Abstract
Vorticella includes more than 100 currently recognized species and represents one of the most taxonomically challenging genera of ciliates. Molecular phylogenetic analysis of Vorticella has been performed so far with only sequences coding for small subunit ribosomal RNA (SSU rRNA); only a few of its species have been investigated using other genetic markers owing to a lack of similar sequences for comparison. Consequently, phylogenetic relationships within the genus remain unclear, and molecular discrimination between morphospecies is often difficult because most regions of the SSU rRNA gene are too highly conserved to be helpful. In this paper, we move molecular systematics for this group of ciliates to the infrageneric level by sequencing additional molecular markers—fast-evolving internal transcribed spacer (ITS) regions—in a broad sample of 66 individual samples of 28 morphospecies of Vorticella collected from Asia, North America and Europe. Our phylogenies all featured two strongly supported, highly divergent, paraphyletic clades (I, II) comprising the morphologically defined genus Vorticella. Three major lineages made up clade I, with a relatively well-resolved branching order in each one. The marked divergence of clade II from clade I confirms that the former should be recognized as a separate taxonomic unit as indicated by SSU rRNA phylogenies. We made the first attempt to elucidate relationships between species in clade II using both morphological and multi-gene approaches, and our data supported a close relationship between some morphospecies of Vorticella and Opisthonecta, indicating that relationships between species in the clade are far more complex than would be expected from their morphology. Different patterns of helix III of ITS2 secondary structure were clearly specific to clades and subclades of Vorticella and, therefore, may prove useful for resolving phylogenetic relationships in other groups of ciliates.
Keywords: ciliate, multi-gene, morphology, phylogeny, peritrich
1. Introduction
Vorticella is the largest genus of sessile peritrich ciliates and its members live in a wide assortment of marine, freshwater and terrestrial environments worldwide [1–3]. Their extraordinary variety of habitats and prodigious abilities as suspension feeders suit them to be used widely as biological indicators for assessing the quality of natural bodies of water [4–7]. Vorticella and other ciliates are also an integral part of the treatment process in sewage treatment systems worldwide [8]. Species of Vorticella are well known as examples of the challenge of identifying ciliates in large genera to the level of species because they have a plastic, highly contractile cell body and show intraspecific variability in size. Also, morphological characters diagnostic for species, including those revealed by protargol and silver nitrate staining, overlap to some degree between putative species. This has resulted in the naming of numerous species and varieties, many of which are of doubtful taxonomic status [1,9].
During the last few decades, several molecular phylogenetic analyses, based especially on sequences of the gene coding for small subunit ribosomal RNA (SSU rRNA), provided resolution of a number of important questions about evolutionary relationships within several taxa of ciliates. These studies demonstrated that traditional taxonomic classifications are coarse—morphospecies can contain several to many cryptic taxa (e.g. Tetrahymena pyriformis [10] and Paramecium aurelia [11]). More recently, both morphological and molecular analyses [9,12] have suggested the existence of cryptic species in the Vorticella group. However, at present, there are no data at all for genes other than SSU rRNA for almost all named Vorticella species. Moreover, genetic distances between SSU rRNA sequences of morphospecies were small [12], and this high degree of conservation makes it difficult to resolve phylogenetic relationships within the genus and make a clear discrimination between morphospecies. Furthermore, the long-standing assumption that the family Vorticellidae was a distinct, stable taxon and that Vorticella was equally distinct and stable at the generic level was upset by convincing molecular evidence [12–15]. Given these important systematic uncertainties and the low resolution at the infrageneric level of phylogenies based on SSU rRNA sequences, molecular phylogenies of Vorticella that include a broader sample of taxa and additional genes are desirable.
The nuclear internal transcribed spacers (ITSs) ITS1 and ITS2 are the most popular markers for phylogenetic inference at the infrageneric level or between closely related genera [16–18]. The primary sequences of these spacers are subject to lower selective constraints compared to coding regions for the ribosomal subunits and, thus, can be highly variable and possibly unalignable between distantly related species owing to intrasequence nucleotide heterogeneity between sampled taxa. With regard to nuclear ITS2, however, these limitations have been overcome by secondary structure analysis that systematically identifies regions of variability as well as areas of substantial conservation [17,19,20]. More and more studies have suggested that phylogenetic analyses incorporating both primary sequences and secondary structures of ITS2 produce the most robust trees and match resolution provided by analyses of COX1 [18,21–23].
Following this lead, we used increased taxon sampling and genetic markers other than the SSU rRNA gene to create a much more comprehensive phylogenetic analysis of the genus Vorticella than has been attempted. In all, samples of 66 populations of Vorticella from three continents were collected and both nuclear ITSs from all of them were sequenced as well as the Histone 4 gene from some. Our goals were: (i) to complete a pilot study of Vorticella and reveal phylogenetic patterns within the genus; (ii) to test whether entities within Vorticella that are extremely similar in morphology are also genetically similar; and (iii) use multi-gene markers to further explore the relationship between morphospecies of Opisthonecta and Vorticella in one divergent clade.
2. Results
(a). Analyses of sequences and secondary structures
GenBank accession numbers of newly obtained sequences are given in the electronic supplementary material, table S1. Lengths of ITS1-5.8S-ITS2 and ITS2 sequences of all samples included in the study were 415–432 and 165–169 nt, and averaged 422 and 167 nt, respectively (see the electronic supplementary material, table S1). The GC content of ITS1-5.8S-ITS2 sequences ranged from 33.65 to 41.83%, with a mean value of 36.47%, and that of ITS2 sequences alone ranged from 28.99 to 47.88%, with a mean value of 35.06% (see the electronic supplementary material, table S1).
