Abstract
A close ortholog to the cytochrome P450cam (CYP101A1) that catalyzes the same hydroxylation of camphor to 5-exo hydroxycamphor is CYP101D1. There are potentially important differences in and around the active site that could contribute to subtle functional differences. Adjacent to the heme iron ligand, Cys357, is Leu358 in P450cam while this residue is Ala in CYP101D1. Leu358 plays a role in binding of the P450cam redox partner, putidaredoxin (Pdx). On the opposite side of the heme about 15 – 20 Å away Asp251 in P450cam plays a critical role in a proton relay network required for O2 activation but forms strong ion pairs with Arg186 and Lys178. In CYP101D1 a Gly replaces Lys178. Thus, the local electrostatic environment and ion pairing is substantially different in CYP101D1. These sites have been systematically mutated in P450cam to the corresponding residues in CYP101D1 and the mutants analyzed by crystallography, kinetics, and UV/Vis spectroscopy. Individually the mutants have little effect on activity or structure but in combination there is a major drop in enzyme activity. This loss in activity is due the mutants being locked in the low-spin state which prevents electron transfer from the P450cam redox partner, Pdx. These studies illustrate the strong synergistic effects on well separated parts of the structure in controlling the equilibrium between the open (low-spin) and closed (high-spin) conformational states.
Cytochromes P450 represent the largest group of heme monooxygenases in biology with over 18,000 distinct P450s identified to date.1 The large number of P450 crystal structures now known make it clear that the overall P450 fold is very conservative. It thus has been possible to use bacterial P450s to establish basic structure-function relationships shared by all P450s. P450cam (CYP101A1) from Pseudomonas putida catalyzes the regio- and stereo-specific hydroxylation of camphor to 5-exo-hydroxy-camphor. It was the first P450 enzyme whose three dimensional structure was determined2 and much of what we currently know about the P450 super-family has been learned using this enzyme.3 More recently a close ortholog to P450cam, CYP101D1, has been characterized including crystal structures.4–6 CYP101D1 catalyzes exactly the same reaction as P450cam at about the same rate. A comparative structural analysis reveals that there are subtle but potentially important differences involving key residues known to be critical for activity in P450cam. Therefore, Nature has provided a natural variant of P450cam to help further probe the role of key active groups.
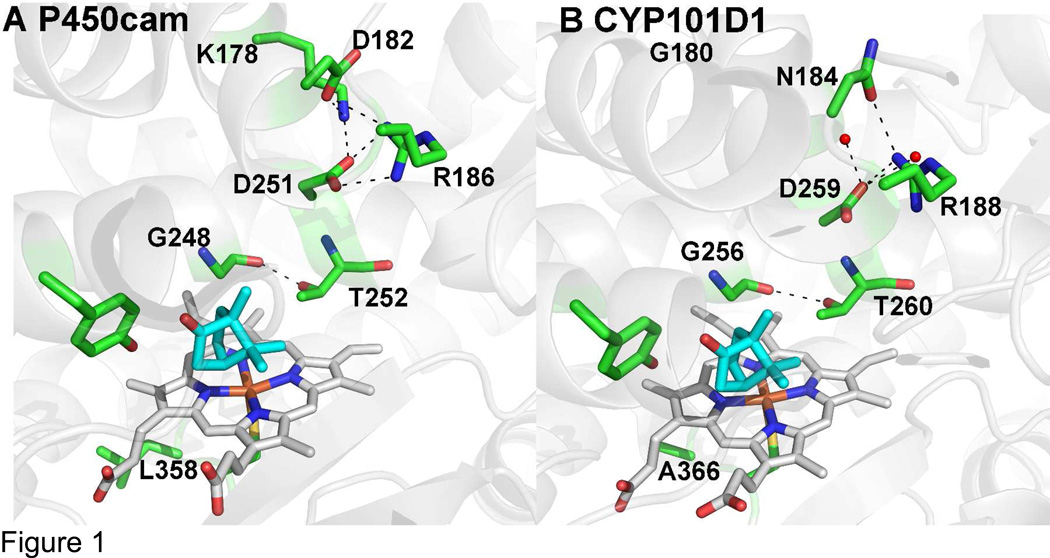
Here we focus on two important differences. Directly adjacent to the thiolate ligand, P450cam has Leu358 while CYP101D1 has Ala366 (Fig. 1). This region is substantially perturbed when the P450cam Fe2S2 electron donor redox partner, putidaredoxin or Pdx, binds and Leu358 moves closer to the heme.7 Changing Leu358 to Pro mimics the effects of Pdx binding since the larger Pro side chain “pushes” on the heme similar to what happens when Pdx binds helping to promote structural changes in the distal pocket required for O2 activation.8,9 These changes are important for the well-known “effector” role of Pdx. That is, P450cam strictly requires Pdx for reduction of oxy- P450cam10 by promoting a structural switch from the closed to the open conformational state.7 This proximal “push” effect is likely to be less important or at least quite different in CYP101D1 owing to the smaller Ala366 at this position. A second key region is Asp251. Asp251 plays a critical role in the delivery of protons to dioxygen, which is required for heterolytic cleavage of the O-O bond.11,12 Asp251 is tied up in strong ion pairs with Arg186 and Lys178, which initially presented problems with how Asp 251 could serve in a proton relay network. In order to serve such a function the Asp251 salt bridges first must be broken and the energetic barrier to rupture these ion pairs is quite high.13 However, it now has been shown that Pdx favors binding the open form of P450cam wherein these salt bridges are broken thereby freeing Asp251 to serve its proposed catalytic function.7 At least part of the effector role of Pdx is to open up the active site to enable a solvent and Asp251 mediated proton relay network to form. The environment around the homologous Asp259 is quite different in CYP101D1. Lys178 in P450cam is replaced by Gly180 in CYP101D1 while Asp182 is replaced by Asn184. To probe the importance of these differences we have systematically converted these residues in P450cam to the corresponding residues in CYP101D1 and have analyzed crystal structures, enzyme activity, electron transfer, and kinetic isotope effects.
