Abstract
Direct reprogramming of one cell type into another provides unprecedented opportunities to study fundamental biology, model disease, and develop regenerative medicine. Different paradigms of reprogramming strategies with different sets of factors have been developed to generate various cell types, including induced pluripotent stem cells, neuronal or neural precursor cells, cardiomyocyte-like cells, endothelial cells, and hepatocyte-like cells. Various exogenous factors, especially small molecules modulating signaling, cellular state, and transcription, have been identified to enhance and enable reprogramming. With an increased understanding of reprogramming mechanisms and discovery of new molecules, it is conceivable that reprogramming can be achieved in a more directed and deterministic manner under entirely chemically defined conditions.
Introduction
Cell fate is controlled by both extrinsic factors (e.g., signaling molecules) and intrinsic factors (e.g., endogenous transcription factors). It has been shown that activation of the LIF-STAT3 and BMP-SMAD signaling pathways are essential for the maintenance of murine embryonic stem cells [1]. Transcription factors (TFs) downstream of the signaling pathways orchestrate with cell type-specific TFs, including Oct4, Sox2 and Nanog that form an auto-regulatory loop, to govern cell fate [1]. Consistent with such mechanism, studies of TF–mediated reprogramming demonstrated that cell fates can be manipulated by exogenous TFs as well. For example, fibroblasts can be induced into pluripotent stem cells (iPSCs) by the Yamanaka factors (Oct4/Sox2/Klf4/c-Myc), or converted to neuronal cells by Brn2/Ascl1/Myt1l [2,3]. Mounting evidence demonstrates that extrinsic factors can functionally mimic reprogramming TFs and/or enhance reprogramming process to facilitate cell fate switching. Here, we review these important extrinsic drivers for somatic cell reprogramming.
Somatic cell reprogramming is regulated by multiple signaling pathways
A successful iPSC reprogramming is to re-establish the intrinsic pluripotency transcriptional network in somatic cells. This network, in which Oct4 plays a pivotal role, involves dozens of pluripotency-associated factors and basal TFs [4].
Several signaling pathways have been reported to regulate the pluripotency of ESCs, indicating that they target certain components of the pluripotency transcriptional network in ESCs. Changes in the chromatin state of pluripotency genes, when driven by transduced factors or other regulators during reprogramming, may allow these signaling pathways to re-establish the pluripotency transcriptional network (Figure 1). We begin this review with a description of some of these key signaling molecules.
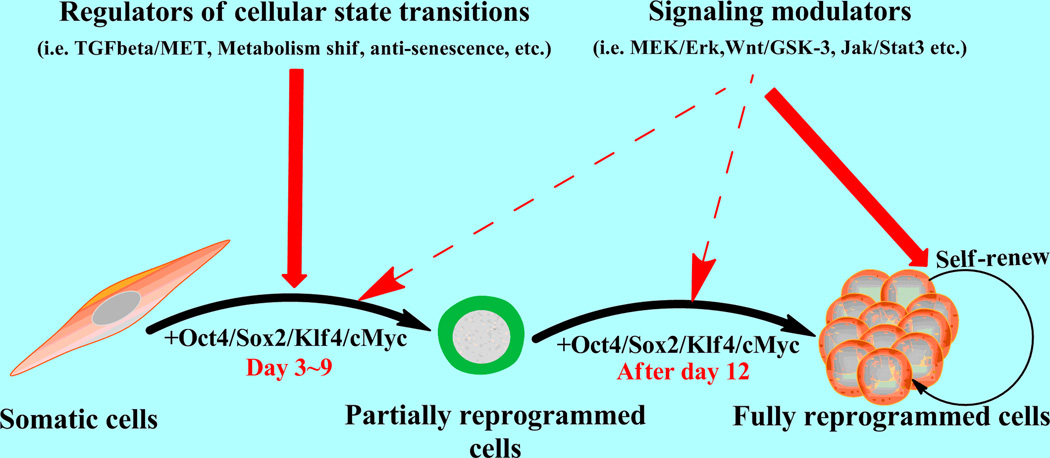
Figure 1. Important roles of signaling modulators and cellular state regulators in somatic cell reprogramming.
Signaling modulators help to re-establish the pluripotency transcriptional network during reprogramming triggered by exogenous key transcription factors, whereas cellular state regulators can facilitate the shift from a somatic to a pluripotent cellular state.
MEK-Erk
Inhibition of MEK and glycogen synthase kinase–3 (GSK-3) by small molecule inhibitors PD0325901 and CHIR99021 (2i) completely eliminated spontaneous differentiation of ESCs in the absence of essential pluripotency signaling pathway activation [5]. During reprogramming, PD0325901 was shown to stabilize and help to select fully reprogrammed iPSCs [6]. Another study suggested that at the late stage of reprogramming, the 2i/LIF condition promoted the transition from partially reprogrammed cells to full iPSCs [7].
