Abstract
Indirect genetic effects (IGEs) describe how an individual's behaviour—which is influenced by his or her genotype—can affect the behaviours of interacting individuals. IGE research has focused on dyads. However, insights from social networks research, and other studies of group behaviour, suggest that dyadic interactions are affected by the behaviour of other individuals in the group. To extend IGE inferences to groups of three or more, IGEs must be considered from a group perspective. Here, I introduce the ‘focal interaction’ approach to study IGEs in groups. I illustrate the utility of this approach by studying aggression among natural genotypes of Drosophila melanogaster. I chose two natural genotypes as ‘focal interactants’: the behavioural interaction between them was the ‘focal interaction’. One male from each focal interactant genotype was present in every group, and I varied the genotype of the third male—the ‘treatment male’. Genetic variation in the treatment male's aggressive behaviour influenced the focal interaction, demonstrating that IGEs in groups are not a straightforward extension of IGEs measured in dyads. Further, the focal interaction influenced male mating success, illustrating the role of IGEs in behavioural evolution. These results represent the first manipulative evidence for IGEs at the group level.
Keywords: aggression, IGEs, fruitfly (Drosophila melanogaster), social behaviour, sexual selection
1. Introduction
Indirect genetic effects (IGEs) describe how an individual's behaviour—which is influenced by his or her genotype—can affect the behaviours of interacting individuals [1]. IGEs can occur when genotypes vary in maternal traits, tendency to cooperate, aggressiveness or in any other trait that affects the phenotypes of conspecifics [1–3]. IGEs are important for understanding individual behaviour and social selection [4], and they can dramatically change the rate and even the direction of behavioural evolution [3,5,6].
IGEs have been identified in a wide range of organisms [7–13], demonstrating that IGEs are common. One limitation of current research is that investigators typically consider IGEs between pairs of individuals [8–10,12,14,15], or in groups containing only two genotypes ([11,16,17]; but see [13,18]). However, many organisms including humans interact in groups that include three or more unrelated individuals, each with its own unique genotype.
To further extend the relevance of IGEs to natural systems, it is important to think about how IGEs influence individual behaviours and fitness in multi-genotype groups. In particular, emerging insights from research on social networks suggest that dyadic interactions may be influenced by the behaviours of other group members [19–21]. The most famous example comes from pigtailed macaques, in which a few adult males are socially dominant and intervene in conflicts between other macaques. Experimental removal of these ‘policing’ males dramatically influences play, grooming and aggressive relationships among individuals in the network [22]. Similar ‘indirect’ effects have been discovered in many organisms including humans [23,24], eusocial insects [25] and guppies [26]; in this context, the term ‘indirect’ refers to the scenario in which dyadic behavioural interactions are influenced by the behaviours of other individuals in the network [21,24].
Experimental progress studying IGEs in groups has been hindered by two problems. In free-living organisms, groups contain genotypes that are not a random sample of the population [27–31], which confounds attempts to measure the effects of the genetic composition of the group on the behaviours within a particular dyad [32]. This problem may be partially overcome by studying organisms in which the social environment can be manipulated (including in the wild, e.g. cross-fostering studies). By contrast, in laboratory studies, factorially manipulating the genotypes of all individuals in the group (i.e. the natural extension of typical IGE studies) requires experimenters to study a very large number of genotypic combinations, even for small groups. Even for most laboratory organisms, this type of experiment is prohibitively large.
To begin to experimentally dissect the role of IGEs in groups, I propose a ‘focal interaction’ approach (figure 1). In this design, two genotypes are chosen as ‘focal interactants’; the interaction between them is the ‘focal interaction’. The term ‘focal interaction’ is intended as an analogy to the more traditional ‘focal individual’ approach [33]. In the focal individual approach, a particular individual is the subject of analysis and interacting conspecifics are considered representatives of the social environment. With the focal interaction approach, the behavioural interaction between the focal interactants is the subject of analysis. To identify ‘second-order’ IGEs, investigators can vary the genotypes of other individuals in the group (i.e. everyone except the focal interactants), and analyse the effects of this manipulation on the focal interaction (figure 1). The advantage of this design is that it dramatically reduces the number of treatments that are necessary, relative to a full factorial design, while still enabling researchers to evaluate the aspects of IGEs in groups that cannot be revealed by studies of dyadic IGEs: the potential effects of the genetic composition of the group on dyadic interactions among individuals [23–26]. Researchers studying eavesdropping [34] and audience effects [35] have used similar designs, demonstrating that the social context can influence dyadic behavioural interactions. However, the role of genetic variation in the social environment—i.e. the potential for second-order IGEs—in shaping dyadic interactions has not yet been considered.
Figure 1.