Figure 1.
Active site models of P450cam (panel A) and CYP10D1 (panel B, PDB code 3LXI).6 In CYP101D1 Lys178 is replaced by a Gly thus leaving room for two water molecules (small red spheres) that H-bond with Asp259.
Experimental procedures
Protein expression and purification
Wild-type and mutant P450cam enzymes along with their redox partners putidaredoxin reductase (Pdr) and putidaredoxin (Pdx) were overexpressed in E. coli and purified as described previously.14–16
Spectroscopic Studies
All UV-visible spectroscopy was performed using a Cary 3 spectrophotometer. P450 content was measured by a reduced CO-difference spectrum using extinction coefficient of 91 mM−1 cm−1 at 450 nm.17 Concentrations of Pdx and Pdr were calculated using extinction coefficients of 5.9 mM−1 cm−1 at 455 nm and 11.0 mM−1 cm−1 at 412 nm, respectively.18,19
Enzyme Assays
Cytochrome P450cam hydroxylation activity was determined in the complete system of three proteins (P450cam, Pdr, Pdx) by measuring rates of camphor-dependent NADH oxidation at 25 °C following previously established protocols.20 Briefly, the reaction mixture of 1.2 mL contained 0.5 µM Pdr, 5 µM Pdx, and 0.5 µM P450cam in 50 mM potassium phosphate buffer, pH 7.4. The rate of NADH oxidation was measured by monitoring the absorbance change at 340 nm using 6.22 mM−1 cm−1. The reaction was initiated by first adding NADH (200 µM final concentration) following which substrate-dependent NADH oxidation was assayed in the presence of 200 µM camphor and was calculated as the difference between the measured rate and the rate of nonspecific NADH oxidation in the absence of camphor. After the completion of the reaction (monitored by UV-Vis), the reaction mixture was removed and organic extraction with dichloromethylene was performed for product formation analysis using GC-MS according to previously established protocol.21 For the kinetic solvent isotope effects, all stock solutions were prepared in 99.9% D2O (Cambridge isotope Laboratories, Inc) and pH meter reading was adjusted by 0.4 units for buffers in D2O. Stock solutions of the proteins were equilibrated prior to each experiment in D2O buffer mixtures before measuring enzyme activities.
Stopped flow
Transfer of the first electron from reduced Pdx to P450cam was measured in carbon monoxide saturated 50 mM phosphate buffer, pH 7.4, containing 3 mM camphor, and glucose oxidase oxygen-scrubbing system, using an SX.18MV stopped-flow spectrophotometer (Applied Photophysics) according to previously published protocols.22 Sample of 1.5 µM of wild type or mutant P450cam enzyme was mixed with excess of Pdx to measure the maximum rate of electron transfer. Reactions were performed with 50-, 70- and 80-fold excess Pdx and did not show any significant difference in rates indicating maximum rates were measured. Formation of the ferrous-CO form of P450cam was monitored at 446 nm at 25 °C.
Crystallization of P450cam mutants
Crystals of the ferric camphor-bound P450cam mutants were grown at room temperature using the hanging drop vapor diffusion method in 50 mM Tris-HCl buffer, pH 7.4, 400 mM KCl, 32% polyethylene glycol 4,000, and 1.2 mM D-camphor as the reservoir solution. The initial droplets containing 2 µL of protein solution at a concentration of 35 mg/mL, and 2 µL of the reservoir solution was equilibrated against a 500 µL of the reservoir solution. To generate the cyanide complex, the ferric camphor-bound crystals were soaked for 10–15 min at room temperature in ~50 mM potassium cyanide in the mother liquor solution and then flash cooled in liquid nitrogen. The brown ferric camphor-bound crystals turned red upon cyanide binding.
All data were collected either in-house using a Rigaku 007HF rotating anode generator with a Saturn 944+ CCD detector or remotely using the Stanford Synchrotron Radiation Lightsource (SSRL) beamline 7-1 as indicated in Table I. Data were indexed, integrated, and scaled with HKL2000.23 Molecular replacement calculations were carried out with Phaser24 through the CCP4i graphic interface25 using ferric camphor-bound wild type P450cam (PDB code 2CPP) as a search model. Further structure refinement were performed by using Phenix.refine.26 Table I lists data collection and refinement statistics.
Table I.