Wnt-GSK-3
Marson et al. showed that Wnt3a-conditioned medium increased reprogramming efficiency in mouse embryonic fibroblasts (MEFs) with ectopic expression of Oct4/Sox2/Klf4 [8]. Similarly, it was demonstrated that CHIR99021 improved the reprogramming in the absence of c-Myc and Sox2 [9]. A Wnt downstream regulator, Tcf3, was reported to occupy the promoter regions of key pluripotency genes, such as Oct4, Nanog and Sox2, to repress their expression [10]. Thus, the positive effects of Wnt pathway in reprogramming may be majorly mediated by reduced Tcf3 activity.
LIF-Stat3
Yang et al. demonstrated that LIF-Stat3 activation increased somatic cell reprogramming efficiency using a system that excluded the possibility of interference by two other LIF-downstream pathways, PI3K-Akt and MEK-Erk [11]. These findings suggest that the role of LIF-Stat3 is to facilitate the transition from incompletely reprogrammed cells (that are Oct4 negative and express retroviral transgenes) into fully reprogrammed iPSCs.
PI3K-Akt
The role of the PI3K-Akt pathway in the reprogramming process has not been fully elucidated. Nakamura et al. showed that activation of Akt promoted reprogramming after cell fusion of ESCs with thymocytes or MEFs [12]. In contrast, it also arrested transition from the two-cell to eight-cell stage after nuclear transfer [12].
Other pathways
Regulation of other pathways, such as the cyclic AMP, Hippo/Yap and Src family kinase pathways, was also reported to increase reprogramming efficiency or functionally replace certain Yamanaka factors [13–16].
Mechanisms that facilitate somatic reprogramming by regulating cellular states
Several mechanisms have been reported to facilitate the reprogramming process without direct activation of pluripotency genes (Figure 1). However, it appears in many cases that the more somatic cells are similar to pluripotent cells, the easier it will be to convert them to pluripotent cells. It is thus plausible that these additional mechanisms facilitate the shift from a somatic to a pluripotent cellular state.
Mesenchymal-to-epithelial transition and TGFβ pathway
During the reprogramming process, fibroblasts lose mesenchymal characteristics and obtain epithelial features, suggesting that the MET process is critical during reprogramming. This is consistent with findings showing that when the TGFβ pathway, which positively regulates the epithelial-to-mesenchymal transition (EMT, a reverse process of MET), was blocked by inhibitor of TGFβ receptor, there was a large increase in iPSC generation [17]. Furthermore, the addition of a specific TGFβ receptor inhibitor could replace Sox2 in reprogramming [13]. Two follow-up studies provided molecular and functional evidence that the MET is necessary for reprogramming [18•,19•].
Metabolic shift
It is evident that, compared with somatic cells, many stem cells (including ESCs) rely more heavily on aerobic glycolysis to support their proliferation [20]. Thus, the transition from oxidative phosphorylation to glycolysis was suggested to be a barrier of somatic reprogramming. The findings showing that hypoxic conditions improved reprogramming support this notion [21]. It was found that PS48, an activator of 3’ phosphoinositide-dependent kinase 1, helped to generate human iPSC with ectopic expression of a single TF (OCT4) by facilitating the metabolic conversion to glycolysis [22]. On the other hand, 2-deoxyglucose, a general inhibitor of glycolysis, greatly impaired iPSC generation [23]. Moreover, the glycolysis transition preceded pluripotency gene expression during reprogramming [23], suggesting that it acts at an early stage.
Anti-senescence pathways
Up-regulation of senescence control genes, including p53, p16INK4a, and p21, was observed as an early event in reprogramming of fibroblasts by the Yamanaka factors [24]. Considering that somatic cells have limited proliferative potential while iPSCs have unlimited capacity for self-renewal, it is likely that cellular senescence is a barrier to reprogramming. This notion is consistent with the observation that fibroblasts from older mice had lower reprogramming efficiency [25]. Several groups pinpointed the p53-p21 pathway as a critical barrier to reprogramming [26]. They showed that knock-down of p53 in human or mouse cells greatly increased iPSC generation.
Somatic cell reprogramming controlled by gene expression regulators
As specific gene expression is central to cell identity, there is no doubt that regulators of gene expression, such as transcription factors, nuclear receptors, epigenetic modifiers and microRNAs, have direct and strong effects on cell fate determination.
Master transcription factors (TFs)
Reprogramming studies have demonstrated that combinations of different cell type-specific TFs could be applied to reprogram somatic cells directly into a variety of cell types, including iPSCs, neuronal cells, cardiomyocyte-like cells, hepatocyte-like cells, and endothelial cells, that are similar to their naturally existing counterparts [2,3,27–31]. In addition, different reprogramming paradigms have been developed. For example, applying transient expression of iPSC factors can reset fibroblasts toward plastic intermediates, which can be redirected by lineage-specific signaling molecules to generate cardiac, neural, or endothelial progenitor cells without passing through the pluripotent state [29,32,33•]. In contrast, neural precursor cells could also be generated using neural-specific TFs, such as Sox2 alone [34].