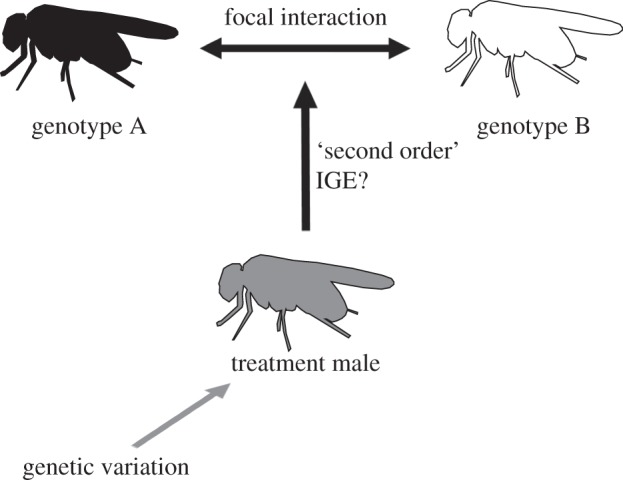
Focal interaction approach. Two male genotypes—genotype A and genotype B—were designated ‘focal interactants’. One male from each of these genotypes was present in every group. The genotype of third male, the ‘treatment male’, varied across groups. Measuring the effect of the behaviour of the treatment male on the focal interaction permits inference about the role of IGEs in groups.
Here, I implement the focal interaction approach by studying aggressive male–male interactions among natural genotypes of the fruitfly, Drosophila melanogaster, and I assess the implications of the resulting second-order IGEs for male–female mating interactions. Flies represent an ideal test case for understanding IGEs. Adults feed on microflora that grows on rotting fruit [36], so social life revolves around these discrete substrates [37]. Flies readily form groups both in the laboratory [38–40] and in the field [37,41]. Thus, social life is integral to fly biology. Of course, flies are also highly amenable to laboratory studies. In particular, heterozygous genotypes—genetically equivalent to identical twins—may be generated repeatedly under controlled conditions [38,40,42]; as heterozygotes, these individuals are genetically indistinguishable from wild flies [43,44]. Using this technique, social groups with specific compositions of natural genotypes may be replicated under controlled conditions [18]. Many studies of dyadic IGEs have been conducted on flies [7,10,13,14,16] including work on IGEs for fly aggressiveness [45].
To extend these findings to a group context, I chose two natural genotypes as focal interactants. One male from each of two focal interactant genotypes was present in every group. The third male in the group—termed the ‘treatment male’—was one of six natural genotypes from the same population as the focal interactants (figure 1). I allowed flies to interact in an environment with a single food patch. By measuring the aggressive behaviour of all individuals, I could determine how the genotype and behaviour of the treatment male influenced the focal interaction, i.e. the number of times that the focal interactants aggressively lunged at each other. By analysing the focal interaction across replicated groups, I could discriminate between random variation in the focal interaction, and variation in the focal interaction that was consistently associated with genetic variation in the behaviour of the treatment male. The latter finding would provide evidence for second-order IGEs, indicating that IGEs in groups are not a straightforward extension of dyadic IGEs.
After observing the behaviour of the three males for 30 min, I added a single virgin female to the group and measured which male she mated with, allowing me to determine whether second-order IGEs for male–male aggressiveness had consequences for male mating success [45].
2. Material and methods
(a). Genotypes
All flies were recurrent F1's made by repeatedly crossing the same inbred parental lines. The parental lines were originally derived from a population in Raleigh, NC, USA, and are numbered arbitrarily (DPGP.org). The direction of the crosses was consistent, to control for maternal effects. For example, genotype 1/2 would be generated by crossing virgin females of genotype 1 to males of genotype 2. The focal interactant crosses were: 208/712 and 707/765. I chose these genotypes as focal interactants based on prior data (J. B. Saltz 2012, unpublished data) which suggested that they show moderate, but different, levels of aggressiveness in a group context. Specifically, I expected 208/712 to be, on average, more aggressive than 707/765. I chose genotypes with moderate levels of aggressiveness because they represent ‘average flies’ from this population. The six treatment genotype crosses were likewise expected to span a range of aggressiveness based on prior data (J. B. Saltz 2012, unpublished data). The treatment genotypes were: 306/391, 304/862, 315/365, 486/380, 637/517 and 732/774. The female genotype was the same in all trials: 303/313. No genotypes shared sires or dams, so relatedness did not vary among groups and could not account for differences in behaviour.
For simplicity, I will refer to focal interactant genotype 208/712 as ‘genotype A’ and focal interactant genotype 707/765 as ‘genotype B.’
(b). Rearing
Rearing vials contained approximately 10 ml of standard fly food and were founded by 10 males and 10 virgin females, to minimize variability in larval density and to ensure a low-competition larval environment. Within 8 h of eclosion, flies were housed in vials (males individually, females in groups of 5) in the experimental room, where they were entrained to a 12 L : 12 D cycle. Trials began when flies were 3 days old; no individual flies were used in more than one trial.
Each fly was carefully marked with a small dab of coloured paint on the day it eclosed [42]. The paint dots allowed me to discriminate between the male genotypes. The paint colour received by the focal interactant genotypes was alternated between trials; as in previous experiments [42], paint colour did not affect behaviour in any of the analyses.