Crystallographic data collection and refinement statistics
| Data set | L358A | CN L358A |
L358A/ K178G |
CN L358A/ K178G |
L358A/ K178G/ D182N |
CN L358A/ K178G/ D182N |
L358P/ K178G |
CN L358P/ K178G |
|---|---|---|---|---|---|---|---|---|
| Data collection | ||||||||
| Space group | P212121 | P212121 | P212121 | P212121 | P212121 | P212121 | P21 | P21 |
| Resolution (Å) | 2.14 | 2.13 | 2.142 | 1.26 | 2.10 | 1.30 | 2.20 | 1.55 |
| Radiation source | Rigaku Saturn |
Rigaku Saturn |
Rigaku Saturn |
SSRL 9-2 | Rigaku Saturn |
SSRL 9-2 | Rigaku Saturn |
SSRL 9-2 |
| Wavelength (Å) | 1.54 | 1.54 | 1.54 | 1.00 | 1.54 | 1.00 | 1.54 | 1.00 |
| Completeness (%) | 90 (89) | 95(85) | 97 (86) | 100(100) | 92 (80) | 95 (94) | 93(68) | 100(100) |
| No of unique reflections |
18,098 | 22,593 | 23,092 | 7895,488 | 22,278 | 95,488 | 37,084 | 53,044 |
| Redundancy | 2.7(2.2) | 3.7(2.5) | 2.7(1.8) | 4.2 (3.8) | 5.1 (2.6) | 3.6 (3.4) | 2.0(1.4) | 3.7(3.6) |
| Rsym or Rmerge | 0.054 | 0.038 | 0.045 | 0.048 | 0.065 | 0.057 | 0.054 | 0.076 |
| (0.07) | (0.07) | (0.13) | (0.64) | (0.22) | (0.67) | (0.16) | (0.43) | |
| I/σ(I) | 34(12) | 30(12) | 33(14) | 39(2) | 34(12) | 35(2) | 11(3.4) | 5.6(2.1) |
| Refinement | ||||||||
| Resolution (Å) | 2.12 | 2.10 | 2.10 | 1.26 | 2.09 | 1.29 | 2.20 | 1.55 |
| B factor (mean) (Å2) |
21.91 | 19.18 | 24.60 | 21.69 | 25.43 | 22.94 | 24.78 | 24.27 |
| Rwork | 0.192 | 0.173 | 0.170 | 0.1920 | 0.176 | 0.195 | 0.170 | 0.173 |
| Rfree | 0.255 | 0.2430 | 0.231 | 0.2150 | 0.235 | 0.215 | 0.243 | 0.212 |
| r.m.s.d bonds (Å) | 0.008 | 0.010 | 0.008 | 0.008 | 0.008 | 0.008 | 0.008 | 0.008 |
| r.m.s.d angles (°) | 1.217 | 1.16 | 1.14 | 1.21 | 1.155 | 1.196 | 1.23 | 1.23 |
| No of atoms | ||||||||
| Protein | 3204 | 3204 | 3199 | 3199 | 3206 | 3199 | 6402 | 3201 |
| Ligand/Ions | 54 | 57 | 55 | 57 | 55 | 57 | 109 | 58 |
| Water | 190 | 415 | 349 | 526 | 294 | 511 | 550 | 447 |
| PDB ID | 4L49 | 4L4D | 4L4A | 4L4E | 4L4B | 4L4F | 4L4C | 4L4G |
Results
Crystal structures
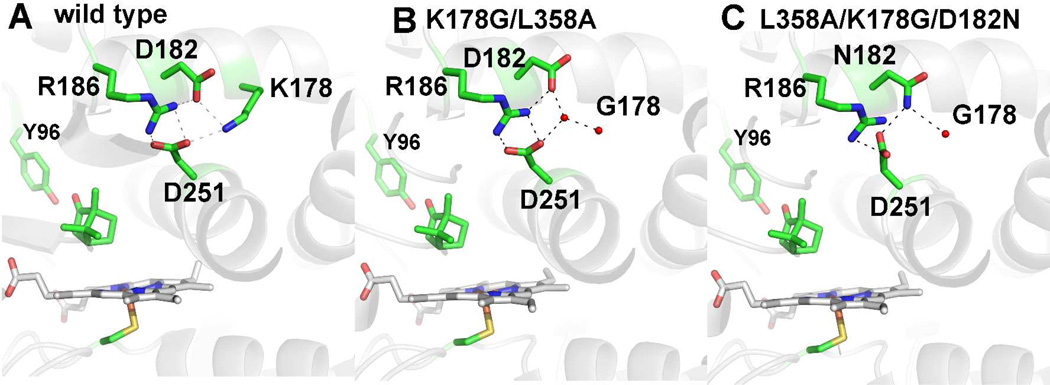
The ferric camphor-bound structures of P450cam are shown in Fig. 2. The L358A mutant is very similar to the wild type enzyme. In L358A/K178G two water molecules replace the missing lysine side chain. Asp182 shifts ~ 1.3 Å away from Asp251 and now H-bonds with one of the water molecules that also H-bonds with side chain of Asp251. When the D182N mutation is added on top of the L358A/K178G mutation to generate the triple mutant, one H-bond with Arg186 is lost and the side chain of Asn182 moves back to its original position where it forms an H-bond with one of the new water molecules substituting for Lys178 and also with the side chain of Asp251 that now adopts a slightly different orientation compared to the WT enzyme. This weakens the salt bridge interactions between the Asp251 with Arg186. In none of the mutants does the peptide of Asp251 display a rotation as seen in CYP101D1 and the H-bond between Gly248 and the side chain of Thr252 remains tight.
Figure 2.
Structures of P450cam wild type (panel A) and the K178G/L358A (panel B), and L358A/K278G/D182N (panel C) mutants. Similar to CYP101D1, replacing Lys178 with Gly leaves room for two water molecules that fill the space left by the missing Lys178 side chain. In the triple mutant where Asp182 is replaced by Asn there is little change other than one less ordered water molecule in the space left by the missing Lys178 side chain. The Asp 251 side adopts a different orientation weakening the H bond with R186
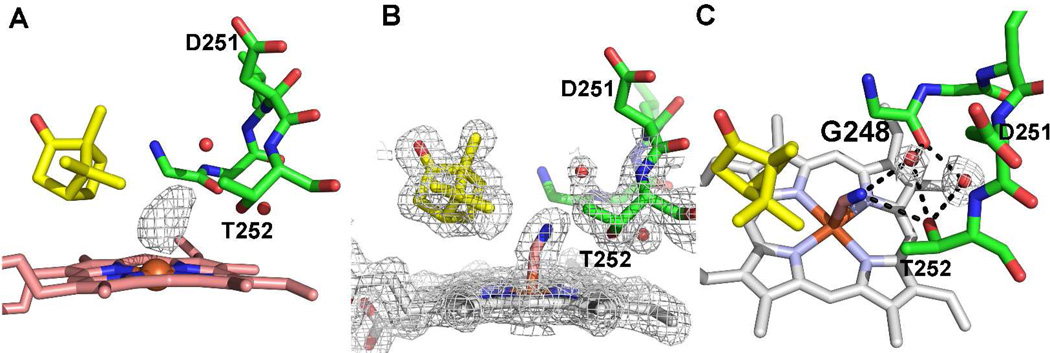
We next solved the structures of the cyanide complexes since it has been shown that cyanide is an excellent mimic for the O2 complex. When O2 binds the I helix opens up enabling key water molecules to enter the active site that form part of the proton relay network required for O2 activation.27–29 CN− coordination to the Fe(III) iron induces the same structural changes in the I helix as does O2 binding to the Fe(II) iron but has the great advantage of being easier to prepare for crystallography.30 In all of the mutants electron density for bound cyanide is clearly observed (Fig. 3). Similar to the wild-type cyanide complex, the cyanide molecule points toward Thr252, which rotates to form a hydrogen bond with the distal nitrogen atom of CN−. This further results in a widening of the groove formed by the stretch of residues encompassing Gly248 to Thr252. Overall, the cyanide complex of all the mutants shows the same conformational change that is observed in the wild type enzyme. This involves (a) breaking of the Thr252-Gly248 H-bond (b) flipping of the Asp251 peptide, and (c) opening of the I helix to accommodate two new water molecules, each H bonded to either Thr252 and Gly248. Since the number and position of the catalytic waters in the active site are the same as the wild type enzyme, we conclude that none of the mutations alter the structure of the proton delivery machinery in the active site. The differences in the orientation of the side chain of Asp251 and Asp182 that were observed in the ferric camphor-bound structure of the mutants as described earlier were maintained in the cyanide structures as well.