Ten species were collected more than once, allowing for some intraspecific comparison (see the electronic supplementary material, table S2). As shown in the electronic supplementary material, table S2, the pairwise distance among the ITS1-5.8S-ITS2 sequences of populations from different localities ranged from 0 to 4.23%. The variation between different populations of these 10 species was usually small. There was no variation between populations of Vorticella convallaria from China and the USA, and the same result was found in Vorticella gracilis-like populations from Austria. Populations of Vorticella aequilata, V. gracilis, Vorticella natans and Vorticella fusca varied slightly more than this, with genetic distances ranging from 0.16 to 1.60%. The pairwise distances among Vorticella similis, Vorticella citrina and Vorticella campanula-like populations were 4.23%, 3.06% and 2.24%, respectively. There was no difference between the three collections of V. campanula from Wuhan (China) and two from Japan, but Austrian collections differed from all of those from Wuhan by 2.89% at most.
We guided alignment of raw ITS2 sequences and identification of genetic characteristics at the species level and below by comparing ITS2 primary sequences to their secondary structures and determining the compensatory base changes (CBCs and hemi-CBCs) for all samples of Vorticella. The general secondary structure of ITS2 in Vorticella included three helices (I, II and III), which is congruent with the putative general model for secondary structure in all sessile peritrich ciliates. Our comparisons show that helix III is the most distinctive region of ITS2 in all samples of Vorticella, and based on its structure, the genus groups into two markedly divergent clades (I and II), with clade I comprising three clearly demarcated subclades (see the electronic supplementary material, figure S1).
(b). Phylogenetic analyses inferred from ITS1-5.8S-ITS2 and ITS2 regions
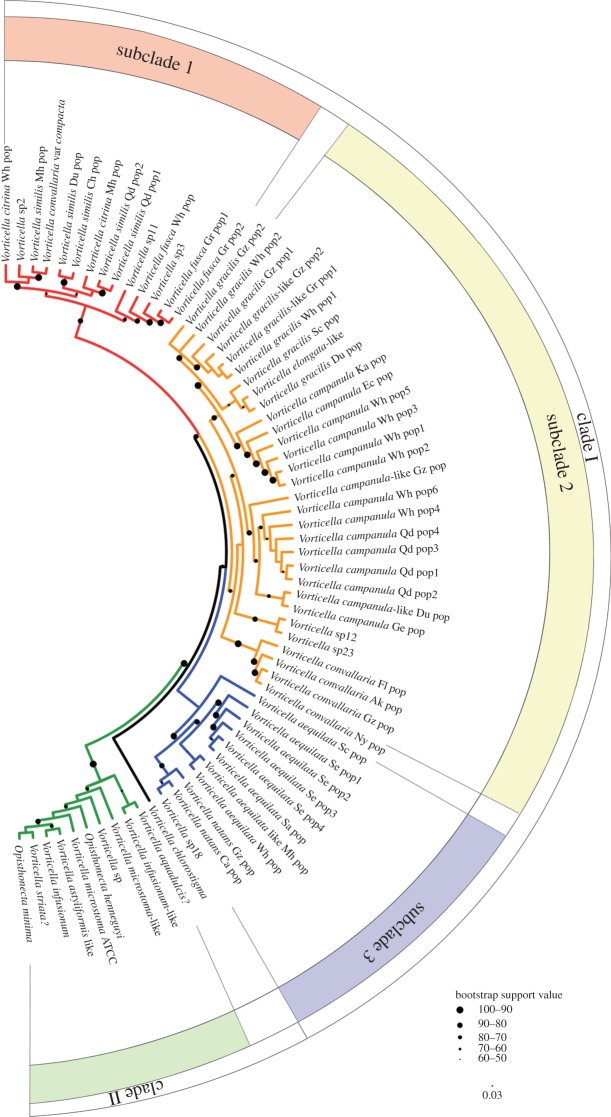
The colour coding of each clade and its subclades in figure 1 is based on the ITS2 tree. All tree topologies provided strong statistical support for monophyly of clade II relative to species of Vorticella s. str. (clade I). Three genetic clusters distinguished by their ITS2 regions corresponded to the three subclades within clade I seen in topologies based on ITS1-5.8S-ITS2 sequences (figures 1 and 2). In subclade 1, morphological characters were consistent with those that define the V. convallaria species complex, which contained the type species of the genus Vorticella. Subclade 2 contains representatives of the morphospecies V. gracilis, Vorticella elongata and V. campanula and two unidentified species. Subclade 3 was basal to the other two and consisted of three identifiable and one unidentified species. Vorticella chlorostigma was divergent from all subclades and branched basally within clade I. Clade II was basal to clade I (and the entire family Vorticellidae; see [12]), comprising morphospecies of the Vorticella microstoma-complexes, Vorticella astyliformis-like, Vorticella infusionum, Vorticella striata?, Vorticella aquadulcis?, one unidentified species and two species of the stalkless, secondarily free-swimming genus Opisthonecta.
Figure 1.
Profile neighbour joining (PNJ) tree constructed with sequence-structure data from the ITS2 region and based on a comprehensive sampling of the genus Vorticella. Major phylogenetic clades are highlighted using differential colour-coding. Scale bar, 0.03 substitutions/site. (Online version in colour.)
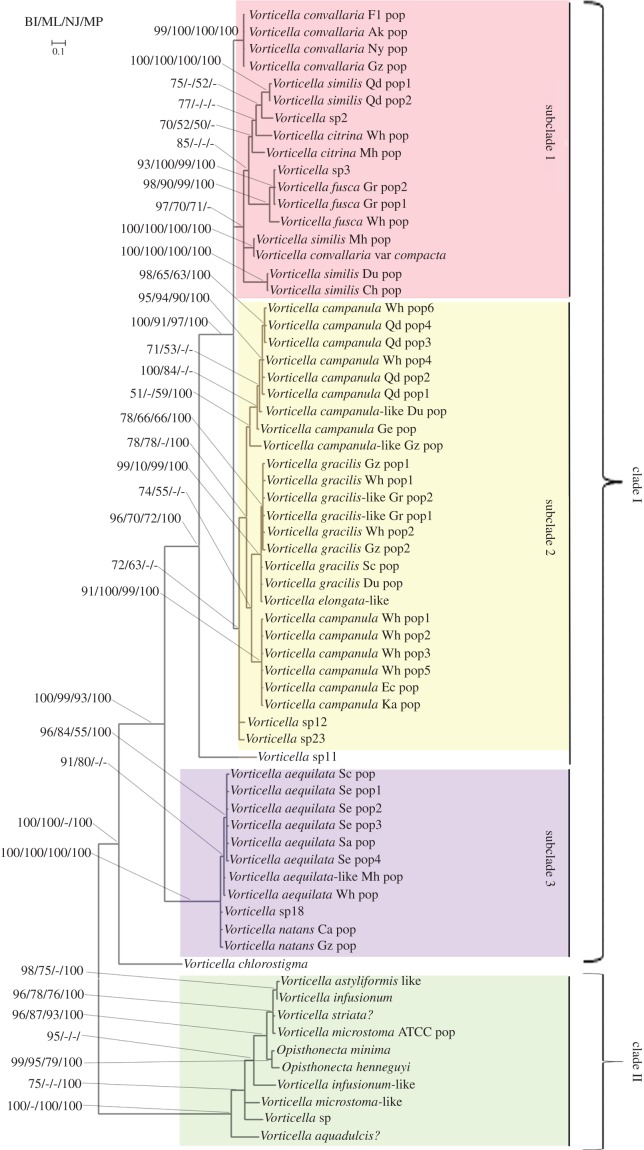
Figure 2.