(c). Environment
To ensure that all flies had the opportunity to encounter each other, groups interacted in relatively small (10 cm diameter, 4 cm high) circular arenas made from Petri dishes. Each arena contained a single (4 cm diameter, 0.5 cm high) patch containing grapefruit food, with a small smear of live yeast on the surface. The grapefruit food was made from a batch containing 9.5 g agar, 18 g yeast extract, 13.5 g malt sugar, 0.25 l water and 0.25 l 100% grapefruit juice [39].
(d). Groups
Each group included one A male, one B male and a male from one of the six treatment genotypes (the treatment male). Each treatment genotype was replicated 21 or 22 times, for a total of 128 trials.
(e). Behavioural assay
The three males were added to the arena without anaesthesia approximately 1 h after lights on. Males were allowed to acclimate for 30 min. Informal observations indicated that aggressive interactions rarely occurred during the acclimation time. After 30 min, I recorded male behaviour for 30 min. I recorded whether each male perched on the food patch, but not the amount of time that the male spent on the patch. I also recorded all instances of lunging behaviour [46], including which male performed the lunge and at whom the lunge was directed.
After 30 min of observation, I added a single virgin female (genotype 303/313; see above) and recorded which male she mated with. Females mated within 30 min in all trials except one, and this trial was removed from the mating analysis. Each female mated with only one male during the 30-min observation period.
(f). Analysis
To describe the focal interaction, I summed the number of times that A males lunged at B males, and the number of times that B males lunged at A males. To appropriately model the data, which were non-normal, generalized linear mixed models (GLMMs) were fitted using SAS PROC GLIMMIX (SAS 2009). I modelled counts of lunges using a Poisson distribution with a logarithm link function, and mating success using a binary distribution with a logit link function. The number of flies that perched on the patch during the 30-min observation period could range from 0 to 3 (the latter if all three males perched on the patch at least once during the 30 min). Changes in behaviour may not be linearly related to the number of flies on the food patch (especially for mating success; [39]); instead of explicitly estimating these nonlinear terms, which were not the focus of this study, I modelled the number of males on that perched on the patch as a classification variable. Because I chose genotypes based on their aggressiveness in prior studies, I modelled the genotypes of the focal interactant and treatment males as fixed predictors. Trial number was modelled as random subject effect to account for repeated measures of the same group (as reflected in the denominator degrees of freedom; SAS 9.2 user's guide). Models included an overdispersion parameter to avoid inflation of F-statistics [47]. Note that parameter estimates from GLMMs do not have intrinsic meaning and therefore can only be meaningfully compared within a given model [48]. I ran six different models to describe aggressive behaviour (described in table 1), so all p-values reported for aggressive behaviour are the nominal p-values multiplied by 6, to correct for multiple testing.
Table 1.
Results of GLMMs describing aggressive behaviour. All models were generalized linear mixed models (see Methods). All p-values were corrected for six tests. Parameter estimates are reported for continuous effects, but note that parameter estimates from GLMMs can only be meaningfully compared within a given model. Not all factors were included in all models (see text); factors not included in a given model are left blank.
| model number | description | response variable | treatment male genotype | males on patch | focal interactant genotype (A or B) | treatment male lunges at focal interactant | focal interactant genotype × treatment male lunges at focal interactant | treatment male lunges at A male | treatment male lunges at B male |
|---|---|---|---|---|---|---|---|---|---|
| 1 | genetic variation in treatment male aggressiveness towards A males | treatment male number of lunges | ** | ** | |||||
| 2 | genetic variation in treatment male aggressiveness towards B males | treatment male number of lunges | ** | * | |||||
| 3 | dyadic IGEs | genotype A or B male number of lunges at treatment male | ** | ** | 0.1035** | * | |||
| 4 | second-order IGEs | focal interaction (number of lunges) | ** | −0.05427 | 0.09834* | ||||
| 5 | second-order IGEs: genotype A | genotype A number of lunges at B | ** | 0.01565 | |||||
| 6 | second-order IGEs: genotype B | genotype B number of lunges at A | n.s. | 0.1048** |
*p < 0.05, **p < 0.001.
3. Results
(a). Dyadic IGEs: aggressive encounters between the treatment male and each focal interactant
In order to generate variation in the social environment of the focal interactants—i.e. to potentially cause IGEs—treatment genotypes must differ in behaviour. I found substantial evidence that the genotype of the treatment male influenced how often he aggressively lunged at A males (aggression model 1, table 1: F5,119 = 100.88, p < 0.0006) and at B males (aggression model 2: F5,119 = 5.31, p = 0.0011; figure 2a).
Figure 2.
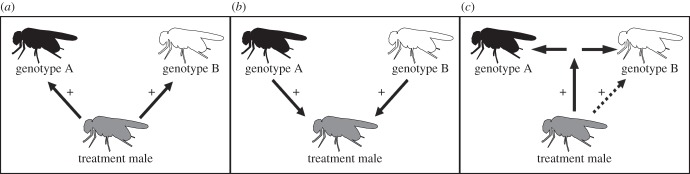
IGEs in groups. Arrows represent statistically significant effects of the treatment male's genotype on aggressive behaviour in the group; plus signs indicate that the coefficient of interaction, psi, was always positive. (a) Treatment male genotypes differ in aggressiveness. (b) More aggressive treatment male genotypes are lunged at more times by the focal interactants, representing dyadic IGEs. (c) Second-order IGEs: genetic variation in treatment male aggressiveness influences the focal interaction (solid arrow); in particular, the treatment male's aggressiveness towards B males affects the focal interaction (dotted arrow).