Figure 3.
A) Omit Fo-Fc electron density map contoured at 3.0 σ showing the CN− electron density. B) Final refined 2Fo-Fc electron density map contoured at 1.0 σ of triple mutant L358A/K178G/D182N cyanide complex. C) The same triple mutant highlighting the H-bonding network. CN− induces the same change in the mutant as in wild type. The Thr252-Gly248 H-bond breaks which widens the I helix groove enabling new waters to enter the active site. Exactly the same change occurs when O2 binds and is considered important to enabling the new waters to participate in a proton relay network required for activity.
Steady state assays and Spectroscopy
NADH oxidation assays were carried out with the wild type and the mutant P450 enzymes as described in the experimental procedures. After the reaction was complete, dichloromethylene was added to extract the product, which was then subjected to GC-MS to measure substrate hydroxylation or actual product formation. All the mutants showed greater than 95% coupling indicating that electrons from NADH are used for actual substrate hydroxylation and not for the wasteful reduction of O2 to peroxide and/or water. The results of the activity assays are shown in Table II. L358A did not have any major effect on activity while K178G shows a modest decrease. However, when present together in L358A/K178G, the mutant enzyme showed a significant decrease in activity compared to the wild type enzyme indicating some synergistic effect of the two mutations. L358P/K178G exhibited a similar decrease in activity. The mutant L358A/K178G/D182N also showed significantly reduced activity compared to the wild type enzyme.
Table II.
NADH turnover rates and Kinetic Solvent Isotope Effects
| P450cam type | Rates at 0 mM KCl (in H2O)(min−1) |
KSIE at 0 mM KCl (in D2O) (min−1) |
Rates at 400 mM KCl (in H2O) (min−1) |
KSIE at 400 mM KCl (in D2O) (min−1) |
|---|---|---|---|---|
| WT | 930 ± 28 | 1.25 ± 0.12 | 230 ± 15 | 1.22 ± 0.09 |
| L358A | 725 ± 17 | 1.06 ± 0.09 | 255 ± 10 | 1.13 ± 0.11 |
| L358P | 610 ± 15 | 1.1 ± 0.10 | 195 ± 9 | 1.29 ± 0.09 |
| K178G | 395 ± 20 | 0.96 ± 0.08 | 206 ± 15 | 0.98 ± 0.07 |
| L358A/K178G | 94 ± 6 | 0.91 ± 0.05 | 165 ± 8 | 1.03 ± 0.06 |
| L358P/K178G | 140 ± 10 | 1.29 ± 0.11 | 130 ± 9 | 1.04 ± 0.09 |
| L358A/K178G/D182N | 150 ± 8 | 0.97 ± 0.09 | 180 ± 9 | 1.02 ± 0.11 |
All rates were measured with 50 mM potassium phosphate buffer at pH 7.4 at room temperature. Rates were measured either in the absence or presence of 400 mM KCl. Under same conditions kinetic solvent isotope effects were also measured and are represented as ratio of the rates in H2O/D2O.
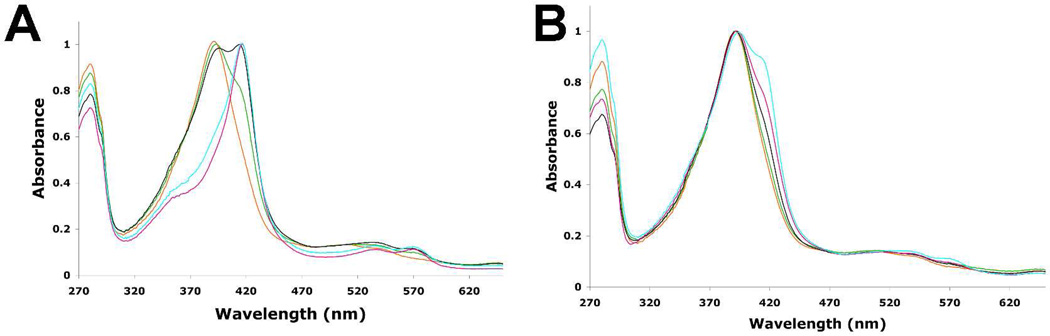
Normally the binding of camphor shifts the spin-state from low (Soret maximum near 417 nm) to high (Soret maximum near 392 nm) which also increases the redox potential.31 This change is required in order for electron transfer from Pdx and thus the low-spin form of P450cam is inactive. Spectroscopic characterization of the mutants in 50 mM potassium phosphate buffer with high concentrations of D-camphor show that mutants that exhibit a significant decrease in activity are mostly low-spin (Figure 4). This is consistent with a previous study on the K178Q mutant, which also did not give a complete spin shift.11 Therefore, the low activity of these mutants is most likely due to a dramatic slowing of the first electron transfer step from Pdx.