Nuclear receptors
Nuclear receptors are transcription factors that can directly bind to DNA and regulate specific gene expression in a ligand-dependent or -independent manner. Like extensively studied master TFs for pluripotency, some nuclear receptors were found to play critical roles in iPSC reprogramming as well as the maintenance of pluripotency. In addition to the well-known core auto-regulatory loop of Oct4-Sox2-Nanog [35], the nuclear receptor Esrrb could form another regulatory circuit with Tbx3 and Tcl1 for the maintenance of ESCs [36]. Furthermore, it was shown that Esrrb, along with Oct4 and Sox2, could convert MEFs to iPSCs [37]. In this process, Esrrb forms a complex with Oct4 and Sox2, and synergistically up-regulates the expression of pluripotency genes in MEFs. Remarkably, with the help of other Yamanaka factors (Klf4/Sox2/c-Myc), another orphan nuclear receptor, Nr5a2, was able to substitute for Oct4 in iPSC generation [38]. In addition, Nr5a2 could greatly enhance iPSC reprogramming in conjunction with activation of another nuclear receptor, RARa/g [39]. The finding that the RARa agonist (CD437) and RARg agonist (AM580) dramatically increased reprogramming efficiency further supports the notion that nuclear receptors play important roles in regulating somatic cell reprogramming.
Epigenetic factors
