both cases, phase transfer removal of SDC significantly contributed to higher protein digestion efficiency and the reduction of variation and bias.
DISCUSSION
We employed qualitative and data-independent quantitative LC-MS/MS to assess efficiency and bias of trypsin-based protein digestion protocols. Mitochondrial protein fractions were used to evaluate protocols based on spin filter-aided as well as standard in-solution digestion methods, previously reported optimal as well as further refined in this study. Our systematic analysis revealed that SDC-assisted in-solution digestion, combined with phase separation, is the most efficient protocol, providing the highest recovery, protein sequence coverage and number of protein identifications, with the least bias toward or against any peptide and protein classes, including membrane proteins. We also demonstrate for the first time that the SDC-assisted protocol is optimal for spin filter-aided digestions, as compared with previously reported methods. Our quantitative workflow thus enabled the objective evaluation of protein sample digestion, identifying two strategies for the quantitative representation of peptides and proteins among all classes, enabling efficient and unbiased protein analysis.
Based on our comparative analysis we concluded that SDC-based digestion protocols are the most efficient compared with other investigated methods using urea or surfactants such as RapiGest. In addition, SDC is inexpensive, aids protein solubility during digestion and enhances trypsin activity, which allows efficient protein digestion in only 5–7 h. Reducing digestion time from the usual overnight incubation was demonstrated to minimize erroneous peptide deamidation by 50% (31).
Our results initially revealed that SDC-based spin-filter protocols slightly surpass the standard ISD protocol in terms of efficiency. More importantly, we demonstrate for the first time that applying SDC as surfactant in spin filter-aided digestion protocols outperforms the standard spin filter-aided digestion protocol that uses urea for removal of SDS (14). SDS as powerful detergent and SDC as efficient surfactant could well be the most optimal combination to date. We however determined that for the effective removal of SDS before digestion, urea remains a necessity and cannot directly be replaced with SDC. For efficient and unbiased sample preparation, urea should however be avoided during digestion, as we clearly demonstrated here. This is in accordance with previous studies, which expressed the preference of using SDC or RapiGest over urea (7, 9, 32). A recent quantitative study showed that the miss-cleavage bias of trypsin in diluted urea can be virtually overcome by sequential Lys-C/trypsin digestion (33). We however demonstrate here that SDC-assisted tryptic digestions are highly efficient, preventing the need for such combinatorial digestions and allowing to avoid urea altogether.
Compared with the SDC-based spin-filter protocol, the SDC-based standard in-solution protocol provided the highest recovery and lowest variation, with the least bias toward generated peptide and protein abundance. Although the SDC-based spin-filter protocol is efficient, the total sample recovery was lower and higher quantitative variation was observed as well as a lower recovery of certain hydrophobic proteins, as compared with the standard ISD:SDC protocol. These observations may in part be because of the filter surface itself or the additional sample handling required for SF-ISD methods, in contrast to the rather straightforward ISD protocol. In general, we consider ISD:SDC to be a very efficient, unbiased, simple and fast protocol, which can generally be applied to many types of protein samples. The use of TEAB instead of ammonium bicarbonate as buffer also ensures that resulting peptide digest are compatible with “downstream” amino reactive reagents, such as TMT and iTRAQ, for labeled quantitative proteomics experiments. The SF-ISD:SDC protocol should be considered when maximum digestion efficiency is needed and it can be applied to almost any type of protein sample. The SF-ISD protocol offers advantages similar to the standard in-gel digestion procedure, including SDS-assisted protein solubilization and removal of interfering substances, but without several of its limitations. We can however only speculate how our findings may relate or apply to gel-based methods as the in-gel digestion workflow was excluded from our comparison.
The in-gel digestion protocol has been extended to the frequently used GeLC-MS workflow, involving protein digestion and LC-MS analysis of peptides recovered from a set of SDS-PAGE gel bands (34, 35). This approach affords effective protein-level fractionation and high proteome coverage (36, 37) but limitations include biased loss and overall lower recovery compared with in-solution digestion and peptide-based fractionation methods (3, 4, 6, 37). Furthermore, GeLC-MS is convenient for metabolic labeling strategies, but presents gel-slice reproducibility issues for chemical labeling and label-free approaches, whereas in-solution digestion can be consistently and widely applied. The necessity to extract peptides from a gel is the prime source of (differential) protein and peptide loss whereas with in-solution digestion essentially all peptides from a given protein have the potential to be detected, provided that efficient and unbiased protein digestion can be achieved using effective in-solution digestion strategies, such as presented here.