Figure 4.
Optical absorption spectra of the WT and mutant P450cam enzymes in 50 mM potassium phosphate buffer , pH 7.4 with 3 mM D-camphor in the absence (A) or presence (B) of 400 mM KCl. WT P450cam is shown in orange, mutants L358A (green), K178G (black) L358A/K178G (magenta) and L358A/K178G/D178N (cyan) are overlaid on the WT spectra.
To further test if the first electron transfer was indeed slowed down in the mutant enzymes stopped flow studies were carried out with the wild type and mutant L358A/K178G to measure the rate of first electron transfer from reduced Pdx (70–80 fold in excess) to ferric P450cam. The mutant L358/K178G was chosen since it showed the most significant reduction in rate in the steady state assays. The rate of reduction for the mutant enzyme (1.9 s−1) was 8–10 fold slower than that of the wild type enzyme thus confirming that reduction of the low-spin mutants is very slow.
The presence of low-spin heme in the mutant enzymes could be due either to the inability of camphor to bind or to the possibility that camphor binds but the water molecule at the sixth coordination site is not expelled. The crystal structures of the mutant enzymes clearly show camphor density but not water coordinated to the iron, so the crystals should be high-spin, which is inconsistent with low-spin spectra in solution. A major difference between the crystallization conditions and steady state assay or UV/Vis measurements was the presence of high concentrations of KCl (400 mM) in the crystallization reservoir solution, which was absent during enzyme assay or spectral studies. Several previous studies on P450cam have shown that K+ ions promote camphor binding and active site dehydration driving the heme toward forming 100% high spin complex upon camphor binding.32–34 Moreover, the crystal structures show that P450cam, but not CYP101D1, has a K+ site very near the key substrate contact residue, Tyr96. K+ thus is important for stabilizing the active site and promotes substrate binding. We therefore recorded spectra under conditions similar to those used in crystallization (400 mM KCl) and found that the mutants now are mostly high-spin (Figure 4B, lower panel). Additional experiments clearly showed that the mutants shift to high-spin as the K+ ion concentration increases and that the effect was specific for K+ ions only. We next measured the activity at high ionic strength (Table II). Although the activity for P450cam decreases with increasing ionic strength20,35 most likely due to poor Pdx binding, the difference in activity between the mutants and wild type at high ionic strength was very modest (Table II). This strongly suggests that the reason the mutants exhibit diminished activity at lower ionic strengths (or lower K+ concentration) is that the fraction of low-spin heme is too high. Since electron transfer to low-spin P450cam is either very slow or does not occur, the loss in activity is due to the inability of Pdx to reduce low-spin P450cam.
Kinetic Solvent isotope effects on NADH oxidation
Kinetic solvent isotope effects (KSIE) are useful probes for determining the presence of proton transfer in the rate-limiting step of enzymatic reactions. Previous studies on P450cam showed that D251N resulted in a drastic increase in the KSIE as compared to the wild type enzyme indicating that this mutation severely affected proton transfer steps.36 This hypothesis was supported by the structure of the oxy complex of this mutant, which showed the absence of catalytic waters in the active site and hence the lack of a proper proton delivery mechanism.37 In the present study, NADH turnover assays for wild type and mutant P450cam enzymes were performed in D2O in presence of either 0 mM or 400 mM KCl. KSIE for the wild type P450cam was in agreement with previous studies of P450cam.36,38,39 In our measurements (Table 2), none of the mutants showed any significant increase in the KSIE. The presence of 400 mM KCl did not alter the isotope effect measurements. These data agree with the structural data on the cyanide complex of the mutant P450 enzymes which show that the proton delivery machinery in the form of the catalytic waters remain intact in the active site.
Discussion
In this study we have probed two regions of P450cam that differ from its close ortholog, CYP101D1. Adjacent to the proximal Cys ligand P450cam has Leu358 while this residue is Ala in CYP101D1. Mutating Leu358 to either Ala or Pro has little effect on activity or structure. On the opposite side of the heme some 15 Å away from Leu358, the critically important Asp251 ion pairs with Arg186 and Lys178 but Lys178 is replaced with Gly in CYP101D1. Conversion of Lys178 to Gly also has little effect on activity or structure. However, mutating both sites results in a large drop in activity owing to the mutants being locked in the low-spin state and low-spin P450cam is not efficiently reduced by Pdx, thus activity decreases. Importantly, the P450cam L358A/K178A mutant mimics exactly the behavior of CYP101D1, with Ala and Gly at the positions corresponding to Leu358 and Lys178 in P450cam, respectively, as being predominantly low-spin5,6 even in the presence of excess camphor. The mutants thus did, indeed, convert P450cam into a more CYP101D1-like P450. The main difference is that CYP101D1 which is about 70% low-spin even in the presence of excess camphor is about as active as fully high-spin P450cam while low-spin P450cam is inactive. The additional complexity with P450cam is that excess K+ can shift the low-activity mutants to the high-spin form and this effect is only marginal in CYP101D1.6 Crystal structures of the CN− complexes in high K+ show the same structural changes as in wild type P450cam including positioning of the key water molecules required for O2 activation. These structures together with our kinetic studies show that the proton relay network is not altered in these mutants.
It is important to note that being predominantly low-spin also means these mutants favor the in the open conformation which differs substantially from the closed high-spin state owing to large movements of the F and G helices which effectively exposes the active site to bulk solvent.40 It is possible that water could coordinate to the heme iron, camphor remains bound, and the active site does not adopt the totally open conformation but, instead, some intermediate state. Indeed, it has been possible to capture intermediate states using substrate analogs with long tethering groups that extend out of the active site.41 However, in the absence of such tethered substrates/ligands these intermediate states are likely to be poorly populated so that the two most stable forms, totally open or closed, dominate. What then requires an explanation is why these mutants are locked in the low-spin open state.