Many studies demonstrated that small-molecule epigenetic modifiers could significantly influence reprogramming process and even substitute for certain reprogramming transcription factors (Figure 2). BIX01294, an inhibitor of G9a histone methyltransferase (HMTase), was shown to enable reprogramming of neural precursor cells or fibroblasts transduced with only two TFs, Oct4 and Klf4 [6]. Besides well-known HDAC inhibitors (e.g., VPA, NaB) that have been demonstrated to facilitate reprogramming in various contexts [40,41], Parnate, an inhibitor of histone demethylase LSD1, was shown to enhance iPSC reprogramming as well [9]. Interestingly, the well-known antioxidant compound vitamin C was recently shown to enhance reprogramming by modulating the activity of the histone demethylases Jhdm1a/1b [42]. These findings highlight the dynamic changes of histone modifications in reprogramming.
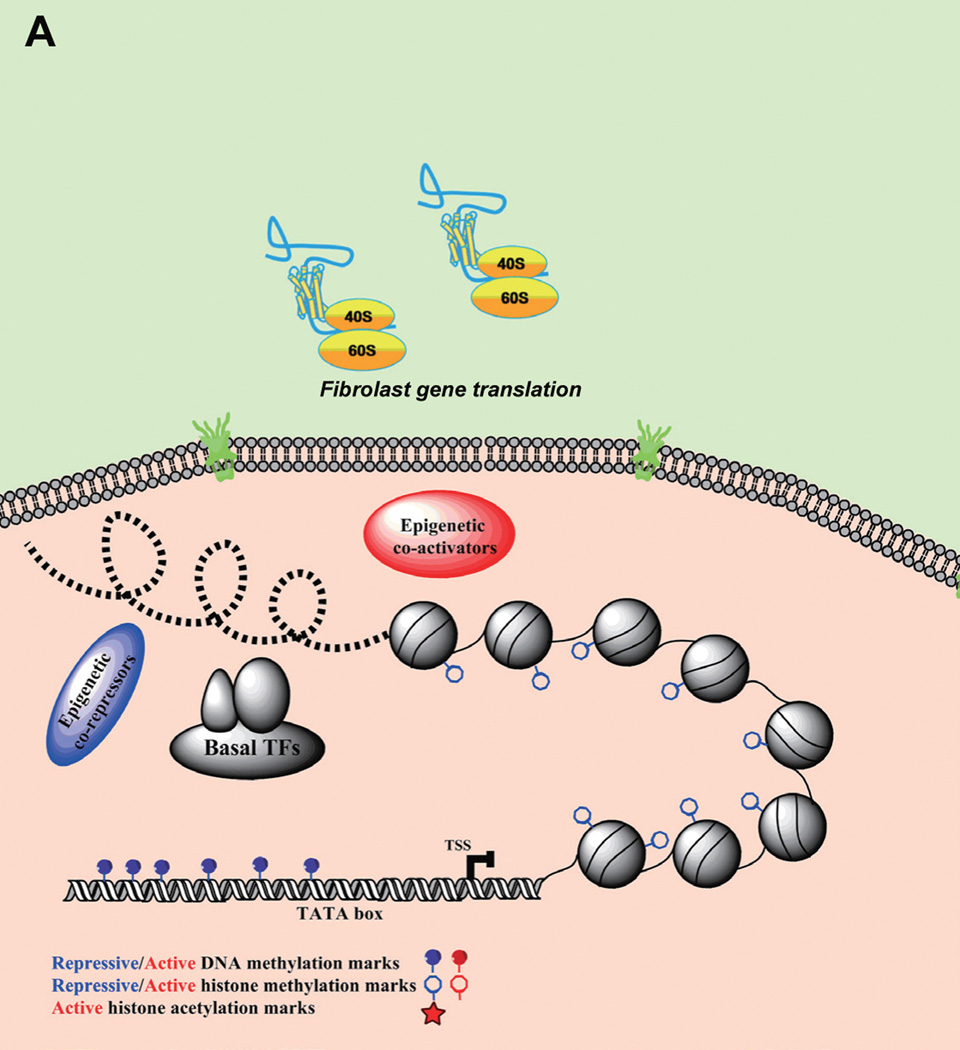
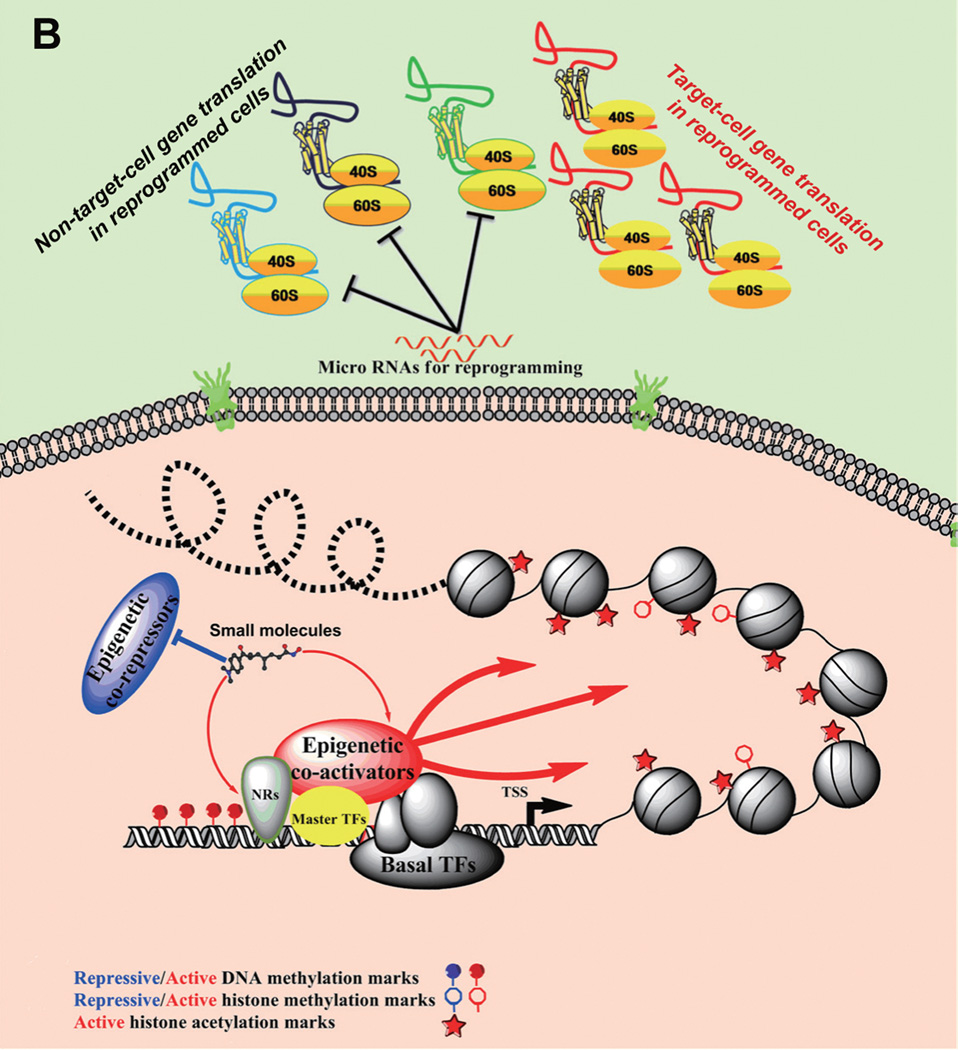
Figure 2. Target-cell genes activated by gene expression regulators and small molecules.
(A) Showing transcriptional silence on target-cell gene loci (i.e. pluripotency genes, neural genes, etc.) in fibroblasts; (B) These genes are activated by exogenous transcriptional regulators, including master transcription factors (TFs), nuclear receptors (NRs), epigenetic co-activator, and related small molecules which can functionally mimic/substitute other exogenous regulators. In addition, microRNAs for reprogramming usually form an inhibitory network to block the path towards other cell fates and thereby facilitate a specific reprogramming process.
