Abstract
The autophagy pathway is an essential component of host defense against viral infection, orchestrating pathogen degradation (xenophagy), innate immune signaling, and certain aspects of adaptive immunity. Single autophagy proteins or cassettes of the core autophagy machinery can also function as antiviral factors independently of the canonical autophagy pathway. Moreover, to survive and propagate within the host, viruses have evolved a variety of strategies to evade autophagic attack and manipulate the autophagy machinery for their own benefit. This review summarizes recent advances in understanding the antiviral and proviral roles of autophagy and previously unappreciated autophagy-independent functions of autophagy-related genes.
Key Words: Autophagy, Virophagy, Antiviral immunity
Introduction
Macroautophagy (herein autophagy) is an evolutionarily ancient pathway that plays essential roles in multiple fundamental physiological processes, including immunity, survival, differentiation, development, and homeostasis [1, 2]. The past few years have witnessed a rapid explosion of studies on autophagy and viral infections [reviewed in [3, 4, 5, 6, 7]]. Indeed, the interplay between autophagy and viruses has been investigated for more than 50 virus species in over 20 families of DNA or RNA viruses. These studies expand our knowledge of the immunological functions of the autophagy machinery as well as the molecular mechanisms of viral life cycles and pathogenesis. Furthermore, modulation of autophagy may represent a novel therapeutic strategy for treating diseases caused by medically important viral pathogens [8].
The autophagy machinery is highly conserved from unicellular eukaryotes to metazoans. The primordial functions of autophagy in unicellular organisms are thought to include energy homeostasis and protein and organelle quality control. Autophagy is likely also one of the most ancient cell-autonomous defense mechanisms to confront microbial invasion. As viruses began co-evolving with eukaryotes as early as 1 billion years ago, long-term mutual adaptations have shaped an extremely complex interplay between autophagy and viruses [3, 5]. For example, the autophagy pathway and/or the autophagy machinery exerts a crucial antiviral role and restricts the virulence of some viruses in vivo in organisms ranging from plants to mammals (table 1). In contrast, many viruses successfully evade autophagy and even exploit the autophagy machinery to propagate viral replication. Recently, increasing evidence indicates that the antiviral and proviral functions of autophagy proteins or the autophagy machinery does not always rely on the canonical autophagic process (fig. 1, 2); rather, in many circumstances, the formation of autophagosomes and their maturation into degradative autolyososmes is not required for such functions of autophagy proteins. In this review, we provide an overview of our current knowledge of the interplay between viruses and the autophagic machinery.
Table 1.
Roles of autophagy (or autophagy-related genes) during viral infection in vivo
| Virus | Host | Autophagy-related genes | Host-virus interplay | Ref. |
|---|---|---|---|---|
| RNA viruses | ||||
| Togaviridae Sindbis virus |
Mouse | beclin 1 | Ectopic Beclin 1 expression in Sindbis virus-infected neurons suppresses viral replication in the brain and reduces mouse mortality | [30] |
| Mouse | Atg5 | Depletion or disruption of Atg5 impairs CNS clearance of Sindbis virus capsid and increases the mortality of infected mice | [14] | |
| Chikungunya virus | Mouse | Atg16L1 | Chikungunya virus induces higher levels of apoptosis and increased lethality in Atg16L1 hypomorphic (Atg16L1HM) mice | [31] |
| Caliciviridae MNV |
Mouse | Atg16L1 | MNV infection in Atg16L1HM mice induces Crohn's disease-like pathological features in the intestine | [41] |
| Dicistroviridae Drosophila C virus |
Drosophila | Atg18 | Atg18 knockdown does not alter Drosophila C virus replication or the survival rate of DCV-infected flies | [10] |
| Orthomyxoviridae Influenza A virus |
Mouse | Atg5 | Atg5 knockdown in the lung reduces influenza virus H5N1-induced inflammation, acute lung injury and mouse mortality | [51] |
| Picornaviridae Coxsackievirus B3 |
Mouse | Atg5 | Pancreatic acinar cell-specific disruption of Atg5 reduces Coxsackievirus B3 replication in the pancreas and pancreatic dysfunction | [80] |
| Rhabdoviridae VSV |
Drosophila | Atg7, Atg12, Atg18 | Silencing these autophagy genes increases VSV replication in vivo and fly mortality after VSV infection | [10] |
| Sigma virus | Drosophila | p62/Ref(2)P | Permissive Ref(2)P alleles increase sigma virus infectivity in flies, whereas restrictive or non-functional alleles decrease sigma virus infectivity | [90] |
| Virgaviridae TMV |
Arabidopsis | ATG3, BECLIN 1, ATG7, VPS34 | Silencing these autophagy genes increases TMV local replication and the spread of hypersensitive-related programmed cell death to uninfected tissues in plants | [29] |
|
DNA viruses Hepadnaviridae HBV |
Mouse | Atg5 | Liver-specific knockout of Atg5 impairs HBV replication in vivo | [81] |
| Herpesviridae HSV-1 |
Mouse | beclin 1 | An HSV-1 mutant lacking the ability to inhibit Beclin 1 (HSV-1 ICP34.5Δ68-87) is attenuated in a intracerabral infection model | [25] |
| Mouse | beclin 1 | HSV-1 ICP34.5?68-87 is attenuated in a corneal infection model in wild-type mice, but not in Rag1 −/− mice lacking B and T cells | [28] | |
| Mouse | Atg5 | CD4+ T cell priming after HSV-1 mucosal infection is impaired in lethally irradiated wild-type mice reconstituted with Atg5–/– hematopoietic cells | [19] | |
| Mouse | beclin 1, Atg5 | HSV-1 ICP34.5Δ68-87 is attenuated in an intravaginal infection model likely due to the antiviral role of autophagy in dorsal root ganglion neurons | [26] | |
| HSV-2 | Mouse | Atg5 | Dendritic cell-conditional deletion of Atg5 impairs CD4+ T cell priming and increases mouse susceptibility to HS-2 | [19] |
| γ-Herpesvirus 68 (γHV68) | Mouse | beclin 1 | γHV68 carrying a vBcl2 variant unable to suppress autophagy fails to efficiently maintain latent infection in the spleen | [27] |
Fig. 1.
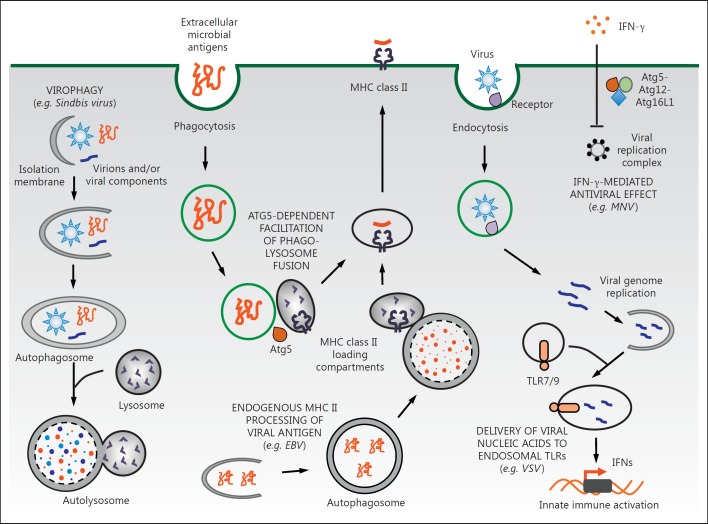
Antiviral functions of autophagy-related genes in viral infection. Shown are selected examples of antiviral functions of autophagy-related genes and representative viruses that may be controlled by such autophagy-related gene antiviral functions. Virions and viral components can be captured and directed to autophagosomes for lysosomal degradation in a process called virophagy (a form of xenophagy). In APCs, autophagy augments adaptive immune responses through promoting viral antigen processing and presentation on MHC molecules. Furthermore, autophagy can restrict viral infection independently of autophagy, such as via promotion of phagosome-to-lysosome fusion (which functions in MHC class II antigen presentation of phagocytosed antigens in dendritic cells) and the direct inhibition of the formation of viral replication complexes (which functions in mediating the antiviral effects of IFN-γ). See text for more details.
Fig. 2.