To obtain optimal results with the presented SDC-based digestion methods, our findings indicate that acid precipitation with phase transfer, as opposed to acid precipitation alone, is required to prevent introducing bias because of the absence or underestimated abundance of particular peptides. A previous study suggested that phase transfer removal of SDC prevents under-representation of hydrophobic peptides (12). That study was purely qualitative and could not distinguish the abundance difference of all peptide and protein classes. Our current quantitative results however indicate that the bias introduced by removal of SDC by acid precipitation is not selective for one particular peptide class. A previous study, using SDC-assisted digestion and phase transfer, reported a lack of negative bias for membrane proteins by using the correlation between mRNA and protein expression in an E. coli lysate (32). The correlation between mRNA and protein expression may however not be reliable enough or practical for comprehensive comparisons of protocol bias in mammalian systems. We now confirmed the lack of bias and further extended these observations by introducing a more straightforward label-free quantitative workflow to determine the most efficient and unbiased digestion protocol on both protein and peptide level for use in peptide-centric proteomics approaches. As novel and updated methods are likely to emerge, our workflow may serve as benchmark for future studies aiming to objectively evaluate digestion of subproteomes as well as more complex samples.
Efficient methods are required to maximize the number of detectable peptides per protein allowing unique proteins and variants to be distinguished and PTMs to be detected. In addition, unbiased generation of peptides is required to most accurately describe the relative and absolute protein levels in a particular sample. This is especially important for label-free (absolute) quantitative approaches, which are becoming increasingly popular. These methods are capable of defining protein abundance as copies per cell and are able to reveal protein stoichiometry in cellular pathways. Recently, estimated absolute protein abundance was achieved in whole proteomes using unlabeled samples and quantitative strategies relying on the most abundant (unique) peptides per protein (38, 39). Efforts to compare digestion protocols using both qualitative and quantitative approaches, such as are reported here, contribute to the improvement of quantitative proteomics workflows used in large-scale studies of cells, tissues, and whole organisms.
Supplementary Material
Acknowledgments
We thank the other members of the Protein Research Group for many helpful discussions and comments.
Footnotes
* This work was supported by grants to O.N.J. from NordForsk Center of Excellence ‘MitoHealth’ and the Danish Ministry for Research, Technology and Innovation.
 This article contains supplemental Figs. S1 to S8 and Tables S1.
This article contains supplemental Figs. S1 to S8 and Tables S1.
1 The abbreviations used are:
- LC-MS/MS
- liquid chromatography tandem mass spectrometry
- AGC
- automatic gain control
- CSV
- comma separated values
- CV
- coefficient of variation
- DIA
- data independent acquisition
- DDA
- data dependent acquisition
- FDR
- false discovery rate
- FPR
- false positive rate
- FWHM
- full width at half maximum
- ISD
- in-solution digestion
- iTRAQ
- isobaric tag for relative and absolute quantitation
- LTQ
- linear trap quadrupole
- PLGS
- ProteinLynx Global Server™
- PT
- phase transfer
- RG
- RapiGest
- SDC
- sodium deoxycholate
- SF-ISD
- spin filter aided in-solution digestion
- TEAB
- triethylammonium bicarbonate
- TM
- transmembrane domain
- TMT
- tandem mass tag
- UPLC
- ultraperformance liquid chromatography.
REFERENCES
- 1. Shevchenko A., Wilm M., Vorm O., Mann M. (1996) Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68, 850–858 [DOI] [PubMed] [Google Scholar]
- 2. Rabilloud T., Vaezzadeh A. R., Potier N., Lelong C., Leize-Wagner E., Chevallet M. (2009) Power and limitations of electrophoretic separations in proteomics strategies. Mass Spectrom. Rev. 28, 816–843 [DOI] [PubMed] [Google Scholar]
- 3. Havlis J., Shevchenko A. (2004) Absolute quantification of proteins in solutions and in polyacrylamide gels by mass spectrometry. Anal. Chem. 76, 3029–3036 [DOI] [PubMed] [Google Scholar]
- 4. Speicher K. D., Kolbas O., Harper S., Speicher D. W. (2000) Systematic analysis of peptide recoveries from in-gel digestions for protein identifications in proteome studies. J. Biomol. Tech. 11, 74–86 [PMC free article] [PubMed] [Google Scholar]