Phylogenetic tree depicting evolutionary relationships among species of Vorticella based on ITS1-5.8S-ITS2 sequences. Branch lengths indicate the inferred number of nucleotide substitutions per site. Nodal support values (posterior probabilities for Bayesian inference (BI) and bootstrap values for other methods) are indicated in the following order: BI/maximum likelihood (ML)/neighbour joining (NJ)/maximum parsimony (MP). Scale bar, 0.1 substitutions/site. (Online version in colour.)
Among samples of all morphospecies of Vorticella individuals, those of V. similis were the most diverse, and representatives could be found in three different clusters within subclade 1. Likewise, the 13 sequences of V. campanula were recovered as a paraphyly, with one sequence being a sister to the V. gracilis cluster within subclade 2. Of the other morphospecies, none had representatives in more than one clade or subclade. The only differences in topology between trees based on the entire ITS1-5.8S-ITS2 region and those based on the ITS2 region alone were placement of Vorticella sp11 and a small cluster of V. convallaria. Vorticella sp11 nested within subclade1 in the ITS2 tree (figure 1) but occupying a position basal to subclade 2 in the ITS1-5.8S-ITS2 tree (figure 2). In the ITS2 tree (figure 1), a small cluster of V. convallaria (Fl, Ak, Gz, Ny) fell within subclade 2; however, this cluster was associated with subclade 1 in an unresolved position in ITS1-5.8S-ITS2 trees (figure 2).
(c). Phylogenetic relationship of species within clade II inferred from both ribosomal and protein-coding gene markers
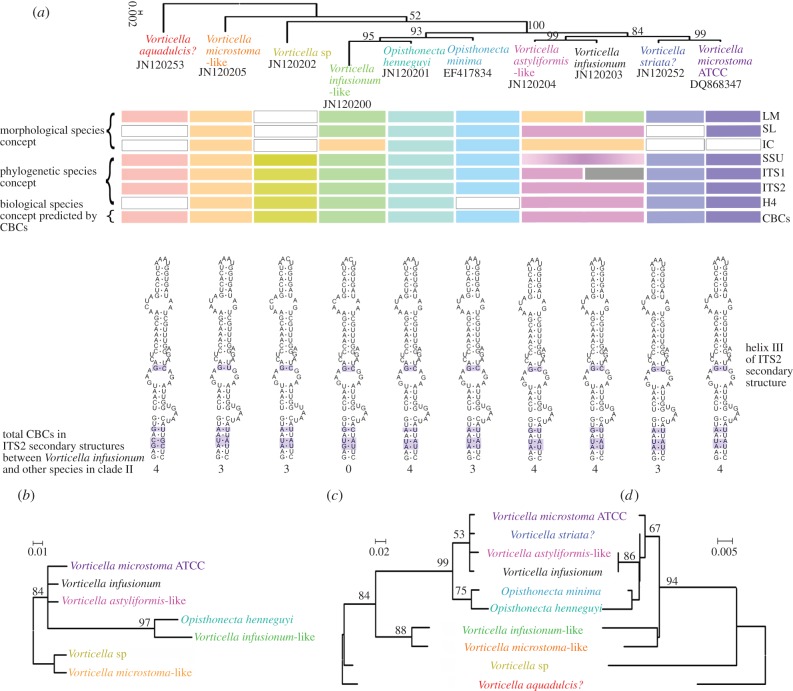
We were able to compare topologies of trees inferred from sets of SSU rRNA, ITS1 and ITS2 data from 10 species in clade II with a tree inferred from Histone H4 data from seven of these species (figure 3). Topologies of Bayesian trees constructed with SSU rRNA, ITS1, and H4 sequences (figure 3a–c) and a profile neighbour joining (PNJ) tree constructed with ITS2 sequences and secondary structures (figure 3d) were almost identical. In all of these trees, a clade containing V. astyliformis-like, V. infusionum, V. microstoma ATCC and V. striata? had a close, well-supported relationship with two members of Opisthonecta. However, the relationship of the species of Opisthonecta to V. infusionum-like was ambiguous. The SSU rRNA phylogeny supported a closer relationship of Opisthonecta henneguyi and V. infusionum-like than between O. henneguyi and Opisthonecta minima. Phylogenies based on ITS1 and ITS2 datasets showed O. henneguyi was sister to O. minima. Vorticella aquadulcis? was the most basal taxon in clade II in trees based on ribosomal markers.
Figure 3.
Bayesian trees inferred from (a) SSU rRNA, (b) Histone 4, and (c) ITS-1 sequences and a PNJ tree inferred from (d) ITS2 primary sequences and secondary structures. Numbers in each node indicate either the posterior probability (Bayesian trees) or bootstrap values (PNJ tree). (a) Horizontal bars show the clustering patterns that can be inferred from gross morphology (LM) and morphology revealed by silver staining (SL and IC), molecular markers (listed on the right side) and sexual reproductive compatibility predicted by CBCs [24]. The blank indicates that corresponding analyses were not carried out. Scale bars: (a) 0.002, (b) 0.01, (c) 0.02 and (d) 0.005 substitutions/site. IC, infraciliature; LM, light microscopy; SL, silverline system. (Online version in colour.)
