Abstract
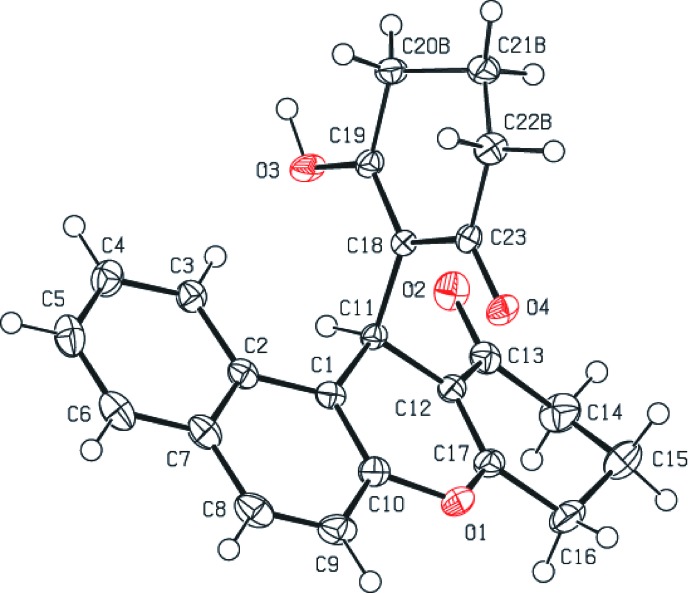
In the xanthenone system of the title compound, C23H20O4, the pyran ring has a maximum deviation of 0.111 (1) Å from planarity and the outer cyclohexene ring exhibits a puckered conformation. The three methylene C atoms of the cyclohexene ring bonded to the pyran unit are disordered over two sets of sites [occupancies = 0.570 (3) and 0.430 (3)]. In the crystal, molecules are linked by C—H⋯O and O—H⋯O hydrogen bonds, forming a two-dimensional network parallel to (110). A C—H⋯π interaction occurs between these networks.
Related literature
For related xanthenone structures, see: Li et al. (2004 ▶); Abdelhamid et al. (2011 ▶); Mohamed et al. (2011 ▶, 2012 ▶). Reddy et al. (2009 ▶); Çelik et al. (2009 ▶). For the industrial and pharmaceutical significance of xanthenes, see: Zare et al. (2012 ▶); Menchen et al. (2003a
▶,b
▶); Sarma & Baruah, (2005 ▶). For ring conformations, see: Cremer & Pople (1975 ▶) and for standard bond lengths, see: Allen et al. (1987 ▶).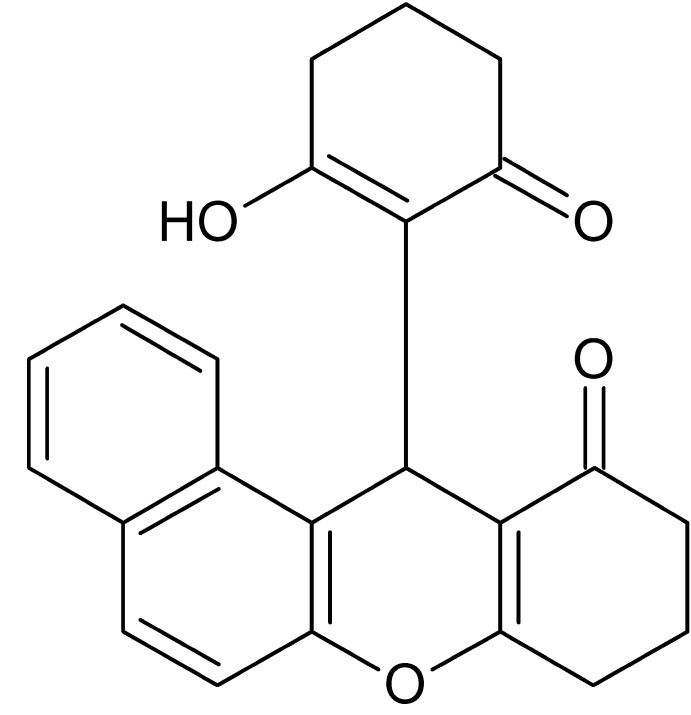
Experimental
Crystal data
C23H20O4
M r = 360.41
Orthorhombic,
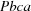
a = 14.2855 (15) Å
b = 13.7461 (12) Å
c = 18.400 (2) Å
V = 3613.2 (6) Å3
Z = 8
Mo Kα radiation
μ = 0.09 mm−1
T = 123 K
0.20 × 0.18 × 0.16 mm
Data collection
Oxford Diffraction Xcalibur, Eos diffractometer
Absorption correction: multi-scan (CrysAlis PRO; Oxford Diffraction, 2010 ▶) T min = 0.994, T max = 1.000
17944 measured reflections
4541 independent reflections
3366 reflections with I > 2σ(I)
R int = 0.042
Refinement
R[F 2 > 2σ(F 2)] = 0.051
wR(F 2) = 0.114
S = 1.04
4541 reflections
258 parameters
8 restraints
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.40 e Å−3
Δρmin = −0.25 e Å−3
Data collection: CrysAlis PRO (Oxford Diffraction, 2010 ▶); cell refinement: CrysAlis PRO; data reduction: CrysAlis PRO; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEP-3 for Windows (Farrugia, 2012 ▶); software used to prepare material for publication: WinGX (Farrugia, 2012 ▶) and PLATON (Spek, 2009 ▶).