IGE theory predicts that the aggressiveness of the focal interactants (whose genotypes are standard in all trials) towards the treatment male might depend on the genetically influenced aggressiveness of that treatment male (aggression model 3, table 1). I found strong support for this prediction. On average, focal interactants lunged more times at treatment males that were more aggressive (parameter estimate = 0.1014, F1,125 = 37.082, p < 0.0006). Genotype A males and genotype B males lunged at treatment males different numbers of times (F1,125 = 18.81, p < 0.0006); means for each focal interactant genotype indicated that A males were more aggressive than B males (A mean = 0.95 lunges, range = 0–27; B mean = 0.28 lunges, range = 0–17). Because the treatment male's aggressiveness differs among treatment genotypes, and this same behaviour affected the aggressiveness of the focal interactant males, this result demonstrates that IGEs influenced focal interactant aggressiveness (figure 2b). Further, there was a significant interaction between these effects: the number of times that a focal interactant male lunged at the treatment male depended on the focal interactant's genotype and also on the number of times he was lunged at by the treatment male (focal interactant genotype × number of treatment male lunges: F1,125 = 7.73, p = 0.02). This interaction suggests that the strength of the IGEs varied across focal interactant genotypes.
(b). Second-order IGEs: effects of treatment male on focal interaction
This experiment was not only designed to measure ‘traditional’ IGEs (as above), but also to evaluate the prediction from social networks research that the treatment male could influence the focal interaction itself. Second-order IGEs would be indicated if a behaviour that varied across treatment genotypes was also associated with differences in the number of times that the focal interactants lunged at each other (labelled ‘IGE?’ in figure 1).
I found strong support for the existence of second-order IGEs (aggression model 4). The number of times that the treatment male lunged at B males significantly affected the focal interaction (parameter estimate = 0.098, F1,122 = 8.22, p = 0.029, figure 3). By contrast, the number of times that the treatment male lunged at A males did not have a discernible effect on the focal interaction (parameter estimate = −0.054, F1,122 = 3.49, p = 0.385). The number of males on the patch was also associated with variation in the focal interaction (F3,122 = 10.57, p < 0.0006). Because the treatment male's aggressiveness towards B males differs among treatment genotypes, this result demonstrates that IGEs significantly affected the focal interaction (figure 2c).
Figure 3.

The focal interaction is affected by genetic variation in treatment male aggressiveness. x-axis describes the number of times that the treatment male lunged at B males. y-axis represents the number of times genotype A males lunged at B males (circles), the number of times genotype B males lunged at A males (crosses), and the total number of times that the focal interactants lunged at each other (i.e. the focal interaction). The dotted line represents a linear regression between the focal interaction and the number of times that the treatment male lunged at B males. N = 128 trials.
To better understand this result, I fitted models of each focal interactant genotype's behaviour towards the other focal interactant (aggression models 5 and 6). I found that the number of times that the treatment male lunged at B males did not significantly affect the number of times that A males lunged at B males (parameter estimate = 0.016, F1,123 = 1.04, p = 1), but the number of males who perched on the patch during the 30-min observation period did affect the number of times that A males lunged at B males (F3,123 = 12.60, p < 0.0006). For B males, I found the opposite result: the number of times that the treatment male lunged at B males was associated with a significantly greater number of times that B males lunged at A males (parameter estimate = 0.1048, F1,123 = 15.94, p < 0.0006), but the number of males who perched on the patch during the 30-min observation period did not significantly affect the number of times that B males lunged at A males (F3,123 = 0.34, p = 1).
(c). Mating consequences of the focal interaction
To determine whether second-order IGEs had consequences for reproductive success, I evaluated whether the mating success of each focal interactant was influenced by the focal interaction and all by the factors affecting the focal interaction, i.e. the number of males who perched on the patch during the 30-min observation period, and the number of times that the treatment male lunged at B males. I also considered two-way interactions between focal interactant genotype and each of these factors.
I found no significant additive effect of the number of males who perched on the patch (F3,120 = 0.05, p = 0.98), the number of times the treatment male lunged at B males (parameter estimate −0.033, F1,120 = 0.94, p = 0.33) or the focal interaction itself (parameter estimate −0.117, F1,120 = 0.09, p = 0.76) on focal interactant mating success. The two focal interactant genotypes did not mate at different frequencies on average (F1,120 = 0.79, p = 0.38).
