Abstract
Pluripotent stem cells can differentiate into nearly all types of cells in the body. This unique potential provides significant promise for cell-based therapies to restore tissues or organs destroyed by injuries, degenerative diseases, aging, or cancer. The discovery of induced pluripotent stem cell (iPSC) technology offers a possible strategy to generate patient-specific pluripotent stem cells. However, because of concerns about the specificity, efficiency, kinetics, and safety of iPSC reprogramming, improvements or fundamental changes in this process are required before their effective clinical use. A chemical approach is regarded as a promising strategy to improve and change the iPSC process. Dozens of small molecules have been identified that can functionally replace reprogramming factors and significantly improve iPSC reprogramming. In addition to the prospect of deriving patient-specific tissues and organs from iPSCs, another attractive strategy for regenerative medicine is transdifferentiation—the direct conversion of one somatic cell type to another. Recent studies revealed a new paradigm of transdifferentiation: using transcription factors (TFs) employed in iPSC generation to induce transdifferentiation, or iPSC TF-based transdifferentiation. This transdifferentiation not only reveals and utilizes the developmentally plastic intermediates generated during iPSC reprogramming, but also produces a very wide range of cells, including expandable tissue-specific precursor cells. Here, we review recent progress of small-molecule approaches in the generation of iPSCs. In addition, we summarize the new concept of iPSC TF–based transdifferentiation and discuss its application in generating various lineage-specific cells, especially cardiovascular cells.
Keywords: Reprogramming, iPSC, small molecule, transdifferentiation and cardiovascular cell
Introduction
Pluripotent stem cells can self-renew indefinitely and undergo differentiation to produce all types of cells in adult bodies. They could be used to generate various cells or even tissues/organs for transplantation. Patients with injuries, degenerative disease, aging, or cancers would benefit from the realization of stem cell–based regenerative medicine. To avoid rejection by the patient’s immune system, cell-based therapies preferably use immune-matched donor cells, or the patient’s own cells, which could be derived from their own pluripotent stem cells.
Reprogramming to pluripotency
Various methods have been developed to reprogram cells to a pluripotent state. In the 1960s, somatic cell nuclear transfer (SCNT) was reported to erase lineage-specific signatures in the nuclei of a somatic cell and reprogram it to a totipotent state1, 2. So far, SCNT approach has been successfully performed in mouse and some other species, but not yet reported in human. Fusion of somatic cells with pluripotent cells was another method proven to enable reprogramming somatic cells to pluripotent cells3, however the utility of this method is limited because the resultant cells are tetraploid. Both methods take advantage of cellular materials to establish pluripotency in somatic cells; SCNT uses material from oocytes, and cell fusion uses material from pluripotent cells. Although SCNT and cell fusion-induced reprogramming are determinative and relatively fast and efficient, significant technical challenges (as well as ethical challenges in the case of SCNT) remain before they can be practical. In addition, they entail complex mixtures of known and undefined factors from oocytes or pluripotent cells to trigger reprogramming, making mechanistic studies more challenging.
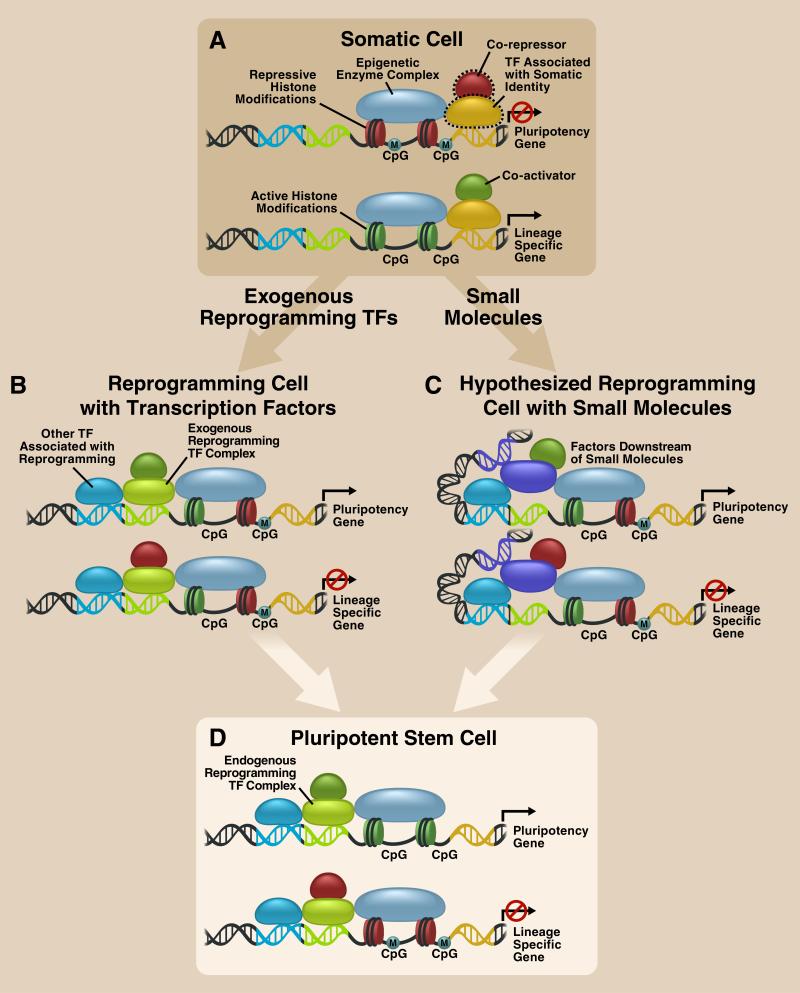
These barriers might be avoided by a new strategy in which mammalian somatic cells are reprogrammed to induced pluripotent stem cells (iPSCs) by ectopic expression of the pluripotent TFs Oct4, Sox2, Klf4, and c-Myc (or Nanog and Lin28 instead of Klf4 and c-Myc) 4-6. In practice, combined with efficient differentiation strategies, iPSCs would be valuable not only to derive functional cells for transplantation, but also to establish patient-specific disease models for drug discovery and development. Similar to the capability of cell-type specific TFs to maintain cell identity by binding to specific DNA sequences across the genome and achieving additional sequence-binding specificity and transcriptional and epigenetic regulation by forming complexes with co-regulatory factors, exogenous iPSC TFs (TFs overexpressed in generation of iPSCs) cooperatively remodel chromatin to activate expression of genes in the pluripotency network and to suppress expression of genes that promote differentiation. Additionally, through both co-occupancy with downstream effectors of various signaling pathways and recruitment of diverse epigenetic enzymes over the whole genome, specific chromosomal binding patterns of exogenous iPSC TFs during the reprogramming process contribute to the establishment of iPSC-specific signal transduction, transcriptional circuitry, and epigenetic pattern. (Figure 1)
Figure 1. Mechanisms of reprogramming to pluripotency induced by exogenous transcription factors and hypothetical mechanisms of small molecule-mediated reprogramming to pluripotency.
A. Somatic cell maintains its identity by transcription factor (TF) mediated activation of lineage specific genes, and TF or heterochromatin mediated silencing of pluripoteny genes.
B. In the paradigm of TF-induced reprogramming to pluripotency, exogenous reprogramming TF complexes (e.g., pluripotency TF complexes coming from oocyte in SCNT, pluripotent cell in cell fusion, or ectopically expressed iPSC TFs), which can recognize and bind to specific sequences across the whole genome, interact with other TFs binding to nearby or distal sites and recruit other transcription cofactors (e.g., activators, repressors, and epigenetic enzyme complexes) to co-occupy and epigenetically modify the genome in a sequence-specific manner. As a result, the gene expression profile and epigenetic pattern of somatic cells gradually becomes iPSC specific. C. Unlike TFs, small molecules do not have the exquisite ability of molecular recognition possessed by TFs, and cannot specifically interact with both DNA and other transcription cofactors. Consequently, small molecules would hypothetically target endogenous components (e.g., TFs in purple and epigenetic enzyme complexes in light blue) in somatic cells to indirectly initiate iPSC reprogramming.
D. Pluripotent stem cell generated by either a TF or small-molecule approach maintains its pluripotency by TF-mediated activation of pluripoteny genes and repression of lineage specific genes.