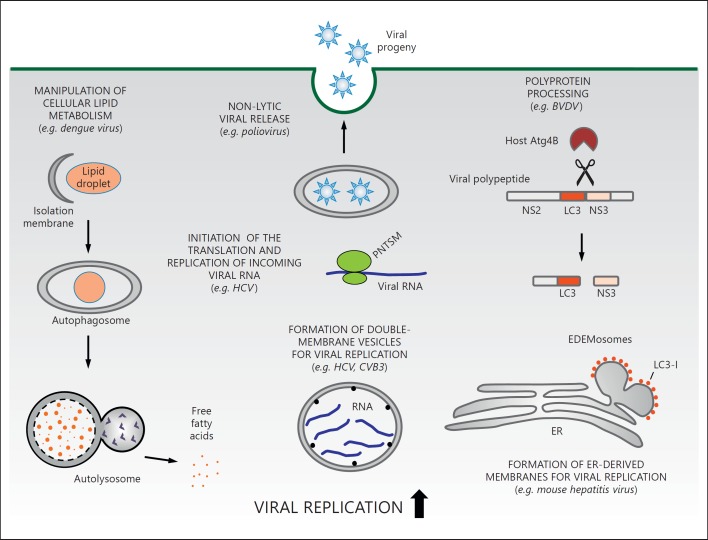
Proviral functions of autophagy-related genes in viral infection. Shown are selected examples of proviral functions of autophagy-related genes and representative viruses that may benefit from such proviral functions. Viruses can exploit autophagy or the autophagy machinery to facilitate their infection. Dengue virus manipulates lipid metabolism for viral replication via lipophagy. Poliovirus utilizes autophagy to promote nonlytic release of virions. HCV employs the autophagy machinery to initiate early viral genome replication and also to conceal viral RNA in autophagosome-like DMVs. Moreover, viruses can exploit autophagy-related proteins in an autophagy-independent manner. For example, MHV employs nonlipidated LC3-I and ER-derived vesicles exporting short-lived ERAD regulators (EDEMosomes) to generate DMVs for viral replication. A cytopathogenic strain of BVDV incorporates an LC3 gene fragment into its genome and utilizes cellular Atg4B to facilitate the processing of the viral polyprotein precursor. CVB3 = Coxsackie virus B3.
The Antiviral Roles of Autophagy and Autophagy Proteins
The canonical function of autophagy, i.e. the formation of double-membrane autophagosomes that deliver cargo to endolysosomal compartments, may exert several different antiviral roles, including: (1) the degradation of intracytoplasmic viral components (a process termed virophagy), (2) the activation of innate immune signaling by delivery of viral nucleic acids to endosomal toll-like receptors (TLRs), (3) the activation of adaptive immunity by presentation of endogenous viral antigens to major histocompatibility complex (MHC) class I and MHC class II molecules, (4) the regulation of innate immune activation by control of mitochondrial quality and reactive oxygen species (ROS) production, and (5) the promotion of cell survival [3, 5]. More broadly, autophagy proteins or cassettes of the autophagy machinery can cooperate with other immune pathways to restrict viral infection in the absence of autophagosome formation and autolysosomal degradation (fig. 1). The signals that trigger virophagy [i.e. viral pathogen-associated molecular patterns (PAMPs) or other cellular signals generated during viral entry or replication such as danger-associated molecular patterns (DAMPs)], the host pattern recognition receptors that sense these PAMPs and DAMPs, the mechanisms by which autophagy functions in the control of virus infection, and the relative contribution of autophagy to the host antiviral response remain incompletely understood.
Signals That Trigger Autophagy during Viral Infection
Potential inducers of autophagy during viral infection include the process of viral entry or viral replication, viral components or assembly intermediates and their derivatives, increased energy requirements, unfolded protein responses, and plasma membrane disturbance. There are several examples of viruses that can induce autophagy independently of viral replication. In Drosophila cells, vesicular stomatitis virus (VSV) induces autophagy via Toll-7 recognition of VSV-G on the surface of virions [9] and both UV-inactivated VSV virions and VSV-G-containing vesicular particles devoid of any other viral components are able to induce autophagy [10]. Measles virus can activate autophagy through a tripartite interaction between its cellular receptor CD46, GOPC, and the Beclin 1-VPS34 complex [11]. Given that CD46 also serves as an entry receptor for human herpesvirus 6, adenovirus, and bovine viral diarrhea virus (BVDV), these viruses may also induce autophagy by a similar mechanism. More generally, the interaction between virion surface protein(s) and host receptors, and/or viral entry-induced internalization of cellular surface molecules may serve as a signal for autophagy initiation.
Autophagy may also be initiated by plasma membrane disturbance independently of the signaling downstream of cellular receptors. The fusogenic function of human immunodeficiency virus type I (HIV-1) envelope glycoprotein gp41 is sufficient to trigger autophagy even in cells expressing mutant CXCR4 molecules defective in signal transduction [12]. Autophagy induction triggered by viral glycoprotein-mediated fusion (which is correlated with syncytium formation) has also been observed during infection with two paromyxoviruses, measles virus and canine distemper virus [13]. Thus, further studies are warranted to examine how plasma membrane disturbance caused by viral entry or syncytium formation between infected cells may function as danger signals capable of inducing autophagy.
For some viruses, such as Sindbis virus (a message sense RNA virus in the alphavirus genus), viral replication is necessary for autophagy induction. In contrast to VSV infection in Drosophila, UV-inactivated Sindbis virus fails to induce autophagy in mammalian cells [14]. It is as of yet unknown whether this is triggered by viral RNA, by the assembled nucleocapsids that are selectively targeted for autophagic degradation [14, 15], and/or by host antiviral molecules activated during viral infection. Recently, the virus-activated host RNAase pathway, RNAase L, was shown to be necessary for autophagy induction by VSV, encephalomyocarditis virus, or 2′,5′-oligoadenylate treatment [16].
Context-Dependent Antiviral Effects of Autophagy
Unlike the proviral activities of autophagy (discussed below), the antiviral activities of autophagy have not been well recapitulated in vitro. To date, at least within tissue culture experimental systems (which are not natural targets for viruses), autophagy deficiency usually does not significantly increase the viral infectivity of either DNA or RNA viruses, such as vaccinia virus [17], herpes simplex virus type I (HSV-1) [18], herpes simplex virus type II (HSV-2) [19], mouse hepatitis virus (MHV) [20], influenza virus [21], Sindbis virus [14], West Nile virus [22], measles virus [13], and canine distemper virus [13]. One possible explanation is that viruses have developed potent strategies to antagonize autophagy. For example, HSV-1 ICP34.5 and viral Bcl-2s encoded by the γ-herpesviruses (γHV68 M11, KSHV vBcl-2) inhibit autophagy through a physical interaction with Beclin 1, and influenza virus M2 inhibits autophagosome maturation [reviewed in [23]]. Additionally, vaccinia virus is thought to disrupt autophagy by inducing aberrant LC3 lipidation and a direct conjugation between the autophagy proteins, Atg3 and Atg12 [24].
However, this scenario fails to explain the observation that viral mutants unable to suppress autophagy do not have replication defects in many cell types in vitro. An HSV-1 mutant virus lacking the Beclin 1-binding domain of ICP34.5 and ability to block autophagy replicates to levels comparable to those of wild-type virus in MEFs and epithelial cells [18, 25, 26]. A γHV68 mutant virus harboring an M11 variant that does not interact with Beclin 1 and inhibit autophagy replicates normally in BHK-21 cells [27].
One important factor underlying these observations appears to be the cell type specificity of the importance of autophagy as an antiviral program. Recently, Yordy et al. [26] demonstrated that dorsal root ganglion neurons do have a significant defect in replication in vitro as well as in vivo of the mutant HSV-1 virus incapable of inhibiting autophagy. Based on this study, as well as previous studies suggesting a crucial role for autophagy in controlling mammalian neuronotropic viruses in vivo [25, 28], an emerging theme is that autophagy, rather than interferon, may play a predominant role in controlling viral infection in postmitotic neurons [6]. However, it is also possible that, even in neurons, autophagy may be insufficient to control robust replication of some viruses. Although not directly examined in vitro, deletion of Atg5 in neurons in vivo increased the susceptibility of mice to lethal central nervous system infection with Sindbis virus but did not increase viral replication [14]. For some viruses such as Sindbis virus, the ratio of replication-defective particles to replication-competent particles may be so high that autophagy deficiency fails to result in a detectable increase in viral replication. While it is intuitive that a noncytolytic mechanisms for controlling viral replication may be particularly important in vital, largely nonrenewable cell populations such as neurons, the exact nature of cellular factors that govern whether autophagy has inhibitory effects on viral replication are poorly understood.