Instead, the effect of the focal interaction depended on the genotype of the focal interactant (focal interaction × focal interactant genotype: F1,120 = 6.15, p = 0.015). Inspection of means for each genotype (figure 4) suggests that this interaction term reflects an increase in the likelihood of mating for A males when the focal interactant males lunge at each other more times; and the opposite for B males. Similarly, the effect of the number of males who perched on the patch (number of males × focal interactant genotype: F3,120 = 3.86, p = 0.011) on focal interactant mating success depended on the genotype of the focal interactant. There was no significant interaction between the number of times the treatment male lunged at B males and the focal interactant genotype (number of times treatment male lunged at B males × focal interactant genotype: F1,120 = 0.64, p = 0.43).
Figure 4.

The focal interaction differentially affects the mating success of the focal interactant genotypes. x-axis describes the number of times that the focal interactants lunged at each other. y-axis represents least-squared mean (adjusted for other factors in the model; see text) probability of mating for A males (circles, solid line) and B males (crosses, dotted line). The total probability of mating for each level of the focal interaction is not constrained to be 1, because the treatment male could also mate. Regression lines illustrate the strength and direction of the relationship between the focal interaction and mating success for each focal interactant genotype. N = 127 trials.
4. Discussion
Extending IGE inferences beyond the level of the dyad is critical to understanding social behaviour and behavioural evolution. Here, I report that dyadic aggressive interactions between a standard pair of D. melanogaster males are influenced by genetic variation in their social group. To my knowledge, this represents the first direct manipulative evidence for second-order IGEs. Further, second-order IGEs had genotype-specific consequences for male mating success, suggesting that second-order IGEs can provide critical insights into social selection.
Second-order IGEs are likely to be common because many animals interact in social groups or networks [19–21]. Second-order IGEs have been hypothesized to contribute to genetic variation in human behaviour, including mental illness [31], but this hypothesis is difficult to test in free-living organisms. The focal interaction approach presented here provides a way to study second-order IGEs using a tractable number of individuals, if the social environment can be experimentally manipulated. The focal interaction approach is amenable to both variance-based and trait-based IGE analyses: variance-partitioning analyses consider the effects of different treatment male genotypes on the focal interaction, and trait-based analyses consider the effects of genetic variation in a particular treatment male trait (in this case, aggressiveness) on the focal interaction [49]. By using the focal interaction approach with both variance-based and trait-based analyses, I found that in groups containing more aggressive treatment genotypes, the focal interactants lunged at each other more times than in groups containing less-aggressive treatment genotypes (figure 3). The effect of genetic variation in treatment, male behaviour on the focal interaction was not revealed by typical IGE analyses, which usually consider dyadic interactions or group averages.
In IGE theory, the coefficient of interaction psi describes the degree to which the behaviour of one individual affects the behaviour of an interacting individual [1–3]. In this experiment, the two focal interactant genotypes exhibited plasticity in aggressiveness in response to different aspects of the social environment. Genotype A males exhibited different levels of aggressiveness towards B males, depending on the number of males on the patch, whereas genotype B males varied their aggressiveness towards A males depending on the aggressive behaviour of the treatment male. The genetic differences in behavioural plasticity evident in this experiment add to the growing literature demonstrating that the coefficient of interaction psi varies across genotypes [9,11,15] which is a necessary condition for plasticity to evolve [11,50–52].
Subtle differences in behavioural plasticity between the focal interactant genotypes highlight the major drawback of the focal interaction approach: that insights garnered from a particular focal interaction may not be generalizable to alternate focal interactions. The focal interaction approach may be most fruitful in cases in which there is some a priori reason to choose a focal interaction. If possible, the experiment may also be replicated with additional sets of focal interactants.
Aggressiveness is heritable in natural populations of fruitflies [45,52,53] and even in laboratory stocks that have presumably undergone long-term directional selection [54]. If females prefer more aggressive males, as is commonly assumed, such directional selection should erode genetic variation in aggressiveness. Indeed, I found in all cases that the sign of the coefficient of interaction, psi, was positive, i.e. higher numbers of 3 aggressive lunges by one individual were associated with more lunges by interacting individuals. Positive values of psi are expected to accelerate the action of directional selection that erodes variation [11].
However, the interpretation of this finding is complicated by the fact that sexual selection on aggressiveness was not directional. Instead, the optimum level of aggressiveness in the group differed among the focal interactant genotypes: genotype A males were more likely to mate in groups in which there were high levels of aggressive interactions, whereas genotype B males showed the opposite pattern (figure 4). An increase in the mating success of A males does not necessarily imply a decrease in the mating success of B males across trials, because the treatment male also mated. Out of the 127 trials in which mating occurred, A males mated 54 times, B males mated 39 times and treatment males mated 34 times. Thus, the opposing effect of the focal interaction on A males and B males is not a statistical artefact, but instead implies that the fitness consequences of aggressive behaviour in the group differed between genotypes. This result is broadly consistent with previous work [39] suggesting that genotypes that are less aggressive on average also have lower mating rates in aggressive social contexts. This type of genotype-by-social environment interaction for mating success can adaptively maintain heritable variation in male aggressiveness [55–57]. Further research is needed to determine how sexual selection generated by second-order IGEs contributes to the depletion and/or adaptive maintenance of genetic variation in social behaviour.