3. Discussion
(a). Phylogenetic and taxonomic relationships within the genus Vorticella s. l.
Traditionally, morphological identification of Vorticella species has been based mostly on characteristics of gross morphology that are visible under the light microscope [1,3]. Morphological variation within species can overlap with variation among species [9], thus, identification using molecular characters is an important alternative for this genus. Information from the ITS1-5.8S-ITS2 region, especially ITS2, has proved to be a source of strong molecular markers for resolving phylogenetic relationships among sessile peritrichs at different taxonomic levels [13,15]. In this work, therefore, we provide a new evaluation of the phylogenetic relationship of Vorticella species using both ITS1-5.8S-ITS2 and ITS2 sequences.
Trees made with both ITS1-5.8S-ITS2 sequences and ITS2 sequences alone featured the same subclades of clade I and, furthermore, showed subclades 1 and 2 as sisters with relatively strong support but with support for monophyly of each one being lower (figures 1 and 2). The large number of specific similarities in helix III of ITS2 that exist between subclades 1 and 2 (see the electronic supplementary material, figure S1) explain this high degree of support for their relationship to one another. By contrast, helix III of subclade 3 is clearly distinct from that of both subclades 1 and 2, confirming it as a divergent lineage within clade I (see the electronic supplementary material, figure S1). The divergent, basal position of Vorticella chlorostigma within clade I in all trees suggests that it might be a representative of yet another subclade. Addition of still more species to analyses is probably the only way to resolve subclades 1–3 and test the hypothesis of existence of a fourth Vorticella chlorostigma clade.
Topological differences did exist between results derived from ITS1-5.8S-ITS2 and ITS2 data. For example, analysis of ITS1-5.8S-ITS2 data places Vorticella sp11 as a basal member of subclade 2 (figure 2), but it is placed within subclade 1 by ITS2 data alone (see the electronic supplementary material, figure S1; figure 1). A possible cause could be the inclusion of phylogenetically distorting characters from the ITS1 region that derive from the relatively high variability in primary sequences among distantly related species of Vorticella.
Previous molecular phylogenetic studies of vorticellid peritrichs based on SSU rRNA data alone and including a relatively small number of sequences [13] indicated that morphospecies of Vorticella in clade II are not members of the genus Vorticella s. str. Sun et al. [12] suggested assigning all species in clade II to the family Astylozoidae s. str. on the basis of distinctive molecular signatures in their SSU rRNA sequences in the absence of unifying morphological characteristics and despite differences in gross morphology resulting from loss of stages from the life cycle by Astylozoon (telotroch lost) and Opisthonecta (trophont lost). In this study, trees based on the ITS1-5.8S-ITS2 and ITS2 regions as well as the distinctive helix 3 of ITS2 in members of clade II provides conclusive support for this taxonomic action and removes any lingering doubt that morphospecies of Vorticella in clade II are not congeneric with species of Vorticella s. str. in clade I.
The V. convallaria species complex includes not only the type species of the genus, but also ciliates that are commonly encountered in both wastewater treatment plants and a variety of natural freshwater habitats worldwide. Historically, this complex consisted of the following taxa: V. convallaria Linnaeus, 1758, V. citrina Müller, 1773, Vorticella nebulifera Müller, 1786, and V. similis Stokes, 1887 [9]. Vorticella nebulifera differs from the other three species by being found in marine habitats but, otherwise, resembles them in terms of basic morphological features. The three freshwater species are morphologically very similar and difficult to differentiate because of their morphological variability. Consequently, their taxonomic status and phylogenetic relationships have posed a problem for more than a century.
In this study, we sequenced the ITS region for 15 samples belonging to the V. convallaria species complex collected from 12 different localities. The relationship between the clade of V. convallaria and the one containing V. similis, V. citrina, V. convallaria var compacta, V. fusca and two unknown species is unresolved (figure 2), but a close relationship between four sequences of V. convallaria and sequences of V. similis, V. citrina and V. convallaria var compacta is indicated by our molecular analyses, confirming that they all belong to the V. convallaria species complex. However, species boundaries within this complex could not be discerned with current data alone—e.g. morphospecies of V. similis were spread over three different clusters within subclade 1 (figure 2). In future studies, sequence data from multiple loci should be combined with morphological and ecological evidence to test species boundaries and assess their evolutionary relationships in this species complex.
(b). Pairwise distances between morphospecies of Vorticella in clade I
Clade I comprised sequences of 46 populations of 10 morphospecies of Vorticella from different localities. The pairwise distance between populations of most of these species was usually small, but V. citrina, V. similis and V. campanula showed a relatively large variation between different populations (see the electronic supplementary material, table S2). Vorticella campanula was the most commonly encountered species in our collections, and a genetic distance of 2.89% between the 13 populations from different localities was observed. In both ITS1-5.8S-ITS2 and ITS2 trees (figures 1 and 2), populations of V. campanula from Austria, China and Japan were recovered as two separate genetic clusters, with one being sister to the V. gracilis cluster. All these results suggest that it is likely that cryptic species exist within the morphospecies V. campanula. Thus, future efforts will focus on testing this hypothesis with broader sampling and inclusion of multiple genes in analyses.
(c). Phylogenetic and taxonomic relationships within clade II inferred from multiple datasets
Vorticella microstoma was the only species other than Opisthonecta and Astylozoon species within clade II that had been included in previous studies, and it appeared to be a phylogenetically isolated organism [15,25]. It is now clear that V. microstoma is merely one representative of a ‘cloud’ of Vorticella morphospecies, all with a relatively small body size and narrow peristome. Results of this study (figures 1 and 2) and previous phylogenetic investigations [12] clearly indicate that Vorticella s. l. is a paraphyletic assemblage; however, relationships within clade II are far from clear. Vorticella morphospecies (see the electronic supplementary material, figure S2a–c,f–h) in this clade share a similar gross morphology and differ only slightly in macronuclear shape/orientation and characters revealed by silver staining that overlap to some degree (see the electronic supplementary material, table S3). Species in clade II also have very similar SSU rRNA sequences, with genetic distances among some morphospecies of Vorticella being larger than those between species of Opisthonecta and other morphospecies of Vorticella (see the electronic supplementary material, table S4). In short, morphology appears to reveal little, if anything about phylogenetic relationships among morphospecies of Vorticella within clade II, and even generic boundaries are unclear in molecular analyses.