Supplementary Material
Crystal structure: contains datablock(s) I. DOI: 10.1107/S1600536813025324/sj5351sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536813025324/sj5351Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536813025324/sj5351Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
Cg3 is the centroid of the C2–C7 benzene ring.
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O3—H3⋯O4i | 0.95 (2) | 1.64 (2) | 2.5793 (15) | 170 (2) |
| C3—H3A⋯O3 | 0.95 | 2.43 | 3.367 (2) | 168 |
| C9—H9⋯O2ii | 0.95 | 2.34 | 3.275 (2) | 170 |
| C14—H14B⋯Cg3iii | 0.99 | 2.85 | 3.750 (2) | 152 |
Symmetry codes: (i) 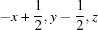 ; (ii)
; (ii) 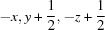 ; (iii)
; (iii) 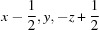 .
.
Acknowledgments
AAA thanks the Ministry of Higher Education in Egypt for a grant to support this collaborative project. Manchester Metropolitan University, Erciyes University and University of Strathclyde are gratefully acknowledged for facilitating this study.
supplementary crystallographic information
1. Comment
Xanthene derivatives have been used as antibacterial, antiviral, antitumor and anti-inflammatory agents (Zare et al., 2012). These compounds also have applications as dyes in laser technology (Menchen et al., 2003a,b), and as pH sensitive fluorescent materials for the visualization of biomolecules (Sarma & Baruah, 2005). Extending our previous studies of xanthenones (Abdelhamid et al., 2011; Mohamed et al., 2011, 2012), we report herein the synthesis and crystal study of a new of xanthenone derivative.
In the title compound shown in Fig. 1, the pyran ring (O1/C1/C10—C12/C17) has a maximum deviation of 0.111 (1) Å from planarity and the outer cyclohexene ring (C12–C17) of the xanthenone moiety is puckered with the puckering parameters (Cremer & Pople, 1975) of QT = 0.455 (2) Å, θ = 124.4 (2) and φ = 352.8 (3)°. The three methylene C atoms (C20/C21/C22) of the other cyclohexene ring attached to the pyran moiety at atom C11 are disordered over two sets of sites with a ratio of refined occupancies of 0.570 (3): 0.430 (3) and both components of the disordered cyclohexene ring are puckered [puckering parameters: QT = 0.445 (6) Å, θ = 48.7 (6) and φ = 183.8 (10)° for major component (C18/C19/C20B–C22B/C23), and QT = 0.471 (8) Å, θ = 130.2 (8) and φ = 353.3 (13)° for minor component (C18/C19/C20A–C22A/C23)].
The bond lengths in the title compound are within normal ranges (Allen et al., 1987) and are comparable those of similar compounds (Li et al., 2004; Abdelhamid et al., 2011; Çelik et al., 2009; Mohamed et al., 2011, 2012; Reddy et al., 2009).
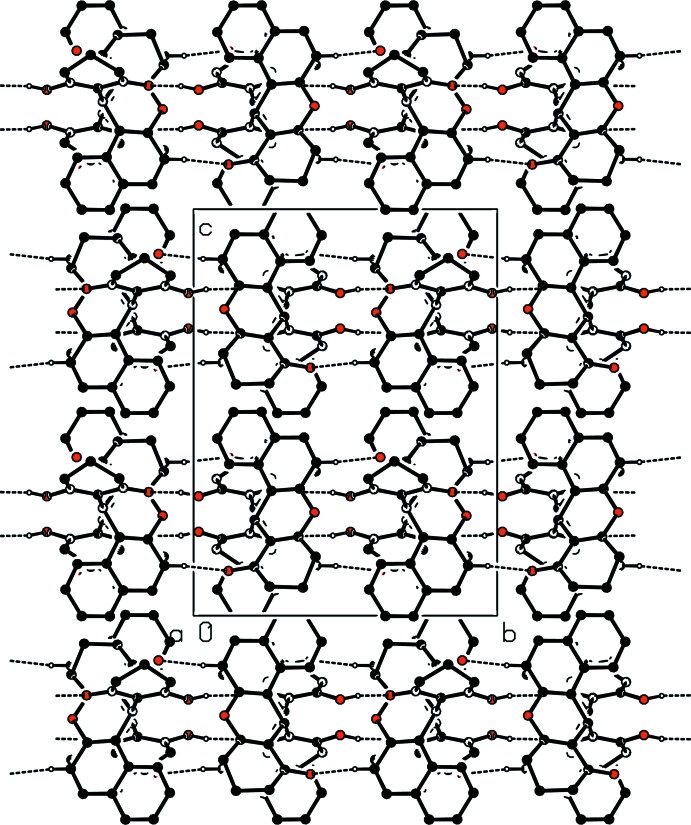
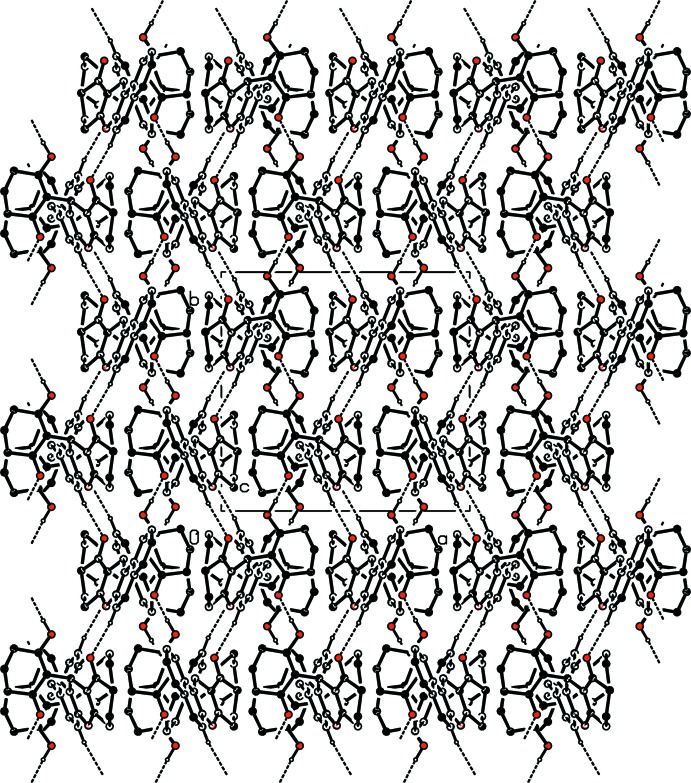
In the crystal structure, C—H···O and O—H···O hydrogen bonds link the neigbouring molecules (Table 1), forming two dimensional networks parallel to the ab-plane (Figs. 2 & 3). A C14—H14B···π interaction also exists between these planes.