Although iPSC reprogramming is technically simpler than SCNT and cell fusion, it only induces a stochastic and nonspecific reprogramming process and is therefore less efficient and slower than SCNT and cell fusion. This difference reflects the possibility that iPSC TFs are core components, but not the complete machinery functional in efficient and specific reprogramming induced by both SCNT and cell fusion. Besides, iPSCs generated by conventional methods raise concerns about their safety (e.g., immunogenicity7 and the risk of tumorigenesis) for clinical applications, as they employ virus-mediated gene delivery, which results in genomic integration of exogenous sequence, and enforced expression of oncogenes.
To increase efficiency, accelerate kinetics, and reduce safety concerns, many improvements in methodology have been achieved by different groups. Several specific cell types were shown to enable reprogramming with higher efficiency and/or less number of exogenous iPSC TFs8. However, nearly all of them rely on overexpression of exogenous iPSC TFs and extra manipulations (e.g., administration of cytokines or small molecules) to get reprogrammed efficiently and rapidly. In addition to reprogramming using different starting cell types, methods using virus-free9-11, removable PiggyBac transposons12, minicircle systems13 or episomal systems14 have been developed. Despite their success in generating iPSCs, often without a genetic footprint, use of DNA constructs leaves the possibility of genomic integration of exogenous sequence. Other attempts to generate iPSCs by nonintegrating virus-mediated gene delivery cannot preclude the safety concerns raised by using viruses15-17. Although recombinant proteins or synthetic mRNAs can produce iPSCs, the protocols are costly and technically challenging18-21. Recently, microRNA was used to generate reprogrammed iPSCs, but the practical utility and robustness of this approach are uncertain22, 23.
Additionally, generation of iPSC with small molecules alone is being attempted. This promising strategy might eliminate many of the drawbacks (e.g., the risk of tumorigenesis from genomic integration of exogenous sequence or overexpression of oncogenes) of conventional and other improved iPSC reprogramming methods. Even if the final outcome of iPSC reprogramming induced by any method is the establishment of pluripotency associated gene expression profile and epigenetic pattern, the small molecule approach would employ a different process/mechanism from other methods to launch reprogramming process. In details, in other methods, exogenously introduced and pluripotency associated elements (e.g., TFs) trigger iPSC reprogramming by directly participating in and directing pluripotency-specific chromatin remodeling in somatic cells, whereas the small molecule approach indirectly initiates iPSC reprogramming by mediating endogenous, non-pluripotency-specific components in somatic cells (Figure 1). Therefore, at this point, successful induction of iPSCs by small molecules would fundamentally change the concept of iPSC reprogramming. Besides, all small molecules identified during development of this method would be possible candidates to further improve iPSC reprogramming and investigate the underlying mechanisms of this process. To date, many small molecules have been identified to modulate the induction of iPSCs, in both functionally replacing some exogenous iPSC TFs and significantly improving the efficiency and quality of iPSC reprogramming (Table 1).
Table 1.
Small molecules identified in iPSC research
| Name & Structure | Target(s)/function | Effect(s) |
|---|---|---|
BIX-01294
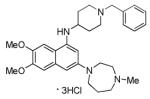
|
methyltransferase G9a inhibitor |
Enables reprogramming of somatic cells by ectopic expression of Oct4 and Klf4 only.46,47 |
RG108
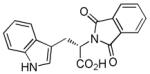
|
DNA methyltransferase inhibitor |
In the presence of BIX-01294, improves reprogramming of MEFs induced by Oct4 and Klf4.46,47 |
Bayk8644 (BayK)
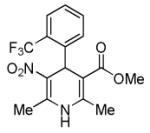
|
L-calcium channel agonist | In the presence of BIX-01294, improves reprogramming of MEFs induced by Oct4 and Klf4.46,47 |
Parnate
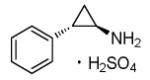
|
lysine-specific demethylase 1 inhibitor |
In combination with CHIR99021, enables reprogramming of human keratinocytes induced by Oct4 and Klf4.50 |
5-Azacytidine (5-azaC)
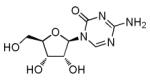
|
DNA methyltransferase inhibitor |
Improves MEF reprogramming efficiency with four iPSC TFs or under c-Myc-free condition.44,54 Facilitates erasure of epigenetic memory retained in established iPSCs.59 |
|
Suberoylanilide hydroxamic acid
(SAHA) |
HDAC inhibitor | Improves MEF reprogramming efficiency with four iPSC TFs.44,54 |
Trichostatin A (TSA)
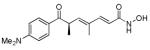
|
HDAC inhibitor | Improves MEF reprogramming efficiency with four iPSC TFs.44,54 Facilitates erasure of epigenetic memory retained in established iPSCs.59 |
Valproic acid (VPA)
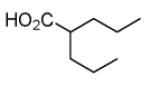
|
HDAC inhibitor | Improves MEF reprogramming efficiency with four iPSC TFs or under c-Myc-free condition.44,54 Enables reprogramming of human fibroblasts induced by Oct4 and Sox2.55 |
CHIR99021
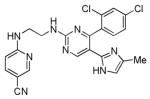
|
Glycogen synthase kinase 3β inhibitor |
Improves reprogramming efficiency of MEFs in the absence of Sox2 and cMyc.50 In combination with Parnate, enables reprogramming of human keratinocytes induced by Oct4 and Klf4.50 |
Kenpaullone
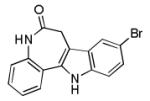
|
Glycogen synthase kinase 3β and cyclin-dependent kinases inhibitor |
Functionally replaces Klf4 in reprogramming of MEFs in the presence of Oct4, Sox2, and cMyc.68 |
SB431542
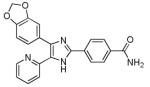
|
TGFβ receptor inhibitor | Enhances MEF reprogramming efficiency in the absence of c-Myc.75, 76 Enhances and accelerates reprogramming of human somatic cells.69 Functionally replaces Sox2 in MEF reprogramming.75,76 |
PD0325901
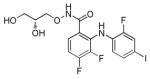
|
MEK inhibitor | Enhances and accelerates reprogramming of somatic cells.69 |
Thiazovivin
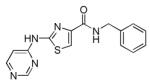
|
ROCK inhibitor | Enhances and accelerates reprogramming of human somatic cells.69 |
A-83-01
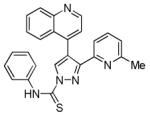
|
TGFβ receptor inhibitor | Enhances MEF reprogramming.70-72 In combination with AMI-5, enables reprogramming of MEFs transduced with Oct4 only.73 |
AMI-5
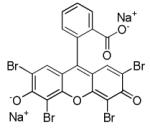
|
Protein arginine methyltransferase inhibitor |
In combination with A-83-01, enables reprogramming of MEFs transduced with Oct4 only.73 |
|
8-Bromoadenosine 3′, 5′-cyclic
monophosphate (8-Br-cAMP) 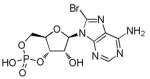
|
Protein kinase A activator | Improves the reprogramming efficiency of human neonatal foreskin fibroblasts transduced with all four iPSC TFs.74 |
Dasatinib
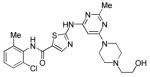
|
Src family kinases Inhibitor | Functionally replaces Sox2 in MEF reprogramming.75,76 |
PP1
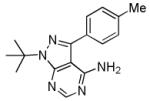
|
Src family kinases Inhibitor | Functionally replaces Sox2 in MEF reprogramming.75,76 |
iPYrazine (iPY)
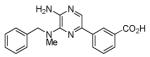
|
Src family kinases Inhibitor | Functionally replaces Sox2 in MEF reprogramming.75,76 |
PS48
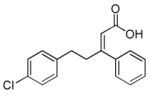
|
3′ phosphoinositide- dependent kinase 1 activator |
In combination with A-83-01, PD0325901 and sodium butyrate, enables reprogramming of human somatic cells transduced with Oct4 only.92 |
|
Sodium butyrate
|
HDAC inhibitor | In combination with A-83-01, PD0325901, and PS48, enables reprogramming of human somatic cells transduced with Oct4 only.92 |
Vitamin C
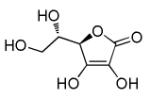
|
Antioxidant and enzyme cofactor |
Promotes iPSC generation from both mouse and human somatic cells.88 |
2, 4-Dinitrophenol
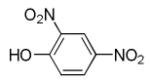
|
Oxidative phosphorylation un-coupler |
Enhances efficiency of reprogramming to iPSC.92 |
Fructose 2, 6-bisphosphate
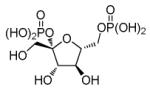
|
Phosphofructokinase 1 activator |
Enhances efficiency of reprogramming to iPSC.92 |
N-oxalylglycine
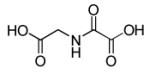
|
Prolyl-4-hydroxylase inhibitor |
Enhances efficiency of reprogramming to iPSC.92 |
Quercetin
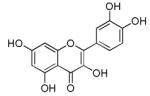
|
Hypoxia-inducible factor pathway activator |
Enhances efficiency of reprogramming to iPSC.92 |
Transdifferentiation
There are two promising strategies to regenerate tissue-specific cell types. In one, pluripotent cells are differentiated into various types of somatic cells. In the other, conventionally known as transdifferentiation, somatic cells are directly reprogrammed to another type of cells. Both approaches could be useful in the field of regenerative medicine and for disease modeling. Transdifferentiation has the advantage of avoiding the use of iPSCs to derive patient-specific cells, making the process faster and more efficient, reducing the risk of pluripotency associated tumorigenesis and probably avoiding immunogenicity identified recently in iPSCs7. In fact, many classic studies of transdifferentiation were thoroughly investigated before or around the time of discovery of iPSCs. For instance, ectopic expression of MyoD induced conversion of fibroblasts to myoblasts24, 25, expression of GATA1 or PU1 promoted the reciprocal transition between myeloid cells and megakaryocyte/erythroid cells26, expression of C/EBPα or C/EBPβ converted pre-B or pre-T cells to macrophages27, 28, and expression of C/EBPβ provoked conversion from pancreatic cells to hepatocytes29. However, most of these transitions were restricted to a relatively narrow lineage (e.g., hematopoietic, mesenchymal, or foregut endodermal system) and were induced by overexpression of a single TF. Success in generating iPSCs suggests constitutive expression of several TFs together might more efficiently induce transdifferentiation between less related cell types. This hypothesis has been verified. Many transdifferentiations induced by a set of lineage-specific TFs have been performed in vitro and in vivo30-40. Additionally, a new concept, iPSC TF–based transdifferentiation, was recently proposed and demonstrated: transient overexpression of iPSC TFs in conjunction with cell-type-specific signals can reprogram somatic cells into diverse lineage-specific cells without going through the pluripotent state. Furthermore, compared with conventional transdifferentiation, this new method has many advantages, such as the use of universal TF system and the ability to generate a multipotent progenitor population.