Close relationships were seen between species of Opisthonecta and some morphospecies of Vorticella in previous studies based only on nuclear SSU rRNA sequences [12]. In this study, we used sets of nuclear SSU rRNA plus the faster evolving genetic markers for H4, ITS1 and ITS2 to assess phylogenetic relationships of the species within clade II. Trees inferred from four different datasets (figure 3) including the same taxa (except the H4 tree, figure 3b) revealed a close, well-supported relationship between a clade comprising two species of Opisthonecta and a clade containing sequences of four morphospecies of Vorticella—V. astyliformis-like, V. infusionum, V. microstoma and V. striata? A monophyletic group comprising species of Opisthonecta and V. infusionum-like was recovered with high support in both SSU rRNA and H4 trees (figure 3a,b), and a sister relationship between species of Opisthonecta and a clade containing V. astyliformis-like, V. infusionum, V. microstoma and V. striata? was supported in ITS1 and ITS2 trees (figure 3c,d).
Morphologically, species of Opisthonecta are markedly different from all morphospecies of Vorticella in clade II, always existing as free-swimming telotrochs (stalkless individuals with a circumferential band of somatic cilia). In most sessile peritrichs, a telotroch is a non-feeding dispersal stage that secretes a stalk and metamorphoses into a trophont by resorbing somatic cilia and reforming oral cilia. In Opisthonecta, by contrast, the somatic ciliature persists and individuals are unable to secrete a stalk; and unlike true telotrochs, zooids are able to develop a normal, ciliated oral complex for suspension feeding (see the electronic supplementary material, figure S2d,e).
At first glance, the loss of the normal trophont morphology appears to be a profound difference between Opisthonecta and other species of sessile peritrichs. In reality, this sort of radical evolutionary loss of a developmental stage has happened in other sessile peritrichs and many kinds of organisms [26–28] and could have resulted from a relatively restricted genetic change in the regulation of development without requiring a large degree of divergence in other morphological or physiological characteristics. For example, Astylozoon is another genus in clade II [12] that is characterized by loss of a developmental stage (lacks telotroch and thus exists as a stalkless, swimming trophont) and also appears to be significantly different in morphology from morphospecies of Vorticella. However, the boundary between them is blurred by the existence of V. astyliformis as a typical stalked trophont at some times and a free-swimming, stalkless trophont at other times [29]. A permanently stalkless trophont (i.e. Astylozoon) could have evolved from such a species by means of a relatively small genetic change (e.g. only 1.976% genetic distance in SSU rRNA sequences exists between A. enriquesi and V. astyliformis), and Opisthonecta may have evolved equally rapidly without experiencing large-scale genetic divergence.
A close relationship between V. astyliformis-like and V. infusionum was supported in all trees (figures 1–3). The two species have similar morphological characteristics (e.g. small size of less than 40 µm, rotund body shape, narrow peristome), and subtle morphological differences in characters revealed by silver staining overlapped (see the electronic supplementary material, figure S2 and table S3). There was a 99.935% similarity in their SSU rRNA sequences (see the electronic supplementary material, table S4). Their ITS2 sequences were identical, hence no CBC was found and there was a genetic distance of only 0.671% between their ITS1 sequences (see the electronic supplementary material, table S4). All of these close genetic and morphological similarities prompt us to conclude that V. astyliformis-like and V. infusionum may be closely related cryptic species or even populations of a single, variable species.
All morphospecies of Vorticella in clade II were placed in a new genus Vorticellides by Foissner et al. [14], partly on the basis of early molecular evidence that one of them—V. astyliformis—was not congeneric with species of Vorticella s. str. and partly on the basis of a morphological characteristic—presence of two epistomial membranes. Results of this study and that of Sun et al. [12] do not support this conclusion. Evolutionary distances between all taxa in clade II (electronic supplementary material, table S4) are small compared with those between taxa in clade I (data were not shown), and there is no support for a single cluster of taxa with stalked trophonts. Moreover, there are not even clear distinctions between species of Opisthonecta and morphospecies of Vorticella in clade II. All morphospecies in clade II appear to have a wide geographical distribution owing to their ability to form cysts and disperse between ephemeral aquatic habitats (temporary pools, wet soil); therefore, a much larger set of samples taken from many geographical regions will be required to reveal evolutionary lineages within clade II with more confidence and resolve the taxonomic status of species and genera within it.
(d). Perspectives
Species of Vorticella, one of the most commonly encountered groups of ciliates in aquatic ecosystems worldwide, are particularly abundant in sewage treatment systems and aquatic farming ponds where organic matter accumulates. Historically, they were considered to be a monophyletic group for over 200 years. However, the molecular divergence observed in the genus Vorticella illustrates that the current catalogue of ciliate genera/classes is incomplete and confirms that it is necessary to recognize a class-level taxon to accommodate all organisms in clade II. Meanwhile, mismatches that were detected between morphological and molecular characters revealed the existence of apparent cryptic species and, possibly, even cryptic genera. Thus, all future workers investigating evolution, biogeography, ecology, conservation, biodiversity and more applied aspects (e.g. water pollution and ecotoxicology) of these Vorticella species should be aware of the necessity to make sure of the specific status of their organisms and their possible biological differences. Finally, our results also underscore the need, in terms of both morphology-based systematics and molecular phylogeny, for multiple markers and a broad set of samples from different geographical areas and types of habitats to understand the evolutionary history of any group of ciliates, which are a major group of microbial eukaryotes.