2. Experimental
The title compound was obtained as the main product during a three component reaction of 1 mmol (206 mg) 4-nitro-2-(trifluoromethyl)aniline, 1 mmol (172 mg) 2-hydroxy-1-naphthaldehyde and 1 mmol (112 mg) 1,3-cyclohexandione in 50 ml ethanol. The reaction mixture was refluxed for 7 h at 351 K. On cooling, the resulting solid was collected, washed with cold ethanol and dried by filtration. The crude product was crystallized by the slow evaporation method over 24 h using ethanol as a solvent. M.p. = 517 K, yield = 95%.
3. Refinement
The hydroxyl H atoms were found from a difference Fourier map and refined freely. The C-bound H-atoms were refined using a riding model with C—H = 0.95 - 1.00 Å and Uĩso(H) = 1.2Ueq(C). The three methylene C atoms (C20/C21/C22) of the other cyclohexene ring bonded to the pyran moiety are disordered over two sets of sites with a ratio of refined occupancies of 0.570 (3): 0.430 (3) [in the refinement, DFIX and EADP instructions were used for the disordered atoms].
Figures
Fig. 1.
The structure of the title compound with the atom numbering scheme. Displacement ellipsoids for non-H atoms are drawn at the 50% probability level. For clarity only atoms of one disorder component of the disordered methylene groups are shown.
Fig. 2.
The packing and hydrogen bonding (dashed lines) of the title compound viewing along the a axis. For clarity only atoms of the major disorder component of the disordered methylene groups are shown.
Fig. 3.
The packing and hydrogen bonding (dashed lines) of the title compound viewing along the c axis. For clarity only atoms of the major disorder component of the disordered methylene groups are shown.
Crystal data
| C23H20O4 | F(000) = 1520 |
| Mr = 360.41 | Dx = 1.325 Mg m−3 |
| Orthorhombic, Pbca | Mo Kα radiation, λ = 0.7107 Å |
| Hall symbol: -P 2ac 2ab | Cell parameters from 3838 reflections |
| a = 14.2855 (15) Å | θ = 3.0–29.5° |
| b = 13.7461 (12) Å | µ = 0.09 mm−1 |
| c = 18.400 (2) Å | T = 123 K |
| V = 3613.2 (6) Å3 | Block, colourless |
| Z = 8 | 0.20 × 0.18 × 0.16 mm |
Data collection
| Oxford Diffraction Xcalibur, Eos diffractometer | 4541 independent reflections |
| Radiation source: Enhance (Mo) X-ray Source | 3366 reflections with I > 2σ(I) |
| Graphite monochromator | Rint = 0.042 |
| Detector resolution: 16.0727 pixels mm-1 | θmax = 29.5°, θmin = 3.0° |
| ω scans | h = −18→18 |
| Absorption correction: multi-scan (CrysAlis PRO; Oxford Diffraction, 2010) | k = −17→17 |
| Tmin = 0.994, Tmax = 1.000 | l = −16→25 |
| 17944 measured reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.051 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.114 | H atoms treated by a mixture of independent and constrained refinement |
| S = 1.04 | w = 1/[σ2(Fo2) + (0.037P)2 + 1.8151P] where P = (Fo2 + 2Fc2)/3 |
| 4541 reflections | (Δ/σ)max = 0.001 |
| 258 parameters | Δρmax = 0.40 e Å−3 |
| 8 restraints | Δρmin = −0.25 e Å−3 |
Special details
