Abstract
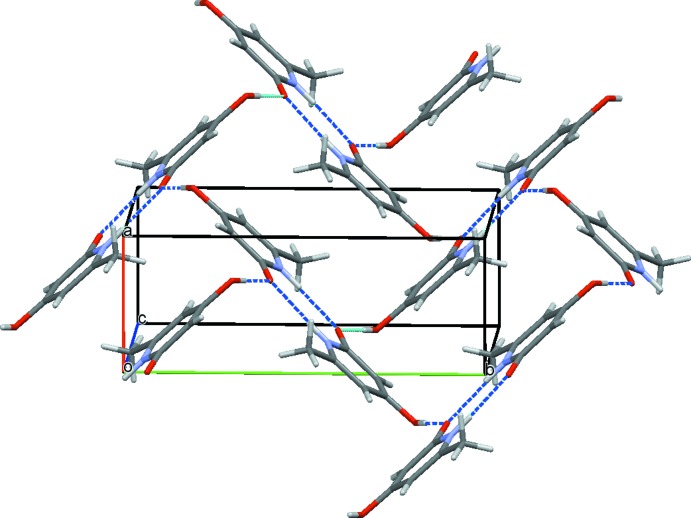
In the crystal structure of the title compound, C6H7NO2, N—H⋯O and O—H⋯O hydrogen bonds link the molecules, forming a zigzag array along [001] and a layer structure parallel to the ab plane.
Related literature
For the potential of related compounds in anti-HIV treatment, see: De Clercq (2005 ▶); Dollé et al. (1995 ▶); Medina-Franco et al. (2007 ▶).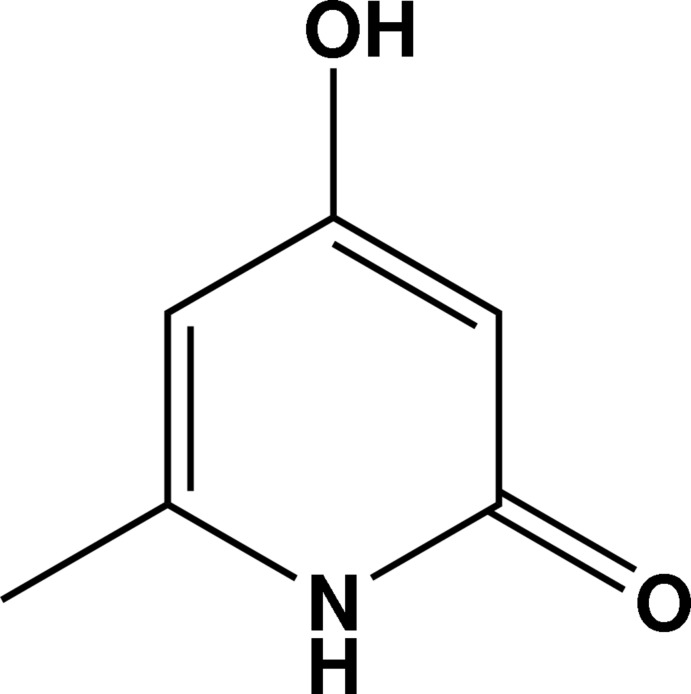
Experimental
Crystal data
C6H7NO2
M r = 125.13
Monoclinic,
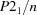
a = 4.7082 (5) Å
b = 12.2988 (8) Å
c = 10.0418 (7) Å
β = 91.303 (7)°
V = 581.32 (8) Å3
Z = 4
Mo Kα radiation
μ = 0.11 mm−1
T = 298 K
0.65 × 0.20 × 0.18 mm
Data collection
Bruker P4 diffractometer
Absorption correction: ψ scan (XSCANS; Siemens, 1996 ▶) T min = 0.216, T max = 0.259
2445 measured reflections
1701 independent reflections
1269 reflections with I > 2σ(I)
R int = 0.026
3 standard reflections every 97 reflections intensity decay: 9.4%
Refinement
R[F 2 > 2σ(F 2)] = 0.050
wR(F 2) = 0.160
S = 1.06
1701 reflections
82 parameters
H-atom parameters constrained
Δρmax = 0.32 e Å−3
Δρmin = −0.25 e Å−3
Data collection: XSCANS (Siemens, 1996 ▶); cell refinement: XSCANS; data reduction: XSCANS; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: SHELXTL (Sheldrick, 2008 ▶); software used to prepare material for publication: SHELXTL.