4. Material and methods
(a). Collection, isolation, fixation and identification of organisms
Sixty-six populations of peritrichs matching the morphological description of the genera Vorticella and Opisthonecta were collected from Europe, Asia and North America and used to obtain sequences of nuclear ITSs. Sequences have been deposited in GenBank with the accession numbers shown in the electronic supplementary material, table S1. Samples were collected from a wide variety of aquatic habitats—streams, irrigation ditches in farmed fields, bogs, lakes, ponds in botanical gardens, ephemeral pools and other areas (see the electronic supplementary material, table S5). Samples were brought directly back to the laboratory, and the cells were confirmed as species of Vorticella with live microscopy and silver staining [30]. Some samples we designated as ‘Vorticella sp-like’, for example V. astyliformis-like, because of some uncertainty in identification with overlapping but not identical morphological characters of known species. Cells were isolated under a stereomicroscope from mixed samples with a micropipette or by clipping off bits of the substrate (e.g. aquatic plants, dead leaves and decomposing plant debris) to which they were attached. Isolated cells were observed in wet mounts under a compound microscope (Olympus/Nikon/Zeiss) and photographed with a digital camera. Some cells were cleaned manually by carrying them through three or more changes of syringe-filtered water and pipetting away contaminants, after which they were fixed with 95% ethanol or transferred to animal tissue lysis buffer (Qiagen, Valencia, CA, USA) for DNA extraction. Some cells were transferred to Petri dishes to start clonal cultures. Cultures of species of Vorticella and Opisthonecta were grown in a medium made with wheat grains or by adding a 5 g l−1 stock solution of boiled and filtered wheat grass powder in small amounts of habitat water to simulate growth of the bacterial community. Cells were picked individually from clonal cultures with a micropipette for DNA extraction. Terminology and systematic classification used in this paper follow Lynn [3].
(b). DNA extraction, gene amplification and gene sequencing
Extraction of DNA from samples was carried out using methods described in [13]. Amplification of the ITS region and a fragment of approximately 150 bp comprising part of the internal H4 region was accomplished according to protocols in [31,32]. Amplicons were cleaned by filtration using a QIAquick PCR Purification Kit (Qiagen Sciences, Germantown, MD, USA) and sequenced in both directions using an ABI 3730-XL DNA analyzer (Applied Biosystems, Foster City, CA, USA). Sequence fragments were assembled into contiguous sequences and edited with the Sequencher v. 4.0 software package (GeneCodes Corp., Ann Arbor, MI, USA). All new sequences have been deposited in the GenBank database (see the electronic supplementary material, table S1 for accession numbers).
(c). Predicting secondary structures of ITS2
We relied primarily on the free-energy minimization approach to secondary structure inference, which assumes that the dominant interactions (H-bonding between bases and stacking between adjacent bases) are local and that conformations adopted by RNA are the lowest free-energy conformations at equilibrium [33]. Consensus structures of ITS2 regions were found using the Alifold Server (http://rna.tbi.univie.ac.at/cgi-bin/alifold.cgi), which predicts structures from an alignment of related RNA sequences [34]. With the guidance of these consensus structures, the secondary structures of ITS2 sequences were predicted with Mfold v. 3.2 (http://mfold.bioinfo.rpi.edu/cgi-bin/rna-form1.cgi) [35] by screening for thermodynamically optimal and suboptimal secondary structures using the default values for folding temperature (37°C), per cent suboptimality (5%), upper bound of computed foldings (50), window parameter (25) and maximum distance between paired bases (no limit). Results for the various species were compared to reveal the folding pattern common to them all, aided by knowledge of the conserved structure of helix II and by the existence of the highly conserved region of DNA on the 5′ side of helix III [20]. This, in turn, established the conserved structural models for sessile peritrichs that revealed evidence of homology useful for phylogenetic analysis [13]. The ITS2 sequences with homologous structures were synchronously aligned using 4SALE [36]. The alignment with structural information was exported. The resultant filename was changed to a filename with the extension ‘.xfasta’ for subsequent analysis.
(d). Phylogenetic analysis
The alignment output file from 4SALE was imported into ProfDistS [37], and ‘RNA/DNA structure Profile Neighbor-Joining’ was selected from the ‘Run’ menu (Bootstraps = 1000, Distance Correction Model = General Time Reversible, Ratematrix Q = Q_ITS2.txt). The resulting tree file with node strengths was viewed in ProfDistS and then visualized and reproduced with FigTree v. 1.2.3 [38] and MEGA v. 4.0 [39].
Phylogenetic analyses of 66 strains were also performed with data from the entire ITS1-5.8S-ITS2 region. Sequences of ITS15.8S-ITS2 regions were aligned using ClustalW as an accessory application of Bioedit v. 7.0.9, then adjusted by eye [40]. The aligned matrix was analysed using four methods (Bayesian inference (BI), maximum likelihood (ML), neighbour joining (NJ) and maximum parsimony (MP)). BI analyses were performed with MrBayes v. 3.1.2 [41] using the GTR + I + G model selected by MrModeltest v. 2 [42] under the Akaike information criterion. Four simultaneous Markov chain Monte Carlo chains were run for 2 500 000 generations sampling every 100 generations. The first 25% trees were discarded as burn-in and the 50% majority-rule consensus tree was determined to calculate the posterior probabilities for each node. The appropriate model GTR + I + G was determined by Modeltest [43] for ML analysis. An ML tree was constructed with PhyML v. 3.0 (via http://www.atgc-montpellier.fr/phyml), which performed ML analysis with heuristic searches and a 1000-fold bootstrap analysis [44]. The NJ and MP analyses were performed with PAUP* 4.0b10 [45], including 1000 bootstrapping replicates. We also performed BI and PNJ analyses of clade II with four datasets and trees based on these sets of sequences constructed as described above.