Here, we will categorize all small molecules identified in iPSC research, based on their function in target cells, and review them separately. Then we will review iPSC TF-induced transdifferentiation and its use to transdifferentiate somatic cells into various linage-specific cells, especially cardiovascular cells.
1. Epigenetic modifiers
During reprogramming, cells not only undergo transcriptional changes but also exhibit epigenetic changes in DNA methylation and histone modifications41-45. These changes convert the epigenetic pattern of somatic cells to an ESC-like state. Several small molecules that target enzymes involved in epigenetic modifications increase the efficiency of cellular reprogramming and sometimes can even functionally replace ectopic expression of certain TFs.
Dramatic changes in the histone methylation pattern are key features of iPSC reprogramming42, 43. Thus, reprogramming would be affected by small molecules that target enzymes involved in histone methylation or demethylation. BIX-01294 (BIX), an inhibitor of histone H3K9 mono- and dimethyltransferase G9a, enables reprogramming by ectopic expression of Oct4 and Klf4 in somatic cells. Moreover, after treatment of neural progenitor cells (NPCs) with BIX-01294 or treatment of mouse embryonic fibrolasts (MEFs) with BIX-01294 combined with a DNA methyltransferase inhibitor RG108 or an L-calcium channel agonist Bayk8644 (BayK), reprogramming with only two TFs (i.e., Oct4 and Klf4) was as efficient as reprogramming with four TFs46, 47. Consistently, G9a mediated H3K9 methylation is necessary for heterochromatinization and silencing of key pluripotency genes, such as Oct4 and Rex1, during early embryogenesis48. Parnate is an inhibitor of lysine-specific demethylase 1 (LSD1) mediated H3K4 demethylation, shown to globally increase H3K4 methylation as well as transcriptional derepression of LSD1 target gene Oct4 in P19 embryonal carcinoma cells49. In line with this observation, Parnate enabled two factors (Oct4 and Klf4) to induce conversion of human keratinocytes to iPSCs, when combined with CHIR99021 (an inhibitor of glycogen synthase kinase 3β )50.
Because inhibition of histone deacetylation and DNA methylation improves the reprogramming efficiency of SCNT51-53, it was hypothesized that such an approach might also aid in the establishment of iPSCs. As predicted, a DNA methyltransferase inhibitor, 5-azacytidine (5-azaC), or three histone deacetylase (HDAC) inhibitors (suberoylanilide hydroxamic acid (SAHA), trichostatin A (TSA) and valproic acid (VPA) ), improves reprogramming efficiency after transduction of the four iPSC TFs in MEFs44, 54. In addition, treatment with 5-azaC or VPA increases reprogramming efficiency under cMyc-free conditions44, 54. Further mechanistic studies showed 5-azaC induced a rapid and stable transition of a fully reprogrammed iPSC state from a partially reprogrammed state with DNA hypermethylation at pluripotency genes and administration of VPA in MEFs upregulated some ESC-specific genes meanwhile downregulated some MEF-specific genes44, 54. Moreover, VPA enables reprogramming of human fibroblasts transduced with only Oct4 and Sox255 and has been used to efficiently generate recombinant protein–induced pluripotent stem cells21.
Although mouse iPSC clones that are functionally identical to ESCs can be generated, subtle differences in gene expression and epigenetic patterns between iPSCs and ESCs could exist in some other iPSC clones41. Such differences could reflect residual expression and epigenetic pattern of original cell-type-specific genes, and/or gained genetic and epigenetic changes, contributed by incomplete epigenetic reprogramming or induced changes imposed by the reprogramming process56-62. To improve upon such incomplete reprogramming, epigenetic memory can be largely erased by treating established iPSCs with 5-azaC and trichostatin A59. On the other hand, such epigenetic memory may favor differentiation of iPSCs toward lineages related to its original cell type59, 61, 62.
In summary, appropriate application of small molecules that function as epigenetic modifiers, either individually or in combination, significantly increases iPSC reprogramming efficiency. Furthermore, administration of epigenetic modifiers can largely diminish epigenetic memory retained in iPSCs, and thereby improve the quality of reprogrammed iPSCs.
2. Signaling modulators
Consistent with the notion that signal transduction pathways mediated by extrinsic factors and intrinsic transcriptional network cooperate to maintain self-renewal and pluripotency of ESCs, signal transduction pathways and TFs act coordinately to reprogram somatic cells to iPSCs63. Therefore, small molecules that target signaling pathways would modulate reprogramming. Some such small molecules have been identified that increase reprogramming efficiency and can functionally replace some TFs in iPSC reprogramming.
Wnt signaling is important for maintaining the pluripotency of ESCs and for self-renewal of adult stem cells in multiple tissues64. In ESCs, TCF3, a downstream effecter of the Wnt signaling pathway, co-occupies genome-wide loci with master pluripotency regulators, such as Oct4 and Nanog, and acts as a transcriptional repressor for targeted genes, competing activity of these master pluripotency regulators. β-Catenin, which is stabilized by activation of the Wnt signaling pathway, directly interacts with TCF3 and reduces inhibitory effect of TCF3 on pluripotency. Therefore, the Wnt signaling pathway is regarded as an integral component of the core transcriptional circuitry in ESCs65. Indeed, β-catenin was involved in initial study of iPSC reprogramming and deletion of TCF3 could strikingly and rapidly enhance the efficiency of NPC reprogramming5, 66. Consistently, Wnt3a enhanced the reprogramming of MEFs to pluripotency in the absence of cMyc67; and CHIR99021, a glycogen synthase kinase 3β inhibitor that activates Wnt signaling, significantly improved reprogramming efficiency in MEFs in the absence of Sox2 and cMyc50. In the same study, treatment with CHIR99021 and Parnate converted human keratinocytes to iPSCs upon ectopic expression of Oct4 and Klf450. Another glycogen synthase kinase 3β inhibitor, kenpaullone, which also inhibits other kinases, could replace Klf4 in reprogramming of MEFs in the presence of Oct4, Sox2, and cMyc. Interestingly, the role of kenpaullone in this process could not be replaced by CHIR99021, and the mechanism is unknown68.