| Geometry. Bond distances, angles etc. have been calculated using the rounded fractional coordinates. All su's are estimated from the variances of the (full) variance-covariance matrix. The cell e.s.d.'s are taken into account in the estimation of distances, angles and torsion angles |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | Occ. (<1) | |
| O1 | 0.03562 (8) | 0.89828 (7) | 0.24583 (6) | 0.0232 (3) | |
| O2 | 0.02801 (9) | 0.61517 (8) | 0.38998 (6) | 0.0269 (4) | |
| O3 | 0.18481 (8) | 0.51704 (7) | 0.29379 (7) | 0.0245 (3) | |
| O4 | 0.22989 (8) | 0.85224 (7) | 0.30414 (6) | 0.0206 (3) | |
| C1 | 0.11053 (11) | 0.75665 (10) | 0.19287 (8) | 0.0170 (4) | |
| C2 | 0.14830 (11) | 0.71387 (10) | 0.12835 (8) | 0.0190 (4) | |
| C3 | 0.18506 (11) | 0.61770 (11) | 0.12698 (9) | 0.0208 (4) | |
| C4 | 0.21973 (12) | 0.57841 (12) | 0.06413 (9) | 0.0259 (5) | |
| C5 | 0.21989 (12) | 0.63171 (13) | −0.00100 (9) | 0.0289 (5) | |
| C6 | 0.18492 (12) | 0.72413 (12) | −0.00158 (9) | 0.0279 (5) | |
| C7 | 0.14894 (12) | 0.76802 (11) | 0.06208 (9) | 0.0233 (5) | |
| C8 | 0.11215 (13) | 0.86400 (11) | 0.06150 (9) | 0.0275 (5) | |
| C9 | 0.07720 (12) | 0.90419 (11) | 0.12294 (9) | 0.0264 (5) | |
| C10 | 0.07683 (11) | 0.84974 (11) | 0.18784 (9) | 0.0206 (4) | |
| C11 | 0.10633 (10) | 0.70142 (9) | 0.26465 (8) | 0.0150 (4) | |
| C12 | 0.04078 (10) | 0.75335 (10) | 0.31646 (8) | 0.0173 (4) | |
| C13 | 0.00454 (11) | 0.69966 (11) | 0.37919 (9) | 0.0212 (5) | |
| C14 | −0.06429 (13) | 0.75053 (13) | 0.42855 (10) | 0.0330 (6) | |
| C15 | −0.04989 (15) | 0.86009 (13) | 0.43098 (11) | 0.0370 (6) | |
| C16 | −0.05019 (13) | 0.90186 (12) | 0.35487 (10) | 0.0288 (5) | |
| C17 | 0.01241 (11) | 0.84547 (11) | 0.30557 (9) | 0.0205 (4) | |
| C18 | 0.20297 (10) | 0.68519 (10) | 0.29769 (8) | 0.0145 (4) | |
| C19 | 0.23694 (11) | 0.59364 (10) | 0.31102 (8) | 0.0180 (4) | |
| C20B | 0.3353 (4) | 0.5759 (6) | 0.3367 (5) | 0.0207 (11) | 0.570 (3) |
| C21B | 0.3740 (2) | 0.66151 (19) | 0.37967 (17) | 0.0236 (7) | 0.570 (3) |
| C22B | 0.3603 (5) | 0.7555 (5) | 0.3376 (5) | 0.0214 (11) | 0.570 (3) |
| C23 | 0.26030 (11) | 0.76828 (10) | 0.31416 (8) | 0.0160 (4) | |
| C22A | 0.3539 (7) | 0.7519 (7) | 0.3496 (7) | 0.0214 (11) | 0.430 (3) |
| C20A | 0.3268 (6) | 0.5728 (8) | 0.3504 (7) | 0.0207 (11) | 0.430 (3) |
| C21A | 0.3963 (3) | 0.6529 (3) | 0.3323 (2) | 0.0236 (7) | 0.430 (3) |
| H3A | 0.18560 | 0.58020 | 0.17030 | 0.0250* | |
| H6 | 0.18470 | 0.75980 | −0.04580 | 0.0330* | |
| H4 | 0.24410 | 0.51410 | 0.06460 | 0.0310* | |
| H5 | 0.24410 | 0.60370 | −0.04430 | 0.0350* | |
| H3 | 0.2178 (17) | 0.4587 (17) | 0.3029 (12) | 0.060 (7)* | |
| H14A | −0.05800 | 0.72400 | 0.47830 | 0.0400* | |
| H14B | −0.12870 | 0.73660 | 0.41160 | 0.0400* | |
| H15A | −0.10050 | 0.89050 | 0.45990 | 0.0440* | |
| H15B | 0.01050 | 0.87490 | 0.45490 | 0.0440* | |
| H16A | −0.11480 | 0.90080 | 0.33540 | 0.0350* | |
| H16B | −0.02910 | 0.97040 | 0.35650 | 0.0350* | |
| H20C | 0.37600 | 0.56360 | 0.29420 | 0.0250* | 0.570 (3) |
| H20D | 0.33630 | 0.51700 | 0.36770 | 0.0250* | 0.570 (3) |
| H21C | 0.34160 | 0.66600 | 0.42710 | 0.0280* | 0.570 (3) |
| H21D | 0.44160 | 0.65130 | 0.38910 | 0.0280* | 0.570 (3) |
