Abstract
Memory consolidation and long-term potentiation require activity-dependent gene transcription, coordinated by an array of transcription factors. Many members of the nuclear receptor superfamily of transcription factors are expressed in the hippocampus immediately after learning, including the Nr4a family of orphan receptors. These activity-dependent transcription factors are critical for hippocampus-dependent contextual fear and object recognition memory, but their role in hippocampal synaptic function is unknown. In this study, we hypothesized that Nr4a transcription factor function is also necessary for hippocampal long-term potentiation. We used a strain of mice expressing a dominant-negative Nr4a transgene. Hippocampal slices from Nr4aDN mutant mice exhibited impairments in transcription-dependent long-term potentiation and were not sensitive to LTP enhancement by the HDAC inhibitor TSA. These results demonstrate that NR4A transcription factor function mediates mechanisms of synaptic plasticity in the hippocampus.
Keywords: Nr4a, Synaptic plasticity, Nuclear receptors, Hippocampus, Long-term potentiation, HDAC inhibitor
1. Introduction
In the hippocampus, the consolidation of long-term memory (LTM) and the maintenance of long-term potentiation (LTP) both depend on cAMP signaling, PKA activity, gene transcription, and de novo protein synthesis (Abel et al., 1997; Huang & Kandel, 1994; Igaz, Vianna, Medina, & Izquierdo, 2002; Nguyen, Abel, & Kandel, 1994; Pittenger et al., 2002). A number of transcription factors are known to support activity-driven transcription in neurons, including well-characterized proteins such as cAMP response element-binding protein (CREB) and the immediate early genes C/EBP and Zif268 (see Alberini, 2009 for review). The large family of nuclear receptor transcription factors is increasingly being investigated for its role in memory and plasticity, and the potential of some nuclear receptors to serve as therapeutic targets in disorders of cognition (Hawk & Abel, 2011; Hawk et al., 2012). For example, agonists for the liver X receptor (LXR), retinoic acid receptor (RAR), retinoid X receptor (RXR), and the peroxisome proliferator-activated receptor gamma (PPARγ) have been found to improve memory and synaptic plasticity in mouse models of disease, most notably in models of Alzheimer’s disease (Cramer et al., 2012; Jiang et al., 2008; Pedersen et al., 2006; Wesson, Borkowski, Landreth, Nixon, & Levy, 2011).
Although most of the nuclear receptors are regulated by lipophilic ligands that readily cross the nuclear membrane, some are considered “orphan” receptors. These proteins either have no known endogenous ligand, or operate independently of ligand binding. One such group of orphan receptors is the Nr4a subfamily, made up of three closely related members (Nr4a1/Nurr1/NGFI-B, Nr4a2/Nur77/HZF-3, and Nr4a3/NOR-1/TEC) (Hawk & Abel, 2011; Maxwell & Muscat, 2006; Paulsen, Weaver, Fahrner, & Milbrandt, 1992; Wansa, Harris, Yan, Ordentlich, & Muscat, 2003). The NR4A proteins are involved in activity-dependent processes in numerous cell types, supporting such phenomena as apoptosis, metabolism, dopaminergic development, and inflammatory response (for review see Hawk & Abel, 2011). The Nr4a genes are immediate-early genes transcribed rapidly in response to external stimuli (Peña de Ortiz & Jamieson, 1996). Mature NR4A proteins are capable of both positively and negatively regulating the transcription of their downstream targets (Johnson, Michelhaugh, Bouhamdan, Schmidt, & Bannon, 2011), which include plasticity-related genes such as Bdnf1 and Fosl2 (Hawk et al., 2012; Volpicelli et al., 2007).
In many systems, Nr4a transcription is controlled by the cAMP/PKA/CREB signaling pathway (Kovalovsky et al., 2002; Lemberger, Parkitna, Chai, Schütz, & Engblom, 2008), which is a cascade critical for transcription of other memory- and plasticity-related genes (Josselyn & Nguyen, 2005). In the hippocampus, training in different behavioral tasks increases the expression of one or all of the Nr4a genes in discrete anatomical regions (Hawk et al., 2012; McNulty et al., 2012; Peña de Ortiz, Maldonado-Vlaar, & Carrasquillo, 2000). The induction of long-term potentiation in vivo by high-frequency stimulation (HFS) increases Nr4a gene expression in hippocampal neurons (Dragunow, Abraham, & Hughes, 1996; Ryan, Mason-Parker, Tate, Abraham, & Williams, 2011), and administration of the GABAA antagonist gabazine enhances excitability and increases Nr4a mRNA levels in hippocampal slices (Pegoraro et al., 2010). Memory enhancement by intrahippocampal administration of the histone deacetylase (HDAC) inhibitor trichostatin A (TSA) further increases the expression of Nr4a1 and Nr4a2 (Vecsey et al., 2007), and knockdown of Nr4a2 attenuates memory enhancement in mice lacking the histone deacetylase HDAC3 (McQuown et al., 2011). Further, disrupting NR4A function by knockout (Rojas, Joodmardi, Hong, Perlmann, & Ogren, 2007), siRNA knockdown (McNulty et al., 2012), antisense oligonucleotides (Colón-Cesario et al., 2006), or expression of a dominant-negative protein (Hawk et al., 2012) impairs learning and long-term memory and blocks memory enhancement by HDAC inhibitors (Hawk et al., 2012). To define the importance of the Nr4a transcription factors in hippocampal synaptic function, we employed a transgenic dominant-negative NR4A protein to inhibit NR4A function. We then measured synaptic function and long-term potentiation at the Schaffer collateral-CA1 synapses. The results presented here support an important role for the NR4A transcription factors in the maintenance of long-term synaptic plasticity, consistent with their role in the consolidation of long-term hippocampus-dependent memory.
2. Methods
2.1. Subjects
Mice were maintained under standard conditions consistent with National Institute of Health guidelines for animal care and use and all experimental procedures were approved by the Institutional Animal Care and Use Committee of the University of Pennsylvania. Mice were maintained on a 12:12 light:dark cycle with lights on at 7 AM. Food and water were provided in their home cages ad libitum. Tissue collection and electrophysiological testing were conducted on male and female adult (2–6 months old) animals during the light portion of the cycle, with animals being sacrificed at approximately 11 AM.