Supplementary Material
Crystal structure: contains datablock(s) I. DOI: 10.1107/S1600536813024240/im2437sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536813024240/im2437Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536813024240/im2437Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H1A⋯O1i | 0.86 | 1.98 | 2.835 (2) | 175 |
| O2—H2B⋯O1ii | 0.82 | 1.79 | 2.609 (2) | 180 |
Symmetry codes: (i) 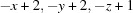 ; (ii)
; (ii) 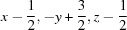 .
.
Acknowledgments
We gratefully acknowledge support for this project by the Dirección General de Educación Superior Tecnológica (DGEST grants 2535.09P and 3604.10-P).
supplementary crystallographic information
1. Comment
The acquired immunodeficiency syndrome (AIDS) is a disease of people who are infected with human immunodeficiency virus (HIV). The use of drugs to fight HIV are called antiretroviral drugs and are characterized by inhibiting essential enzymes for virus replication, such as reverse transcriptase (De Clercq, 2005).
Pyridin-2 (1H)-one hybrids are a kind of compounds that inhibit the reverse transcription process and have been shown, by molecular modeling, their good performance as non-nucleoside inhibitors of HIV-1 reverse transcriptase. Recent design of the pyridin-2(1H)-one hybrids has generated active molecules against wild type and mutant strains of HIV, as in the case of second-generation hybrid pyridinone-UC781 (Medina-Franco et al.., 2007). In this work, and as part of our ongoing research, we have synthesized pyridin-2 (1H)-one hybrids (Dollé et al., 1995) of second generation with different polar groups at C-3 and also with different olefinic groups at C-4, similar to pyridinone-UC781. The compound 4-hydroxy-6-methylpyridin-2 (1H)-one is an intermediate in the synthesis of second-generation hybrids with a polar nitro group at C-3.
We have synthesized the title compound (I) and report its crystal structure here (Fig. 1). In the crystal structure adjacent networks are linked together via intermolecular hydrogen bond interactions (table 1) [N1—H1A···O1i (2.8349 Å), symmetry codes: (i) –x + 2, –y + 2, - z + 1] and [O2—H2B···O1ii (2.6086 Å), symmetry codes: (ii) x – 1/2, - y + 3/2, z – 1/2] to form a zigzag array along the [001] direction and molecules are forming a layer structure parallel to the ab plane (Fig. 2).
2. Experimental
The synthesis of 4-hydroxy-6-methylpyridin-2(1H)-one includes reagents and reagent grade solvents, which were used without further purification. In a round bottom flask of 500 ml equipped with a magnetic stirrer was placed 10.0 g of ethyl 4-hydroxy-6-methyl-2-oxo-1,2-dihydropyridine-3-carboxylate (0.05 mol) in 350 ml of hydrochloric acid 1 N. The mixture was stirred at reflux for 72 h. 4-Hydroxy-6-methylpyridin-2(1H)-one precipitated as a white solid (6.2 g, 99%, m. p. 273–275 °C, for analytical data, see _exptl_special_details section). Crystals of the title compound suitable for Xray diffraction were obtained by dissolving 100 mg of 4-hydroxy-6-methyl-pyridine-2 (1H)-one in 10 ml of methanol-diethylether (1:1, v / v) and placing the solution in a glass vial. The solution was allowed to stand at room temperature for 7 days and the crystals formed were filtered.
3. Refinement
All H atoms were positioned geometrically and refined using a riding model, with C—H = 0.93 Å for aryl and 0.96 Å for methyl H atoms. Isotropic thermal parameters were fixed to Uiso(H) = 1.2 Ueq(C) for aryl and Uiso(H) = 1.5 Ueq(C) for methyl H atoms.
Figures
Fig. 1.
Molecular structure of the title compound with displacement ellipsoids drawn at the 30% probability level.
Fig. 2.
Hydrogen-bond (indicated as dashed lines) network of the title compound leading to a two dimensional network along the ab plane.