During reprogramming, mesenchymal-type fibroblasts undergo dramatic epithelial-like morphological changes and correlated gene expression changes, such as upregulation of E-cadherin (which is highly expressed in pluripotent cells) and concomitant downregulation of Snail. This so-called mesenchymal-to epithelial-transition (MET) inevitably occurs during successful reprogramming of cells. Therefore, small molecules that facilitate the MET process were hypothesized to enhance reprogramming69. Indeed, small molecules that target three known MET mechanisms (for derepression of epithelial phenotype with upregulation and stabilization of E-cadherin), including inhibition of TGFβ receptor (by SB431542), MEK (by PD0325901), or ROCK (by thiazovivin), individually or in combination significantly enhanced reprogramming of human somatic cells and increased reprogramming speed69. This MET mechanism was further characterized in three subsequent studies, where additional small molecules that inhibit the TGFβ pathway or upregulate E-cadherin were used70-72. More recently, another TGFβ receptor inhibitor, A-83-01, combined with a protein arginine methyltransferase inhibitor, AMI-5, enabled reprogramming of MEFs transduced with Oct4 alone73.
A few other small molecules that affect many other signaling pathways also facilitate iPSC reprogramming. A cyclic AMP analog, 8-bromoadenosine 3′, 5′-cyclic monophosphate (8-Br-cAMP), improved the reprogramming efficiency of human neonatal foreskin fibroblasts transduced with all four iPSC TFs74. Inhibitors of Src family kinases, including Dasatinib, PP1, and iPYrazine (iPY), sufficiently supported reprogramming of MEFs in the absence of exogenous Sox2, as efficiently as TGFβ inhibitors75, 76. All these observations are consistent with previously reports in which the PKA pathway and Src family kinases were shown to influence ESC self-renewal and/or differentiation77-79.
In summary, small-molecule modulators of signaling pathways, individually or in combination, and sometimes even together with epigenetic modifiers, induce reprogramming with higher efficiency and/or fewer exogenous TFs, by affecting the integrated cellular network consisting of both signaling pathways and transcriptional circuitry.
3. Cell senescence alleviators
Cell senescence is typically seen during cellular reprogramming and is considered one of the barriers of reprogramming that account for the slow kinetics and low efficiency of this process80. Indeed, the early stage of iPSC reprogramming entails a stress response with characteristics of cell senescence, including upregulated expression of p53, p21CIP1, and p16Ink4a/p19Arf 81. Furthermore, downregulating the expression of any of these genes increased the efficiency and speed of iPSC reprogramming82-87. A natural antioxidant, vitamin C, promoted iPSC generation from both mouse and human somatic cells through indirect reduction of p53 and p21CIP1 expression and partial alleviation of cell senescence88.
4. Metabolism regulators
Differentiated adult somatic cells typically use mitochondrial oxidative phosphorylation for cell growth, whereas pluripotent stem cells mainly use glycolytic metabolism89-91. In rapidly proliferating pluripotent stem cells, glycolytic metabolism more effectively produces various macromolecular precursors to meet the metabolic/energy demand and generates fewer reactive oxygen species, which induce oxidative stress89, 91. Therefore, transition from mitochondrial oxidative phosphorylation to glycolytic metabolism would be a crucial step in iPSC reprogramming91, 92. This metabolic reprogramming is further supported by the ability of hypoxia-inducible factor 1α and cMyc to promote glycolytic metabolism and to improve the efficiency of iPSC reprogramming90, 93. Consistently, a recent study showed PS48, a small-molecule activator of 3′ phosphoinositide-dependent kinase 1, combined with A-83-01, PD0325901, and sodium butyrate (a HDAC inhibitor), enabled reprogramming of human adult keratinocytes, umbilical vein endothelial cells, or amniotic fluid derived cells transduced with only Oct4, in which PS48 activated phosphatidylinositol 3-kinase (PI3K)/Akt pathway was shown to upregulate expression of several key glycolytic genes and facilitate the conversion from mitochondrial oxidative phosphorylation to glycolytic metabolism92. Remarkably, this study also revealed iPSC reprogramming efficiency was significantly increased by several other small molecules that enhance glycolytic metabolism and/or depress mitochondrial oxidative phosphorylation, including 2, 4-dinitrophenol (an oxidative phosphorylation uncoupler), fructose 2, 6-bisphosphate (an activator of phosphofructokinase 1), N-oxalylglycine (an inhibitor of prolyl-4-hydroxylase), and quercetin (an activator of hypoxia-inducible factor pathway)92. In a word, all evidences associated with these small molecules confirm the notion of metabolic reprogramming.
5. Trandifferentiation
Built upon iPSC reprogramming strategy and previous studies on cellular transdifferentiation restricted within several lineages, functional neurons, cardiomyocytes, hepatocytes, or macrophage-like cells can be converted from fibroblasts by ectopic expression of multiple lineage-specific TFs or microRNAs30-38. Furthermore, transdifferentiation has also been induced with lineage-specific TFs in vivo39, 40. However, transdifferentiated cells generated by ectopic expression of lineage-specific TFs were mostly one type of terminally differentiated cells. Therefore, conventional transdifferentiation typically would not allow isolation, expansion, and characterization of the reprogrammed cells, all of which are prerequisite for clinical application. In a word, the restricted proliferative capacity and limited cell type diversity of these transdifferentiated cells may substantially compromise their potential application in regenerative therapy.
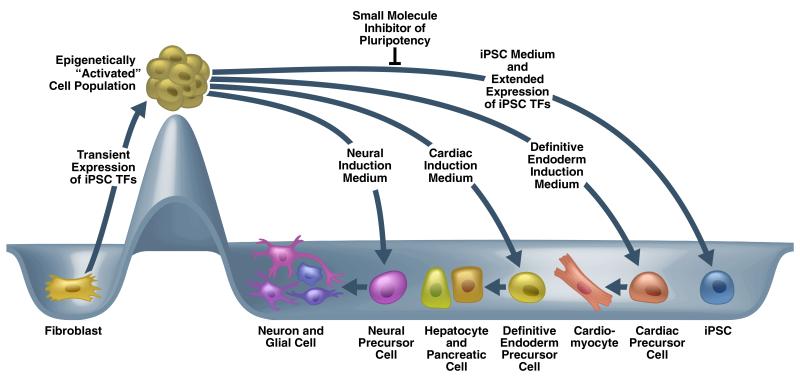
Recently, a new paradigm of transdifferentiation was devised to generate various lineage-specific precursor cells by combining transient overexpression of the iPSC TFs and treatment with soluble signaling molecules. The basis of this universal transdifferentiation strategy is that conventional iPSC reprogramming proceeds as a slow, step-wise, and nondeterminative process that gives rise to iPSCs only at the late stage and with low efficiency and to populations of epigenetically unstable/plastic cells at early and intermediate stages. In addition, generating iPSCs requires an extended period (e.g., 8–12 days) of iPSC TF expression and a specific signaling environment (e.g., leukemia inhibitory factor). These observations suggest that temporal iPSC TF expression combined with different signaling environments (e.g., growth factors/cytokines, and small-molecule modulators of signaling pathways) would dictate reprogrammed cell fate. This would be consistent with the notion that cell-type-specific TFs direct different cell fates cooperatively with interacted signaling downstream TFs. Indeed, this concept and strategy was demonstrated by direct conversion of fibroblasts to cardiac94, neural95, or definitive endoderm (unpublished data) precursor cells using transient expression of iPSC TFs (e.g., 4 days), followed by treatment with BMP4, FGF4, or Activin-A, respectively. (Figure 2) Compared with conventional transdifferentiation, in which different cell specifications are determined by ectopic expression of different sets of lineage specific TFs, all iPSC TF-based transdifferentiations share the same TFs in the initial step: transient overexpression of iPSC TFs. Such transient gene expression might be more easily replaced with safer and more convenient methods without genetic modifications. Next, we will separately review iPSC TF–induced trandifferentiation toward different lineages.
Figure 2. A simplified and conceptual paradigm of iPSC TF–based transdifferentiation.
Temporally restricted overexpression of iPSC TFs in fibroblasts leads to the rapid generation of epigenetically “activated” cells, which can be further reprogrammed to iPSC by both extended expression of iPSC TFs and culture of iPSC medium, and parallelly can be coaxed by other signals and small molecule inhibitor of pluripotency to “relax” back into various differentiated state(s), ultimately giving rise to somatic cells entirely distinct from the starting population.