The dominant-negative NR4A construct (NR4ADN) is a form of NR4A1 that contains the dimerization domain and DNA-binding domain but lacks the transactivation domain, and instead contains an HA and an YFP tag (Hawk et al., 2012; Robert, Martin, & Tremblay, 2006). The Nr4aDN mice were generated as previously described (see Hawk et al., 2012). Briefly, the transgenic Nr4aDN mice express a dominant-negative NR4A construct under the control of the tetracycline operator (tetO). This Nr4aDN transgene was microinjected into C57BL/6 zygotes, and founders were then crossed with C57BL/6 mice bearing the CaMKII-tTA (line B) transgene (Mayford et al., 1996) to produce double-transgenic mice that expressed the Nr4aDN construct postnatally in excitatory forebrain neurons (Hawk et al., 2012). Double-transgenic (CaMKII-tTA+; Nr4aDN+) males were bred to C57BL/6 females. From the resulting litters experimental Nr4aDN mutant mice were double-transgenic (CaMKII-tTA+; Nr4aDN+), while non-transgenic and single-transgenic littermates served as controls. Mice were raised in the absence of doxycycline, but were placed on doxycycline chow after weaning to suppress transgene expression in some control experiments.
2.2. Electrophysiological recordings
To assess the effects of Nr4a dominant-negative transgene expression on hippocampal LTP, mice were killed by cervical dislocation and their hippocampi were quickly dissected in ice-cold oxygenated artificial CSF (aCSF; 124 mM NaCl, 4.4 mM KCl, 1.3 mM MgSO4·7H2O, 1 NaH2PO4·H2O, 26.2 mM NaHCO3, 2.5 mM CaCl2·2H2O, 10 mM glucose). Transverse hippocampal slices were cut 400 μm thick using a Stoelting tissue chopper, placed in an interface recording chamber, and perfused with oxygenated aCSF at 28.0 °C. Slices were allowed to recover for at least 2 h before beginning electrophysiological recordings. Single-pathway recordings were made using a single bipolar stimulating electrode made from nichrome wire (A–M Systems) placed in the stratum radiatum of the CA1 subfield and used to elicit action potentials in the axons of CA3 pyramidal neurons. Field potentials (fEPSPs) were recorded using an aCSF-filled glass microelectrode (A–M Systems) with a resistance between 0.5 and 5 MΩ placed in the stratum radiatum region of CA1. Data collection was handled by Clampex software (Molecular Devices, Palo Alto, CA) and was analyzed using Clampfit (Molecular Devices). The peak fEPSP amplitude induced by the stimulating electrode was required to be at least 5 mV, and stimulus intensity during the recording was set to produce a response of 40% of the maximum fEPSP amplitude. Test stimulation occurred once every minute. Baseline responses were recorded for 20 min before LTP induction or drug application. To examine early-phase LTP (E-LTP) one train of stimuli at 100 Hz for 1 s was applied through the stimulating electrode. To examine late-phase LTP (L-LTP) four trains of stimuli at 100 Hz for 1 s were delivered 5 min apart (spaced 4-train HFS), or a theta-burst (TBS) protocol consisting of 15 pulses of 40 ms duration delivered at a rate of 5 Hz was used. Recordings continued for 160 min after LTP induction. The initial slopes of the recorded fEPSPs were normalized to the averaged slope of the 20 baseline traces and expressed as a percentage of this baseline. Input–output characteristics in area CA1 were investigated by recording the fEPSPs elicited by stimuli of decreasing intensity. The initial fEPSP slopes were plotted against the amplitudes of corresponding presynaptic fiber volleys and fit with linear regressions. The maximum elicited fEPSP slope was also recorded as a measure of synaptic strength. Paired-pulse facilitation, a short-term form of synaptic plasticity and a measure of presynaptic function, was measured in slices from control and Nr4aDN mutant mice. Paired stimuli were delivered with varying interpulse intervals (300, 200, 100, 50 and 25 ms) and the initial fEPSP slope from the second stimulus was plotted relative to the slope from the first stimulus to give the facilitation ratio.
2.3. Drugs
The HDAC inhibitor Trichostatin A (TSA; AG Scientific) was dissolved at 32 mM in 100% ethanol and diluted to a 16.5 mM stock solution in 50% ethanol. Aliquots of the TSA stock solution were stored at −20 °C. TSA was diluted to a final concentration of 1.65 μM in oxygenated aCSF and slices were perfused with drug or vehicle (0.005% ethanol in aCSF) for 20 min prior to stimulation and throughout the remainder of the recording (Vecsey et al., 2007).
2.4. Statistical analysis
The initial slope of the recorded fEPSP was used to quantify synaptic potentiation, normalized to the averaged 20-min baseline value. Between-group differences in the maintenance of LTP were analyzed by performing a one-way repeated measures ANOVA on the final 20-min epoch of the recordings (Vecsey et al., 2007, 2009). When multiple comparisons were made to a control group, a two-way repeated measures ANOVA with Dunnett’s post hoc test was used with genotype and doxycycline treatment as factors and fEPSP slope as the dependent variable. To evaluate potential differences in paired-pulse facilitation a two-way repeated measures ANOVA was used with genotype and inter-stimulus interval as factors and the facilitation ratio as the dependent variable. For evaluation of input–output characteristics a t-test was performed comparing the average linear regression slopes for control mice and Nr4aDN mutants. Synaptic strength was analyzed with a t-test comparing the maximum elicited fEPSP slopes in control and mutant mice. All statistical analyses were performed using STATISTICA 7 software (StatSoft Inc.; Tulsa, OK). Significance for all tests was set at p < 0.05.