In contrast to in vitro experimental systems, the protective role of autophagy against viral infection is strongly supported by in vivo studies using viral variants deficient in autophagy inhibition or model organisms deficient in autophagy function (table 1). In Arabidopsis, disruption of BECLIN 1 or other autophagy genes dramatically increased local replication of tobacco mosaic virus (TMV) and resulted in the uncontrolled spread of programmed cell death [29]. In Drosophila, silencing of the autophagy genes, Atg5,Atg8, and Atg18, increased VSV replication in vivo and decreased fly survival [10]. In mammals, ectopic expression of Beclin 1 in neurons decreased Sindbis virus replication in brain, virus-induced neuronal apoptosis, and animal lethality [30]. Conversely, deletion of Atg5 in virally infected neurons led to increased virus-induced apoptosis and mortality during central nervous system Sindbis virus infection [14]. Hypomorphic expression of Atg16L1 resulted in increased mortality of mice infected with another alphavirus, chikungunya virus [31]. Compared to wild-type HSV-1, the HSV-1 mutant lacking the ability to bind to Beclin 1 and inhibit autophagy resulted in decreased mortality after intracerebral infection [25], decreased ocular disease after intracorneal infection [28], and decreased mortality after intravaginal infection [26]. These findings collectively demonstrate that the autophagy machinery functions as an essential component of host defense against many viral infections in vivo.
In in vivo studies, the specific nature of the protective effects of autophagy also appears to be context dependent. Dorsal root ganglion cells appear to rely on autophagy to control HSV-1 replication in a cell-autonomous manner [26], whereas in the cornea, autophagy seems to be used primarily to promote viral clearance by increasing CD4 T-cell responses [28]. Although ‘supraphysiological’ levels of autophagy can decrease Sindbis virus replication in neurons [30], deletion of endogenous autophagy does not affect levels of infectious virus in mouse brain, but does impede the clearance of viral nucleocapsids in neurons, resulting in increased neuronal death [14]. Thus, autophagy can function in antiviral defense in vivo through the direct control of viral replication, through the clearance of ‘toxic’ viral nucleocapsids, and/or through enhancing adaptive immune responses. It is also likely that other functions of autophagy proteins, such as the regulation of innate immunity and inflammatory signaling, may also contribute to protective effects during viral infection.
Virophagy Is a Form of Selective Autophagy
The autophagy pathway is known to selectively target unwanted cellular cargo to autophagosomes, including intracellular bacteria, damaged mitochondria, peroxisomes, lipid droplets, and protein aggregates. Classically, this process is thought to involve the labeling of such cargo with ubiquitin and an adaptor protein that binds to both ubiquitin and LC3. However, it has been unknown whether virions can be selectively targeted for autophagy and, if so, whether this process is also ubiquitin dependent. Recent studies have established that Sindbis virus nucleocapsids are selectively targeted for autophagic degradation by a mechanism that involves the well-known adaptor protein p62/SQSTM1, as well as a newly identified adaptor protein, SMURF1, an HECT domain containing E3 ligase that interacts with p62/SQSTM1 [15]. p62/SQSTM1 and SMURF1 both interact biochemically with Sindbis virus nucleocapsid in a ubiquitin-independent manner. Moreover, the ubiquitin E3 ligase domain of Smurf1 is dispensable for its function in selective autophagy. Therefore, the p62/SQSTM1-Smurf1 pathway represents a novel ubiquitin-independent mechanism of selective autophagy during viral infection. It will be interesting to determine what signals earmark unwanted viral nucleocapsids for virophagy, and whether similar mechanisms are used to target other families of viruses.
Autophagy Proteins Function in Adaptive Immunity in Viral Infection
Accumulating evidence demonstrates that the autophagy machinery promotes MHC class II presentation of viral (and other) endogenously synthesized antigens in different antigen-presenting cells (APCs), such as B cells, macrophages, dendritic cells, and fibroblasts. This concept was first demonstrated in studies with the Epstein-Barr virus (EBV) nuclear antigen 1 (EBNA1) [32]. Subsequently, an artificial fusion protein of influenza matrix protein 1 (MP1) and LC3 was shown to increase MP1 localization to autophagosomes and enhance antigen presentation to CD4+ T cells by dendritic cells, B cells, and epithelial cells [33]. A recent study of respiratory syncytial virus infection found that disruption of autophagy significantly inhibited the maturation of dendritic cells and their antigen presentation to CD4+ T cells, resulting in decreased cytokine production in response to respiratory syncytial virus infection [34].
Autophagy may also promote the presentation of endogenous viral antigens on MHC class I molecules and the activation of CD8+ T cells [35]. During late, but not early, stages of HSV-1 infection, autophagy was found to contribute to viral glycoprotein gB presentation to MHC class I molecules in macrophages and antigen presentation efficiency was higher in macrophages infected by the HSV-1.ΔICP34.5 mutant lacking the viral autophagy inhibitor ICP34.5 than in cells infected by wild-type HSV-1 [35]. In three other reports, autophagy was found to be dispensable for cross-presentation of viral antigens by MHC class I molecules, including in HSV-1-, EBV-, or HIV-1-infected dendritic cells or lymphoblast cells [19, 32, 36]. It is not yet known whether autophagy plays a more general role in class I MHC antigen presentation in viral infection or whether its role is dependent on the pathogen, the type of APC, and/or the route of antigen presentation. The contribution of autophagy to CD8+ T-cell activation in vivo also needs to be further explored.
Given the role of autophagy in adaptive immunity, it is not surprising that viruses employ multiple strategies to antagonize this function of autophagy. As noted above, the HSV-1 encoded virulence protein ICP34.5, that interacts with Beclin 1, limits CD4 T-cell responses during corneal infection [28]. More recently, Leib et al. [28] also showed that ICP34.5 has a unique function in dendritic cells that is not observed in fibroblasts or neurons; rather than blocking autophagosome formation (which it does in other cell types), ICP34.5 blocks autophagosomal maturation in dendritic cells, leading to impaired viral antigen presentation to MHC class II loading compartments and decreased CD4 T-cell activation [37]. In a somewhat similar fashion, influenza A virus matrix protein 2 (M2) interacts with Beclin 1, blocks autophagosome fusion with lysosomes [21], and restricts antigen presentation by MHC class II-loading compartments [33]. HIV-1 shuts down autophagy and immunoamphisome formation through rapid envelope-mediated mTOR activation, which impairs HIV-1 antigen processing and presentation [36]. EBNA1 is thought to shelter itself from being targeted by autophagy by having predominant nuclear localization [38].
The function of autophagy in endogenous viral antigen presentation is thought to represent ‘canonical’ autophagy, at least in the sense that the process involves the formation of double-membrane autophagosomes that deliver cytosolic components to lysosome-like compartments. In contrast, a canonical autophagy-independent role has been shown for Atg5 in dendritic cell MHC class II antigen presentation. Mice with dendritic cell depletion of Atg5 show defective CD4 T-cell priming and increased mortality after HSV-2 infection. Atg5−/− dendritic cells are defective in the processing and presentation of extracellular microbial antigens due to an impairment in phagosome-to-lysosome fusion rather than a defect in general autophagy [19]. These observations suggest that autophagy proteins may function in distinct cellular membrane trafficking events to contribute to effective MHC class II presentation of both endogenous and exogenous viral antigens.
Autophagy Proteins Function in the Regulation of Innate Immune Signaling
During viral infection, the fine-tuning of immune activation is beneficial to the host [1, 3, 5]; indeed, the termination of virus-triggered immune responses is often equally if not more critical than the initiation of such responses. Autophagy (or specific functions of autophagy proteins) can provide a negative feedback mechanism that reduces the magnitude and duration of innate responses to viral infection, thereby protecting the host from abnormalities caused by sustained immune activation. One interesting example of this is the inhibitory role of ATG16L1 in Crohn's disease. Crohn's disease is a common type of inflammatory bowel disease involving mucosal ulceration and inflammation. A polymorphism in the autophagy-related gene, ATG16L1 (T300A), is one of the Crohn's disease susceptibility alleles [39]. Compared with wild-type mice, mice lacking Atg16L1 in hematopoietic cells are highly susceptible to dextran sulfate sodium-induced acute colitis and show higher mortality due to severe intestinal inflammation [40]. Persistent infection with murine norovirus (MNV) results in Crohn's disease-like pathologic abnormalities of Paneth cells in mice with an Atg16L1 hypomorphic allele (Atg16L1HM) without enhancing viral replication [41]. However, this intestinal abnormality relies on TNF-α, IFN-γ, and commensal bacteria - three important factors associated with Crohn's disease pathogenesis in humans. This suggests that MNV-induced Paneth cell abnormalities in Atg16L1HM mice are likely triggered by an altered cytokine response and increased inflammation in vivo. Since the ATG16L1 T300A mutation associated with Crohn's disease does not alter canonical autophagy [42], this phenotype is most likely due to autophagy-independent functions of Atg16L1. Of note, several autophagy-related proteins have been shown to negatively regulate innate immune activation through a direct interaction with essential signaling molecules. For example, the conjugation of Atg5-Atg12 to CARD domains of RIG-I and MAVS exhibits an inhibitory effect on RIG-I-MAVS-mediated type I interferon production [43]. Atg9a reduces dsDNA-induced innate immune responses by inhibiting STING translocation and the assembly of STING and TBK1 [44].