5.1 iPSC TF–based trandifferentiation of fibroblasts into cardiac cells
The mammalian heart lacks significant regenerative capacity, so transplantation of autologous cardiac cells generated in vitro is considered a possible therapy for heart disease. In a previous report, three cardiac TFs, Gata4, Mef2c, and Tbx5, enabled transdifferentiation of mouse postnatal cardiac or dermal fibroblasts directly into cardiomyocyte-like cells31. However, it took several weeks to generate beating cardiomyocytes, and the efficiency was very low. As an alternative method, temporal expression of iPSC TFs (Oct4/Sox2/Klf4/cMyc) in mouse fibroblasts for only 4–6 days with a JAK inhibitor and without LIF, followed by treatment with BMP4 in a chemically defined medium for additional 5 days, resulted in a large number of spontaneously contracting cardiomyocytes94. This process was further characterized as a direct transdifferentiation, during which no pluripotent intermediates arose. Furthermore, an inverse relationship was demonstrated between induction of cardiomyocytes and generation of iPSCs, which could explain why cardiogenic transdifferentiation was facilitated by small-molecule inhibition of pluripotency but was impaired by prolonged overexpression of iPSC TFs94.
Compared with the cardiac TF induced transdifferentiation, cardiogenic transdifferentiation triggered by transient expression of iPSC TFs followed by treatment with BMP4 is much faster (first spontaneous contractions beginning after 11 days versus 4–5 weeks) and more efficient.31, 94 Moreover, multipotent cardiovascular precursor cells arose at an intermediate stage, as suggested by the pattern of gene expression during iPSC TF induced transdifferentiation94. If such cells could be isolated and expanded in vitro, they would eventually be a renewable source for unlimited amounts of cardiomyocytes and other terminally differentiated cardiovascular cells as well. Therefore, relative to cardiac TF induced terminally differentiated cardiomyocytes, which are restricted in cell type and limited in capacity to propagate after transplantation in vivo31, these multipotent cardiovascular precursor cells may be a more versatile cell source for modeling cardiovascular disease and for cell-based therapy.
For ultimate clinical applications, it would be highly desirable to develop a more robust condition for human system without any genetic modifications and safety concerns. Knowledge and techniques gained from development of iPSC reprogramming might be especially helpful to improve this new method of cardiogenic transdifferentiation.
5.2 iPSC TF–based trandifferentiation of fibroblasts toward other lineages
In addition to cardiac cells, other lineage-specific precursor cells, such as neural and definitive endoderm precursor cells, have also been generated from mouse and human fibroblasts by the same paradigm with different signaling molecules95. In these studies, transient expression (4–6 days) of iPSC TFs in fibroblasts was the initial step shared by all. Another shared step was treatment with a JAK inhibitor to suppress the LIF-STAT3 pathway during iPSC reprogramming, which would prevent establishment of pluripotency in reprogrammed cells and facilitate the generation of developmentally plastic intermediate cells. The epigenetically activated cells were treated with FGF4 to generate neural precursor cells or with Activin-A to generate definitive endoderm precursor cells. Importantly, the efficiently and fast converted neural precursor cells could be expanded for serial passages and then differentiated into mature and subtype-specific neuronal cells and glial cells95. Additionally, human fibroblasts have been directly converted to multipotent blood progenitors by prolonged ectopic expression of Oct4 and treatment with IGFII, bFGF, Flt3L and SCF96.
In summary, cardiovascular cells and other lineage-specific precursor cells can be directly converted from iPSC TF-induced transdifferentiation, which implies that such strategy would provide a general platform to induce a broad range of cells for various applications.
Perspectives
The stem cell field has embarked on exciting discoveries that both iPSCs and lineage specific cells can be reprogrammed from somatic cells by ectopic expression of iPSC TFs. Because of the close relationship between these two types of reprogramming, they share some technical challenges and safety considerations that need to be addressed before their clinical applications. To date, several strategies have overcome these hurdles in respect of iPSC reprogramming, and some may also work well in iPSC TF–based transdifferentiation. As reviewed above, regarding iPSC reprogramming, small molecules are not only valuable to significantly promote cellular reprogramming and functionally substitute ectopic expression of TFs, but also provide insights into molecular mechanism underlying this process. Even though pleiotropy of small molecules may result in some side effects compromising the desired reprogramming process, appropriate employment of small molecule combinations would largely diminish these side effects and potentially have synergistic effects on reprogramming. In the future, complete small-molecule-based reprogramming will fundamentally change the reprogramming paradigm through a mechanism that involves activation of endogenous TFs by small molecules rather than by exogenously provided reprogramming TFs. Moreover, the success of small-molecule based transdifferentiation would greatly reduce safety concerns in the clinical application of reprogrammed cells by avoiding issues caused both by the generation of pluripotent cells (e.g., the risk of pluripotency associated tumorigenesis) and the introduction of exogenous TFs. Therefore, better understanding of these reprogramming processes and further development of these small molecules may ultimately be useful for in vivo stem cell biology and therapy.
Acknowledgements
We thank Saiyong Zhu, Mingliang Zhang, Baoming Nie, Yu Zhang, Chen Yu, and other members in Ding lab for their constructive discussions, Stephen Ordway, and Gary Howard for editing the manuscript, and the Gladstone Graphics Department for assistance on figures.
Sources of Funding Sheng Ding is supported by funding from NICHD, NHLBI, NEI, and NIMH/NIH, California Institute for Regenerative Medicine, Prostate Cancer Foundation, and the Gladstone Institute.
Nonstandard abbreviations
- iPSC
induced pluripotent stem cell
- ESC
embryonic stem cell
- NPC
neural progenitor cell
- MEF
mouse embryonic fibroblast
- TF
transcription factor
- SCNT
somatic cell nuclear transfer
- MET
mesenchymal-to-epithelial-transition
- LSD1
lysine-specific demethylase 1
- PI3K
phosphatidylinositol 3-kinase
- BIX
BIX-01294
- BayK
Bayk8644
- 5-azaC
5-azacytidine
- HDAC
histone deacetylase
- SAHA
suberoylanilide hydroxamic acid
- TSA
trichostatin A
- VPA
valproic acid
- 8-Br-cAMP
8-bromoadenosine 3′, 5′-cyclic monophosphate
- iPY
iPYrazine
Footnotes
Disclosures None
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference
- 1.Gurdon JB, Wilmut I. Nuclear transfer to eggs and oocytes. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a002659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jullien J, Pasque V, Halley-Stott RP, Miyamoto K, Gurdon JB. Mechanisms of nuclear reprogramming by eggs and oocytes: a deterministic process? Nat Rev Mol Cell Biol. 2011;12:453–459. doi: 10.1038/nrm3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yamanaka S, Blau HM. Nuclear reprogramming to a pluripotent state by three approaches. Nature. 2010;465:704–712. doi: 10.1038/nature09229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 5.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 6.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, Slukvin, Thomson JA. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 7.Zhao T, Zhang ZN, Rong Z, Xu Y. Immunogenicity of induced pluripotent stem cells. Nature. 2011;474:212–215. doi: 10.1038/nature10135. [DOI] [PubMed] [Google Scholar]