3. Results
3.1. Nr4aDN expression does not change basal synaptic properties
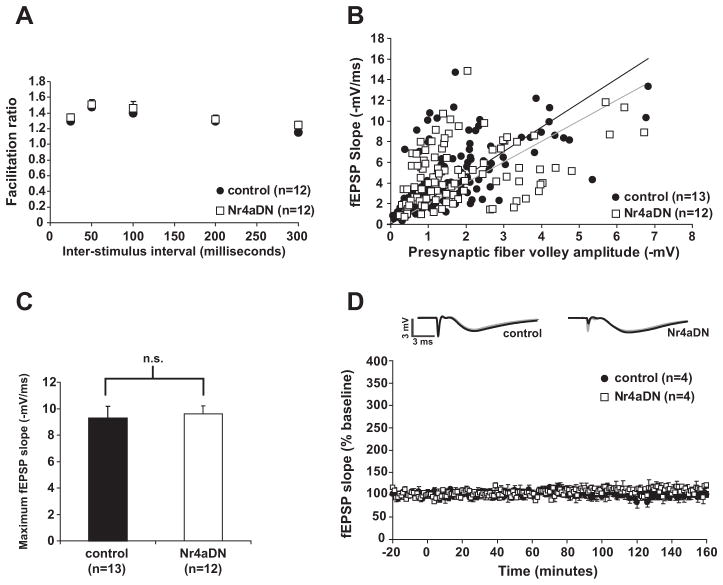
The Nr4a genes are transcribed as immediate-early genes in response to external stimuli such as synaptic activation and in turn they regulate a number of downstream genes, including some involved in synaptic plasticity (Hawk & Abel, 2011). The dominant-negative form of Nr4a expressed in the mutant mice blocks the function of the NR4A proteins, so we began by examining basal synaptic properties to determine if this attenuation of NR4A-mediated transcription caused any disturbances in normal synaptic function at the Schaffer collateral synapses. Paired-pulse facilitation (Fig. 1A), a form of short-term plasticity and a measure of presynaptic release mechanisms, was unchanged by Nr4aDN expression (p = 0.267). The input–output relationship (Fig. 1B) in the Nr4aDN mutants was not altered compared to control mice (p = 0.573), indicating that the relationship between presynaptic release and postsynaptic response was unchanged. Synaptic strength, measured as the maximum fEPSP slope induced by stimulation (Fig. 1C), was comparable between genotypes (control = 9.25 ± 0.91 mV/ms, Nr4aDN = 9.62 ± 0.6 mV/ms; p = 0.791). Further, field recordings in the absence of LTP-inducing stimulation (Fig. 1D) did not significantly deviate from control baseline responses over time (p = 0.256) indicating that transgene expression does not affect synaptic stability or the health of hippocampal slices.
Fig. 1.
Basal transmission is unaffected by Nr4aDN transgene expression. (A) Paired-pulse facilitation, a measure of presynaptic release, was not affected by Nr4aDN expression (p = 0.267). (B) Input–output curves, plotted with fEPSP slope as function of the presynaptic fiber volley, are not affected by Nr4aDN expression (p = 0.573). (C) The maximum fEPSP slopes from Nr4aDN mutants and controls were not significantly different (p = 0.791). (D) Baseline synaptic response in the absence of stimulation is not altered in Nr4aDN mutants when compared to controls (p = 0.256).
3.2. Expression of the dominant-negative Nr4a transgene impairs transcription-dependent hippocampal LTP
Previously, our lab has found that Nr4aDN mutant mice have deficits in hippocampus-dependent long-term contextual fear memory but not in short-term memory (Hawk et al., 2012). Other labs have found that interfering with hippocampal Nr4a2 function by heterozygous deletion (Rojas et al., 2007) or by antisense injection (Colón-Cesario et al., 2006) impairs some forms of long-term memory, while siRNA knockdown of Nr4a1 or Nr4a2 impairs specific forms of object memory (McNulty et al., 2012). Hippocampal LTP, which is defined as an activity-dependent change in synaptic strength between neurons, is a long-lasting form of synaptic plasticity that shares many of its underlying molecular mechanisms with long-term memory (Huang, Nguyen, Abel, & Kandel, 1996; Martin, Grimwood, & Morris, 2000). LTP can be divided into two distinct categories: transient early LTP (E-LTP) that only lasts for 1–2 h and is independent of transcription, and late LTP (L-LTP) which lasts for several hours and requires both transcription and translation (Abel & Nguyen, 2008; Huang & Kandel, 1994; Nguyen et al., 1994).
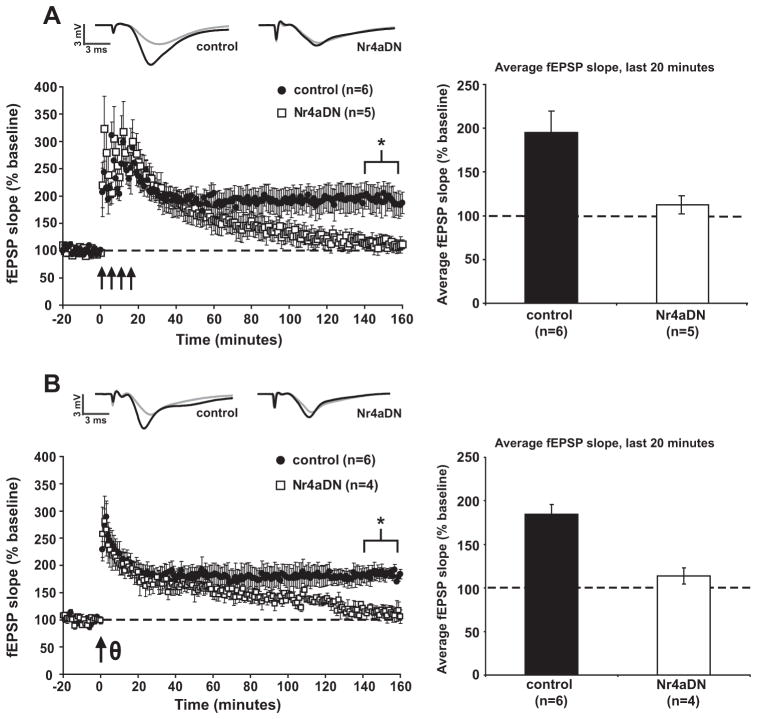
To further explore the role of the Nr4a family in activity-dependent hippocampal function and long-term changes in synaptic strength, we examined two forms of transcription-dependent L-LTP in slices from Nr4aDN mutant mice (Fig. 2). In control animals, the delivery of four 1 s, 100 Hz stimuli spaced 5 min apart induces long-lasting long-term potentiation (Fig. 2A; control, avg. of fEPSP slope over final 20 min = 195.6 ± 23.9%). In hippocampal slices from Nr4aDN mutant animals the same stimulation protocol induced potentiation initially but failed to produce maintained L-LTP (Fig. 2A; Nr4aDN, avg. of fEPSP slope over final 20 min = 112.5 ± 10.5%; p = 0.016). Theta-burst stimulation, which mimics the natural pattern of input from the axons of CA3 pyramidal neurons to the dendrites of CA1 neurons (Nguyen & Kandel, 1997), also elicited L-LTP in control mice but not in Nr4aDN mutant littermates (Fig. 2B; avg. of fEPSP slope over final 20 min for controls = 184.4 ± 11.6%, for Nr4aDN = 113.7 ± 9.4%; control vs. Nr4aDN p = 0.002). These data suggest that transcription-dependent forms of plasticity in the hippocampus depend on the normal activity of the NR4A transcription factors and their downstream target genes.