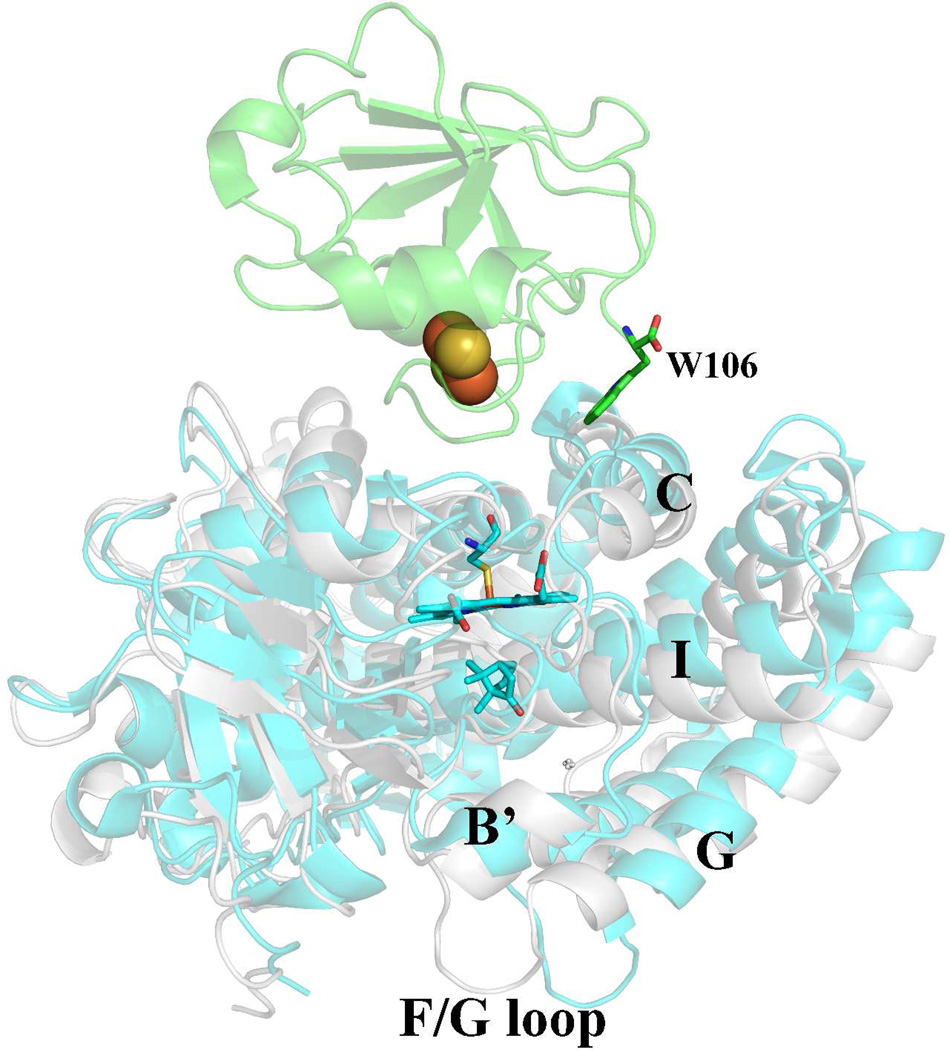
The P450cam-Pdx crystal structure7 provides a structural explanation. The structure shows that Pdx favors binding to the open form of P450cam. The switch between the open and closed forms is complex and involves a large part of the protein.40 Pdx binds very near Leu358 and this residue adopts a new rotamer conformation which moves the Leu side chain closer to the heme. The Pro side chain in the L358P mutant also is closer to the heme8 and thus partially mimics9 the “push” effect of Pdx binding. The P450cam C helix moves “up” (Fig. 5) to contact Trp106 in Pdx and this change is coupled to a movement of the I helix toward the conformation required for O2 activation as well as large movements in the F and G helices which expose the active site. Lys178 and Arg186 are on the F/G helical segments that change the most and hence, the Asp251 salt bridges with these residues are broken. This frees Asp251 to serve its catalytic function11,12 in shuttling protons to dioxygen. Thus, the changes near the proximal face of the heme are coupled to changes in the substrate access channel and especially the local environment of Asp251 ~15–20 Å away and so it is not surprising that mutations in regions that are involved in the open/close transition are synergistic and shift P450cam to the open low-spin state.
Figure 5.
Structure of the P450cam-Pdx complex (PDB code 4JX1).7 The C helix moves “up” to better interact with Trp106 in Pdx. This movement is coupled to a shift of P450cam from the closed (gray) to open (cyan) state. Shifting to the open state results in large movements of the F and G helices and F/G loop which breaks the Asp251 salt bridges. Asp251 thus is free to serve its catalytic function.
Overall these results illustrate the delicate balance between the open and closed conformational states as well as a strong structural connection between the Pdx binding site (Leu358) and the region around Asp251 that undergoes a large structural change in the open/closed transition. It appears that these complex interactions between spin-state, redox partner binding, and conformational changes are less stringent in CYP101D1 primarily because this P450 is predominantly low-spin even in the presence of substrate5 but still is as active as P450cam. This opens up some interesting questions on how two such similar P450s can behave so differently and what, if any, the connection is between these various levels of conformational dynamics, activity, and biological function.
Acknowledgments
We are grateful to Joumana Jamal for making the P450cam mutants L358A and L358A/K178G. We would also like to acknowledge Irina Sevrioukova and Sarvind Tripathi for helpful discussions. This paper involves research carried out at the Stanford Synchrotron Radiation Laboratory, a national user facility operated by Stanford University on behalf of the U.S. Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program, and the National Institute of General Medical Sciences.