The autophagy machinery acts at different steps to limit the magnitude of inflammation. Mitophagy, the selective autophagic targeting of damaged mitochondria for lysosomal degradation, prevents the cytosolic accumulation of ROS and mitochondrial DNA, and thereby restricts inflammasome activation [45, 46]. Assembled inflammasomes undergo ubiquitination and are targeted to autophagosomes via the autophagy adaptor p62 [47]. Furthermore, autophagy also controls IL-1β secretion by targeting pro-IL-1β for lysosomal degradation [48].
In some contexts, autophagy or the autophagy machinery can also stimulate innate immune or inflammatory signaling. Autophagy-related proteins participate in type I interferon production by plasmacytoid dendritic cells exposed to VSV [49] and HIV-1 [50], likely through promoting the delivery of viral ssRNA to endosomal Toll-like receptors. Moreover, siRNA knockdown of Atg5 in the lung reduced inflammation, acute lung injury, and the mortality of influenza virus H5N1-infected mice without suppression of viral replication [51]. Thus, depending on the specific context, autophagy may either enhance or reduce inflammatory signaling. Further studies are necessary to understand the determinants of such responses and how they can be modified to benefit the host in the treatment of viral infections.
Autophagy-Independent Antiviral Activities of Autophagy-Related Genes (ATGs)
In complex multicellular eukaryotes, ATGs orchestrate diversified functions of cellular and organismal responses to a variety of dangerous stimuli including viral infection [5]. Over the past 5 years, autophagy-independent antiviral functions of ATGs have been revealed in different experimental systems. These include the promotion of phagosome-to-lysosome fusion (discussed above) and mediation of the effects of IFN-γ on the disruption of MNV replication complexes. IFN-γ treatment triggers the recruitment of the autophagy proteins Atg5, Atg12, and Atg16L1 to the MNV polymerase, and these autophagy proteins (as well as Atg7 but not Atg4B or the digestive activity of autophagy) are required for IFN-γ-induced inhibition of the formation of cytoplasmic MNV replication complexes and MNV replication [52]. Although not yet studied in the context of viral infection, other recently identified autophagy-independent functions of certain autophagy proteins that may prove relevant to viral control include a role in the recruitment of immunity-related GTPases to pathogen-containing vacuoles [53] as well as in the unconventional secretion of IL-1β [54].
Autophagy Induction as a Potential Antiviral Strategy
The antiviral function of autophagy suggests that autophagy induction may be a feasible strategy to treat certain viral infections. In support of this concept, autophagy induction by either rapamycin or vitamin D treatment has been shown to restrict HIV-replication in vitro in primary human macrophages [55, 56]. In addition, a cell-permeable autophagy-inducing peptide (derived from a region of Beclin 1 targeted by HIV-1 Nef) restricts the in vitro replication of several viruses, including HIV-1, Sindbis virus, chikungunya virus, and West Nile virus, and also reduces the mortality of neonatal mice infected with either chikungunya virus or West Nile virus [57]. Therefore, autophagy upregulation may be a novel promising approach to broad-spectrum antiviral therapies.
The Proviral Roles of Autophagy and Autophagy Proteins
Autophagy has likely maintained a selective pressure on viruses throughout millions years of co-evolution with their eukaryotic hosts. Consequently, different viruses have developed diversified strategies to avoid and/or suppress canonical autophagic degradation and autophagy-mediated immune activation. These viral antagonism strategies include: (1) avoidance of autophagic capture, (2) suppression of autophagy initiation, and (3) suppression of autophagosomal maturation [3]. The successful evasion of the antiviral effects of autophagy allows viruses to manipulate and exploit the autophagy pathway and/or components of the autophagy machinery to their own advantage. To date, the autophagy pathway and/or components of the autophagy machinery have been implicated in proviral roles for several RNA and DNA viruses (fig. 2). However, the precise molecular mechanisms of most proviral activities have not been completely defined.
Viral Exploitation of the Autophagy Pathway and/or the Autophagy Machinery
The proviral activity was first discovered in poliovirus-infected cells, in which autophagy promotes both viral replication and nonlytic release [58]. There are several possible ways in which intact or incomplete autophagy might benefit viral infection. As an essential mechanism to maintain nutrient and energy homeostasis in response to stress, autophagy may help supply building blocks for viral replication. As a prosurvival mechanism, autophagy may help keep virally infected cells alive, allowing more prolonged replication. As a pathway that mediates dynamic membrane rearrangements, autophagy (and/or specific autophagy proteins) may facilitate the formation of intracellular membranes required for viral replication. As a process that involves many enzymatic complexes, including the ubiquitin ligase protein conjugation systems and the class III phosphatidylinositol-3 kinase complex, some of the biochemical functions of these complexes might be co-opted for use in viral replication. Many of these, and likely other, features of autophagy may be utilized to foster viral replication. In this review, we focus on selected recent studies that have not previously been discussed in other published reviews [for a more complete discussion of this topic, we refer readers to [3, 4, 5, 6, 7]].
Dengue Virus Manipulates Lipid Metabolism for Replication
In mammals, lipid droplets can be selectively degraded by lysosomes via autophagy to release free fatty acids (FFAs) in a selective autophagy process referred to as lipophagy [59]. Dengue virus infection induces autophagy-dependent processing of lipid droplets and triglycerides to generate FFAs, resulting in an increase in cellular β-oxidation and ATP generation [60]. Autophagy inhibition greatly reduces lipophagy and Dengue virus yield without affecting viral entry or translation, and this defect in viral replication is completely rescued by exogenous FFAs [60]. These findings provide the first example of viral exploitation of lipophagy to enhance infection by modulating host lipid metabolism.
Autophagy May Enhance Kaposi's Sarcoma-Associated Herpesvirus Lytic Reactivation from Latency
Kaposi's sarcoma-associated herpesvirus (KSHV) is a human γ-herpesvirus associated with multiple cancers, including lymphoproliferative disorders and Kaposi's sarcoma. Two autophagy suppressors in the KSHV genome have been identified, including viral Bcl-2 and v-FLIP [61, 62]. KSHV v-FLIP blockade of autophagy (via an interaction with Atg3) is thought to enhance the proliferation of KSHV-infected cells by blocking autophagy-dependent senescence induced by the viral oncogenic protein v-cyclin [63]. In contrast to this role of autophagy as an anti-KSHV host defense mechanism (by promoting oncogene-induced senescence), another study reported that autophagy may have a proviral function during KSHV infection. Genetic or pharmacological inhibition of autophagy was shown to block KSHV replication and transcription activator (RTA)-mediated lytic gene expression or DNA replication, respectively [64].
The Autophagy Machinery Is Exploited for Hepatitis C Virus Infection
Hepatitis C virus (HCV) is one of best-studied viruses that is reported to exploit components of the autophagy pathway to promote its replication. Different subtypes of HCV (1a, 1b, and 2a) induce the apparent accumulation of autophagosomes (i.e. vacuoles that have autophagic markers such as LC3 and ATG5) [65, 66], and NS4B, an HCV nonstructural protein, interacts with Rab5 and Beclin 1/Vps34 and functions in autophagy initiation [67]. Of note, HCV infection triggers an incomplete autophagy response likely by blocking autophagosome maturation [66, 68]. The activation of early autophagy is important for the HCV life cycle, as knockdown of several different autophagy genes (LC3, ATG4B, ATG7, ATG12, and beclin 1) significantly reduces HCV RNA replication and yield [66, 69, 70, 71, 72].