- 5. Stewart II, Thomson T., Figeys D. (2001) 18O labeling: a tool for proteomics. Rapid Commun. Mass Spectrom. 15, 2456–2465 [DOI] [PubMed] [Google Scholar]
- 6. Wiśniewski J. R., Ostasiewicz P., Mann M. (2011) High recovery FASP applied to the proteomic analysis of microdissected formalin fixed paraffin embedded cancer tissues retrieves known colon cancer markers. J. Proteome Res. 10, 3040–3049 [DOI] [PubMed] [Google Scholar]
- 7. Chen E. I., Cociorva D., Norris J. L., Yates J. R., 3rd (2007) Optimization of mass spectrometry-compatible surfactants for shotgun proteomics. J. Proteome Res. 6, 2529–2538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chen E. I., McClatchy D., Park S. K., Yates J. R., 3rd (2008) Comparisons of mass spectrometry compatible surfactants for global analysis of the mammalian brain proteome. Anal. Chem. 80, 8694–8701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Proc J. L., Kuzyk M. A., Hardie D. B., Yang J., Smith D. S., Jackson A. M., Parker C. E., Borchers C. H. (2010) A quantitative study of the effects of chaotropic agents, surfactants, and solvents on the digestion efficiency of human plasma proteins by trypsin. J. Proteome Res. 9, 5422–5437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Coleman R., Iqbal S., Godfrey P. P., Billington D. (1979) Membranes and bile formation. Composition of several mammalian biles and their membrane-damaging properties. Biochem. J. 178, 201–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhou J., Zhou T., Cao R., Liu Z., Shen J., Chen P., Wang X., Liang S. (2006) Evaluation of the application of sodium deoxycholate to proteomic analysis of rat hippocampal plasma membrane. J. Proteome Res. 5, 2547–2553 [DOI] [PubMed] [Google Scholar]
- 12. Masuda T., Tomita M., Ishihama Y. (2008) Phase transfer surfactant-aided trypsin digestion for membrane proteome analysis. J. Proteome Res. 7, 731–740 [DOI] [PubMed] [Google Scholar]
- 13. Manza L. L., Stamer S. L., Ham A. J., Codreanu S. G., Liebler D. C. (2005) Sample preparation and digestion for proteomic analyses using spin filters. Proteomics 5, 1742–1745 [DOI] [PubMed] [Google Scholar]
- 14. Wisniewski J. R., Zougman A., Nagaraj N., Mann M. (2009) Universal sample preparation method for proteome analysis. Nat. Methods 6, 359–362 [DOI] [PubMed] [Google Scholar]
- 15. Newmeyer D. D., Ferguson-Miller S. (2003) Mitochondria: releasing power for life and unleashing the machineries of death. Cell 112, 481–490 [DOI] [PubMed] [Google Scholar]
- 16. Pagliarini D. J., Dixon J. E. (2006) Mitochondrial modulation: reversible phosphorylation takes center stage? Trends Biochem. Sci. 31, 26–34 [DOI] [PubMed] [Google Scholar]
- 17. Taylor S. W., Fahy E., Zhang B., Glenn G. M., Warnock D. E., Wiley S., Murphy A. N., Gaucher S. P., Capaldi R. A., Gibson B. W., Ghosh S. S. (2003) Characterization of the human heart mitochondrial proteome. Nat. Biotechnol. 21, 281–286 [DOI] [PubMed] [Google Scholar]
- 18. Vigerust N. F., Cacabelos D., Burri L., Berge K., Wergedahl H., Christensen B., Portero-Otin M., Viste A., Pamplona R., Berge R. K., Bjørndal B. (2012) Fish oil and 3-thia fatty acid have additive effects on lipid metabolism but antagonistic effects on oxidative damage when fed to rats for 50 weeks. J. Nutr. Biochem. 23, 1384–1393 [DOI] [PubMed] [Google Scholar]
- 19. Martens L., Vandekerckhove J., Gevaert K. (2005) DBToolkit: processing protein databases for peptide-centric proteomics. Bioinformatics 21, 3584–3585 [DOI] [PubMed] [Google Scholar]
- 20. Reisinger F., Martens L. (2009) Database on Demand - an online tool for the custom generation of FASTA-formatted sequence databases. Proteomics 9, 4421–4424 [DOI] [PubMed] [Google Scholar]
- 21. Silva J. C., Gorenstein M. V., Li G. Z., Vissers J. P., Geromanos S. J. (2006) Absolute quantification of proteins by LCMSE: a virtue of parallel MS acquisition. Mol. Cell. Proteomics 5, 144–156 [DOI] [PubMed] [Google Scholar]
- 22. Gobom J., Nordhoff E., Mirgorodskaya E., Ekman R., Roepstorff P. (1999) Sample purification and preparation technique based on nano-scale reversed-phase columns for the sensitive analysis of complex peptide mixtures by matrix-assisted laser desorption/ionization mass spectrometry. J. Mass Spectrom. 34, 105–116 [DOI] [PubMed] [Google Scholar]