Abbreviations
- CYP
Cytochrome P450
- Pdx
Putidaredoxin
- Pdr
Putidaredoxin reductase
- NADH
Nicotinamide adenine dinucleotide hydride
- GC-MS
Gas chromatography-mass spectrometry
- KSIE
Kinetic solvent isotope effect
References
- 1.Guengerich FP, Munro AW. Unusual Cytochrome P450 Enzymes and Reactions. J. Biol. Chem. 2013 doi: 10.1074/jbc.R113.462275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Poulos TL, Finzel BC, Gunsalus IC, Wagner GC, Kraut J. The 2.6-Å crystal structure of Pseudomonas putida cytochrome P-450. J. Biol. Chem. 1985;260:16122–16130. [PubMed] [Google Scholar]
- 3.Poulos TL. Structural biology of heme monooxygenases. Biochem. Biophys. Res. Commun. 2005;338:337–345. doi: 10.1016/j.bbrc.2005.07.204. [DOI] [PubMed] [Google Scholar]
- 4.Bell SG, Dale A, Rees NH, Wong L-L. A cytochrome P450 class I electron transfer system from Novosphingobium aromaticivorans. Appl. Microbiol. Biotechnol. 2010;86:163–175. doi: 10.1007/s00253-009-2234-y. [DOI] [PubMed] [Google Scholar]
- 5.Bell SG, Wong L-L. P450 enzymes from the bacterium Novosphingobium aromaticivorans. Biochrm. Biophys. Res. Comm. 2007;360:666–672. doi: 10.1016/j.bbrc.2007.06.119. [DOI] [PubMed] [Google Scholar]
- 6.Yang W, Bell SG, Wang H, Zhou W, Hoskins N, Dale A, Bartlam M, Wong LL, Rao Z. Molecular characterization of a class I P450 electron transfer system from Novosphingobium aromaticivorans DSM12444. J. Biol. Chem. 2010;285:27372–27384. doi: 10.1074/jbc.M110.118349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tripathi S, Li H, Poulos TL. Structural basis for effector control and redox partner recognition in cytochrome P450. Science. 2013 doi: 10.1126/science.1235797. in press. [DOI] [PubMed] [Google Scholar]
- 8.Nagano S, Tosha T, Ishimori K, Morishima I, Poulos TL. Crystal structure of the cytochrome P450cam mutant that exhibits the same spectral perturbations induced by putidaredoxin binding. J. Biol. Chem. 2004;279:42844–42849. doi: 10.1074/jbc.M404217200. [DOI] [PubMed] [Google Scholar]
- 9.Tosha T, Yoshioka S, Ishimori K, Morishima I. L358P mutation on cytochrome P450cam simulates structural changes upon putidaredoxin binding - The structural changes trigger electron transfer to oxy-P450cam from electron donors. J. Biol. Chem. 2004;279:42836–42843. doi: 10.1074/jbc.M404216200. [DOI] [PubMed] [Google Scholar]
- 10.Lipscomb JD, Sligar SG, Namtvedt MJ, Gunsalus IC. Autooxidation and hydroxylation reactions of oxygenated cytochrome P-450cam. J. Biol. Chem. 1976;251:1116–1124. [PubMed] [Google Scholar]
- 11.Gerber NC, Sligar SG. A role for Asp-251 in cytochrome P-450cam oxygen activation. J. Biol. Chem. 1994;269:4260–4266. [PubMed] [Google Scholar]
- 12.Gerber NS, Sligar SG. Catalytic mechanism of cytrochrome P450 - evidence for a distal charge relay system. J. Amer. Chem .Soc. 1992;174:725–735. [Google Scholar]
- 13.Lounnas V, Wade RC. Exceptionally stable salt bridges in cytochrome P450cam have functional roles. Biochemistry. 1997;36:5402–5417. doi: 10.1021/bi9622940. [DOI] [PubMed] [Google Scholar]
- 14.Sevrioukova I, Gracia C, Li H, Bhaskar B, Poulos TL. Crystal structure of putidaredoxin, the [2Fe-2S] component of the P450cam monooxygenase system from Pseudomonas putida. J. Molec. Biol. 2003;333:377–392. doi: 10.1016/j.jmb.2003.08.028. [DOI] [PubMed] [Google Scholar]
- 15.Sevrioukova I, Li H, Poulos TL. Crystal structure of putidaredoxin reductase from Pseudomonas putida, the final structural component of the P450cam monooxygenase system. J. Mol. Biol. 2004;236:889–902. doi: 10.1016/j.jmb.2003.12.067. [DOI] [PubMed] [Google Scholar]
- 16.Yoshioka S, Takahashi S, Ishimori K, Morishima I. Roles of the axial push effect in cytochrome P450cam studied with the site-directed mutagenesis at the heme proximal site. J. Inorg. Biochem. 2000;81:141–151. doi: 10.1016/s0162-0134(00)00097-0. [DOI] [PubMed] [Google Scholar]
- 17.Omura T, Sato R. The Carbon Monoxide-Binding Pigment of Liver MicrosomesIEvidence for Its Hemoprotein Nature. J. Biol. Chem. 1964;239:2370–2378. [PubMed] [Google Scholar]
- 18.Kuznetsov VY, Blair E, Farmer PJ, Poulos TL, Pifferitti A, Sevrioukova IF. The putidaredoxin reductase-putidaredoxin electron transfer complex: theoretical and experimental studies. J. Biol. Chem. 2005;280:16135–16142. doi: 10.1074/jbc.M500771200. [DOI] [PubMed] [Google Scholar]
- 19.Gunsalus IC, Wagner GC. Bacterial P-450cam methylene monooxygenase components: cytochrome m, putidaredoxin, and putidaredoxin reductase. Methods Enzymol. 1978;52:166–188. doi: 10.1016/s0076-6879(78)52019-3. [DOI] [PubMed] [Google Scholar]
- 20.Sevrioukova I, Hazzard JT, Tollin G, Poulos TL. Laser flash induced electron transfer in P450cam monoxygenase: putidaredoxin reductase-putidaredoxin interaction. Biochemistry. 2001;40:10592–10600. doi: 10.1021/bi010874d. [DOI] [PubMed] [Google Scholar]