Crystal data
| C6H7NO2 | F(000) = 264 |
| Mr = 125.13 | Dx = 1.430 Mg m−3 |
| Monoclinic, P21/n | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2yn | Cell parameters from 51 reflections |
| a = 4.7082 (5) Å | θ = 6.6–12.3° |
| b = 12.2988 (8) Å | µ = 0.11 mm−1 |
| c = 10.0418 (7) Å | T = 298 K |
| β = 91.303 (7)° | Prismatic, colorless |
| V = 581.32 (8) Å3 | 0.65 × 0.20 × 0.18 mm |
| Z = 4 |
Data collection
| Bruker P4 diffractometer | 1269 reflections with I > 2σ(I) |
| Radiation source: fine-focus sealed tube | Rint = 0.026 |
| Graphite monochromator | θmax = 30.0°, θmin = 2.6° |
| 2θ/ω scans | h = −1→6 |
| Absorption correction: ψ scan (XSCANS; Siemens, 1996) | k = −1→17 |
| Tmin = 0.216, Tmax = 0.259 | l = −14→14 |
| 2445 measured reflections | 3 standard reflections every 97 reflections |
| 1701 independent reflections | intensity decay: 9.4% |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.050 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.160 | H-atom parameters constrained |
| S = 1.06 | w = 1/[σ2(Fo2) + (0.1P)2] where P = (Fo2 + 2Fc2)/3 |
| 1701 reflections | (Δ/σ)max < 0.001 |
| 82 parameters | Δρmax = 0.32 e Å−3 |
| 0 restraints | Δρmin = −0.25 e Å−3 |
Special details
| Experimental. IR: 3296, 3094, 2891, 1640 cm^-1. 1Ĥ NMR (CDCl~3~): δ 10.99 (s, NH-1), 10.40 (s, OH), 5.59 (s, H-3), 5.34 (s, H-5) 2.07 (s, 3H, CH~3~—C-6). ^13Ĉ NMR (CDCl~3~): δ 167.6, 164.8, 145.9, 98.2, 95.7, 18.5. MS m/e (int. rel): [M]^+^ 125 (100), 97 (16). |
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| O1 | 0.7844 (3) | 0.91089 (9) | 0.60053 (9) | 0.0364 (3) | |
| O2 | 0.2045 (2) | 0.68616 (10) | 0.32761 (10) | 0.0396 (3) | |
| H2B | 0.2292 | 0.6556 | 0.2562 | 0.059* | |
| N1 | 0.7786 (3) | 0.93124 (10) | 0.37596 (11) | 0.0301 (3) | |
| H1A | 0.9049 | 0.9813 | 0.3858 | 0.036* | |
| C1 | 0.6811 (3) | 0.88072 (12) | 0.48782 (12) | 0.0288 (3) | |
| C2 | 0.4786 (3) | 0.79830 (13) | 0.46911 (13) | 0.0326 (3) | |
| H2A | 0.4026 | 0.7638 | 0.5426 | 0.039* | |
| C3 | 0.3912 (3) | 0.76793 (12) | 0.34229 (13) | 0.0303 (3) | |
| C4 | 0.4958 (3) | 0.82420 (12) | 0.23072 (14) | 0.0321 (3) | |
| H4A | 0.4353 | 0.8053 | 0.1450 | 0.038* | |
| C5 | 0.6858 (3) | 0.90624 (12) | 0.25012 (13) | 0.0293 (3) | |
| C6 | 0.8049 (4) | 0.97389 (15) | 0.14088 (15) | 0.0403 (4) | |
| H6D | 0.7267 | 0.9501 | 0.0567 | 0.060* | |
| H6A | 1.0078 | 0.9662 | 0.1411 | 0.060* | |
| H6B | 0.7566 | 1.0488 | 0.1548 | 0.060* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0491 (7) | 0.0370 (6) | 0.0227 (5) | −0.0064 (5) | −0.0062 (4) | 0.0009 (4) |
| O2 | 0.0483 (7) | 0.0407 (6) | 0.0297 (5) | −0.0133 (5) | 0.0024 (5) | −0.0057 (4) |
| N1 | 0.0360 (6) | 0.0315 (6) | 0.0227 (5) | −0.0037 (5) | −0.0032 (5) | 0.0022 (4) |
| C1 | 0.0352 (7) | 0.0297 (7) | 0.0214 (6) | 0.0034 (6) | −0.0013 (5) | 0.0010 (5) |