Fig. 2.
L-LTP is impaired in Nr4aDN mutant mice. (A) Four trains of stimulation spaced 5 min apart (indicated by arrows) induces L-LTP in control slices but not slices taken from Nr4aDN mutant mice (p = 0.016). (B) Theta-burst LTP (TBS; indicated by arrow) produced less potentiation in the late maintenance phase in Nr4aDN mutant mice compared to control animals (p = 0.002).
3.3. LTP impairment in Nr4aDN mutant mice depends on transgene expression
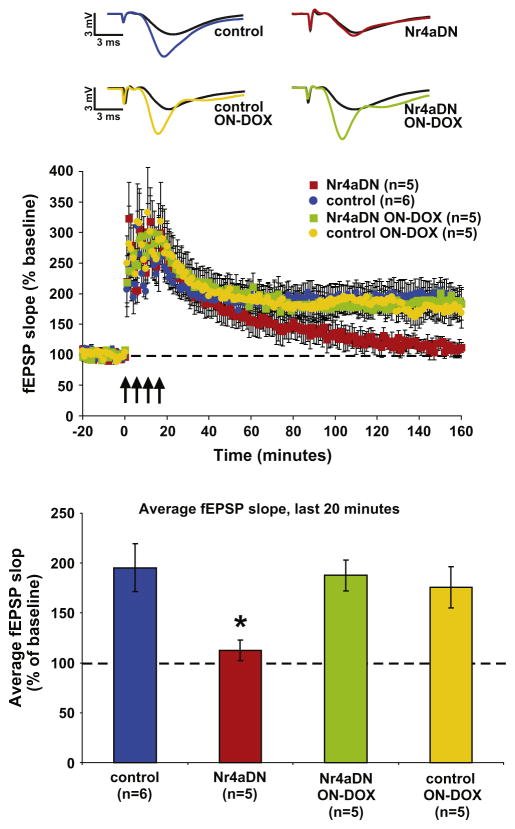
The deficits in long-term contextual memory that were reported in Nr4aDN transgenic mice were rescued by transgene suppression (Hawk et al., 2012), indicating that observed impairments were not due to development effects of blocking NR4A protein function. We sought to address this issue at the synaptic level in Nr4aDN transgenic mice. Nr4aDN mice and littermate controls were reared in the absence of doxycycline, and then placed on a doxycycline diet from the time of weaning until they were 2 months old, a 4-week treatment that is sufficient to suppress expression of the transgene and restore contextual fear memory in Nr4aDN mutant mice (Hawk et al., 2012). A spaced 4-train stimulation protocol was applied to slices from both the doxycycline-treated Nr4aDN mice and doxycycline-treated control animals and found to produce long-lasting LTP in both groups (Fig. 3; avg. fEPSP slope over final 20 min in ON-DOX controls = 175.6 ± 20.8%, in ON-DOX Nr4aDN = 187.7 ± 15.4%). To determine the effects of transgene suppression on LTP, we included our previously collected off-doxycycline spaced 4-train LTP data (Fig. 2A) in our analyses. We compared on- and off-doxycycline LTP using a two-way ANOVA (factors: genotype, doxycycline treatment) and found no significant main effects, but a significant interaction between genotype and doxycycline treatment (p = 0.03). Post-hoc comparisons revealed that although Nr4aDN mutant L-LTP was impaired with respect to controls (p = 0.02), this deficit was rescued by transgene suppression with doxycycline (p = 0.984). In contrast, doxycycline did not affect L-LTP in control mice (p = 0.814). The rescue of LTP impairment by transgene suppression indicates that the loss of NR4A-mediated transcription does not permanently alter synaptic function and demonstrates that the observed deficits in L-LTP are due to transgene expression and not attributable to potential insertional or developmental effects of the transgene.
Fig. 3.
The Nr4aDN LTP impairment can be rescued by transgene suppression. Off-doxycycline Nr4aDN and control data reproduced here from Fig. 2A for comparison. After 4 weeks on doxycyline chow to suppress the tTA; Tet-O system, spaced 4-train stimulation produces sustained L-LTP in the ON-DOX control and Nr4aDN mice that does not differ from controls (Dunnett’s post hoc, control vs. Nr4aDN ON-DOX, p = 0.984). Doxycycline did not affect LTP in control mice (Dunnett’s post hoc, control vs. control ON-DOX, p = 0.814). [*p < 0.05, two-way repeated measures ANOVA followed by Dunnett’s post hoc, control vs. Nr4aDN; p > 0.05 for control vs. Nr4aDN ON-DOX and control vs. control ON-DOX]
3.4. Nr4aDN expression blocks LTP enhancement by HDAC inhibition
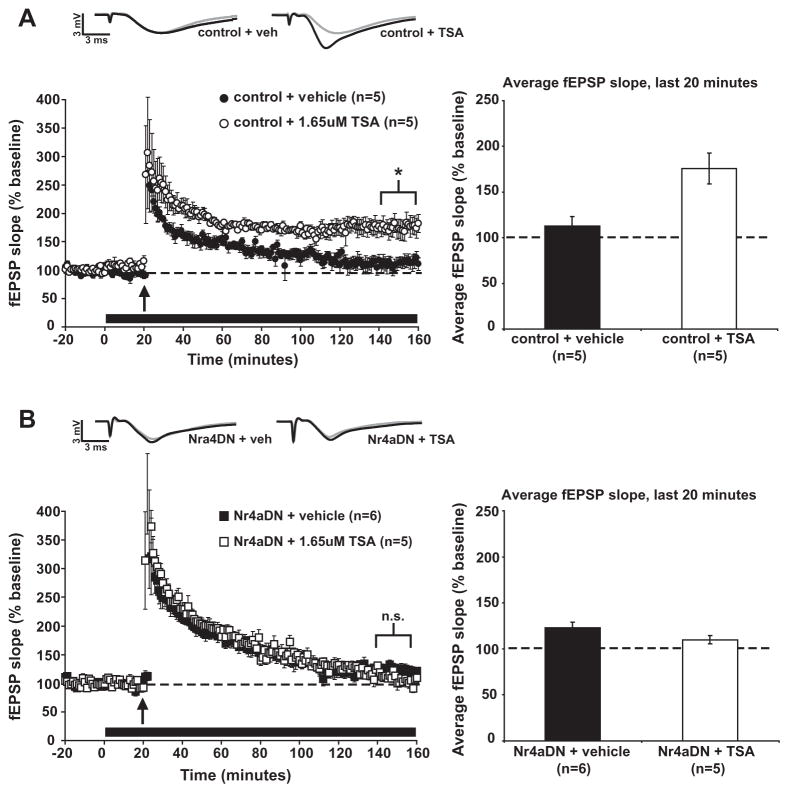
Long-term memory and LTP are both enhanced by the pharmacological inhibition of histone deacetylase enzymes, which produces increases in histone acetylation (Levenson et al., 2004; Vecsey et al., 2007). These enhancements are dependent on the transcription factor CREB, and hippocampal administration of the HDAC inhibitor TSA increases the expression of Nr4a1 and Nr4a2, both Cre-regulated genes (Vecsey et al., 2007). Conditional knockout of the class I histone deacetylase HDAC3 in mice enhances memory and increases the expression of Nr4a2, while siRNA knockdown of Nr4a2 abolishes the memory enhancement observed in HDAC3 mutant mice (McQuown et al., 2011). Further, the expression of a dominant-negative NR4Aprotein blocks memory enhancement by the HDAC inhibitor TSA (Hawk et al., 2012). These results indicate that the NR4A proteins are critical in mediating the memory-enhancing effects of HDAC inhibition. We explored the possibility that NR4A proteins may also be critical for the LTP-enhancing effects of HDAC inhibition by testing the ability of TSA to enhance 1-train E-LTP in Nr4aDN mutant mice. A single 1 s, 100 Hz tetanus induces short-lived E-LTP in both control mice (Fig. 4A; WT VEH, avg. of fEPSP slope over final 20 min = 112.6 ± 10.5%) and Nr4aDN mutant mice (Fig. 4B; Nr4aDN VEH avg. fEPSP slope overfinal 20 min = 122.8 ± 6.0%). When acute slices from control animals were exposed to 1.65 μMTSA, the same stimulus induced significantly enhanced long-lasting L-LTP compared to vehicle-treated slices (Fig. 4A; avg. fEPSP slope over final 20 min = 175.7 ± 16.8%, TSA vs. VEH within control slices, p = 0.016). In slices from Nr4aDN mutant animals, HDAC inhibitor treatment failed to enhance LTP (Fig. 4B; avg. fEPSP slope over final 20 min = 109.9 ± 4.5%, TSA vs. VEH within Nr4aDN slices, p = 0.11). Our results support previous findings that blocking NR4A function prevents memory enhancement by acute TSA treatment (Hawk et al., 2012), and indicate that the NR4A transcription factors are necessary for mediating the effects of HDAC inhibition on synaptic plasticity.
Fig. 4.
Nr4aDN expression blocks LTP enhancement by the HDAC inhibitor TSA. (A) Perfusion of control slices with the HDAC inhibitor TSA enhances potentiation following a single 1s, 100 Hz train of stimulation (indicated by arrow) when compared to 1-train LTP induced in the presence of vehicle (p = 0.016). (B) In slices from Nr4aDN mutants a single train of stimulation (indicated by arrow) produces short-term E-LTP in the presence of both vehicle and 1.65 μM TSA (p = 0.11). [Black bar indicates drug/vehicle treatment.]
4. Discussion
The present study investigated the role of NR4A transcription factor function in LTP in the hippocampus using a dominant-negative NR4A construct to block transactivation by native NR4A proteins (Hawk et al., 2012; Robert et al., 2006). We found that transcription-dependent LTP induced by either spaced high-frequency stimulation or TBS was greatly attenuated in the transgenic mice. However, Nr4aDN expression had no effect on the basal properties of the Schaffer collateral synapses or the stability of synaptic transmission. In addition, we demonstrated that the enhancement of LTP by the HDAC inhibitor TSA (Vecsey et al., 2007) also requires NR4A transcription factor function. Previous studies from our lab (Hawk et al., 2012; Vecsey et al., 2007) and others (Colón-Cesario et al., 2006; McNulty et al., 2012; McQuown et al., 2011; Peña de Ortiz et al., 2000; Rojas et al., 2007) have implicated the Nr4a family in the formation and expression of several types of memory. Here, we provide evidence that these transcription factors are involved in mediating activity-dependent changes in synaptic strength, which is thought to underlie memory formation, supporting the idea that the NR4A proteins are important regulators of plastic and cognitive processes.
We find that two forms of transcription-dependent L-LTP, one induced by spaced 4-train HFS and the other induced by theta-burst stimulation, were both disrupted by Nr4aDN expression. Mutant slices responded to stimulation with an initial potentiation, but were unable to sustain that potentiation through the subsequent transcription- and translation-dependent phase of LTP (Huang et al., 1996; Nguyen et al., 1994). The experience-dependent changes in synaptic strength that occur during LTP share many of the same pathways and mechanisms as long-term memory formation, and some of these (for example, PKA signaling and the activation of CREB) are known to initiate the transcription of the Nr4a genes (Lemberger et al., 2008). Contextual fear conditioning is accompanied by increased hippocampal expression of all three Nr4a family genes in the 2 h following training (Hawk et al., 2012). Other groups have reported upregulation of Nr4a1 in CA1 (Von Hertzen & Giese, 2005) and Nr4a2 in both CA1 and CA3 (Peña de Ortiz et al., 2000) after hippocampus-dependent memory tasks, while Colón-Cesario et al. (2006) demonstrated that the injection of antisense oligonucleotides directed against Nr4a2 attenuated learning on a spatial discrimination task. Inducing synaptic plasticity also increases Nr4a expression; the induction of LTP in vivo by high-frequency stimulation has been found to increase Nr4a mRNA levels in hippocampal neurons (Dragunow et al., 1996; Ryan et al., 2011), and treating cultured hippocampal slices with the GABAA antagonist gabazine increases neuronal excitability and increases expression of all three Nr4a genes (Pegoraro et al., 2010). Taken together with our results, these studies suggest that the activity-dependent transcription of the Nr4a genes and subsequent changes in NR4A-regulated transcription are critical for memory consolidation and the underlying changes in synaptic plasticity.