Recent mechanistic studies of iPSC reprogramming further illustrated how epigenetic changes are orchestrated in the early and late stages of reprogramming. Koche et al. showed that activated chromatin marks (e.g., H3K4 methylation) were targeted to promoters of pluripotency and developmentally regulated genes (e.g., Fgf4 and Lin28) prior to transcriptional activation during the early phase of iPSC reprogramming [43]. It was also reported that two epigenetic factors, Parp1 and Tet2, were recruited to pluripotency loci (e.g., Nanog and Esrrb) and established early epigenetic marks by converting 5-methylcytosine (5mC) into 5-hydroxymethylcytosine (5hmC) during reprogramming [44]. Interestingly, it was reported that along with Klf4, Sox2, and c-Myc, another Tet family protein Tet1 could enable somatic cell reprogramming in the absence of the key transcription factor Oct4 or nuclear receptors Esrrb and Nr5a2 [45], highlighting the important role of DNA demethylation (through hydroxymethylation) in reprogramming. Furthermore, other specific histone modifications were identified to occur in reprogramming. For example, inhibition of the H3K79 histone methyltransferase DOT1L (e.g., by a small molecule inhibitor) and the H3K9 methyltransferase Setdb1 (e.g., by RNAi) was shown to enhance iPSC generation [46,47]. These findings indicate that histone methylations of both H3K79 and H3K9 are barriers to iPSC reprogramming.
In addition, a recent study provided additional details of certain epigenetic changes during reprogramming [48••]. As Thy1 (a fibroblast marker) is linearly down-regulated and SSEA1 and Oct4 are linearly up-regulated during reprogramming, the reprogramming process in this study was roughly divided into three stages: early (day 3, Thy1-), intermediate (day 6–9, SSEA1+), and late (day 12, Oct4+). To determine certain epigenetic profiles in the different stages of reprogramming, ChIP-seq analyses were performed using antibodies against H3K4me3 (an active histone mark) and H3K27me3 (a repressive histone mark) in cells undergoing reprogramming. It was found that the genes carrying H3K4me3 marks were activated early or gradually (e.g. Fbx15, Cdc25c), whereas genes that were activated late (e.g. Oct4, Nanog) were often either unmarked with H3K4me3 or marked with both H3K4me3 and K3K27me3 in fibroblasts. It was also found that the demethylation of DNA did not happen until the late stage of reprogramming.
MicroRNAs
It was demonstrated that some mouse ESC–specific, cell-cycle-regulating (ESCC) microRNAs, including miR-291-3p, miR-294, and miR-295, could substitute c-Myc and enhance iPSC reprogramming with Oct4/Sox2/Klf4 [49]. Moreover, Subramanyam et al. showed that human ESCC miRNA orthologs hsa-miR-302b and hsa-miR-372 promoted human somatic cell reprogramming through multiple targets, including cell cycle regulators, epigenetic modifiers, and MET regulators [50]. In addition to iPSC generation, microRNAs were also shown as powerful regulator for lineage-specific reprogramming. It was reported that miR-9* and miR-124 were found to directly induce human fibroblasts into neurons with NeuroD2, Ascl1, and Myt1l [51•]. It was also demonstrated that miR-124 in conjunction with Brn2 and Mytl1 could convert human adult fibroblasts into mature neurons, suggesting that miR-124 plays an important role in neuronal specification [52•]. This finding also was supported by recent studies in which knocking down a single RNA-binding, polypyrimidine-tract-binding (PTB) protein could generate mature neurons from mouse fibroblasts via the action of miR-124 [53].
Perspectives
Among these exogenously delivered factors, small molecules and microRNAs, which can be chemically synthesized and do not modify target cell genome, have emerged as powerful tools to manipulate cell fate. While microRNAs offer the advantage of specifically targeting a large number of genes, small molecules provide precise temporal and tunable control over protein function, including rapid and reversible activation and inhibition. With an increased understanding of reprogramming mechanisms and discovery of new molecules, it is conceivable that reprogramming can be achieved in a more efficient and deterministic manner under entirely chemically defined conditions.