| C2 | 0.0399 (8) | 0.0340 (7) | 0.0238 (6) | −0.0026 (6) | 0.0015 (5) | 0.0009 (5) |
| C3 | 0.0324 (7) | 0.0311 (7) | 0.0273 (6) | 0.0002 (6) | 0.0000 (5) | −0.0014 (5) |
| C4 | 0.0380 (8) | 0.0357 (8) | 0.0224 (6) | 0.0007 (6) | −0.0021 (5) | −0.0023 (5) |
| C5 | 0.0341 (7) | 0.0319 (7) | 0.0218 (6) | 0.0027 (6) | −0.0004 (5) | 0.0016 (5) |
| C6 | 0.0534 (10) | 0.0420 (9) | 0.0255 (6) | −0.0049 (8) | 0.0007 (6) | 0.0068 (6) |
Geometric parameters (Å, º)
| O1—C1 | 1.2768 (16) | C2—H2A | 0.9300 |
| O2—C3 | 1.3418 (18) | C3—C4 | 1.415 (2) |
| O2—H2B | 0.8200 | C4—C5 | 1.359 (2) |
| N1—C5 | 1.3629 (17) | C4—H4A | 0.9300 |
| N1—C1 | 1.3719 (17) | C5—C6 | 1.496 (2) |
| N1—H1A | 0.8600 | C6—H6D | 0.9600 |
| C1—C2 | 1.401 (2) | C6—H6A | 0.9600 |
| C2—C3 | 1.3809 (19) | C6—H6B | 0.9600 |
| C3—O2—H2B | 109.5 | C5—C4—C3 | 119.30 (13) |
| C5—N1—C1 | 123.42 (13) | C5—C4—H4A | 120.4 |
| C5—N1—H1A | 118.3 | C3—C4—H4A | 120.4 |
| C1—N1—H1A | 118.3 | C4—C5—N1 | 119.76 (13) |
| O1—C1—N1 | 117.79 (14) | C4—C5—C6 | 124.34 (13) |
| O1—C1—C2 | 124.99 (13) | N1—C5—C6 | 115.90 (14) |
| N1—C1—C2 | 117.21 (12) | C5—C6—H6D | 109.5 |
| C3—C2—C1 | 120.46 (13) | C5—C6—H6A | 109.5 |
| C3—C2—H2A | 119.8 | H6D—C6—H6A | 109.5 |
| C1—C2—H2A | 119.8 | C5—C6—H6B | 109.5 |
| O2—C3—C2 | 119.01 (13) | H6D—C6—H6B | 109.5 |
| O2—C3—C4 | 121.24 (12) | H6A—C6—H6B | 109.5 |
| C2—C3—C4 | 119.74 (14) | ||
| C5—N1—C1—O1 | 179.84 (14) | O2—C3—C4—C5 | −179.71 (13) |
| C5—N1—C1—C2 | 1.0 (2) | C2—C3—C4—C5 | 1.3 (2) |
| O1—C1—C2—C3 | −176.62 (15) | C3—C4—C5—N1 | 1.8 (2) |
| N1—C1—C2—C3 | 2.2 (2) | C3—C4—C5—C6 | −178.05 (15) |
| C1—C2—C3—O2 | 177.69 (13) | C1—N1—C5—C4 | −3.0 (2) |
| C1—C2—C3—C4 | −3.3 (2) | C1—N1—C5—C6 | 176.88 (14) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1A···O1i | 0.86 | 1.98 | 2.835 (2) | 175 |
| O2—H2B···O1ii | 0.82 | 1.79 | 2.609 (2) | 180 |
Symmetry codes: (i) −x+2, −y+2, −z+1; (ii) x−1/2, −y+3/2, z−1/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: IM2437).
References
- De Clercq, E. (2005). J. Med. Chem. 48, 1297–1313. [DOI] [PubMed]
- Dollé, V., Fan, E., Nguyen, C. H., Aubertin, A.-M., Kirn, A., Andreola, M. L., Jamieson, G., Tarrago-Litvak, L. & Bisagni, E. (1995). J. Med. Chem. 38, 4679–4686. [DOI] [PubMed]
- Medina-Franco, J. L., Martinez-Mayorga, K., Juárez-Gordiano, C. & Castillo, R. (2007). ChemMedChem, 2, 1141–1147. [DOI] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Siemens (1996). XSCANS Siemens Analytical X-ray Instruments Inc., Madison, Wisconsin, USA.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I. DOI: 10.1107/S1600536813024240/im2437sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536813024240/im2437Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536813024240/im2437Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report