Acute administration of HDAC inhibitors has repeatedly been shown to enhance long-term memory (Haettig et al., 2011; Levenson et al., 2004; Stefanko et al., 2009; Vecsey et al., 2007) and extinction (Lattal, Barrett, & Wood, 2007; Stafford, Raybuck, Ryabinin, & Lattal, 2012) in vivo, and can transform short-term E-LTP into transcription-dependent L-LTP in ex vivo hippocampal slices. Accumulated research indicates that the Nr4a nuclear receptors are important regulators of HDAC inhibitor-mediated memory enhancement. Vecsey et al. (2007) reported that Nr4a1 and Nr4a2 mRNA levels were increased during memory consolidation in mice administered TSA, while Hawk and colleagues (2012) found that contextual fear memory in Nr4aDN mice was not enhanced by TSA treatment. Knocking out HDAC3 enhances memory, mimicking the effects of HDAC inhibitor treatment, and this effect is reversed by siRNA knockdown of Nr4a2 (McQuown et al., 2011). Here, we replicated the finding that acute TSA treatment enhances LTP in hippocampal slices from wild-type mice, and we further demonstrated that HDAC inhibition is unable to enhance LTP in slices lacking NR4A activity. These data suggest that the NR4A transcription factors mediate the memory- and plasticity-enhancing effects of HDAC inhibitors. As histone acetylation-regulated and activity-dependent genes, the Nr4a family is rapidly transcribed in response to neuronal activity (Maxwell & Muscat, 2006; McQuown et al., 2011; Peña de Ortiz & Jamieson, 1996; Vecsey et al., 2007). The putative gene targets of NR4A include Bdnf and Fosl2, both of which are upregulated by HDAC inhibitor treatment (Hawk et al., 2012). Bdnf is of particular interest as a candidate by which the Nr4a family influences neuronal plasticity and memory and mediates the effects of histone deacetylase inhibition, because it is an acetylation-regulated gene and its expression coincides with a later wave of transcription (Cunha, Brambilla, & Thomas, 2010) that could be controlled by newly translated NR4A proteins. An important future direction will be to identify those NR4A target genes important for synaptic plasticity and memory.
Research into the pharmacological manipulation of nuclear receptors has shown that altering the function of RXR, LXR, and PPAR can rescue memory and memory deficits in mouse models of Alzheimer’s disease and neurodegeneration (Cramer et al., 2012; Jiang et al., 2008; Pedersen et al., 2006). RXR is able to form transcriptionally active heterodimers with LXR and PPAR, as well as the NR4A nuclear receptors; this partnering facilitates the transcription of the gene targets of either partner and could link the function of these receptors in the brain (Mangelsdorf & Evans, 1995; Maxwell & Muscat, 2006). Although the NR4A proteins are orphan receptors and lack the canonical nuclear receptor ligand-binding domain (Wansa et al., 2003), a number of small-molecule activators exist that target one or more of the NR4A transcription factors (Dubois, Hengerer, & Mattes, 2006; Inamoto et al., 2008; Li, Lee, & Safe, 2012). Impairing the function of the NR4A nuclear receptors attenuates memory, so could pharmacologically increasing their function enhance it? Such research would have important practical applications in studies of human aging and neuropsychiatric illness. In some human patients, Nr4a2 mutations have been identified and associated with schizophrenia (Buervenich et al., 2000; Xing, Zhang, Russell, & Post, 2006), while Nr4a2+/− mutant mice display schizophrenia-like behaviors (Rojas et al., 2007). In humans, decreased Nr4a2 gene expression has also been reported to accompany aging (Chu, Kompoliti, Cochran, Mufson, & Kordower, 2002). Pharmacological activation of NR4A could be used to compensate for transcriptional deficiency caused by mutation or loss of these transcription factors and could be utilized to counter the effects of age-related cognitive decline or NR4A dysfunction in schizophrenia.
Acknowledgments
We would like to thank Shane Poplawski, Alan Park, Michelle Dumoulin, and Josh Hawk for their comments on the article and the experimental design. This work was supported by predoctoral NRSA fellowship 1F31NS079019 to M. Bridi and NIH Grant R01-MH087463 to T. Abel.
References
- Abel T, Nguyen PV, Barad M, Deuel TA, Kandel ER, Bourtchouladze R. Genetic demonstration of a role for PKA in the late phase of LTP and in hippocampus-based long-term memory. Cell. 1997;88(5):615–626. doi: 10.1016/s0092-8674(00)81904-2. [DOI] [PubMed] [Google Scholar]
- Abel T, Nguyen PV. Regulation of hippocampus-dependent memory by cyclic AMP-dependent protein kinase. Progress in Brain Research. 2008;169:97–115. doi: 10.1016/S0079-6123(07)00006-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberini CM. Transcription factors in long-term memory and synaptic plasticity. Physiological Reviews. 2009;89(1):121–145. doi: 10.1152/physrev.00017.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buervenich S, Carmine A, Arvidsson M, Xiang F, Zhang Z, Sydow O, et al. NURR1 mutations in cases of schizophrenia and manic-depressive disorder. American Journal of Medical Genetics. 2000;96(6):808–813. doi: 10.1002/1096-8628(20001204)96:6<808::aid-ajmg23>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- Chu Y, Kompoliti K, Cochran EJ, Mufson EJ, Kordower JH. Age-related decreases in Nurr1 immunoreactivity in the human substantia nigra. The Journal of Comparative Neurology. 2002;450(3):203–214. doi: 10.1002/cne.10261. [DOI] [PubMed] [Google Scholar]