Acknowledgements
Sheng Ding is supported by funding from NICHD, NHLBI, NEI, NIMH/NIH, California Institute for Regenerative Medicine, and the Gladstone Institute. We apologize to all scientists whose work could not be properly discussed and cited here due to limited space.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. Integration of External Signaling Pathways with the Core Transcriptional Network in Embryonic Stem Cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 2.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 3.Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Sudhof TC, Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463:1035–1041. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ng H-H, Surani MA. The transcriptional and signalling networks of pluripotency. Nature Cell Biology. 2011;13:490–496. doi: 10.1038/ncb0511-490. [DOI] [PubMed] [Google Scholar]
- 5.Ying Q-L, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, Cohen P, Smith A. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–523. doi: 10.1038/nature06968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shi Y, Do JT, Desponts C, Hahm HS, Scholer HR, Ding S. A combined chemical and genetic approach for the generation of induced pluripotent stem cells. Cell Stem Cell. 2008;2:525–528. doi: 10.1016/j.stem.2008.05.011. [DOI] [PubMed] [Google Scholar]
- 7.Silva J, Barrandon O, Nichols J, Kawaguchi J, Theunissen TW, Smith A. Promotion of reprogramming to ground state pluripotency by signal inhibition. PLoS Biol. 2008;6:e253. doi: 10.1371/journal.pbio.0060253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marson A, Foreman R, Chevalier B, Bilodeau S, Kahn M, Young RA, Jaenisch R. Wnt Signaling Promotes Reprogramming of Somatic Cells to Pluripotency. Cell Stem Cell. 2008;3:132–135. doi: 10.1016/j.stem.2008.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li W, Zhou H, Abujarour R, Zhu S, Young Joo J, Lin T, Hao E, Scholer HR, Hayek A, Ding S. Generation of human-induced pluripotent stem cells in the absence of exogenous Sox2. STEM CELLS. 2009;27:2992–3000. doi: 10.1002/stem.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cole MF, Johnstone SE, Newman JJ, Kagey MH, Young RA. Tcf3 is an integral component of the core regulatory circuitry of embryonic stem cells. Genes & Development. 2008;22:746–755. doi: 10.1101/gad.1642408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang J, van Oosten AL, Theunissen TW, Guo G, Silva JCR, Smith A. Stat3 Activation Is Limiting for Reprogramming to Ground State Pluripotency. Cell Stem Cell. 2010;7:319–328. doi: 10.1016/j.stem.2010.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nakamura T, Inoue K, Ogawa S, Umehara H, Ogonuki N, Miki H, Kimura T, Ogura A, Nakano T. Effects of Akt signaling on nuclear reprogramming. Genes Cells. 2008;13:1269–1277. doi: 10.1111/j.1365-2443.2008.01243.x. [DOI] [PubMed] [Google Scholar]
- 13.Ichida JK, Blanchard J, Lam K, Son EY, Chung JE, Egli D, Loh KM, Carter AC, Di Giorgio FP, Koszka K, et al. A Small-Molecule Inhibitor of Tgf-β Signaling Replaces Sox2 in Reprogramming by Inducing Nanog. Cell Stem Cell. 2009;5:491–503. doi: 10.1016/j.stem.2009.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lian I, Kim J, Okazawa H, Zhao J, Zhao B, Yu J, Chinnaiyan A, Israel MA, Goldstein LSB, Abujarour R, et al. The role of YAP transcription coactivator in regulating stem cell self-renewal and differentiation. Genes & Development. 2010;24:1106–1118. doi: 10.1101/gad.1903310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Staerk J, Lyssiotis CA, Medeiro LA, Bollong M, Foreman RK, Zhu S, Garcia M, Gao Q, Bouchez LC, Lairson LL, et al. Pan-Src Family Kinase Inhibitors Replace Sox2 during the Direct Reprogramming of Somatic Cells. Angewandte Chemie International Edition. 2011;50:5734–5736. doi: 10.1002/anie.201101042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Y, Adjaye J. A Cyclic AMP Analog, 8-Br-cAMP, Enhances the Induction of Pluripotency in Human Fibroblast Cells. Stem Cell Reviews and Reports. 2010;7:331–341. doi: 10.1007/s12015-010-9209-3. [DOI] [PubMed] [Google Scholar]
- 17.Lin T, Ambasudhan R, Yuan X, Li W, Hilcove S, Abujarour R, Lin X, Hahm HS, Hao E, Hayek A, et al. A chemical platform for improved induction of human iPSCs. Nature Methods. 2009;6:805–808. doi: 10.1038/nmeth.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.•Li R, Liang J, Ni S, Zhou T, Qing X, Li H, He W, Chen J, Li F, Zhuang Q, et al. A Mesenchymal-to-Epithelial Transition Initiates and Is Required for the Nuclear Reprogramming of Mouse Fibroblasts Cell Stem Cell 2010751–63. [DOI] [PubMed] [Google Scholar]
- 19. Samavarchi-Tehrani P, Golipour A, David L, Sung H-k, Beyer TA, Datti A, Woltjen K, Nagy A, Wrana JL. Functional Genomics Reveals a BMP-Driven Mesenchymal-to-Epithelial Transition in the Initiation of Somatic Cell Reprogramming. Cell Stem Cell. 2010;7:64–77. doi: 10.1016/j.stem.2010.04.015. This paper and ref. [18] mechanistically characterized MET as a key early step in iPSC reprogramming process.