Genetic variation in the behaviour of the treatment male significantly affected the focal interaction (figure 3, aggression model 4) but I did not detect a direct effect of the treatment male's behaviour on focal interactant mating success. This result suggests that a simple analysis of mating success, without considering second-order IGEs, would not have revealed the complexity of male–male interactions that ultimately influenced mating success. Thus, consideration of IGEs is important for understanding sexual selection in groups. Creative application of the focal individual approach across species will doubtless provide novel insights into the expression and evolution of social behaviours.
Acknowledgements
Thanks to Judy Stamps, Andy Sih, Brad Foley and Sergey Nuzhdin for productive discussion. Thanks to Judy Stamps, Allen Moore and two anonymous reviewers for insightful comments on the manuscript.
Data accessibility
Data deposited in the dryad repository: http://doi.org/10.5061/dryad.3ds26.
Funding statement
Research reported in this publication was supported by the National Institute of Mental Health of the National Institutes of Health under Award nos. R01MH100879 and MH091561; and by a DDIG from the National Science Foundation under Award no. IOS-1110371.
References
- 1.Moore AJ, Brodie ED, III, Wolf JB. 1997. Interacting phenotypes and the evolutionary process: I. Direct and indirect genetic effects of social interactions. Evolution 51, 1352–1362 (doi:10.2307/2411187) [DOI] [PubMed] [Google Scholar]
- 2.Moore AJ, Pizzari T. 2005. Quantitative genetic models of sexual conflict based on interacting phenotypes. Am. Nat. 165, S88–S97 (doi:10.1086/429354) [DOI] [PubMed] [Google Scholar]
- 3.McGlothlin JW, Moore AJ, Wolf JB, Brodie ED. 2010. Interacting phenotypes and the evolutionary process. III. Social evolution. Evolution 64, 2558–2574 (doi:10.1111/j.1558-5646.2010.01012.x) [DOI] [PubMed] [Google Scholar]
- 4.Wolf JB, Brodie ED, III, Moore AJ. 1999. Interacting phenotypes and the evolutionary process. II. Selection resulting from social interactions. Am. Nat. 153, 254–266 (doi:10.1086/303168) [DOI] [PubMed] [Google Scholar]
- 5.Agrawal AF, Brodie ED, III, Wade MJ. 2001. On indirect effects in structured populations. Am. Nat. 158, 308–323 (doi:10.1086/321324) [DOI] [PubMed] [Google Scholar]
- 6.Bijma P, Muir WM, Van Arendonk JAM. 2007. Multilevel selection 1: quantitative genetics of inheritance and response to selection. Genetics 175, 277–288 (doi:10.1534/genetics.106.062711) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lewontin RC. 1954. The effects of population density and composition on viability in Drosophila melanogaster. Evolution 9, 27–41 (doi:10.2307/2405355) [Google Scholar]
- 8.Griffing B, Zsiros E. 1971. Heterosis associated with genotype–environment interactions. Genetics 68, 443–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ayre DJ, Grosberg RK. 1995. Aggression, habituation, and clonal coexistence in the sea anemone Anthopleura elegantissima. Am. Nat. 146, 427–453 (doi:10.1086/285808) [Google Scholar]
- 10.Wolf JB. 2003. Genetic architecture and evolutionary constraint when the environment contains genes. Proc. Natl Acad. Sci. USA 100, 4655–4660 (doi:10.1073/pnas.0635741100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bleakely BH, Brodie ED., III 2009. Indirect genetic effects influence antipredator behavior in guppies: estimates of the coefficient of interaction psi and the inheritance of reciprocity. Evolution 63, 1796–1806 (doi:10.1111/j.1558-5646.2009.00672.x) [DOI] [PubMed] [Google Scholar]
- 12.Wilson AJ, Gelin U, Perron M-C, Réale D. 2009. Indirect genetic effects and the evolution of aggression in a vertebrate system. Proc. R. Soc. B 276, 533–541 (doi:10.1098/rspb.2008.1193) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Billeter J-C, Jagadeesh S, Stepek N, Azanchi R, Levine JD. 2012. Drosophila melanogaster females change mating behaviour and offspring production based on social context. Proc. R. Soc. B 279, 2417–2425 (doi:10.1098/rspb.2011.2676) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pischedda A, Stewart AD, Little MK, Rice WR. 2011. Male genotype influences female reproductive investment in Drosophila melanogaster. Proc. R. Soc. B 278, 2165–2172 (doi:10.1098/rspb.2010.2272) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sartori C, Mantovani R. 2013. Indirect genetic effects and the genetic basis of social dominance: evidence from cattle. Heredity 110, 3–9 (doi:10.1038/hdy.2012.56) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kent C, Azanchi R, Smith B, Formosa A, Levine JD. 2008. Social context influences chemical communication in D. melanogaster males. Curr. Biol. 18, 1–6 (doi:10.1016/j.cub.2007.11.056) [DOI] [PubMed] [Google Scholar]