The core autophagy machinery may participate in different facets of HCV infection. Through mechanisms that are not yet understood, the autophagy machinery was shown to be required for the initiation of translation and replication of the incoming HCV genome (but not for subsequent rounds of replication) [69]. ATG5 transiently interacts with the HCV RNA polymerase NS5B, leading to the speculation that ATG5 might function as a proviral factor during the onset of viral infection [73]. Some studies suggest an association between HCV replication (HCV RNA and components of the HCV replicase complex) and double-membrane vesicles (DMVs) that have autophagosomal markers [72, 74]. Additional studies will be needed to determine whether these DMVs represent true autophagosomes or whether multiple components of the canonical autophagy machinery may also be used to generate double-membrane structures used as scaffolds in RNA virus replication.
One advantage of the utilization of DMVs is that this may allow HCV to conceal its viral RNA from cytosolic RNA sensors and thereby evade host antiviral responses. This strategy is not a unique feature of viruses in the family Flaviviridae. Porcine reproductive and respiratory syndrome virus, a member of the family Arteriviridae, also induces an incomplete autophagy response (i.e. induction of early autophagy with a block in autophagosome-lysosome fusion) that is required for viral replication, and the viral replicase NSP2 partially co-localizes with LC3 [75]. In addition, Coxsackievirus B3 and other picornaviruses have been shown to induce the formation of DMVs that contain autophagosomal markers, block autophagosome-lysosome fusion, and display reduced viral replication with pharmacological or genetic inhibition of autophagy [58, 76]. Therefore, viral hijacking of autophagy protein-mediated DMV formation may be a common replication strategy for RNA viruses.
Autophagy May Promote DNA Virus Replication
Hepatitis B virus (HBV) likely utilizes the core autophagic machinery to promote viral DNA replication [77] and viral envelopment [78]. Promotion of viral DNA genome replication by autophagy or the autophagy machinery is also observed during infection with two other DNA viruses, KSHV [64] and porcine circovirus type 2 [79]. It will be interesting to explore whether there is a common molecular mechanism underlying the involvement of the autophagy machinery in the genome replication of DNA viruses.
Atg5 Is Required for Viral Replication and Pathogenesis in vivo
Two recent studies on Coxackievirus B3 [80] and HBV [81] provide in vivo evidence that the Atg5 autophagy protein exerts proviral functions. Pancreatic acinar cell-specific disruption of Atg5 dramatically reduces Coxsackievirus B3 replication in the pancreas and decreases viral-induced pancreatic dysfunction [80]. The precise mechanism of decreased viral replication is not fully understood but is postulated to result from reduced synthesis of viral genomic RNA due to a deficiency in the formation of compound membrane vesicles and paracrystalline arrays (which are believed to be viral replication platforms). Liver-specific knockout of Atg5 reduces HBV DNA replicative intermediates in the liver to nearly undetectable levels and circulating HBV DNA in sera by more than 90% [81]. These in vivo studies suggest that for certain viruses, such as picornaviruses and HBV, pharmacological disruption of autophagy might exert antiviral effects. However, it is not yet known whether the in vivo phenotypes of Atg5 deficiency in Coxsackievirus B and HBV infection are due to defective autophagy or the lack of other Atg5-dependent functions.
Immunity-Related GTPase Family M May Be a Common Viral Target to Modulate Autophagy or Exploit the Autophagy Machinery
Human immunity-related GTPase family M (IRGM) and the mouse ortholog Irgm1 were originally linked to autophagy by virtue of their requirement in IFN-γ-mediated autophagy induction and elimination of intracellular mycobacteria [82]. A recent study suggests that IRGM may be a common target of RNA viruses that subvert autophagy [83]. Using yeast two-hybrid and bioinformatics analyses, human IRGM was shown to interact with several core autophagy proteins, including ATG5, ATG10, and LC3B, as well as with several viral proteins, including measles virus capsid, HCV NS3, and HIV Nef. Expression of these viral proteins is sufficient to induce autophagy through an IRGM-dependent mechanism, and the genetic silencing of IRGM decreases measles-, HCV-, and HIV-1-induced autophagy and particle production [83]. The precise mechanisms by which viral targeting of IRGM promotes viral replication remain to be determined.
Murine Cytomegalovirus Inhibits the Inflammatory Cascade by Targeting the NF-κB Essential Modulator NEMO/IKKγ for Autophagic Degradation
The inflammatory signaling cascade activated during viral infection is thought to be key to pathogen elimination [84]. Thus, another potential strategy that viruses may utilize to co-opt autophagy for their own benefit is to dampen the inflammatory cascade by directing the selective autophagic degradation of modulators of inflammatory signaling. The murine cytomegalovirus protein M45 interacts with NEMO/IKKγ, an essential modulator of NF-κB signaling, and directs it to the autophagosome for lysosome-mediated degradation, thereby blunting the host cytokine response [85]. It will be interesting to determine whether this type of selective targeting of an essential host defense component for autophagic degradation is a more general strategy of viral immune evasion.
Viral Exploitation of ATGs in an Autophagy-Independent Manner
There is increasing evidence that individual autophagy proteins may be utilized in viral replication in a manner that is independent of the autophagy pathway. The best-studied demonstration of this concept is with the murine coronavirus, MHV [86]. MHV employs LC3-I (the nonlipidated, nonautophagosome-associated form of LC3) and ER-derived vesicles exporting short-lived ERAD regulators (EDEMosomes) to generate the DMVs required for viral replication [86]. The knockdown of LC3 reduces DMV formation and viral replication; however, neither Atg5 nor Atg7 is required for MHV replication [20, 86]. Therefore, the role of nonlipidated LC3 in MHV infection represents a novel autophagy-independent function in membrane trafficking and viral replication. This finding underscores the need for caution in interpreting whether the autophagy pathway functions in viral replication based solely on observations of LC3 and viral protein colocalization.
Another interesting example of viral exploitation of LC3 in an autophagy-independent manner is provided by studies of the pestivirus BVDV. The induction of lethal mucosal disease in cattle is usually associated with the generation of a cytopathogenic BVDV strain in animals persistently infected with a noncytopathogenic strain. Sequence analysis of a naturally occurring recombinant cytopathogenic BVDV strain, JaCP, from an animal with mucosal disease showed that JaCP and the variants in vivo all carry the full-length cellular LC3 gene insertion in the viral genome [87, 88]. The LC3 insertion in the JaCP genome is in frame of the viral polyprotein precursor, and the LC3-specific processing of the viral polyprotein by the host LC3-specific protease Atg4B is essential for the processing of the NS3 viral protein and for BVDV replication [89]. Thus, BVDV co-opts the enzymatic function of host LC3 processing by Atg4B to optimize viral replication. Further studies are required to determine whether the biochemical functions of autophagy proteins are utilized in the replication strategies of other viruses.
Conclusion
In the past several years, considerable advances have been made in understanding the complex interplay between autophagy and viruses. Autophagy is an ancient conserved cell-autonomous defense mechanism and the autophagy-related genes (ATGs) orchestrate diverse aspects of host antiviral responses. During co-evolution with their hosts, many viruses have developed a variety of strategies to subvert and/or manipulate the autophagy pathway. Some viruses may also exploit components of the autophagy machinery in a ‘modular’ manner to promote their replication. Moreover, even for the same virus, the interaction between autophagy and viral replication may vary widely depending on the specific cellular target.
While considerable advances have been made in understanding the interplay between autophagy and viruses, many important questions remain to be answered. It is still largely unclear how autophagy is activated during viral infection, how autophagy functions to restrict the replication of some viruses, how autophagy functions to promote the replication of some viruses, and how autophagy proteins are utilized by the host and by the virus in noncanonical autophagy-independent fashions to negatively or positively regulate viral replication. Unraveling the answers to these questions will enhance our understanding of virology, autophagy, and novel autophagy-independent functions of autophagy proteins, as well as potentially identify additional targets that can be used to therapeutically modulate autophagy in the treatment of viral diseases.
Acknowledgements
We thank Drs. Matthew Leveno and Sanae Shoji-Kawata for critical reading of the manuscript. The work in the authors' laboratory was supported by NIH grants RO1 AI151267 and U54AI057156 to B.L. We apologize to those authors whose work could not be cited due to space limitations.