- 23. Geromanos S. J., Vissers J. P., Silva J. C., Dorschel C. A., Li G. Z., Gorenstein M. V., Bateman R. H., Langridge J. I. (2009) The detection, correlation, and comparison of peptide precursor and product ions from data independent LC-MS with data dependant LC-MS/MS. Proteomics 9, 1683–1695 [DOI] [PubMed] [Google Scholar]
- 24. Silva J. C., Denny R., Dorschel C. A., Gorenstein M., Kass I. J., Li G. Z., McKenna T., Nold M. J., Richardson K., Young P., Geromanos S. (2005) Quantitative proteomic analysis by accurate mass retention time pairs. Anal. Chem. 77, 2187–2200 [DOI] [PubMed] [Google Scholar]
- 25. Li G. Z., Vissers J. P., Silva J. C., Golick D., Gorenstein M. V., Geromanos S. J. (2009) Database searching and accounting of multiplexed precursor and product ion spectra from the data independent analysis of simple and complex peptide mixtures. Proteomics 9, 1696–1719 [DOI] [PubMed] [Google Scholar]
- 26. Schwämmle V., Jensen O. N. (2010) A simple and fast method to determine the parameters for fuzzy c-means cluster analysis. Bioinformatics 26, 2841–2848 [DOI] [PubMed] [Google Scholar]
- 27. Xie X. L., Beni G. (1991) A validity measure for fuzzy clustering. IEEE Trans. Pattern Anal. Mach. Intell. 13, 841–847 [Google Scholar]
- 28. Tabb D. L., Vega-Montoto L., Rudnick P. A., Variyath A. M., Ham A. J., Bunk D. M., Kilpatrick L. E., Billheimer D. D., Blackman R. K., Cardasis H. L., Carr S. A., Clauser K. R., Jaffe J. D., Kowalski K. A., Neubert T. A., Regnier F. E., Schilling B., Tegeler T. J., Wang M., Wang P., Whiteaker J. R., Zimmerman L. J., Fisher S. J., Gibson B. W., Kinsinger C. R., Mesri M., Rodriguez H., Stein S. E., Tempst P., Paulovich A. G., Liebler D. C., Spiegelman C. (2010) Repeatability and reproducibility in proteomic identifications by liquid chromatography-tandem mass spectrometry. J. Proteome Res. 9, 761–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Silva J. C., Denny R., Dorschel C., Gorenstein M. V., Li G. Z., Richardson K., Wall D., Geromanos S. J. (2006) Simultaneous qualitative and quantitative analysis of the Escherichia coli proteome: a sweet tale. Mol. Cell. Proteomics 5, 589–607 [DOI] [PubMed] [Google Scholar]
- 30. Chou M. F., Schwartz D. (2011) Biological sequence motif discovery using motif-x. Curr. Protoc. Bioinformatics Chapter 13, Unit 13 15–24 [DOI] [PubMed] [Google Scholar]
- 31. Hao P., Ren Y., Alpert A. J., Sze S. K. (2011) Detection, evaluation and minimization of nonenzymatic deamidation in proteomic sample preparation. Mol. Cell. Proteomics 10, O111.009381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Masuda T., Saito N., Tomita M., Ishihama Y. (2009) Unbiased quantitation of Escherichia coli membrane proteome using phase transfer surfactants. Mol. Cell. Proteomics 8, 2770–2777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Glatter T., Ludwig C., Ahrné E., Aebersold R., Heck A. J., Schmidt A. (2012) Large-scale quantitative assessment of different in-solution protein digestion protocols reveals superior cleavage efficiency of tandem Lys-C/trypsin proteolysis over trypsin digestion. J. Proteome Res. 11, 5145–5156 [DOI] [PubMed] [Google Scholar]
- 34. Schirle M., Heurtier M. A., Kuster B. (2003) Profiling core proteomes of human cell lines by one-dimensional PAGE and liquid chromatography-tandem mass spectrometry. Mol. Cell. Proteomics 2, 1297–1305 [DOI] [PubMed] [Google Scholar]
- 35. Simpson R. J., Connolly L. M., Eddes J. S., Pereira J. J., Moritz R. L., Reid G. E. (2000) Proteomic analysis of the human colon carcinoma cell line (LIM 1215): development of a membrane protein database. Electrophoresis 21, 1707–1732 [DOI] [PubMed] [Google Scholar]
- 36. de Godoy L. M., Olsen J. V., Cox J., Nielsen M. L., Hubner N. C., Fröhlich F., Walther T. C., Mann M. (2008) Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature 455, 1251–1254 [DOI] [PubMed] [Google Scholar]
- 37. Fang Y., Robinson D. P., Foster L. J. (2010) Quantitative analysis of proteome coverage and recovery rates for upstream fractionation methods in proteomics. J. Proteome Res. 9, 1902–1912 [DOI] [PubMed] [Google Scholar]
- 38. Schmidt A., Beck M., Malmstrom J., Lam H., Claassen M., Campbell D., Aebersold R. (2011) Absolute quantification of microbial proteomes at different states by directed mass spectrometry. Mol. Syst. Biol. 7, 510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ludwig C., Claassen M., Schmidt A., Aebersold R. (2012) Estimation of absolute protein quantities of unlabeled samples by selected reaction monitoring mass spectrometry. Mol. Cell. Proteomics 11, M111.013987. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.