- 21.Churbanova IY, Poulos TL, Sevrioukova IF. Production and characterization of a functional putidaredoxin reductase-putidaredoxin covalent complex. Biochemistry. 2010;49:58–67. doi: 10.1021/bi901872s. [DOI] [PubMed] [Google Scholar]
- 22.Kuznetsov VY, Poulos TL, Sevrioukova IF. Putidaredoxin-to-cytochrome P450cam electron transfer: Differences between the two reductive steps required for catalysis. Biochemistry. 2006;45:11934–11944. doi: 10.1021/bi0611154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Otwinowski Z, Minor W. Macromolecular Crystallography, Pt A. 1997;Vol. 276:307–326. [Google Scholar]
- 24.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AGW, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. Overview of the CCP4 suite and current developments. Acta Crystallogr. D. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schlichting I, Berendzen J, Chu K, Stock AM, Maves SA, Benson DE, Sweet RM, Ringe D, Petsko GA, Sligar SG. The catalytic pathway of cytochrome P450cam at atomic resolution. Science. 2000;287:1615–1622. doi: 10.1126/science.287.5458.1615. [DOI] [PubMed] [Google Scholar]
- 28.Denisov IG, Makris TM, Sligar SG, Schlichting I. Structure and chemistry of cytochrome P450. Chem Rev. 2005;105:2253–2277. doi: 10.1021/cr0307143. [DOI] [PubMed] [Google Scholar]
- 29.Nagano S, Poulos TL. Crystallographic study on the dioxygen complex of wild-type and mutant cytochrome P450cam - Implications for the dioxygen activation mechanism. J. Biol. Chem. 2005;280:31659–31663. doi: 10.1074/jbc.M505261200. [DOI] [PubMed] [Google Scholar]
- 30.Fedorov R, Ghosh DK, Schlichting I. Crystal structures of cyanide complexes of P450cam and the oxygenase domain of inducible nitric oxide synthase-structural models of the short-lived oxygen complexes. Arch Biochem Biophys. 2003;409:25–31. doi: 10.1016/s0003-9861(02)00555-6. [DOI] [PubMed] [Google Scholar]
- 31.Sligar SG. Coupling of spin, substrate, and redox equilibria in cytochrome P450. Biochemistry. 1976;15:5399–5406. doi: 10.1021/bi00669a029. [DOI] [PubMed] [Google Scholar]
- 32.Di Primo C, Hui Bon Hoa G, Douzou P, Sligar S. Mutagenesis of a single hydrogen bond in cytochrome P-450 alters cation binding and heme solvation. J. Biol. Chem. 1990;265:5361–5363. [PubMed] [Google Scholar]
- 33.Westlake AC, Harford-Cross CF, Donovan J, Wong LL. Mutations of glutamate-84 at the putative potassium-binding site affect camphor binding and oxidation by cytochrome p450cam. Eur. J. Biochem. 1999;265:929–935. doi: 10.1046/j.1432-1327.1999.00793.x. [DOI] [PubMed] [Google Scholar]
- 34.Deprez E, Gill E, Helms V, Wade RC, Hui Bon Hoa G. Specific and non-specific effects of potassium cations on substrate-protein interactions in cytochromes P450cam and P450lin. J. Inorg. Biochem. 2002;91:597–606. doi: 10.1016/s0162-0134(02)00467-1. [DOI] [PubMed] [Google Scholar]
- 35.Unno M, Shimada H, Toba Y, Makino R, Ishimura Y. Role of Arg112 of cytochrome p450cam in the electron transfer from reduced putidaredoxin. Analyses with site-directed mutants. J. Biol. Chem. 1996;271:17869–17874. doi: 10.1074/jbc.271.30.17869. [DOI] [PubMed] [Google Scholar]
- 36.Vidakovic M, Sligar SG, Li H, Poulos TL. Understanding the role of the essential Asp251 in cytochrome p450cam using site-directed mutagenesis, crystallography, and kinetic solvent isotope effect. Biochemistry. 1998;37:9211–9219. doi: 10.1021/bi980189f. [DOI] [PubMed] [Google Scholar]
- 37.Nagano S, Cupp-Vickery JR, Poulos TL. Crystal structures of the ferrous dioxygen complex of wild-type cytochrome P450eryF and its mutants, A245S and A245T - Investigation of the proton transfer system in P450eryF. J. Biol. Chem. 2005;280:22102–22107. doi: 10.1074/jbc.M501732200. [DOI] [PubMed] [Google Scholar]
- 38.Purdy MM, Koo LS, de Montellano PR, Klinman JP. Mechanism of O2 activation by cytochrome P450cam studied by isotope effects and transient state kinetics. Biochemistry. 2006;45:15793–15806. doi: 10.1021/bi061726w. [DOI] [PubMed] [Google Scholar]
- 39.Makris TM, von Koenig K, Schlichting I, Sligar SG. Alteration of P450 distal pocket solvent leads to impaired proton delivery and changes in heme geometry. Biochemistry. 2007;46:14129–14140. doi: 10.1021/bi7013695. [DOI] [PubMed] [Google Scholar]
- 40.Lee YT, Wilson RF, Rupniewski I, Goodin DB. P450cam visits an open conformation in the absence of substrate. Biochemistry. 2010;49:3412–3419. doi: 10.1021/bi100183g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee YT, Glazer EC, Wilson RF, Stout CD, Goodin DB. Three clusters of conformational states in p450cam reveal a multistep pathway for closing of the substrate access channel. Biochemistry. 2011;50:693–703. doi: 10.1021/bi101726d. [DOI] [PMC free article] [PubMed] [Google Scholar]