- 8.Gonzalez F, Boue S, Izpisua Belmonte JC. Methods for making induced pluripotent stem cells: reprogramming a la carte. Nat Rev Genet. 2011;12:231–242. doi: 10.1038/nrg2937. [DOI] [PubMed] [Google Scholar]
- 9.Kaji K, Norrby K, Paca A, Mileikovsky M, Mohseni P, Woltjen K. Virus-free induction of pluripotency and subsequent excision of reprogramming factors. Nature. 2009;458:771–775. doi: 10.1038/nature07864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okita K, Hong H, Takahashi K, Yamanaka S. Generation of mouse-induced pluripotent stem cells with plasmid vectors. Nat Protoc. 2010;5:418–428. doi: 10.1038/nprot.2009.231. [DOI] [PubMed] [Google Scholar]
- 11.Okita K, Nakagawa M, Hyenjong H, Ichisaka T, Yamanaka S. Generation of mouse induced pluripotent stem cells without viral vectors. Science. 2008;322:949–953. doi: 10.1126/science.1164270. [DOI] [PubMed] [Google Scholar]
- 12.Woltjen K, Michael IP, Mohseni P, Desai R, Mileikovsky M, Hamalainen R, Cowling R, Wang W, Liu P, Gertsenstein M, Kaji K, Sung HK, Nagy A. piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature. 2009;458:766–770. doi: 10.1038/nature07863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jia F, Wilson KD, Sun N, Gupta DM, Huang M, Li Z, Panetta NJ, Chen ZY, Robbins RC, Kay MA, Longaker MT, Wu JC. A nonviral minicircle vector for deriving human iPS cells. Nat Methods. 2010;7:197–199. doi: 10.1038/nmeth.1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yu J, Hu K, Smuga-Otto K, Tian S, Stewart R, Slukvin, Thomson JA. Human induced pluripotent stem cells free of vector and transgene sequences. Science. 2009;324:797–801. doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seki T, Yuasa S, Oda M, Egashira T, Yae K, Kusumoto D, Nakata H, Tohyama S, Hashimoto H, Kodaira M, Okada Y, Seimiya H, Fusaki N, Hasegawa M, Fukuda K. Generation of induced pluripotent stem cells from human terminally differentiated circulating T cells. Cell Stem Cell. 2010;7:11–14. doi: 10.1016/j.stem.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 16.Stadtfeld M, Nagaya M, Utikal J, Weir G, Hochedlinger K. Induced pluripotent stem cells generated without viral integration. Science. 2008;322:945–949. doi: 10.1126/science.1162494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou W, Freed CR. Adenoviral gene delivery can reprogram human fibroblasts to induced pluripotent stem cells. Stem Cells. 2009;27:2667–2674. doi: 10.1002/stem.201. [DOI] [PubMed] [Google Scholar]
- 18.Kim D, Kim CH, Moon JI, Chung YG, Chang MY, Han BS, Ko S, Yang E, Cha KY, Lanza R, Kim KS. Generation of human induced pluripotent stem cells by direct delivery of reprogramming proteins. Cell Stem Cell. 2009;4:472–476. doi: 10.1016/j.stem.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Warren L, Manos PD, Ahfeldt T, Loh YH, Li H, Lau F, Ebina W, Mandal PK, Smith ZD, Meissner A, Daley GQ, Brack AS, Collins JJ, Cowan C, Schlaeger TM, Rossi DJ. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7:618–630. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yakubov E, Rechavi G, Rozenblatt S, Givol D. Reprogramming of human fibroblasts to pluripotent stem cells using mRNA of four transcription factors. Biochem Biophys Res Commun. 2010;394:189–193. doi: 10.1016/j.bbrc.2010.02.150. [DOI] [PubMed] [Google Scholar]
- 21.Zhou H, Wu S, Joo JY, Zhu S, Han DW, Lin T, Trauger S, Bien G, Yao S, Zhu Y, Siuzdak G, Scholer HR, Duan L, Ding S. Generation of induced pluripotent stem cells using recombinant proteins. Cell Stem Cell. 2009;4:381–384. doi: 10.1016/j.stem.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anokye-Danso F, Trivedi CM, Juhr D, Gupta M, Cui Z, Tian Y, Zhang Y, Yang W, Gruber PJ, Epstein JA, Morrisey EE. Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell. 2011;8:376–388. doi: 10.1016/j.stem.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miyoshi N, Ishii H, Nagano H, Haraguchi N, Dewi DL, Kano Y, Nishikawa S, Tanemura M, Mimori K, Tanaka F, Saito T, Nishimura J, Takemasa I, Mizushima T, Ikeda M, Yamamoto H, Sekimoto M, Doki Y, Mori M. Reprogramming of mouse and human cells to pluripotency using mature microRNAs. Cell Stem Cell. 2011;8:633–638. doi: 10.1016/j.stem.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 24.Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51:987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- 25.Schafer BW, Blakely BT, Darlington GJ, Blau HM. Effect of cell history on response to helix-loop-helix family of myogenic regulators. Nature. 1990;344:454–458. doi: 10.1038/344454a0. [DOI] [PubMed] [Google Scholar]
- 26.Graf T. Differentiation plasticity of hematopoietic cells. Blood. 2002;99:3089–3101. doi: 10.1182/blood.v99.9.3089. [DOI] [PubMed] [Google Scholar]
- 27.Laiosa CV, Stadtfeld M, Xie H, de Andres-Aguayo L, Graf T. Reprogramming of committed T cell progenitors to macrophages and dendritic cells by C/EBP alpha and PU.1 transcription factors. Immunity. 2006;25:731–744. doi: 10.1016/j.immuni.2006.09.011. [DOI] [PubMed] [Google Scholar]
- 28.Xie H, Ye M, Feng R, Graf T. Stepwise reprogramming of B cells into macrophages. Cell. 2004;117:663–676. doi: 10.1016/s0092-8674(04)00419-2. [DOI] [PubMed] [Google Scholar]
- 29.Shen CN, Slack JM, Tosh D. Molecular basis of transdifferentiation of pancreas to liver. Nat Cell Biol. 2000;2:879–887. doi: 10.1038/35046522. [DOI] [PubMed] [Google Scholar]
- 30.Feng R, Desbordes SC, Xie H, Tillo ES, Pixley F, Stanley ER, Graf T. PU.1 and C/EBPalpha/beta convert fibroblasts into macrophage-like cells. Proc Natl Acad Sci U S A. 2008;105:6057–6062. doi: 10.1073/pnas.0711961105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ieda M, Fu JD, Delgado-Olguin P, Vedantham V, Hayashi Y, Bruneau BG, Srivastava D. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pfisterer U, Kirkeby A, Torper O, Wood J, Nelander J, Dufour A, Bjorklund A, Lindvall O, Jakobsson J, Parmar M. Direct conversion of human fibroblasts to dopaminergic neurons. Proc Natl Acad Sci U S A. 2011;108:10343–10348. doi: 10.1073/pnas.1105135108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Sudhof TC, Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Caiazzo M, Dell’Anno MT, Dvoretskova E, Lazarevic D, Taverna S, Leo D, Sotnikova TD, Menegon A, Roncaglia P, Colciago G, Russo G, Carninci P, Pezzoli G, Gainetdinov RR, Gustincich S, Dityatev A, Broccoli V. Direct generation of functional dopaminergic neurons from mouse and human fibroblasts. Nature. 2011;476:224–227. doi: 10.1038/nature10284. [DOI] [PubMed] [Google Scholar]
- 35.Huang P, He Z, Ji S, Sun H, Xiang D, Liu C, Hu Y, Wang X, Hui L. Induction of functional hepatocyte-like cells from mouse fibroblasts by defined factors. Nature. 2011;475:386–389. doi: 10.1038/nature10116. [DOI] [PubMed] [Google Scholar]
- 36.Pang ZP, Yang N, Vierbuchen T, Ostermeier A, Fuentes DR, Yang TQ, Citri A, Sebastiano V, Marro S, Sudhof TC, Wernig M. Induction of human neuronal cells by defined transcription factors. Nature. 2011;476:220–223. doi: 10.1038/nature10202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sekiya S, Suzuki A. Direct conversion of mouse fibroblasts to hepatocyte-like cells by defined factors. Nature. 2011;475:390–393. doi: 10.1038/nature10263. [DOI] [PubMed] [Google Scholar]