References
- 1.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deretic V, Levine B. Autophagy, immunity, and microbial adaptations. Cell Host Microbe. 2009;5:527–549. doi: 10.1016/j.chom.2009.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dreux M, Chisari FV. Viruses and the autophagy machinery. Cell Cycle. 2010;9:1295–1307. doi: 10.4161/cc.9.7.11109. [DOI] [PubMed] [Google Scholar]
- 5.Levine B, Mizushima N, Virgin HW. Autophagy in immunity and inflammation. Nature. 2011;469:323–335. doi: 10.1038/nature09782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yordy B, Iwasaki A. Autophagy in the control and pathogenesis of viral infection. Curr Opin Virol. 2011;1:196–203. doi: 10.1016/j.coviro.2011.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jordan TX, Randall G. Manipulation or capitulation: virus interactions with autophagy. Microbes Infect. 2012;14:126–139. doi: 10.1016/j.micinf.2011.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rubinsztein DC, Codogno P, Levine B. Autophagy modulation as a potential therapeutic target for diverse diseases. Nat Rev Drug Discov. 2012;11:709–730. doi: 10.1038/nrd3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakamoto M, Moy RH, Xu J, Bambina S, Yasunaga A, Shelly SS, Gold B, Cherry S. Virus recognition by Toll-7 activates antiviral autophagy in Drosophila. Immunity. 2012;36:658–667. doi: 10.1016/j.immuni.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shelly S, Lukinova N, Bambina S, Berman A, Cherry S. Autophagy is an essential component of Drosophila immunity against vesicular stomatitis virus. Immunity. 2009;30:588–598. doi: 10.1016/j.immuni.2009.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Joubert PE, Meiffren G, Grégoire IP, Pontini G, Richetta C, Flacher M, Azocar O, Vidalain PO, Vidal M, Lotteau V, Codogno P, Rabourdin-Combe C, Faure M. Autophagy induction by the pathogen receptor CD46. Cell Host Microbe. 2009;6:354–366. doi: 10.1016/j.chom.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 12.Denizot M, Varbanov M, Espert L, Robert-Hebmann V, Sagnier S, Garcia E, Curriu M, Mamoun R, Blanco J, Biard-Piechaczyk M. HIV-1 gp41 fusogenic function triggers autophagy in uninfected cells. Autophagy. 2008;4:998–1008. doi: 10.4161/auto.6880. [DOI] [PubMed] [Google Scholar]
- 13.Delpeut S, Rudd PA, Labonté P, von Messling., V Membrane fusion-mediated autophagy induction enhances morbillivirus cell-to-cell spread. J Virol. 2012;86:8527–8535. doi: 10.1128/JVI.00807-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Orvedahl A, MacPherson S, Sumpter R, Jr, Tallóczy Z, Zou Z, Levine B. Autophagy protects against Sindbis virus infection of the central nervous system. Cell Host Microbe. 2010;7:115–127. doi: 10.1016/j.chom.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Orvedahl A, Sumpter R, Jr, Xiao G, Ng A, Zou Z, Tang Y, Narimatsu M, Gilpin C, Sun Q, Roth M, Forst CV, Wrana JL, Zhang YE, Luby-Phelps K, Xavier RJ, Xie Y, Levine B. Image-based genome-wide siRNA screen identifies selective autophagy factors. Nature. 2011;480:113–117. doi: 10.1038/nature10546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chakrabarti A, Ghosh PK, Banerjee S, Gaughan C, Silverman RH. RNase L triggers autophagy in response to viral infections. J Virol. 2012;86:11311–11321. doi: 10.1128/JVI.00270-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang H, Monken CE, Zhang Y, Lenard J, Mizushima N, Lattime EC, Jin S. Cellular autophagy machinery is not required for vaccinia virus replication and maturation. Autophagy. 2006;2:91–95. doi: 10.4161/auto.2.2.2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alexander DE, Ward SL, Mizushima N, Levine B, Leib DA. Analysis of the role of autophagy in replication of herpes simplex virus in cell culture. J Virol. 2007;81:12128–12134. doi: 10.1128/JVI.01356-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee HK, Mattei LM, Steinberg BE, Alberts P, Lee YH, Chervonsky A, Mizushima N, Grinstein S, Iwasaki A. In vivo requirement for Atg5 in antigen presentation by dendritic cells. Immunity. 2010;32:227–239. doi: 10.1016/j.immuni.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao Z, Thackray LB, Miller BC, Lynn TM, Becker MM, Ward E, Mizushima NN, Denison MR, Virgin HW., 4th Coronavirus replication does not require the autophagy gene ATG5. Autophagy. 2007;3:581–585. doi: 10.4161/auto.4782. [DOI] [PubMed] [Google Scholar]
- 21.Gannagé M, Dormann D, Albrecht R, Dengjel J, Torossi T, Rämer PC, Lee M, Strowig T, Arrey F, Conenello G, Pypaert M, Andersen J, García-Sastre A, Münz C. Matrix protein 2 of influenza A virus blocks autophagosome fusion with lysosomes. Cell Host Microbe. 2009;6:367–380. doi: 10.1016/j.chom.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beatman E, Oyer R, Shives KD, Hedman K, Brault AC, Tyler KL, David Beckham., J West Nile virus growth is independent of autophagy activation. Virology. 2012;433:262–272. doi: 10.1016/j.virol.2012.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He C, Levine B. The Beclin 1 interactome. Curr Opin Cell Biol. 2010;22:140–149. doi: 10.1016/j.ceb.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moloughney JG, Monken CE, Tao H, Zhang H, Thomas JD, Lattime EC, Jin SV. Vaccinia virus leads to ATG12-ATG3 conjugation and deficiency in autophagosome formation. Autophagy. 2011;7:1434–1447. doi: 10.4161/auto.7.12.17793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Orvedahl A, Alexander D, Tallóczy Z, Sun Q, Wei Y, Zhang W, Burns D, Leib DA, Levine B. HSV-1 ICP34.5 confers neurovirulence by targeting the Beclin 1 autophagy protein. Cell Host Microbe. 2007;1:23–35. doi: 10.1016/j.chom.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 26.Yordy B, Iijima N, Huttner A, Leib D, Iwasaki A. A neuron-specific role for autophagy in antiviral defense against herpes simplex virus. Cell Host Microbe. 2012;12:334–345. doi: 10.1016/j.chom.2012.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.E X, Hwang S, Oh S, Lee JS, Jeong JH, Gwack Y, Kowalik TF, Sun R, Jung JU, Liang C. Viral Bcl-2-mediated evasion of autophagy aids chronic infection of gammaherpesvirus 68. PloS Pathog. 2009;5:e1000609. doi: 10.1371/journal.ppat.1000609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leib DA, Alexander DE, Cox D, Yin J, Ferguson TA. Interaction of ICP34.5 with Beclin 1 modulates herpes simplex virus type 1 pathogenesis through control of CD4+ T-cell responses. J Virol. 2009;83:12164–12171. doi: 10.1128/JVI.01676-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu Y, Schiff M, Czymmek K, Tallóczy Z, Levine B, Dinesh-Kumar SP. Autophagy regulates programmed cell death during the plant innate immune response. Cell. 2005;121:567–577. doi: 10.1016/j.cell.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 30.Liang XH, Kleeman LK, Jiang HH, Gordon G, Goldman JE, Berry G, Herman B, Levine B. Protection against fatal Sindbis virus encephalitis by beclin, a novel Bcl-2-interacting protein. J Virol. 1998;72:8586–8596. doi: 10.1128/jvi.72.11.8586-8596.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Joubert PE, Werneke SW, de la Calle C, Guivel-Benhassine F, Giodini A, Peduto L, Levine B, Schwartz O, Lenschow DJ, Albert ML. Chikungunya virus-induced autophagy delays caspase-dependent cell death. J Exp Med. 2012;209:1029–1047. doi: 10.1084/jem.20110996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paludan C, Schmid D, Landthaler M, Vockerodt M, Kube D, Tuschl T, Münz C. Endogenous MHC class II processing of a viral nuclear antigen after autophagy. Science. 2005;307:593–596. doi: 10.1126/science.1104904. [DOI] [PubMed] [Google Scholar]