- 20.Folmes Clifford DL, Dzeja Petras P, Nelson Timothy J, Terzic A. Metabolic Plasticity in Stem Cell Homeostasis and Differentiation. Cell Stem Cell. 2012;11:596–606. doi: 10.1016/j.stem.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoshida Y, Takahashi K, Okita K, Ichisaka T, Yamanaka S. Hypoxia Enhances the Generation of Induced Pluripotent Stem Cells. Cell Stem Cell. 2009;5:237–241. doi: 10.1016/j.stem.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 22.Zhu S, Li W, Zhou H, Wei W, Ambasudhan R, Lin T, Kim J, Zhang K, Ding S. Reprogramming of human primary somatic cells by OCT4 and chemical compounds. Cell Stem Cell. 2010;7:651–655. doi: 10.1016/j.stem.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Folmes CD, Nelson TJ, Martinez-Fernandez A, Arrell DK, Lindor JZ, Dzeja PP, Ikeda Y, Perez-Terzic C, Terzic A. Somatic oxidative bioenergetics transitions into pluripotency-dependent glycolysis to facilitate nuclear reprogramming. Cell Metab. 2011;14:264–271. doi: 10.1016/j.cmet.2011.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Banito A, Rashid ST, Acosta JC, Li S, Pereira CF, Geti I, Pinho S, Silva JC, Azuara V, Walsh M, et al. Senescence impairs successful reprogramming to pluripotent stem cells. Genes & Development. 2009;23:2134–2139. doi: 10.1101/gad.1811609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li H, Collado M, Villasante A, Strati K, Ortega S, Canamero M, Blasco MA, Serrano M. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature. 2009;460:1136–1139. doi: 10.1038/nature08290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Menendez S, Camus S, Izpisua Belmonte JC. p53: guardian of reprogramming. Cell Cycle. 2010;9:3887–3891. doi: 10.4161/cc.9.19.13301. [DOI] [PubMed] [Google Scholar]
- 27.Huang P, He Z, Ji S, Sun H, Xiang D, Liu C, Hu Y, Wang X, Hui L. Induction of functional hepatocyte-like cells from mouse fibroblasts by defined factors. Nature. 2011;475:386–389. doi: 10.1038/nature10116. [DOI] [PubMed] [Google Scholar]
- 28.Ieda M, Fu JD, Delgado-Olguin P, Vedantham V, Hayashi Y, Bruneau BG, Srivastava D. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li J, Huang NF, Zou J, Laurent TJ, Lee JC, Okogbaa J, Cooke JP, Ding S. Conversion of Human Fibroblasts to Functional Endothelial Cells by Defined Factors. Arterioscler Thromb Vasc Biol. 2013 doi: 10.1161/ATVBAHA.112.301167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qian L, Huang Y, Spencer CI, Foley A, Vedantham V, Liu L, Conway SJ, Fu JD, Srivastava D. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature. 2012;485:593–598. doi: 10.1038/nature11044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sekiya S, Suzuki A. Direct conversion of mouse fibroblasts to hepatocyte-like cells by defined factors. Nature. 2011;475:390–393. doi: 10.1038/nature10263. [DOI] [PubMed] [Google Scholar]
- 32.Efe JA, Hilcove S, Kim J, Zhou H, Ouyang K, Wang G, Chen J, Ding S. Conversion of mouse fibroblasts into cardiomyocytes using a direct reprogramming strategy. Nat Cell Biol. 2011;13:215–222. doi: 10.1038/ncb2164. [DOI] [PubMed] [Google Scholar]
- 33. Kim J, Efe JA, Zhu S, Talantova M, Yuan X, Wang S, Lipton SA, Zhang K, Ding S. Direct reprogramming of mouse fibroblasts to neural progenitors. Proc Natl Acad Sci U S A. 2011;108:7838–7843. doi: 10.1073/pnas.1103113108. This paper reported the first case of reprogramming fibroblasts into induced neural progenitors by transient Yamanaka factors.
- 34.Ring KL, Tong LM, Balestra ME, Javier R, Andrews-Zwilling Y, Li G, Walker D, Zhang WR, Kreitzer AC, Huang Y. Direct reprogramming of mouse and human fibroblasts into multipotent neural stem cells with a single factor. Cell Stem Cell. 2012;11:100–109. doi: 10.1016/j.stem.2012.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ivanova N, Dobrin R, Lu R, Kotenko I, Levorse J, DeCoste C, Schafer X, Lun Y, Lemischka IR. Dissecting self-renewal in stem cells with RNA interference. Nature. 2006;442:533–538. doi: 10.1038/nature04915. [DOI] [PubMed] [Google Scholar]
- 37.Feng B, Jiang J, Kraus P, Ng JH, Heng JC, Chan YS, Yaw LP, Zhang W, Loh YH, Han J, et al. Reprogramming of fibroblasts into induced pluripotent stem cells with orphan nuclear receptor Esrrb. Nat Cell Biol. 2009;11:197–203. doi: 10.1038/ncb1827. [DOI] [PubMed] [Google Scholar]