| H22C | 0.37900 | 0.81120 | 0.36840 | 0.0260* | 0.570 (3) |
| H22D | 0.40120 | 0.75510 | 0.29410 | 0.0260* | 0.570 (3) |
| H8 | 0.11210 | 0.90030 | 0.01760 | 0.0330* | |
| H9 | 0.05310 | 0.96860 | 0.12230 | 0.0320* | |
| H11 | 0.07880 | 0.63600 | 0.25440 | 0.0180* | |
| H20A | 0.35200 | 0.50900 | 0.33500 | 0.0250* | 0.430 (3) |
| H20B | 0.31560 | 0.57080 | 0.40350 | 0.0250* | 0.430 (3) |
| H21A | 0.41270 | 0.64980 | 0.28010 | 0.0280* | 0.430 (3) |
| H21B | 0.45420 | 0.64360 | 0.36100 | 0.0280* | 0.430 (3) |
| H22A | 0.34670 | 0.75800 | 0.40290 | 0.0260* | 0.430 (3) |
| H22B | 0.39770 | 0.80330 | 0.33340 | 0.0260* | 0.430 (3) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| O1 | 0.0271 (6) | 0.0151 (5) | 0.0274 (6) | 0.0044 (4) | −0.0008 (5) | 0.0007 (4) |
| O2 | 0.0325 (7) | 0.0199 (6) | 0.0284 (6) | −0.0053 (5) | 0.0035 (5) | 0.0024 (5) |
| O3 | 0.0248 (6) | 0.0096 (5) | 0.0392 (7) | 0.0003 (4) | −0.0094 (5) | −0.0003 (4) |
| O4 | 0.0222 (6) | 0.0106 (5) | 0.0290 (6) | −0.0015 (4) | 0.0001 (5) | 0.0000 (4) |
| C1 | 0.0156 (7) | 0.0143 (7) | 0.0212 (8) | −0.0028 (6) | −0.0029 (6) | 0.0018 (6) |
| C2 | 0.0161 (7) | 0.0197 (7) | 0.0213 (8) | −0.0045 (6) | −0.0029 (6) | 0.0004 (6) |
| C3 | 0.0208 (8) | 0.0196 (7) | 0.0220 (8) | −0.0029 (6) | −0.0009 (7) | −0.0006 (6) |
| C4 | 0.0242 (9) | 0.0240 (8) | 0.0295 (9) | −0.0024 (7) | 0.0017 (7) | −0.0052 (7) |
| C5 | 0.0264 (9) | 0.0385 (10) | 0.0219 (8) | −0.0062 (7) | 0.0028 (7) | −0.0075 (7) |
| C6 | 0.0267 (9) | 0.0370 (9) | 0.0201 (8) | −0.0090 (7) | −0.0026 (7) | 0.0037 (7) |
| C7 | 0.0213 (8) | 0.0263 (8) | 0.0222 (8) | −0.0070 (6) | −0.0033 (7) | 0.0027 (7) |
| C8 | 0.0309 (10) | 0.0265 (8) | 0.0251 (9) | −0.0054 (7) | −0.0053 (8) | 0.0098 (7) |
| C9 | 0.0290 (9) | 0.0170 (7) | 0.0333 (10) | −0.0004 (6) | −0.0064 (8) | 0.0069 (7) |
| C10 | 0.0197 (8) | 0.0173 (7) | 0.0249 (8) | −0.0014 (6) | −0.0026 (7) | 0.0000 (6) |
| C11 | 0.0158 (7) | 0.0105 (6) | 0.0186 (7) | −0.0009 (5) | −0.0010 (6) | −0.0002 (5) |
| C12 | 0.0129 (7) | 0.0171 (7) | 0.0220 (8) | −0.0009 (6) | −0.0013 (6) | −0.0028 (6) |
| C13 | 0.0168 (8) | 0.0231 (8) | 0.0236 (8) | −0.0040 (6) | −0.0013 (7) | −0.0018 (6) |
| C14 | 0.0263 (10) | 0.0400 (10) | 0.0326 (10) | 0.0039 (8) | 0.0110 (8) | 0.0010 (8) |
| C15 | 0.0332 (11) | 0.0389 (10) | 0.0390 (11) | 0.0124 (8) | 0.0068 (9) | −0.0085 (8) |
| C16 | 0.0237 (9) | 0.0242 (8) | 0.0386 (10) | 0.0067 (7) | 0.0018 (8) | −0.0061 (7) |
| C17 | 0.0161 (8) | 0.0192 (7) | 0.0261 (8) | 0.0008 (6) | −0.0027 (7) | −0.0023 (6) |
| C18 | 0.0147 (7) | 0.0138 (6) | 0.0150 (7) | 0.0005 (5) | 0.0004 (6) | −0.0003 (5) |
| C19 | 0.0189 (8) | 0.0137 (7) | 0.0213 (8) | 0.0003 (6) | −0.0010 (6) | −0.0011 (6) |
| C20B | 0.0199 (13) | 0.0172 (8) | 0.025 (3) | 0.0027 (9) | −0.0037 (16) | 0.0015 (15) |
| C21B | 0.0171 (12) | 0.0240 (11) | 0.0297 (13) | 0.0017 (9) | −0.0052 (11) | −0.0040 (12) |
| C22B | 0.0152 (11) | 0.0190 (9) | 0.030 (3) | −0.0042 (9) | 0.0005 (15) | −0.0012 (14) |
| C23 | 0.0166 (7) | 0.0153 (7) | 0.0161 (7) | −0.0005 (5) | 0.0026 (6) | −0.0006 (6) |