- 33.Comber JD, Robinson TM, Siciliano NA, Snook AE, Eisenlohr LC. Functional macroautophagy induction by influenza A virus without a contribution to major histocompatibility complex class II-restricted presentation. J Virol. 2011;85:6453–6463. doi: 10.1128/JVI.02122-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morris S, Swanson MS, Lieberman A, Reed M, Yue Z, Lindell DM, Lukacs NW. Autophagy-mediated dendritic cell activation is essential for innate cytokine production and APC function with respiratory syncytial virus responses. J Immunol. 2011;187:3953–3961. doi: 10.4049/jimmunol.1100524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.English L, Chemali M, Duron J, Rondeau C, Laplante A, Gingras D, Alexander D, Leib D, Norbury C, Lippé R, Desjardins M. Autophagy enhances the presentation of endogenous viral antigens on MHC class I molecules during HSV-1 infection. Nat Immunol. 2009;10:480–487. doi: 10.1038/ni.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blanchet FP, Moris A, Nikolic DS, Lehmann M, Cardinaud S, Stalder R, Garcia E, Dinkins C, Leuba F, Wu L, Schwartz O, Deretic V, Piguet V. Human immunodeficiency virus-1 inhibition of immunoamphisomes in dendritic cells impairs early innate and adaptive immune responses. Immunity. 2010;32:654–569. doi: 10.1016/j.immuni.2010.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gobeil PA, Leib DA. Herpes simplex virus γ34.5 interferes with autophagosome maturation and antigen presentation in dendritic cells. MBio. 2012;3:e00267–e00312. doi: 10.1128/mBio.00267-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leung CS, Haigh TA, Mackay LK, Rickinson AB, Taylor GS. Nuclear location of an endogenously expressed antigen, EBNA1, restricts access to macroautophagy and the range of CD4 epitope display. Proc Natl Acad Sci USA. 2010;107:2165–2170. doi: 10.1073/pnas.0909448107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cadwell K, Liu JY, Brown SL, Miyoshi H, Loh J, Lennerz JK, Kishi C, Kc W, Carrero JA, Hunt S, Stone CD, Brunt EM, Xavier RJ, Sleckman BP, Li E, Mizushima N, Stappenbeck TS, Virgin HW., 4th A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456:259–263. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, Satoh T, Omori H, Noda T, Yamamoto N, Komatsu M, Tanaka K, Kawai T, Tsujimura T, Takeuchi O, Yoshimori T, Akira S. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature. 2008;456:264–268. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- 41.Cadwell K, Patel KK, Maloney NS, Liu TC, Ng AC, Storer CE, Head RD, Xavier R, Stappenbeck TS, Virgin HW. Virus-plus-susceptibility gene interaction determines Crohn's disease gene Atg16L1 phenotypes in intestine. Cell. 2010;141:1135–1145. doi: 10.1016/j.cell.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kuballa P, Huett A, Rioux JD, Daly MJ, Xavier RJ. Impaired autophagy of an intracellular pathogen induced by a Crohn's disease associated ATG16L1 variant. PLoS One. 2008;3:e3391. doi: 10.1371/journal.pone.0003391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jounai N, Takeshita F, Kobiyama K, Sawano A, Miyawaki A, Xin KQ, Ishii KJ, Kawai T, Akira S, Suzuki K, Okuda K. The Atg5 Atg12 conjugate associates with innate antiviral immune responses. Proc Natl Acad Sci USA. 2007;104:14050–14055. doi: 10.1073/pnas.0704014104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saitoh T, Fujita N, Hayashi T, Takahara K, Satoh T, Lee H, Matsunaga K, Kageyama S, Omori H, Noda T, Yamamoto N, Kawai T, Ishii K, Takeuchi O, Yoshimori T, Akira S. Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc Natl Acad Sci USA. 2009;106:20842–20846. doi: 10.1073/pnas.0911267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou R, Yazdi AS, Menu P, Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011;469:221–225. doi: 10.1038/nature09663. [DOI] [PubMed] [Google Scholar]
- 46.Nakahira K, Haspel JA, Rathinam VA, Lee SJ, Dolinay T, Lam HC, Englert JA, Rabinovitch M, Cernadas M, Kim HP, Fitzgerald KA, Ryter SW, Choi AM. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat Immunol. 2011;12:222–230. doi: 10.1038/ni.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shi CS, Shenderov K, Huang NN, Kabat J, Abu-Asab M, Fitzgerald KA, Sher A, Kehrl JH. Activation of autophagy by inflammatory signals limits IL-1β production by targeting ubiquitinated inflammasomes for destruction. Nat Immunol. 2012;13:255–263. doi: 10.1038/ni.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Harris J, Hartman M, Roche C, Zeng SG, O'Shea A, Sharp FA, Lambe EM, Creagh EM, Golenbock DT, Tschopp J, Kornfeld H, Fitzgerald KA, Lavelle EC. Autophagy controls IL-1beta secretion by targeting pro-IL-1beta for degradation. J Biol Chem. 2011;286:9587–9597. doi: 10.1074/jbc.M110.202911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee HK, Lund JM, Ramanathan B, Mizushima N, Iwasaki A. Autophagy-dependent viral recognition by plasmacytoid dendritic cells. Science. 2007;315:1398–1401. doi: 10.1126/science.1136880. [DOI] [PubMed] [Google Scholar]
- 50.Zhou D, Kang KH, Spector SA. Production of interferon α by human immunodeficiency virus type 1 in human plasmacytoid dendritic cells is dependent on induction of autophagy. J Infect Dis. 2012;205:1258–1267. doi: 10.1093/infdis/jis187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sun Y, Li C, Shu Y, Ju X, Zou Z, Wang H, Rao S, Guo F, Liu H, Nan W, Zhao Y, Yan Y, Tang J, Zhao C, Yang P, Liu K, Wang S, Lu H, Li X, Tan L, Gao R, Song J, Gao X, Tian X, Qin Y, Xu KF, Li D, Jin N, Jiang C. Inhibition of autophagy ameliorates acute lung injury caused by avian influenza A H5N1 infection. Sci Signal. 2012;5:ra16. doi: 10.1126/scisignal.2001931. [DOI] [PubMed] [Google Scholar]
- 52.Hwang S, Maloney NS, Bruinsma MW, Goel G, Duan E, Zhang L, Shrestha B, Diamond MS, Dani A, Sosnovtsev SV, Green KY, Lopez-Otin C, Xavier RJ, Thackray LB, Virgin HW. Nondegradative role of Atg5-Atg12/Atg16L1 autophagy protein complex in antiviral activity of interferon gamma. Cell Host Microbe. 2012;11:397–409. doi: 10.1016/j.chom.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhao Z, Fux B, Goodwin M, Dunay IR, Strong D, Miller BC, Cadwell K, Delgado MA, Ponpuak M, Green KG, Schmidt RE, Mizushima N, Deretic V, Sibley LD, Virgin HW. Autophagosome-independent essential function for the autophagy protein Atg5 in cellular immunity to intracellular pathogens. Cell Host Microbe. 2008;4:458–469. doi: 10.1016/j.chom.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dupont N, Jiang S, Pilli M, Ornatowski W, Bhattacharya D, Deretic V. Autophagy-based unconventional secretory pathway for extracellular delivery of IL-1β. EMBO J. 2011;30:4701–4711. doi: 10.1038/emboj.2011.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Campbell GR, Spector SA. Hormonally active vitamin D3 (1alpha,25-dihydroxycholecalciferol) triggers autophagy in human macrophages that inhibits HIV-1 infection. J Biol Chem. 2011;286:18890–18902. doi: 10.1074/jbc.M110.206110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Campbell GR, Spector SA. Vitamin D inhibits human immunodeficiency virus type 1 and Mycobacterium tuberculosis infection in macrophages through the induction of autophagy. PLoS Pathog. 2012;8:e1002689. doi: 10.1371/journal.ppat.1002689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shoji-Kawata S, Sumpter R, Jr, Leveno M, Campbell G, Zou Z, Kinch L, Wilkins A, Sun Q, Pallauf K, MacDuff D, Huerta C, Virgin HW IV, Helms JB, Eerland R, Tooze SA, Xavier R, Lenschow DJ, Yamamoto A, King D, Lichtarge O, Grishin NK, Spector SA, Kaloyanova DV, Levine B. Identification of a candidate therapeutic autophagy-inducing peptide. Nature. 2013 doi: 10.1038/nature11866. DOI:10.1038/nature11866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jackson WT, Giddings TH, Jr, Taylor MP, Mulinyawe S, Rabinovitch M, Kopito RR, Kirkegaard K. Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol. 2005;3:e156. doi: 10.1371/journal.pbio.0030156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Singh R, Kaushik S, Wang Y, Xiang Y, Novak I, Komatsu M, Tanaka K, Cuervo AM, Czaja MJ. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Heaton NS, Randall G. Dengue virus-induced autophagy regulates lipid metabolism. Cell Host Microbe. 2010;8:422–432. doi: 10.1016/j.chom.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, Packer M, Schneider MD, Levine B. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–939. doi: 10.1016/j.cell.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 62.Lee JS, Li Q, Lee JY, Lee SH, Jeong JH, Lee HR, Chang H, Zhou FC, Gao SJ, Liang C, Jung JU. FLIP-mediated autophagy regulation in cell death control. Nat Cell Biol. 2009;11:1355–1362. doi: 10.1038/ncb1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Leidal AM, Cyr DP, Hill RJ, Lee PW, McCormick C. Subversion of autophagy by Kaposi's sarcoma-associated Herpesvirus impairs oncogene-induced senescence. Cell Host Microbe. 2012;11:167–180. doi: 10.1016/j.chom.2012.01.005. [DOI] [PubMed] [Google Scholar]