- 17.Wang J, Ross KG, Keller L. 2008. Genome-wide expression patterns and the genetic architecture of a fundamental social trait. PLoS Genet. 4, e1000127 (doi:10.1371/journal.pgen.1000127) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saltz JB, Alicuben ET, Grubman J, Harkenrider M, Megowan N, Nuzhdin SV. 2012. Non-additive indirect effects of group genetic diversity on larval viability in D. melanogaster imply key role of maternal decision-making. Mol. Ecol. 21, 2270–2281 (doi:10.1111/j.1365-294X.2012.05518.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krause J, Croft DP, James R. 2007. Social network theory in the behavioural sciences: potential applications. Behav. Ecol. Sociobiol. 62, 15–27 (doi:10.1007/s00265-007-0445-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wey T, Blumstein DT, Shen W, Jordán F. 2008. Social network analysis of animal behaviour: a promising tool for the study of sociality. Anim. Behav. 75, 333–344 (doi:10.1016/j.anbehav.2007.06.020) [Google Scholar]
- 21.Sih A, Hanser SF, McHugh KA. 2009. Social network theory: new insights and issues for behavioral ecologists. Behav. Ecol. Sociobiol. 63, 975–988 (doi:10.1007/s00265-009-0725-6) [Google Scholar]
- 22.Flack JC, Girvan M, de Waal FBM, Krakauer DC. 2006. Policing stabilizes construction of social niches in primates. Nature 439, 426–429 (doi:10.1038/nature04326) [DOI] [PubMed] [Google Scholar]
- 23.Fowler JH, Christakis NA. 2010. Cooperative behavior cascades in human social networks. Proc. Natl Acad. Sci. USA 107, 5334–5448 (doi:10.1073/pnas.0913149107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ferrin DL, Dirks KT, Shah PP. 2006. Direct and indirect effects of third-party relationships on interpersonal trust. J. Appl. Psychol. 91, 870–883 (doi:10.1037/0021-9010.91.4.870) [DOI] [PubMed] [Google Scholar]
- 25.Fewell JH. 2003. Social insect networks. Science 301, 1867–1870 (doi:10.1126/science.1088945) [DOI] [PubMed] [Google Scholar]
- 26.Darden SK, Watts L. 2012. Male sexual harassment alters female social behavior towards other females. Biol. Lett. 8, 186–188 (doi:10.1098/rsbl.2011.0807) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Godfrey SS, Bradley JK, Sih A, Bull MC. 2012. Lovers and fighters in sleepy lizard land: where do aggressive males fit into a social network? Anim. Behav. 83, 209–215 (doi:10.1016/j.anbehav.2011.10.028) [Google Scholar]
- 28.Beaver KM, Wright JP, DeLisi M. 2008. Delinquent peer group formation: evidence of a gene × environment correlation. J. Genet. Psychol. 169, 227–244 (doi:10.3200/GNTP.169.3.227-244) [DOI] [PubMed] [Google Scholar]
- 29.Fowler JH, Dawes CT, Christakis NA. 2009. Model of genetic variation in human social networks. Proc. Natl Acad. Sci. USA 106, 1720–7124 (doi:10.1073/pnas.0806746106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lea AJ, Blumstein DT, Wey TW, Martin JGA. 2010. Heritable victimization and the benefits of agonistic relationships. Proc. Natl Acad. Sci. USA 107, 21 587–21 592 (doi:10.1073/pnas.1009882107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fowler JH, Settle JE, Christakis NA. 2011. Correlated genotypes in friendship networks. Proc. Natl Acad. Sci. USA 108, 1993–1997 (doi:10.1073/pnas.1011687108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Falconer DS, Mackay TFC. 1996. Introduction to quantitative genetics, 4th edn London, UK: Longman [Google Scholar]
- 33.Altmann J. 1974. Observational study of behavior: sampling methods. Behaviour 49, 227–267 (doi:10.1163/156853974X00534) [DOI] [PubMed] [Google Scholar]
- 34.Earley RL. 2010. Social eavesdropping and the evolution of conditional cooperation and cheating strategies. Phil. Trans. R. Soc. B 365, 2675–2686 (doi:10.1098/rstb.2010.0147) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dzieweczynski TL, Earley RL, Green TM, Rowland WJ. 2005. Audience effect is context dependent in Siamese fighting fish, Betta splendens. Behav. Ecol. 16, 1025–1030 (doi:10.1093/beheco/ari088) [Google Scholar]
- 36.Powell JR. 1997. Progress and prospects in evolutionary biology: the Drosophila model. New York, NY: Oxford University Press [Google Scholar]
- 37.Wertheim B, Allemand R, Vet L, Dicke M. 2006. Effects of aggregation pheromone on individual and food web interactions: a field study on Drosophila. Ecol. Entomol. 31, 216–226 (doi:10.1111/j.1365-2311.2006.00757.x) [Google Scholar]