- Colón-Cesario WI, Martínez-Montemayor MM, Morales S, Félix J, Cruz J, Adorno M, et al. Knockdown of Nurr1 in the rat hippocampus: Implications to spatial discrimination learning and memory. Learning & Memory. 2006;13(6):734–744. doi: 10.1101/lm.407706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer PE, Cirrito JR, Wesson DW, Lee CYD, Karlo JC, Zinn AE, et al. ApoE-directed therapeutics rapidly clear β-amyloid and reverse deficits in AD mouse models. Science. 2012;335(6075):1503–1506. doi: 10.1126/science.1217697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha C, Brambilla R, Thomas KL. A simple role for BDNF in learning and memory? Frontiers in Molecular Neuroscience. 2010;3(1) doi: 10.3389/neuro.02.001.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragunow M, Abraham W, Hughes P. Activation of NMDA and muscarinic receptors induces nur-77 mRNA in hippocampal neurons. Molecular Brain Research. 1996;36:349–356. doi: 10.1016/0169-328x(95)00294-3. [DOI] [PubMed] [Google Scholar]
- Dubois C, Hengerer B, Mattes H. Identification of a potent agonist of the orphan nuclear receptor Nurr1. ChemMedChem. 2006;1(9):955–958. doi: 10.1002/cmdc.200600078. [DOI] [PubMed] [Google Scholar]
- Haettig J, Stefanko DP, Multani ML, Figueroa DX, McQuown SC, Wood MA. HDAC inhibition modulates hippocampus-dependent long-term memory for object location in a CBP-dependent manner. Learning & Memory. 2011;18:71–79. doi: 10.1101/lm.1986911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawk JD, Abel T. The role of NR4A transcription factors in memory formation. Brain Research Bulletin. 2011;85(1–2):21–29. doi: 10.1016/j.brainresbull.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawk JD, Bookout AL, Poplawski SG, Bridi M, Rao AJ, Sulewski ME, et al. NR4A nuclear receptors support memory enhancement by histone deacetylase inhibitors. The Journal of Clinical Investigation. 2012;122(10):3593–3602. doi: 10.1172/JCI64145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Kandel ER. Recruitment of long-lasting and protein kinase A-dependent long-term potentiation in the CA1 region of hippocampus requires repeated tetanization. Learning & Memory. 1994;1(1):74–82. [PubMed] [Google Scholar]
- Huang YY, Nguyen PV, Abel T, Kandel ER. Long-lasting forms of synaptic potentiation in the mammalian hippocampus. Learning & Memory. 1996;3(2–3):74–85. doi: 10.1101/lm.3.2-3.74. [DOI] [PubMed] [Google Scholar]
- Igaz LM, Vianna MRM, Medina JH, Izquierdo I. Two time periods of hippocampal mRNA synthesis are required for memory consolidation of fear-motivated learning. The Journal of Neuroscience. 2002;22(15):6781–6789. doi: 10.1523/JNEUROSCI.22-15-06781.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inamoto T, Papineni S, Chintharlapalli S, Cho SD, Safe S, Kamat AM. 1,1-Bis(3′-indolyl)-1-(p-chlorophenyl)methane activates the orphan nuclear receptor Nurr1 and inhibits bladder cancer growth. Molecular Cancer Therapeutics. 2008;7(12):3825–3833. doi: 10.1158/1535-7163.MCT-08-0730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Q, Lee CYD, Mandrekar S, Wilkinson B, Cramer P, Zelcer N, et al. ApoE promotes the proteolytic degradation of Abeta. Neuron. 2008;58(5):681–693. doi: 10.1016/j.neuron.2008.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MM, Michelhaugh SK, Bouhamdan M, Schmidt CJ, Bannon MJ. The transcription factor NURR1 exerts concentration-dependent effects on target genes mediating distinct biological processes. Frontiers in Neuroscience. 2011;5:135. doi: 10.3389/fnins.2011.00135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josselyn SA, Nguyen PV. CREB, synapes, and memory disorders: Past progress and future challenges. Current Drug Targets – CNS & Neurological Disorders. 2005;4(5):481–497. doi: 10.2174/156800705774322058. [DOI] [PubMed] [Google Scholar]
- Kovalovsky D, Refojo D, Liberman AC, Hochbaum D, Pereda MP, Coso OA, et al. Activation and induction of NUR77/NURR1 in corticotrophs by CRh/cAMP: Involvement of calcium, protein kinase A, and MAPK pathways. Molecular Endocrinology. 2002;16(7):1638–1651. doi: 10.1210/mend.16.7.0863. [DOI] [PubMed] [Google Scholar]
- Lattal KM, Barrett RM, Wood MA. Systemic or intrahippocampal delivery of histone deacetylase inhibitors facilitates fear extinction. Behavioral Neuroscience. 2007;121(5):1125–1131. doi: 10.1037/0735-7044.121.5.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemberger T, Parkitna JR, Chai M, Schütz G, Engblom D. CREB has a context-dependent role in activity-regulated transcription and maintains neuronal cholesterol homeostasis. The FASEB Journal. 2008;22(8):2872–2879. doi: 10.1096/fj.08-107888. [DOI] [PubMed] [Google Scholar]
- Levenson JM, O’Riordan KJ, Brown KD, Trinh MA, Molfese DL, Sweatt JD. Regulation of histone acetylation during memory formation in the hippocampus. The Journal of Biological Chemistry. 2004;279(39):40545–40559. doi: 10.1074/jbc.M402229200. [DOI] [PubMed] [Google Scholar]
- Li X, Lee SO, Safe S. Structure-dependent activation of NR4A2 (Nurr1) by 1,1-bis(3′-indolyl)-1-(aromatic)methane analogs in pancreatic cancer cells. Biochemical Pharmacology. 2012;83(10):1445–1455. doi: 10.1016/j.bcp.2012.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangelsdorf DJ, Evans RM. The RXR heterodimers and orphan receptors. Cell. 1995;83(6):841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- Martin SJ, Grimwood PD, Morris RGM. Synaptic plasticity and memory: An evaluation of the hypothesis. Annual Review of Neuroscience. 2000;23:649–711. doi: 10.1146/annurev.neuro.23.1.649. [DOI] [PubMed] [Google Scholar]
- Maxwell M, Muscat GEO. The NR4A subgroup: Immediate early response genes with pleiotropic physiological roles. Nuclear Receptor Signaling. 2006;4:e002. doi: 10.1621/nrs.04002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayford M, Bach ME, Huang YY, Wang L, Hawkins RD, Kandel ER. Control of memory formation through regulated expression of a CaMKII transgene. Science. 1996;274(5293):1678–1683. doi: 10.1126/science.274.5293.1678. [DOI] [PubMed] [Google Scholar]