- 64.Wen HJ, Yang Z, Zhou Y, Wood C. Enhancement of autophagy during lytic replication by the Kaposi's sarcoma-associated herpesvirus replication and transcription activator. J Virol. 2010;84:7448–7458. doi: 10.1128/JVI.00024-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ait-Goughoulte M, Kanda T, Meyer K, Ryerse JS, Ray RB, Ray R. Hepatitis C virus genotype 1a growth and induction of autophagy. J Virol. 2008;82:2241–2249. doi: 10.1128/JVI.02093-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sir D, Chen WL, Choi J, Wakita T, Yen TS, Ou JH. Induction of incomplete autophagic response by hepatitis C virus via the unfolded protein response. Hepatology. 2008;48:1054–1061. doi: 10.1002/hep.22464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Su WC, Chao TC, Huang YL, Weng SC, Jeng KS, Lai MM. Rab5 and class III phosphoinositide 3-kinase Vps34 are involved in hepatitis C virus NS4B-induced autophagy. J Virol. 2011;85:10561–10571. doi: 10.1128/JVI.00173-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Taguwa S, Kambara H, Fujita N, Noda T, Yoshimori T, Koike K, Moriishi K, Matsuura Y. Dysfunction of autophagy participates in vacuole formation and cell death in cells replicating hepatitis C virus. J Virol. 2011;85:13185–13194. doi: 10.1128/JVI.06099-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dreux M, Gastaminza P, Wieland SF, Chisari FV. The autophagy machinery is required to initiate hepatitis C virus replication. Proc Natl Acad Sci USA. 2009;106:14046–14051. doi: 10.1073/pnas.0907344106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tanida I, Fukasawa M, Ueno T, Kominami E, Wakita T, Hanada K. Knockdown of autophagy-related gene decreases the production of infectious hepatitis C virus particles. Autophagy. 2009;5:937–945. doi: 10.4161/auto.5.7.9243. [DOI] [PubMed] [Google Scholar]
- 71.Shrivastava S, Raychoudhuri A, Steele R, Ray R, Ray RB. Knockdown of autophagy enhances the innate immune response in hepatitis C virus-infected hepatocytes. Hepatology. 2011;53:406–414. doi: 10.1002/hep.24073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sir D, Kuo CF, Tian Y, Liu HM, Huang EJ, Jung JU, Machida K, Ou JH. Replication of hepatitis C virus RNA on autophagosomal membranes. J Biol Chem. 2012;287:18036–18043. doi: 10.1074/jbc.M111.320085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Guévin C, Manna D, Bélanger C, Konan KV, Mak P, Labonté P. Autophagy protein ATG5 interacts transiently with the hepatitis C virus RNA polymerase (NS5B) early during infection. Virology. 2010;405:1–7. doi: 10.1016/j.virol.2010.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ferraris P, Blanchard E, Roingeard P. Ultrastructural and biochemical analyses of hepatitis C virus-associated host cell membranes. J Gen Virol. 2010;91:2230–2237. doi: 10.1099/vir.0.022186-0. [DOI] [PubMed] [Google Scholar]
- 75.Sun MX, Huang L, Wang R, Yu YL, Li C, Li PP, Hu XC, Hao HP, Ishag HA, Mao X. Porcine reproductive and respiratory syndrome virus induces autophagy to promote virus replication. Autophagy. 2012;8:1434–1447. doi: 10.4161/auto.21159. [DOI] [PubMed] [Google Scholar]
- 76.Wong J, Zhang J, Si X, Gao G, Mao I, McManus BM, Luo H. Autophagosome supports coxsackievirus B3 replication in host cells. J Virol. 2008;82:9143–9153. doi: 10.1128/JVI.00641-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sir D, Tian Y, Chen WL, Ann DK, Yen TS, Ou JH. The early autophagic pathway is activated by hepatitis B virus and required for viral DNA replication. Proc Natl Acad Sci USA. 2010;107:4383–4388. doi: 10.1073/pnas.0911373107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li J, Liu Y, Wang Z, Liu K, Wang Y, Liu J, Ding H, Yuan Z. Subversion of cellular autophagy machinery by hepatitis B virus for viral envelopment. J Virol. 2011;85:6319–6333. doi: 10.1128/JVI.02627-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhu B, Xu F, Li J, Shuai J, Li X, Fang W. Porcine circovirus type 2 explores the autophagic machinery for replication in PK-15 cells. Virus Res. 2012;163:476–485. doi: 10.1016/j.virusres.2011.11.012. [DOI] [PubMed] [Google Scholar]
- 80.Alirezaei M, Flynn CT, Wood MR, Whitton JL. Pancreatic acinar cell-specific autophagy disruption reduces Coxsackievirus replication and pathogenesis in vivo. Cell Host Microbe. 2012;11:298–305. doi: 10.1016/j.chom.2012.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tian Y, Sir D, Kuo CF, Ann DK, Ou JH. Autophagy required for hepatitis B virus replication in transgenic mice. J Virol. 2011;85:13453–13456. doi: 10.1128/JVI.06064-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Singh SB, Davis AS, Taylor GA, Deretic V. Human IRGM induces autophagy to eliminate intracellular mycobacteria. Science. 2006;313:1438–1441. doi: 10.1126/science.1129577. [DOI] [PubMed] [Google Scholar]
- 83.Grégoire IP, Richetta C, Meyniel-Schicklin L, Borel S, Pradezynski F, Diaz O, Deloire A, Azocar O, Baguet J, Le Breton M, Mangeot PE, Navratil V, Joubert PE, Flacher M, Vidalain PO, André P, Lotteau V, Biard-Piechaczyk M, Rabourdin-Combe C, Faure M. IRGM is a common target of RNA viruses that subvert the autophagy network. PLoS Pathog. 2011;7:e1002422. doi: 10.1371/journal.ppat.1002422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Medzhitov R. Origin and physiological roles of inflammation. Nature. 2008;454:428–435. doi: 10.1038/nature07201. [DOI] [PubMed] [Google Scholar]
- 85.Fliss PM, Jowers TP, Brinkmann MM, Holstermann B, Mack C, Dickinson P, Hohenberg H, Ghazal P, Brune W. Viral mediated redirection of NEMO/IKKγ to autophagosomes curtails the inflammatory cascade. PLoS Pathog. 2012;8:e1002517. doi: 10.1371/journal.ppat.1002517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Reggiori F, Monastyrska I, Verheije MH, Calì T, Ulasli M, Bianchi S, Bernasconi R, de Haan CA, Molinari M. Coronaviruses Hijack the LC3-I-positive EDEMosomes, ER-derived vesicles exporting short-lived ERAD regulators, for replication. Cell Host Microbe. 2010;7:500–508. doi: 10.1016/j.chom.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Meyers G, Stoll D, Gunn M. Insertion of a sequence encoding light chain 3 of microtubule-associated proteins 1A and 1B in a pestivirus genome: connection with virus cytopathogenicity and induction of lethal disease in cattle. J Virol. 1998;72:4139–4148. doi: 10.1128/jvi.72.5.4139-4148.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fricke J, Gunn M, Meyers G. A family of closely related bovine viral diarrhea virus recombinants identified in an animal suffering from mucosal disease: new insights into the development of a lethal disease in cattle. Virology. 2001;291:77–90. doi: 10.1006/viro.2001.1170. [DOI] [PubMed] [Google Scholar]
- 89.Fricke J, Voss C, Thumm M, Meyers G. Processing of a pestivirus protein by a cellular protease specific for light chain 3 of microtubule-associated proteins. J Virol. 2004;78:5900–5912. doi: 10.1128/JVI.78.11.5900-5912.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Carré-Mlouka A, Gaumer S, Gay P, Petitjean AM, Coulondre C, Dru P, Bras F, Dezélée S, Contamine D. Control of sigma virus multiplication by the ref(2)P gene of Drosophila melanogaster: an in vivo study of the PB1 domain of Ref(2)P. Genetics. 2007;176:409–419. doi: 10.1534/genetics.106.063826. [DOI] [PMC free article] [PubMed] [Google Scholar]