| C22A | 0.0152 (11) | 0.0190 (9) | 0.030 (3) | −0.0042 (9) | 0.0005 (15) | −0.0012 (14) |
| C20A | 0.0199 (13) | 0.0172 (8) | 0.025 (3) | 0.0027 (9) | −0.0037 (16) | 0.0015 (15) |
| C21A | 0.0171 (12) | 0.0240 (11) | 0.0297 (13) | 0.0017 (9) | −0.0052 (11) | −0.0040 (12) |
Geometric parameters (Å, º)
| O1—C10 | 1.3894 (19) | C20B—C21B | 1.522 (9) |
| O1—C17 | 1.3584 (19) | C21A—C22A | 1.523 (11) |
| O2—C13 | 1.2250 (19) | C21B—C22B | 1.519 (8) |
| O3—C19 | 1.3281 (18) | C22A—C23 | 1.505 (11) |
| O4—C23 | 1.2469 (17) | C22B—C23 | 1.503 (8) |
| O3—H3 | 0.95 (2) | C3—H3A | 0.9500 |
| C1—C10 | 1.370 (2) | C4—H4 | 0.9500 |
| C1—C11 | 1.525 (2) | C5—H5 | 0.9500 |
| C1—C2 | 1.431 (2) | C6—H6 | 0.9500 |
| C2—C3 | 1.423 (2) | C8—H8 | 0.9500 |
| C2—C7 | 1.429 (2) | C9—H9 | 0.9500 |
| C3—C4 | 1.369 (2) | C11—H11 | 1.0000 |
| C4—C5 | 1.405 (2) | C14—H14A | 0.9900 |
| C5—C6 | 1.365 (2) | C14—H14B | 0.9900 |
| C6—C7 | 1.414 (2) | C15—H15A | 0.9900 |
| C7—C8 | 1.420 (2) | C15—H15B | 0.9900 |
| C8—C9 | 1.354 (2) | C16—H16A | 0.9900 |
| C9—C10 | 1.409 (2) | C16—H16B | 0.9900 |
| C11—C18 | 1.525 (2) | C20A—H20B | 0.9900 |
| C11—C12 | 1.515 (2) | C20A—H20A | 0.9900 |
| C12—C13 | 1.465 (2) | C20B—H20D | 0.9900 |
| C12—C17 | 1.345 (2) | C20B—H20C | 0.9900 |
| C13—C14 | 1.510 (2) | C21A—H21B | 0.9900 |
| C14—C15 | 1.521 (3) | C21A—H21A | 0.9900 |
| C15—C16 | 1.514 (3) | C21B—H21D | 0.9900 |
| C16—C17 | 1.491 (2) | C21B—H21C | 0.9900 |
| C18—C23 | 1.438 (2) | C22A—H22B | 0.9900 |
| C18—C19 | 1.371 (2) | C22A—H22A | 0.9900 |
| C19—C20B | 1.502 (6) | C22B—H22D | 0.9900 |
| C19—C20A | 1.502 (10) | C22B—H22C | 0.9900 |
| C20A—C21A | 1.520 (11) | ||
| C10—O1—C17 | 117.92 (11) | C5—C4—H4 | 120.00 |
| C19—O3—H3 | 110.5 (14) | C4—C5—H5 | 120.00 |
| C2—C1—C11 | 121.94 (12) | C6—C5—H5 | 120.00 |
| C10—C1—C11 | 120.64 (13) | C5—C6—H6 | 119.00 |
| C2—C1—C10 | 117.41 (13) | C7—C6—H6 | 119.00 |
| C1—C2—C7 | 119.77 (13) | C7—C8—H8 | 120.00 |
| C3—C2—C7 | 117.82 (14) | C9—C8—H8 | 120.00 |
| C1—C2—C3 | 122.41 (13) | C8—C9—H9 | 120.00 |
| C2—C3—C4 | 121.00 (15) | C10—C9—H9 | 120.00 |
| C3—C4—C5 | 121.03 (15) | C1—C11—H11 | 107.00 |
| C4—C5—C6 | 119.44 (15) | C12—C11—H11 | 107.00 |
| C5—C6—C7 | 121.57 (15) | C18—C11—H11 | 107.00 |
| C2—C7—C6 | 119.14 (14) | C13—C14—H14A | 109.00 |
| C6—C7—C8 | 121.64 (15) | C13—C14—H14B | 109.00 |
| C2—C7—C8 | 119.21 (14) | C15—C14—H14A | 109.00 |
| C7—C8—C9 | 120.62 (15) | C15—C14—H14B | 109.00 |
| C8—C9—C10 | 119.49 (14) | H14A—C14—H14B | 108.00 |
| O1—C10—C9 | 113.41 (13) | C14—C15—H15A | 110.00 |
| C1—C10—C9 | 123.49 (15) | C14—C15—H15B | 110.00 |
| O1—C10—C1 | 123.06 (14) | C16—C15—H15A | 110.00 |
| C1—C11—C18 | 112.50 (12) | C16—C15—H15B | 110.00 |
| C12—C11—C18 | 112.19 (12) | H15A—C15—H15B | 108.00 |
| C1—C11—C12 | 109.56 (11) | C15—C16—H16A | 109.00 |
| C11—C12—C17 | 122.43 (13) | C15—C16—H16B | 109.00 |
| C13—C12—C17 | 119.04 (14) | C17—C16—H16A | 109.00 |
| C11—C12—C13 | 118.49 (12) | C17—C16—H16B | 109.00 |
| O2—C13—C14 | 121.31 (15) | H16A—C16—H16B | 108.00 |
| C12—C13—C14 | 118.09 (13) | C19—C20A—H20A | 110.00 |