- McNulty SE, Barrett RM, Vogel-Ciernia A, Malvaez M, Hernandez N, Davatolhagh MF, et al. Differential roles for Nr4a1 and Nr4a2 in object location vs. object recognition long-term memory. Learning & Memory. 2012;19(12):588–592. doi: 10.1101/lm.026385.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuown SC, Barrett RM, Matheos DP, Post RJ, Rogge GA, Alenghat T, et al. HDAC3 is a critical negative regulator of long-term memory formation. The Journal of Neuroscience. 2011;31(2):764–774. doi: 10.1523/JNEUROSCI.5052-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen PV, Abel T, Kandel ER. Requirement of a critical period of transcription for induction of a late phase of LTP. Science. 1994;265(5175):1104–1107. doi: 10.1126/science.8066450. [DOI] [PubMed] [Google Scholar]
- Nguyen PV, Kandel ER. Brief theta-burst stimulation induces a transcription-dependent late phase of LTP requiring cAMP in area CA1 of the mouse hippocampus. Learning & Memory. 1997;4(2):230–243. doi: 10.1101/lm.4.2.230. [DOI] [PubMed] [Google Scholar]
- Paulsen RE, Weaver CA, Fahrner TJ, Milbrandt J. Domains regulating transcriptional activity of the inducible orphan receptor NGFI-B. The Journal of Biological Chemistry. 1992;267(23):16491–19496. [PubMed] [Google Scholar]
- Pedersen WA, McMillan PJ, Kulstad JJ, Leverenz JB, Craft S, Haynatzki GR. Rosiglitazone attenuates learning and memory deficits in Tg2576 Alzheimer mice. Experimental Neurology. 2006;199(2):265–273. doi: 10.1016/j.expneurol.2006.01.018. [DOI] [PubMed] [Google Scholar]
- Pegoraro S, Broccard FD, Ruaro ME, Bianchini D, Avossa D, Pastore G, et al. Sequential steps underlying neuronal plasticity induced by a transient exposure to gabazine. The Journal of Cellular Physiology. 2010;222:713–728. doi: 10.1002/jcp.21998. [DOI] [PubMed] [Google Scholar]
- Peña de Ortiz S, Jamieson GA. HZF-3, an immediate-early orphan receptor homologous to NURR1/NOT: Induction upon membrane depolarization and seizures. Molecular Brain Research. 1996;38(1):1–13. doi: 10.1016/0169-328x(95)00263-r. [DOI] [PubMed] [Google Scholar]
- Peña de Ortiz S, Maldonado-Vlaar CS, Carrasquillo Y. Hippocampal expression of the orphan nuclear receptor gene hzf-3/nurr1 during spatial discrimination learning. Neurobiology of Learning and Memory. 2000;74(2) doi: 10.1006/nlme.1999.3952. [DOI] [PubMed] [Google Scholar]
- Pittenger C, Huang YY, Paletzki RF, Bourtchouladze R, Scanlin H, Vronskaya S, et al. Reversible inhibition of CREB/ATF transcription factors in region CA1 of the dorsal hippocampus disrupts hippocampus-dependent spatial memory. Neuron. 2002;34(3):447–462. doi: 10.1016/s0896-6273(02)00684-0. [DOI] [PubMed] [Google Scholar]
- Robert NM, Martin LJ, Tremblay JJ. The orphan nuclear receptor NR4A1 regulates insulin-like 3 gene transcription in Leydig cells. Biology of Reproduction. 2006;74(2):322–330. doi: 10.1095/biolreprod.105.044560. [DOI] [PubMed] [Google Scholar]
- Rojas P, Joodmardi E, Hong Y, Perlmann T, Ogren SO. Adult mice with reduced Nurr1 expression: An animal model for schizophrenia. Molecular Psychiatry. 2007;12(8):756–766. doi: 10.1038/sj.mp.4001993. [DOI] [PubMed] [Google Scholar]
- Ryan MM, Mason-Parker SE, Tate WP, Abraham WC, Williams JM. Rapidly induced gene networks following induction of long-term potentiation at perforant path synapses in vivo. Hippocampus. 2011;21:541–553. doi: 10.1002/hipo.20770. [DOI] [PubMed] [Google Scholar]
- Stafford JM, Raybuck JD, Ryabinin AE, Lattal KM. Increasing histone acetylation in the hippocampus-infralimbic network enhances fear extinction. Biological Psychiatry. 2012;72(1):25–33. doi: 10.1016/j.biopsych.2011.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanko DP, Barrett RM, Ly AR, Reolon GK, Wood MA. Modulation of long-term memory for object recognition via HDAC inhibition. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(23):9447–9452. doi: 10.1073/pnas.0903964106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vecsey CG, Baillie GS, Jaganath D, Havekes R, Daniels A, Wimmer M, et al. Sleep deprivation impairs cAMP signalling in the hippocampus. Nature. 2009;461(7267):1122–1125. doi: 10.1038/nature08488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vecsey CG, Hawk JD, Lattal KM, Stein JM, Fabian SA, Attner MA, et al. Histone deacetylase inhibitors enhance memory and synaptic plasticity via CREB:CBP-dependent transcriptional activation. The Journal of Neuroscience. 2007;27(23):6128–6140. doi: 10.1523/JNEUROSCI.0296-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpicelli F, Caiazzo M, Greco D, Consales C, Leone L, Perrone-Capano C, et al. Bdnf gene is a downstream target of Nurr1 transcription factor in rat midbrain neurons in vitro. Journal of Neurochemistry. 2007;102(2):441–453. doi: 10.1111/j.1471-4159.2007.04494.x. [DOI] [PubMed] [Google Scholar]
- Von Hertzen LSJ, Giese KP. Memory reconsolidation engages only a subset of immediate-early genes induced during consolidation. The Journal of Neuroscience. 2005;25(8):1935–1942. doi: 10.1523/JNEUROSCI.4707-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wansa KDSA, Harris JM, Yan G, Ordentlich P, Muscat GEO. The AF-1 domain of the orphan nuclear receptor NOR-1 mediates trans-activation, coactivator recruitment, and activation by the purine anti-metabolite 6-mercaptopurine. The Journal of Biological Chemistry. 2003;278:24776–24790. doi: 10.1074/jbc.M300088200. [DOI] [PubMed] [Google Scholar]
- Wesson DW, Borkowski AH, Landreth GE, Nixon RA, Levy E. Sensory network dysfunction, behavioral impairments, and their reversibility in an Alzheimer’s β-amyloidosis mouse model. The Journal of Neuroscience. 2011;31(44):15962–15971. doi: 10.1523/JNEUROSCI.2085-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing G, Zhang L, Russell S, Post R. Reduction of dopamine-related transcription factors Nurr1 and NGFI-B in the prefrontal cortex in schizophrenia and bipolar disorders. Schizophrenia Research. 2006;84(1):36–56. doi: 10.1016/j.schres.2005.11.006. [DOI] [PubMed] [Google Scholar]