- 38.Heng JC, Feng B, Han J, Jiang J, Kraus P, Ng JH, Orlov YL, Huss M, Yang L, Lufkin T, et al. The nuclear receptor Nr5a2 can replace Oct4 in the reprogramming of murine somatic cells to pluripotent cells. Cell Stem Cell. 2010;6:167–174. doi: 10.1016/j.stem.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 39.Wang W, Yang J, Liu H, Lu D, Chen X, Zenonos Z, Campos LS, Rad R, Guo G, Zhang S, et al. Rapid and efficient reprogramming of somatic cells to induced pluripotent stem cells by retinoic acid receptor gamma and liver receptor homolog 1. Proc Natl Acad Sci U S A. 2011;108:18283–18288. doi: 10.1073/pnas.1100893108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mali P, Chou BK, Yen J, Ye Z, Zou J, Dowey S, Brodsky RA, Ohm JE, Yu W, Baylin SB, et al. Butyrate greatly enhances derivation of human induced pluripotent stem cells by promoting epigenetic remodeling and the expression of pluripotency-associated genes. STEM CELLS. 2010;28:713–720. doi: 10.1002/stem.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huangfu D, Osafune K, Maehr R, Guo W, Eijkelenboom A, Chen S, Muhlestein W, Melton DA. Induction of pluripotent stem cells from primary human fibroblasts with only Oct4 and Sox2. Nat Biotechnol. 2008;26:1269–1275. doi: 10.1038/nbt.1502. [DOI] [PubMed] [Google Scholar]
- 42.Esteban MA, Wang T, Qin B, Yang J, Qin D, Cai J, Li W, Weng Z, Chen J, Ni S, et al. Vitamin C enhances the generation of mouse and human induced pluripotent stem cells. Cell Stem Cell. 2010;6:71–79. doi: 10.1016/j.stem.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 43.Koche RP, Smith ZD, Adli M, Gu H, Ku M, Gnirke A, Bernstein BE, Meissner A. Reprogramming factor expression initiates widespread targeted chromatin remodeling. Cell Stem Cell. 2011;8:96–105. doi: 10.1016/j.stem.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Doege CA, Inoue K, Yamashita T, Rhee DB, Travis S, Fujita R, Guarnieri P, Bhagat G, Vanti WB, Shih A, et al. Early-stage epigenetic modification during somatic cell reprogramming by Parp1 and Tet2. Nature. 2012;488:652–655. doi: 10.1038/nature11333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gao Y, Chen J, Li K, Wu T, Huang B, Liu W, Kou X, Zhang Y, Huang H, Jiang Y, et al. Replacement of Oct4 by Tet1 during iPSC Induction Reveals an Important Role of DNA Methylation and Hydroxymethylation in Reprogramming. Cell Stem Cell. 2013;12:453–469. doi: 10.1016/j.stem.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 46.Chen J, Liu H, Liu J, Qi J, Wei B, Yang J, Liang H, Chen Y, Wu Y, Guo L, et al. H3K9 methylation is a barrier during somatic cell reprogramming into iPSCs. Nat Genet. 2013;45:34–42. doi: 10.1038/ng.2491. [DOI] [PubMed] [Google Scholar]
- 47.Onder TT, Kara N, Cherry A, Sinha AU, Zhu N, Bernt KM, Cahan P, Marcarci BO, Unternaehrer J, Gupta PB, et al. Chromatin-modifying enzymes as modulators of reprogramming. Nature. 2012;483:598–602. doi: 10.1038/nature10953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Polo JM, Anderssen E, Walsh RM, Schwarz BA, Nefzger CM, Lim SM, Borkent M, Apostolou E, Alaei S, Cloutier J, et al. A molecular roadmap of reprogramming somatic cells into iPS cells. Cell. 2012;151:1617–1632. doi: 10.1016/j.cell.2012.11.039. This paper reveals a molecular roadmap of reprogramming and provides insights into somatic cell reprogramming.
- 49.Judson RL, Babiarz JE, Venere M, Blelloch R. Embryonic stem cell-specific microRNAs promote induced pluripotency. Nat Biotechnol. 2009;27:459–461. doi: 10.1038/nbt.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Subramanyam D, Lamouille S, Judson RL, Liu JY, Bucay N, Derynck R, Blelloch R. Multiple targets of miR-302 and miR-372 promote reprogramming of human fibroblasts to induced pluripotent stem cells. Nat Biotechnol. 2011;29:443–448. doi: 10.1038/nbt.1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yoo AS, Sun AX, Li L, Shcheglovitov A, Portmann T, Li Y, Lee-Messer C, Dolmetsch RE, Tsien RW, Crabtree GR. MicroRNA-mediated conversion of human fibroblasts to neurons. Nature. 2011;476:228–231. doi: 10.1038/nature10323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ambasudhan R, Talantova M, Coleman R, Yuan X, Zhu S, Lipton SA, Ding S. Direct reprogramming of adult human fibroblasts to functional neurons under defined conditions. Cell Stem Cell. 2011;9:113–118. doi: 10.1016/j.stem.2011.07.002. In this paper, miR124 along with Brn2 and Myt1l could enable human neuronal reprogramming in the absence of the essential factor, Ascl1, for neuronal reprogramming.
- 53. Xue Y, Ouyang K, Huang J, Zhou Y, Ouyang H, Li H, Wang G, Wu Q, Wei C, Bi Y, et al. Direct Conversion of Fibroblasts to Neurons by Reprogramming PTB-Regulated MicroRNA Circuits. Cell. 2013;152:82–96. doi: 10.1016/j.cell.2012.11.045. In this paper, the miR124 downstream target protein PTB was identified as a key player during neuronal reprogramming.