| O2—C13—C12 | 120.58 (14) | C19—C20A—H20B | 110.00 |
| C13—C14—C15 | 112.84 (15) | C21A—C20A—H20A | 110.00 |
| C14—C15—C16 | 110.37 (15) | C21A—C20A—H20B | 110.00 |
| C15—C16—C17 | 111.34 (15) | H20A—C20A—H20B | 108.00 |
| O1—C17—C16 | 111.15 (13) | H20C—C20B—H20D | 108.00 |
| C12—C17—C16 | 125.43 (15) | C19—C20B—H20D | 109.00 |
| O1—C17—C12 | 123.39 (14) | C21B—C20B—H20C | 109.00 |
| C11—C18—C19 | 121.74 (13) | C19—C20B—H20C | 109.00 |
| C19—C18—C23 | 119.33 (13) | C21B—C20B—H20D | 109.00 |
| C11—C18—C23 | 118.92 (12) | C20A—C21A—H21B | 110.00 |
| O3—C19—C18 | 119.12 (14) | C22A—C21A—H21A | 110.00 |
| O3—C19—C20A | 116.3 (4) | C20A—C21A—H21A | 110.00 |
| C18—C19—C20B | 122.4 (3) | H21A—C21A—H21B | 108.00 |
| C18—C19—C20A | 124.3 (4) | C22A—C21A—H21B | 110.00 |
| O3—C19—C20B | 118.1 (3) | C20B—C21B—H21D | 110.00 |
| C19—C20A—C21A | 108.3 (7) | C20B—C21B—H21C | 110.00 |
| C19—C20B—C21B | 112.2 (5) | H21C—C21B—H21D | 108.00 |
| C20A—C21A—C22A | 110.0 (6) | C22B—C21B—H21C | 110.00 |
| C20B—C21B—C22B | 110.3 (5) | C22B—C21B—H21D | 110.00 |
| C21A—C22A—C23 | 113.4 (7) | C23—C22A—H22B | 109.00 |
| C21B—C22B—C23 | 111.6 (5) | H22A—C22A—H22B | 108.00 |
| C18—C23—C22A | 118.6 (4) | C21A—C22A—H22A | 109.00 |
| O4—C23—C22A | 120.8 (4) | C21A—C22A—H22B | 109.00 |
| C18—C23—C22B | 120.6 (3) | C23—C22A—H22A | 109.00 |
| O4—C23—C18 | 120.38 (14) | C21B—C22B—H22C | 109.00 |
| O4—C23—C22B | 118.8 (3) | C21B—C22B—H22D | 109.00 |
| C2—C3—H3A | 120.00 | C23—C22B—H22C | 109.00 |
| C4—C3—H3A | 119.00 | C23—C22B—H22D | 109.00 |
| C3—C4—H4 | 119.00 | H22C—C22B—H22D | 108.00 |
| C17—O1—C10—C1 | −12.4 (2) | C18—C11—C12—C13 | −72.26 (16) |
| C17—O1—C10—C9 | 165.31 (14) | C18—C11—C12—C17 | 110.02 (16) |
| C10—O1—C17—C12 | 11.4 (2) | C1—C11—C18—C19 | −120.34 (15) |
| C10—O1—C17—C16 | −166.72 (13) | C1—C11—C18—C23 | 58.68 (17) |
| C10—C1—C2—C3 | −179.85 (14) | C12—C11—C18—C19 | 115.59 (15) |
| C10—C1—C2—C7 | −0.5 (2) | C12—C11—C18—C23 | −65.40 (17) |
| C11—C1—C2—C3 | −0.6 (2) | C11—C12—C13—O2 | 1.1 (2) |
| C11—C1—C2—C7 | 178.76 (14) | C11—C12—C13—C14 | −177.18 (14) |
| C2—C1—C10—O1 | 177.82 (14) | C17—C12—C13—O2 | 178.93 (15) |
| C2—C1—C10—C9 | 0.4 (2) | C17—C12—C13—C14 | 0.6 (2) |
| C11—C1—C10—O1 | −1.5 (2) | C11—C12—C17—O1 | 3.6 (2) |
| C11—C1—C10—C9 | −178.93 (15) | C11—C12—C17—C16 | −178.61 (15) |
| C2—C1—C11—C12 | −164.88 (14) | C13—C12—C17—O1 | −174.15 (14) |
| C2—C1—C11—C18 | 69.61 (17) | C13—C12—C17—C16 | 3.7 (2) |
| C10—C1—C11—C12 | 14.38 (19) | O2—C13—C14—C15 | 152.03 (16) |
| C10—C1—C11—C18 | −111.14 (16) | C12—C13—C14—C15 | −29.7 (2) |
| C1—C2—C3—C4 | 179.23 (15) | C13—C14—C15—C16 | 53.5 (2) |
| C7—C2—C3—C4 | −0.1 (2) | C14—C15—C16—C17 | −48.5 (2) |
| C1—C2—C7—C6 | −178.84 (15) | C15—C16—C17—O1 | −160.62 (14) |
| C1—C2—C7—C8 | 0.2 (2) | C15—C16—C17—C12 | 21.3 (2) |
| C3—C2—C7—C6 | 0.5 (2) | C11—C18—C19—O3 | 0.9 (2) |
| C3—C2—C7—C8 | 179.57 (15) | C11—C18—C19—C20B | 173.9 (4) |
| C2—C3—C4—C5 | −0.2 (2) | C23—C18—C19—O3 | −178.09 (14) |
| C3—C4—C5—C6 | 0.0 (3) | C23—C18—C19—C20B | −5.1 (5) |
| C4—C5—C6—C7 | 0.4 (3) | C11—C18—C23—O4 | 2.7 (2) |