- 38.Stamps J, McElreath R, Eason P. 2005. Alternative models of conspecific attraction in flies and crabs. Behav. Ecol. 16, 974–980 (doi:10.1093/beheco/ari083) [Google Scholar]
- 39.Saltz JB, Foley BR. 2011. Natural genetic variation in social niche construction: social effects of aggression drive disruptive sexual selection in Drosophila melanogaster. Am. Nat 177, 645–654 (doi:10.1086/659631) [DOI] [PubMed] [Google Scholar]
- 40.Saltz JB. 2011. Natural genetic variation in social environment choice: context-dependent gene–environment correlation in Drosophila melanogaster. Evolution 65, 2325–2334 (doi:10.1111/j.1558-5646.2011.01295.x) [DOI] [PubMed] [Google Scholar]
- 41.Wertheim B, Dicke M, Vet LEM. 2002. Behavioural plasticity in support of a benefit for aggregation pheromone use in Drosophila melanogaster. Entomol. Exp. et Applicata 103, 61–71 (doi:10.1046/j.1570-7458.2002.00954.x) [Google Scholar]
- 42.Stamps J, Buechner M, Alexander K, Davis J, Zuniga N. 2005. Genotypic differences in space use and movement patterns in Drosophila melanogaster. Anim. Behav. 70, 609–618 (doi:10.1016/j.anbehav.2004.11.018) [Google Scholar]
- 43.Wahlsten D. 2001. Standardizing tests of mouse behavior: reasons, recommendations, and reality. Physiol. Behav. 73, 695–704 (doi:10.1016/S0031-9384(01)00527-3) [DOI] [PubMed] [Google Scholar]
- 44.Brakefield PM. 2003. Artificial selection and the development of ecologically relevant phenotypes. Ecology 84, 1661–1671 (doi:10.1890/0012-9658(2003)084[1661:ASATDO]2.0.CO;2) [Google Scholar]
- 45.Cabral LG, Foley BR, Nuzhdin SV. 2008. Does sex trade with violence among genotypes in Drosophila melanogaster? PLoS ONE 3, e1986 (doi:10.1371/journal.pone.0001986) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nilsen SP, Chan Y-B, Huber R, Kravitz EA. 2004. Gender-selective patterns of aggressive behavior in Drosophila melanogaster. Proc. Natl Acad. Sci. USA 101, 12 342–12 347 (doi:10.1073/pnas.0404693101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bolker BM, Brooks ME, Clark CJ, Geange SW, Poulsen JR, Stevens MHH, White J-S. 2009. Generalized linear mixed models: a practical guide for ecology and evolution. Trends Ecol. Evol. 24, 127–135 (doi:10.1016/j.tree.2008.10.008) [DOI] [PubMed] [Google Scholar]
- 48.Littel RC, Milliken GA, Stroup WW, Wolfinger RG, Schabenberger O. 2006. SAS for mixed models, 2nd edn Cary, NC: SAS Publishing [Google Scholar]
- 49.McGlothlin JW, Brodie ED., III 2009. How to measure indirect genetic effects: the congruence of trait-based and variance-partitioning approaches. Evolution 63, 1785–1795 (doi:10.1111/j.1558-5646.2009.00676.x) [DOI] [PubMed] [Google Scholar]
- 50.Bailey NW, Zuk M. 2012. Socially flexible female choice differs among populations of the Pacific field cricket: geographical variation in the interaction coefficient psi (Ψ). Proc. R. Soc. B 279, 3589–3596 (doi:10.1098/rspb.2012.0631) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kazancıoğlu E, Klug H, Alonzo SH. 2012. The evolution of social interactions changes predictions about interacting phenotypes. Evolution 66, 2056–2064 (doi:10.1111/j.1558-5646.2012.01585.x) [DOI] [PubMed] [Google Scholar]
- 52.Hoffmann AA. 1988. Heritable variation for territorial success in two Drosophila melanogaster populations. Anim. Behav. 36, 1180–1189 (doi:10.1016/S0003-3472(88)80077-0) [Google Scholar]
- 53.Edwards AC, Rollmann SM, Morgan TJ, Mackay TFC. 2006. Quantitative genomics of aggressive behavior in Drosophila melanogaster. PLoS Genet. 2, e154 (doi:10.1371/journal.pgen.0020154) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dierick HA, Greenspan RJ. 2006. Molecular analysis of flies selected for aggressive behavior. Nat. Genet. 38, 1023–1031 (doi:10.1038/ng1864) [DOI] [PubMed] [Google Scholar]
- 55.Gillespie JH, Turelli M. 1989. Genotype–environment interactions and the maintenance of polygenic variation. Genetics 121, 129–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Harris E, McKane A, Wolf J. 2008. The maintenance of heritable variation through social competition. Evolution 62, 337–347 (doi:10.1111/j.1558-5646.2007.00302.x) [DOI] [PubMed] [Google Scholar]
- 57.Nonacs P, Kapheim KM. 2007. Social heterosis and the maintenance of genetic diversity. J. Evol. Biol. 20, 2253–2265 (doi:10.1111/j.1420-9101.2007.01418.x) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data deposited in the dryad repository: http://doi.org/10.5061/dryad.3ds26.