Acknowledgements
Much appreciation is expressed to Drs Wilhelm Foissner (University of Salzburg), Weibo Song (Ocean University of China), Xiaofeng Lin (South China Normal University), and Wei Miao and Yingchun Gong (Institute of Hydrobiology, Chinese Academy of Sciences) for their institutional support. We are grateful to the Institute of Marine Sciences, University of North Carolina at Chapel Hill for providing facilities for our work in the vicinity of Morehead City, NC, USA. Dr Yasushi Kusuoka (Lake Biwa Museum), Dr Chengjie Fu (Uppsala University), Dr Jie Xiong (Institute of Hydrobiology, Chinese Academy of Sciences), and Mr Xiaokun Shi and Mr Bo Li (South China Normal University) are thanked for their invaluable help with collecting samples and taking photomicrographs.
Funding statement
This work was supported by the Natural Science Foundation of China (no. 31372167, P.S.; no. 41306125, D.X.), National Science Foundation grant (DEB-0716348, to J.C.C.) and Basic Science Research Program through the NRF funded by the Government of Korea (MEST) (no. 2012R1A1A2005751, to M.K.S.).
References
- 1.Warren A. 1986. A revision of the genus Vorticella (Ciliophora: Peritrichida). Bull. Br. Mus. Nat. Hist. 50, 1–57 [Google Scholar]
- 2.Corliss JO. 1979. The ciliated protozoa: characterization, classification and guide to the literature, 2nd edn Oxford, UK: Pergamon Press [Google Scholar]
- 3.Lynn DH. 2008. The ciliated protozoa. Characterization, classification, and guide to the literature, 3rd edn Dordrecht, The Netherlands: Springer [Google Scholar]
- 4.Azam F, Fenchel T, Field JG, Gray JS, Meyerreil LA, Thingstad F. 1983. The ecological role of water-column microbes in the sea. Mar. Ecol. Progr. Ser. 10, 257–263 (doi:10.3354/meps010257) [Google Scholar]
- 5.Mahadevan L, Matsudaira P. 2000. Motility powered by supramolecular springs and ratchets. Science 288, 95–99 (doi:10.1126/science.288.5463.95) [DOI] [PubMed] [Google Scholar]
- 6.Sherr BF, Sherr EB, Rassoulzadegan F. 1988. Rates of digestion of bacteria by marine phagotrophic protozoa: temperature-dependence. Appl. Environ. Microbiol. 54, 1091–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lloyd B. 1973. The construction of a sand profile sampler: its use in the study of the Vorticella populations and the general interstitial microfauna of slow sand filters. Water Resour. 7, 963–973 (doi:10.1016/0043-1354(73)90178-4) [Google Scholar]
- 8.Berger H, Foissner W.2003. Illustrated guide and ecological notes to ciliate indicator species (Protozoa, Ciliophora) in running waters, lakes, and sewage plants. Handbuch Angewandte Limnologie 17, 160 p. Ecomed, Landsberg, Germany.
- 9.Foissner W, Berger H, Kohmann F. 1992. Taxonomische und ökologische Revision der Ciliaten des Saprobiensystems-Band II: Peritrichida, Heterotrichida, Odontostomatida. München, Germany: Informationsberichte des Bayer, Landesamtes für Wasserwirtschaft 5/92, 1–311 [Google Scholar]
- 10.Simon EM, Nanney DL, Doerder FP. 2008. The ‘Tetrahymena pyriformis’ complex of cryptic species. Biodivers. Conserv. 17, 365–380 (doi:10.1007/s10531-007-9255-6) [Google Scholar]
- 11.Catania F, Wurmser F, Potekhin AA, Przyboś E, Lynch M. 2009. Genetic diversity in the Paramecium aurelia species complex. Mol. Biol. Evol. 26, 421–431 (doi:10.1093/molbev/msn266) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun P, Clamp JC, Xu D, Kusuoka Y, Miao W. 2012. Vorticella Linnaeus, 1767 (Ciliophora, Oligohymenophora, Peritrichia) is a grade not a clade: redefinition of Vorticella and the families Vorticellidae and Astylozoidae using molecular characters derived from the gene coding for small subunit ribosomal RNA. Protist 163, 129–142 (doi:10.1016/j.protis.2011.06.005) [DOI] [PubMed] [Google Scholar]
- 13.Sun P, Clamp JC, Xu D. 2010. Analysis of the secondary structure of ITS transcripts in peritrich ciliates (Ciliophora, Oligohymenophorea): Implications for structural evolution and phylogenetic reconstruction. Mol. Phylogenet. Evol. 56, 242–251 (doi:10.1016/j.ympev.2010.02.030) [DOI] [PubMed] [Google Scholar]
- 14.Foissner W, Blake N, Wolf K, Breiner HW, Stoeck T. 2010. Morphological and molecular characterization of some peritrichs (Ciliophora: Peritrichida) from tank bromeliads, including two new genera: Orborhabdostyla and Vorticellides. Acta. Protozool. 48, 291–319 [PMC free article] [PubMed] [Google Scholar]
- 15.Sun P, Clamp JC, Xu D, Kusuoka Y, Hori M. 2011. Molecular phylogeny of the family Vorticellidae (Ciliophora, Peritrichia) using combined datasets with a special emphasis on the three morphologically similar genera Carchesium, Epicarchesium and Apocarchesium. Int. J. Syst. Evol. Microbiol. 61, 1001–1010 (doi:10.1099/ijs.0.020255-0) [DOI] [PubMed] [Google Scholar]
- 16.Young I, Coleman AW. 2004. The advantages of the ITS2 region of the nuclear rDNA cistron for analysis of phylogenetic relationships of insects: a Drosophila example. Mol. Phylogenet. Evol. 30, 236–242 (doi:10.1016/S1055-7903(03)00178-7) [DOI] [PubMed] [Google Scholar]
- 17.Schultz J, Wolf M. 2009. ITS2 sequence-structure analysis in phylogenetics: a how-to manual for molecular systematics. Mol. Phylogenet. Evol. 52, 520–523 (doi:10.1016/j.ympev.2009.01.008) [DOI] [PubMed] [Google Scholar]