| C5—C6—C7—C2 | −0.7 (3) | C11—C18—C23—C22B | −172.0 (4) |
| C5—C6—C7—C8 | −179.71 (17) | C19—C18—C23—O4 | −178.22 (14) |
| C2—C7—C8—C9 | 0.3 (3) | C19—C18—C23—C22B | 7.0 (4) |
| C6—C7—C8—C9 | 179.32 (17) | O3—C19—C20B—C21B | −159.1 (4) |
| C7—C8—C9—C10 | −0.5 (3) | C18—C19—C20B—C21B | 27.9 (7) |
| C8—C9—C10—O1 | −177.54 (15) | C19—C20B—C21B—C22B | −50.8 (7) |
| C8—C9—C10—C1 | 0.1 (3) | C20B—C21B—C22B—C23 | 52.4 (6) |
| C1—C11—C12—C13 | 162.05 (13) | C21B—C22B—C23—O4 | 153.7 (4) |
| C1—C11—C12—C17 | −15.67 (19) | C21B—C22B—C23—C18 | −31.5 (7) |
Hydrogen-bond geometry (Å, º)
Cg3 is the centroid of the C2–C7 benzene ring.
| D—H···A | D—H | H···A | D···A | D—H···A |
| O3—H3···O4i | 0.95 (2) | 1.64 (2) | 2.5793 (15) | 170 (2) |
| C3—H3A···O3 | 0.95 | 2.43 | 3.367 (2) | 168 |
| C9—H9···O2ii | 0.95 | 2.34 | 3.275 (2) | 170 |
| C11—H11···O3 | 1.00 | 2.34 | 2.8228 (17) | 108 |
| C14—H14B···Cg3iii | 0.99 | 2.85 | 3.750 (2) | 152 |
Symmetry codes: (i) −x+1/2, y−1/2, z; (ii) −x, y+1/2, −z+1/2; (iii) x−1/2, y, −z+1/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: SJ5351).
References
- Abdelhamid, A. A., Mohamed, S. K., Allahverdiyev, M. A., Gurbanov, A. V. & Ng, S. W. (2011). Acta Cryst. E67, o785. [DOI] [PMC free article] [PubMed]
- Allen, F. H., Kennard, O., Watson, D. G., Brammer, L., Orpen, A. G. & Taylor, R. (1987). J. Chem. Soc. Perkin Trans. 2, pp. S1–19.
- Çelik, Í., Akkurt, M., Jarrahpour, A., Ebrahimi, E. & Büyükgüngör, O. (2009). Acta Cryst. E65, o2522–o2523. [DOI] [PMC free article] [PubMed]
- Cremer, D. & Pople, J. A. (1975). J. Am. Chem. Soc. 97, 1354–1358.
- Farrugia, L. J. (2012). J. Appl. Cryst. 45, 849–854.
- Li, Y.-L., Wang, X.-S., Shi, D.-Q., Tu, S.-J. & Zhang, Y. (2004). Acta Cryst. E60, o1439–o1441.
- Menchen, S. M., Benson, S. C., Lam, J. Y. L., Zhen, W., Sun, D., Rosenblum, B. B., Khan, S. H. & Taing, M. (2003a). US Patent, US 6583168.
- Menchen, S. M., Benson, S. C., Lam, J. Y. L., Zhen, W., Sun, D., Rosenblum, B. B., Khan, S. H. & Taing, M. (2003b). Chem Abstr 139, 54287f.
- Mohamed, S. K., Abdelhamid, A. A., Khalilov, A. N., Gurbanov, A. V. & Ng, S. W. (2011). Acta Cryst. E67, o850–o851. [DOI] [PMC free article] [PubMed]
- Mohamed, S. K., Akkurt, M., Abdelhamid, A. A., Fanwick, P. E. & Potgeiter, H. (2012). Acta Cryst. E68, o1710. [DOI] [PMC free article] [PubMed]
- Oxford Diffraction (2010). CrysAlis PRO Oxford Diffraction Ltd, Yarnton, England.
- Reddy, B. P., Vijayakumar, V., Narasimhamurthy, T., Suresh, J. & Lakshman, P. L. N. (2009). Acta Cryst. E65, o916. [DOI] [PMC free article] [PubMed]
- Sarma, R. J. & Baruah, J. B. (2005). Dyes Pigm 64, 91–92.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
- Zare, A., Mokhlesi, M., Hasaninejad, A. & Hekmat-Zadehk, T. (2012). E-J. Chem. 9, 1854–1863.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I. DOI: 10.1107/S1600536813025324/sj5351sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536813025324/sj5351Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536813025324/sj5351Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report