- 38.Yoo AS, Sun AX, Li L, Shcheglovitov A, Portmann T, Li Y, Lee-Messer C, Dolmetsch RE, Tsien RW, Crabtree GR. MicroRNA-mediated conversion of human fibroblasts to neurons. Nature. 2011;476:228–231. doi: 10.1038/nature10323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takeuchi JK, Bruneau BG. Directed transdifferentiation of mouse mesoderm to heart tissue by defined factors. Nature. 2009;459:708–711. doi: 10.1038/nature08039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou Q, Brown J, Kanarek A, Rajagopal J, Melton DA. In vivo reprogramming of adult pancreatic exocrine cells to beta-cells. Nature. 2008;455:627–632. doi: 10.1038/nature07314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chin MH, Mason MJ, Xie W, Volinia S, Singer M, Peterson C, Ambartsumyan G, Aimiuwu O, Richter L, Zhang J, Khvorostov I, Ott V, Grunstein M, Lavon N, Benvenisty N, Croce CM, Clark AT, Baxter T, Pyle AD, Teitell MA, Pelegrini M, Plath K, Lowry WE. Induced pluripotent stem cells and embryonic stem cells are distinguished by gene expression signatures. Cell Stem Cell. 2009;5:111–123. doi: 10.1016/j.stem.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hawkins RD, Hon GC, Lee LK, Ngo Q, Lister R, Pelizzola M, Edsall LE, Kuan S, Luu Y, Klugman S, Antosiewicz-Bourget J, Ye Z, Espinoza C, Agarwahl S, Shen L, Ruotti V, Wang W, Stewart R, Thomson JA, Ecker JR, Ren B. Distinct epigenomic landscapes of pluripotent and lineage-committed human cells. Cell Stem Cell. 2010;6:479–491. doi: 10.1016/j.stem.2010.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maherali N, Sridharan R, Xie W, Utikal J, Eminli S, Arnold K, Stadtfeld M, Yachechko R, Tchieu J, Jaenisch R, Plath K, Hochedlinger K. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell. 2007;1:55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 44.Mikkelsen TS, Hanna J, Zhang X, Ku M, Wernig M, Schorderet P, Bernstein BE, Jaenisch R, Lander ES, Meissner A. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sridharan R, Tchieu J, Mason MJ, Yachechko R, Kuoy E, Horvath S, Zhou Q, Plath K. Role of the murine reprogramming factors in the induction of pluripotency. Cell. 2009;136:364–377. doi: 10.1016/j.cell.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shi Y, Desponts C, Do JT, Hahm HS, Scholer HR, Ding S. Induction of pluripotent stem cells from mouse embryonic fibroblasts by Oct4 and Klf4 with small-molecule compounds. Cell Stem Cell. 2008;3:568–574. doi: 10.1016/j.stem.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 47.Shi Y, Do JT, Desponts C, Hahm HS, Scholer HR, Ding S. A combined chemical and genetic approach for the generation of induced pluripotent stem cells. Cell Stem Cell. 2008;2:525–528. doi: 10.1016/j.stem.2008.05.011. [DOI] [PubMed] [Google Scholar]
- 48.Feldman N, Gerson A, Fang J, Li E, Zhang Y, Shinkai Y, Cedar H, Bergman Y. G9a-mediated irreversible epigenetic inactivation of Oct-3/4 during early embryogenesis. Nat Cell Biol. 2006;8:188–194. doi: 10.1038/ncb1353. [DOI] [PubMed] [Google Scholar]
- 49.Lee MG, Wynder C, Schmidt DM, McCafferty DG, Shiekhattar R. Histone H3 lysine 4 demethylation is a target of nonselective antidepressive medications. Chem Biol. 2006;13:563–567. doi: 10.1016/j.chembiol.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 50.Li W, Zhou H, Abujarour R, Zhu S, Young Joo J, Lin T, Hao E, Scholer HR, Hayek A, Ding S. Generation of human-induced pluripotent stem cells in the absence of exogenous Sox2. Stem Cells. 2009;27:2992–3000. doi: 10.1002/stem.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Blelloch R, Wang Z, Meissner A, Pollard S, Smith A, Jaenisch R. Reprogramming efficiency following somatic cell nuclear transfer is influenced by the differentiation and methylation state of the donor nucleus. Stem Cells. 2006;24:2007–2013. doi: 10.1634/stemcells.2006-0050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kishigami S, Mizutani E, Ohta H, Hikichi T, Thuan NV, Wakayama S, Bui HT, Wakayama T. Significant improvement of mouse cloning technique by treatment with trichostatin A after somatic nuclear transfer. Biochem Biophys Res Commun. 2006;340:183–189. doi: 10.1016/j.bbrc.2005.11.164. [DOI] [PubMed] [Google Scholar]
- 53.Rybouchkin A, Kato Y, Tsunoda Y. Role of histone acetylation in reprogramming of somatic nuclei following nuclear transfer. Biol Reprod. 2006;74:1083–1089. doi: 10.1095/biolreprod.105.047456. [DOI] [PubMed] [Google Scholar]
- 54.Huangfu D, Maehr R, Guo W, Eijkelenboom A, Snitow M, Chen AE, Melton DA. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat Biotechnol. 2008;26:795–797. doi: 10.1038/nbt1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huangfu D, Osafune K, Maehr R, Guo W, Eijkelenboom A, Chen S, Muhlestein W, Melton DA. Induction of pluripotent stem cells from primary human fibroblasts with only Oct4 and Sox2. Nat Biotechnol. 2008;26:1269–1275. doi: 10.1038/nbt.1502. [DOI] [PubMed] [Google Scholar]
- 56.Ghosh Z, Wilson KD, Wu Y, Hu S, Quertermous T, Wu JC. Persistent donor cell gene expression among human induced pluripotent stem cells contributes to differences with human embryonic stem cells. PLoS One. 2010;5:e8975. doi: 10.1371/journal.pone.0008975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Marchetto MC, Yeo GW, Kainohana O, Marsala M, Gage FH, Muotri AR. Transcriptional signature and memory retention of human-induced pluripotent stem cells. PLoS One. 2009;4:e7076. doi: 10.1371/journal.pone.0007076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stadtfeld M, Apostolou E, Akutsu H, Fukuda A, Follett P, Natesan S, Kono T, Shioda T, Hochedlinger K. Aberrant silencing of imprinted genes on chromosome 12qF1 in mouse induced pluripotent stem cells. Nature. 2010;465:175–181. doi: 10.1038/nature09017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim K, Doi A, Wen B, Ng K, Zhao R, Cahan P, Kim J, Aryee MJ, Ji H, Ehrlich LI, Yabuuchi A, Takeuchi A, Cunniff KC, Hongguang H, McKinney-Freeman S, Naveiras O, Yoon TJ, Irizarry RA, Jung N, Seita J, Hanna J, Murakami P, Jaenisch R, Weissleder R, Orkin SH, Weissman IL, Feinberg AP, Daley GQ. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467:285–290. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ohi Y, Qin H, Hong C, Blouin L, Polo JM, Guo T, Qi Z, Downey SL, Manos PD, Rossi DJ, Yu J, Hebrok M, Hochedlinger K, Costello JF, Song JS, Ramalho-Santos M. Incomplete DNA methylation underlies a transcriptional memory of somatic cells in human iPS cells. Nat Cell Biol. 2011;13:541–549. doi: 10.1038/ncb2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Polo JM, Liu S, Figueroa ME, Kulalert W, Eminli S, Tan KY, Apostolou E, Stadtfeld M, Li Y, Shioda T, Natesan S, Wagers AJ, Melnick A, Evans T, Hochedlinger K. Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nat Biotechnol. 2010;28:848–855. doi: 10.1038/nbt.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bar-Nur O, Russ HA, Efrat S, Benvenisty N. Epigenetic memory and preferential lineage-specific differentiation in induced pluripotent stem cells derived from human pancreatic islet Beta cells. Cell Stem Cell. 2011;9:17–23. doi: 10.1016/j.stem.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 63.Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, Loh YH, Yeo HC, Yeo ZX, Narang V, Govindarajan KR, Leong B, Shahab A, Ruan Y, Bourque G, Sung WK, Clarke ND, Wei CL, Ng HH. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 64.Reya T, Clevers H. Wnt signalling in stem cells and cancer. Nature. 2005;434:843–850. doi: 10.1038/nature03319. [DOI] [PubMed] [Google Scholar]
- 65.Niwa H. Wnt: what’s needed to maintain pluripotency? Nat Cell Biol. 2011;13:1024–1026. doi: 10.1038/ncb2333. [DOI] [PubMed] [Google Scholar]