- 18.Wiemers M, Keller A, Wolf M. 2009. ITS2 secondary structure improves phylogeny estimation in a radiation of blue butterflies of the subgenus Agrodiaetus (Lepidoptera: Lycaenidae: Polyommatus). BMC Evol. Biol. 9, 300 (doi:10.1186/1471-2148-9-300) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coleman AW. 2003. ITS2 is a double-edged tool for eukaryote evolutionary comparisons. Trends Genet. 19, 370–375 (doi:10.1016/S0168-9525(03)00118-5) [DOI] [PubMed] [Google Scholar]
- 20.Coleman AW. 2007. Pan-eukaryote ITS2 homologies revealed by RNA secondary structure. Nucleic Acids Res. 35, 3322–3329 (doi:10.1093/nar/gkm233) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keller A, Förster F, Müller T, Dandekar T, Schultz J. 2010. Including RNA secondary structures improves accuracy and robustness in reconstruction of phylogenetic trees. Biol. Direct 5, 4 (doi:10.1186/1745-6150-5-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moniz MBJ, Kaczmarska I. 2009. Barcoding diatoms: is there a good marker? Mol. Ecol. Resour. 9(Suppl. s1), 65–74 (doi:10.1111/j.1755-0998.2009.02633.x) [DOI] [PubMed] [Google Scholar]
- 23.Yao H, et al. 2010. Use of ITS2 region as the universal DNA barcode for plants and animals. PLoS ONE 5, e13102 (doi:10.1371/journal.pone.0013102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coleman AW. 2009. Is there a molecular key to the level of ‘biological species’ in eukaryotes? A DNA guide. Mol. Phylogenet. Evol. 50, 197–203 (doi:10.1016/j.ympev.2008.10.008) [DOI] [PubMed] [Google Scholar]
- 25.Miao W, Feng W, Yu Y, Zhang X, Shen Y. 2004. Phylogenetic relationships of the subclass Peritrichia (Oligohymenophorea, Ciliophora) inferred from small subunit rRNA gene sequences. J. Eukaryot. Microbiol. 51, 180–186 (doi:10.1111/j.1550-7408.2004.tb00543.x) [DOI] [PubMed] [Google Scholar]
- 26.Strathmann RR. 1978. The evolution and loss of feeding larval stages of marine invertebrates. Evolution 32, 894–906 (doi:10.2307/2407502) [DOI] [PubMed] [Google Scholar]
- 27.Wake DB, Hanken J. 1996. Direct development in the lungless salamanders: what are the consequences for developmental biology, evolution and phylogenesis? Int. J. Dev. Biol. 40, 859–869 [PubMed] [Google Scholar]
- 28.Callery EM, Fang H, Elinson RP. 2001. Frogs without polliwogs: evolution of anuran direct development. BioEssays 23, 233–241 (doi:10.1002/1521-1878(200103)23:3<233::AID-BIES1033>3.0.CO;2-Q) [DOI] [PubMed] [Google Scholar]
- 29.Foissner W. 1981. Morphologie und taxonomie einiger heterotricher und peritricher ciliaten (Protozoa: Ciliophora) aus alpinen boden. Protistologica 17, 29–43 [Google Scholar]
- 30.Wilbert N. 1975. Eine verbesserte Technik der Protargolimprägnation für Ciliaten. Mikrokosmos 64, 171–179 [Google Scholar]
- 31.Goggin CL, Murphy NE. 2000. Conservation of sequence in the internal transcribed spacers and 5.8S ribosomal RNA among geographically separated isolates of parasitic scuticociliates (Ciliophora, Orchitophryidae). Dis. Aquat. Organ. 40, 79–83 (doi:10.3354/dao040079) [DOI] [PubMed] [Google Scholar]
- 32.Bernhard D, Schlegel M. 1998. Evolution of histone H4 and H3 genes in different ciliate lineages. J. Mol. Evol. 46, 344–354 (doi:10.1007/PL00006311) [DOI] [PubMed] [Google Scholar]
- 33.Xia T, Mathews DH, Turner DH. 2001. Thermodynamics of RNA secondary structure formation. In RNA (eds Soll D, Nishimura S, Moore P.), pp. 21–48 Amsterdam, The Netherlands: Pergamon Press [Google Scholar]
- 34.Hofacker IL, Fekete M, Stadler PF. 2002. Secondary structure prediction for aligned RNA sequences. J. Mol. Biol. 319, 1059–1066 (doi:10.1016/S0022-2836(02)00308-X) [DOI] [PubMed] [Google Scholar]
- 35.Zuker M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31, 3406–3415 (doi:10.1093/nar/gkg595) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seibel PN, Müller T, Dandekar T, Wolf M. 2008. Synchronous visual analysis and editing of RNA sequence and secondary structure alignments using 4SALE BMC Res . Notes 1, 91 (doi:10.1186/1756-0500-1-91) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wolf M, Ruderisch B, Dandekar T, Müller T. 2008. ProfdistS: (Profile-) distance based phylogeny on sequence-structure alignments. Bioinformatics 24, 2401–2402 (doi:10.1093/bioinformatics/btn453) [DOI] [PubMed] [Google Scholar]
- 38.Rambaut A.2009. FigTree v 1.2.3. Program distributed by the author, available from http://tree.bio.ed.ac.uk/software/figtree/
- 39.Tamura K, Dudley J, Nei M, Kumar S. 2007. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24, 1596–1599 (doi:10.1093/molbev/msm092) [DOI] [PubMed] [Google Scholar]
- 40.Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98 [Google Scholar]
- 41.Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574 (doi:10.1093/bioinformatics/btg180) [DOI] [PubMed] [Google Scholar]
- 42.Nylander J.2004. MrModeltest version 2. Distributed by the author. Department of Systematic Zoology, Evolutionary Biology Centre. Uppsala University, Uppsala, Sweden.
- 43.Posada D, Crandall KA. 1998. Modeltest: testing the model of DNA substitution. Bioinformatics 14, 817–818 (doi:10.1093/bioinformatics/14.9.817) [DOI] [PubMed] [Google Scholar]
- 44.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321 (doi:10.1093/sysbio/syq010) [DOI] [PubMed] [Google Scholar]
- 45.Swofford DL. 2002. PAUP*: phylogenetic analysis using parsimony (*and other methods), v. 4 Sunderland, MA: Sinauer Associates [Google Scholar]