- 66.Lluis F, Ombrato L, Pedone E, Pepe S, Merrill BJ, Cosma MP. T-cell factor 3 (Tcf3) deletion increases somatic cell reprogramming by inducing epigenome modifications. Proc Natl Acad Sci U S A. 2011;108:11912–11917. doi: 10.1073/pnas.1017402108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Marson A, Foreman R, Chevalier B, Bilodeau S, Kahn M, Young RA, Jaenisch R. Wnt signaling promotes reprogramming of somatic cells to pluripotency. Cell Stem Cell. 2008;3:132–135. doi: 10.1016/j.stem.2008.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lyssiotis CA, Foreman RK, Staerk J, Garcia M, Mathur D, Markoulaki S, Hanna J, Lairson LL, Charette BD, Bouchez LC, Bollong M, Kunick C, Brinker A, Cho CY, Schultz PG, Jaenisch R. Reprogramming of murine fibroblasts to induced pluripotent stem cells with chemical complementation of Klf4. Proc Natl Acad Sci U S A. 2009;106:8912–8917. doi: 10.1073/pnas.0903860106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lin T, Ambasudhan R, Yuan X, Li W, Hilcove S, Abujarour R, Lin X, Hahm HS, Hao E, Hayek A, Ding S. A chemical platform for improved induction of human iPSCs. Nat Methods. 2009;6:805–808. doi: 10.1038/nmeth.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen T, Yuan D, Wei B, Jiang J, Kang J, Ling K, Gu Y, Li J, Xiao L, Pei G. E-cadherin-mediated cell-cell contact is critical for induced pluripotent stem cell generation. Stem Cells. 2010;28:1315–1325. doi: 10.1002/stem.456. [DOI] [PubMed] [Google Scholar]
- 71.Li R, Liang J, Ni S, Zhou T, Qing X, Li H, He W, Chen J, Li F, Zhuang Q, Qin B, Xu J, Li W, Yang J, Gan Y, Qin D, Feng S, Song H, Yang D, Zhang B, Zeng L, Lai L, Esteban MA, Pei D. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell. 2010;7:51–63. doi: 10.1016/j.stem.2010.04.014. [DOI] [PubMed] [Google Scholar]
- 72.Samavarchi-Tehrani P, Golipour A, David L, Sung HK, Beyer TA, Datti A, Woltjen K, Nagy A, Wrana JL. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell. 2010;7:64–77. doi: 10.1016/j.stem.2010.04.015. [DOI] [PubMed] [Google Scholar]
- 73.Yuan X, Wan H, Zhao X, Zhu S, Zhou Q, Ding S. Brief report: combined chemical treatment enables Oct4-induced reprogramming from mouse embryonic fibroblasts. Stem Cells. 2011;29:549–553. doi: 10.1002/stem.594. [DOI] [PubMed] [Google Scholar]
- 74.Wang Y, Adjaye J. A cyclic AMP analog, 8-Br-cAMP, enhances the induction of pluripotency in human fibroblast cells. Stem Cell Rev. 2011;7:331–341. doi: 10.1007/s12015-010-9209-3. [DOI] [PubMed] [Google Scholar]
- 75.Ichida JK, Blanchard J, Lam K, Son EY, Chung JE, Egli D, Loh KM, Carter AC, Di Giorgio FP, Koszka K, Huangfu D, Akutsu H, Liu DR, Rubin LL, Eggan K. A small-molecule inhibitor of TGF-beta signaling replaces Sox2 in reprogramming by inducing Nanog. Cell Stem Cell. 2009;5:491–503. doi: 10.1016/j.stem.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Staerk J, Lyssiotis CA, Medeiro LA, Bollong M, Foreman RK, Zhu S, Garcia M, Gao Q, Bouchez LC, Lairson LL, Charette BD, Supekova L, Janes J, Brinker A, Cho CY, Jaenisch R, Schultz PG. Pan-Src family kinase inhibitors replace Sox2 during the direct reprogramming of somatic cells. Angew Chem Int Ed Engl. 2011;50:5734–5736. doi: 10.1002/anie.201101042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Anneren C, Cowan CA, Melton DA. The Src family of tyrosine kinases is important for embryonic stem cell self-renewal. J Biol Chem. 2004;279:31590–31598. doi: 10.1074/jbc.M403547200. [DOI] [PubMed] [Google Scholar]
- 78.Faherty S, Fitzgerald A, Keohan M, Quinlan LR. Self-renewal and differentiation of mouse embryonic stem cells as measured by Oct4 expression: the role of the cAMP/PKA pathway. In Vitro Cell Dev Biol Anim. 2007;43:37–47. doi: 10.1007/s11626-006-9001-5. [DOI] [PubMed] [Google Scholar]
- 79.Meyn MA, 3rd, Schreiner SJ, Dumitrescu TP, Nau GJ, Smithgall TE. SRC family kinase activity is required for murine embryonic stem cell growth and differentiation. Mol Pharmacol. 2005;68:1320–1330. doi: 10.1124/mol.104.010231. [DOI] [PubMed] [Google Scholar]
- 80.Zhao R, Daley GQ. From fibroblasts to iPS cells: induced pluripotency by defined factors. J Cell Biochem. 2008;105:949–955. doi: 10.1002/jcb.21871. [DOI] [PubMed] [Google Scholar]
- 81.Banito A, Rashid ST, Acosta JC, Li S, Pereira CF, Geti I, Pinho S, Silva JC, Azuara V, Walsh M, Vallier L, Gil J. Senescence impairs successful reprogramming to pluripotent stem cells. Genes Dev. 2009;23:2134–2139. doi: 10.1101/gad.1811609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hanna J, Saha K, Pando B, van Zon J, Lengner CJ, Creyghton MP, van Oudenaarden A, Jaenisch R. Direct cell reprogramming is a stochastic process amenable to acceleration. Nature. 2009;462:595–601. doi: 10.1038/nature08592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hong H, Takahashi K, Ichisaka T, Aoi T, Kanagawa O, Nakagawa M, Okita K, Yamanaka S. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature. 2009;460:1132–1135. doi: 10.1038/nature08235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kawamura T, Suzuki J, Wang YV, Menendez S, Morera LB, Raya A, Wahl GM, Belmonte JC. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature. 2009;460:1140–1144. doi: 10.1038/nature08311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Marion RM, Strati K, Li H, Murga M, Blanco R, Ortega S, Fernandez-Capetillo O, Serrano M, Blasco MA. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature. 2009;460:1149–1153. doi: 10.1038/nature08287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Utikal J, Polo JM, Stadtfeld M, Maherali N, Kulalert W, Walsh RM, Khalil A, Rheinwald JG, Hochedlinger K. Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature. 2009;460:1145–1148. doi: 10.1038/nature08285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhao Y, Yin X, Qin H, Zhu F, Liu H, Yang W, Zhang Q, Xiang C, Hou P, Song Z, Liu Y, Yong J, Zhang P, Cai J, Liu M, Li H, Li Y, Qu X, Cui K, Zhang W, Xiang T, Wu Y, Zhao Y, Liu C, Yu C, Yuan K, Lou J, Ding M, Deng H. Two supporting factors greatly improve the efficiency of human iPSC generation. Cell Stem Cell. 2008;3:475–479. doi: 10.1016/j.stem.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 88.Esteban MA, Wang T, Qin B, Yang J, Qin D, Cai J, Li W, Weng Z, Chen J, Ni S, Chen K, Li Y, Liu X, Xu J, Zhang S, Li F, He W, Labuda K, Song Y, Peterbauer A, Wolbank S, Redl H, Zhong M, Cai D, Zeng L, Pei D. Vitamin C enhances the generation of mouse and human induced pluripotent stem cells. Cell Stem Cell. 2010;6:71–79. doi: 10.1016/j.stem.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 89.Kondoh H, Lleonart ME, Nakashima Y, Yokode M, Tanaka M, Bernard D, Gil J, Beach D. A high glycolytic flux supports the proliferative potential of murine embryonic stem cells. Antioxid Redox Signal. 2007;9:293–299. doi: 10.1089/ars.2006.1467. [DOI] [PubMed] [Google Scholar]
- 90.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Prigione A, Fauler B, Lurz R, Lehrach H, Adjaye J. The senescence-related mitochondrial/oxidative stress pathway is repressed in human induced pluripotent stem cells. Stem Cells. 2010;28:721–733. doi: 10.1002/stem.404. [DOI] [PubMed] [Google Scholar]
- 92.Zhu S, Li W, Zhou H, Wei W, Ambasudhan R, Lin T, Kim J, Zhang K, Ding S. Reprogramming of human primary somatic cells by OCT4 and chemical compounds. Cell Stem Cell. 2010;7:651–655. doi: 10.1016/j.stem.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yoshida Y, Takahashi K, Okita K, Ichisaka T, Yamanaka S. Hypoxia enhances the generation of induced pluripotent stem cells. Cell Stem Cell. 2009;5:237–241. doi: 10.1016/j.stem.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 94.Efe JA, Hilcove S, Kim J, Zhou H, Ouyang K, Wang G, Chen J, Ding S. Conversion of mouse fibroblasts into cardiomyocytes using a direct reprogramming strategy. Nat Cell Biol. 2011;13:215–222. doi: 10.1038/ncb2164. [DOI] [PubMed] [Google Scholar]
- 95.Kim J, Efe JA, Zhu S, Talantova M, Yuan X, Wang S, Lipton SA, Zhang K, Ding S. Direct reprogramming of mouse fibroblasts to neural progenitors. Proc Natl Acad Sci U S A. 2011;108:7838–7843. doi: 10.1073/pnas.1103113108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Szabo E, Rampalli S, Risueno RM, Schnerch A, Mitchell R, Fiebig-Comyn A, Levadoux-Martin M, Bhatia M. Direct conversion of human fibroblasts to multilineage blood progenitors. Nature. 2010;468:521–526. doi: 10.1038/nature09591. [DOI] [PubMed] [Google Scholar]