Abstract
Recombination allows faithful chromosomal segregation during meiosis and contributes to the production of new heritable allelic variants that are essential for the maintenance of genetic diversity. Therefore, an appreciation of how this variation is created and maintained is of critical importance to our understanding of biodiversity and evolutionary change. Here, we analysed the recombination features from species representing the major eutherian taxonomic groups Afrotheria, Rodentia, Primates and Carnivora to better understand the dynamics of mammalian recombination. Our results suggest a phylogenetic component in recombination rates (RRs), which appears to be directional, strongly punctuated and subject to selection. Species that diversified earlier in the evolutionary tree have lower RRs than those from more derived phylogenetic branches. Furthermore, chromosome-specific recombination maps in distantly related taxa show that crossover interference is especially weak in the species with highest RRs detected thus far, the tiger. This is the first example of a mammalian species exhibiting such low levels of crossover interference, highlighting the uniqueness of this species and its relevance for the study of the mechanisms controlling crossover formation, distribution and resolution.
Keywords: phylogenetic analysis, meiosis, selection, divergence time, tiger, elephant shrew
1. Introduction
Most eukaryotes exchange genetic information through recombination during meiosis. This is a process that increases genetic diversity in haploid genotypes and provides physical connections between homologues during the first meiotic division, thus contributing to correct chromosomal segregation. Although recombination can occur at the somatic level, only recombination in the germline can produce new heritable chromosomal variants and hence contribute to the possible formation of new species ([1] and references therein).
Recombination occurs early in meiosis and is triggered by programmed double-stranded breaks (DSBs) [2]. The broken ends are processed with approximately half of them producing double Holliday junctions and crossovers (COs), whereas the remainders are resolved as non-crossovers (NCOs) [3]. CO assurance is carefully controlled by homeostasis, which in turn regulates the CO–NCO ratio [4,5]. It is generally accepted that COs exhibit three principal characteristics: (i) they take place in discrete regions of the genome, (ii) there is at least one crossover per pair of chromosomes (which ensures the proper disjunction of homologous chromosomes) and (iii) they show ‘interference’ (COs tend to follow an evenly spaced distribution) [3]. However, it has been recently demonstrated that not all COs are subject to interference, leading to recognition of interfering (class I) and non-interfering (class II) COs in different organisms [6,7]. Non-interfering COs are Mus81–Mms4-dependent and distributed randomly along the chromosomes independently of each other; the interfering COs are distributed according to a gamma distribution and are Msh4–Msh5-dependent [6,7]. The choice of pathway followed appears species-specific. In fissioning yeast, for example, nearly all COs are dependent on Mus81–Eme1, whereas in worms most COs are subjected to interference [8]. Budding yeast, in turn, occupies an intermediate position where the majority of COs occur through the class I pathway, and the class II pathway may exist primarily as a back-up [6]. It is likely that the pattern in other mammals follows that identified in the mouse (the only mammalian species in which this phenomenon has been studied in any detail). Here, most COs manifest interference and are controlled by the proteins Msh4–Msh5 [9], although some Mus81 activity has been detected during meiosis [10]. In fact, in Mlh1-deficient mice chiasmata are reduced, but not entirely removed [11], suggesting the possible presence of class II COs in mammals.
Only very recently has progress been made in identifying the mechanisms that control CO formation and distribution. In mammals, the conventional argument has been that although recombination rates (RRs) may vary considerably between species when comparing high-resolution (kb) recombination maps (in primates for example, [12]), these differences disappear when comparisons are carried out at a broader scale (Mb) [13,14]. In fact, recent studies in mice [15] and primates [16] have suggested that closely related species tend to have similar average rates of recombination, but whether these observations hold for all mammalian species remains to be tested. Here, we have analysed RRs in phylogenetically diverse species selected from the mammalian tree of life (ToL), including the major eutherian taxonomic groups represented by marsupials, afrotherians, rodents, eulipotyphlans, primates, ungulates and carnivores, in an attempt to better understand its dynamics and impact on mammalian evolution. Our results suggest that phylogeny affects the RRs of eutherian mammals, with species that diversified earlier in the evolutionary tree displaying lower RRs than those from more derived phylogenetic branches. Moreover, analyses of recombination maps that include the widely divergent elephant shrew (Afrotheria) and tiger (Laurasiatheria) show that interference among MLH1 foci shows different levels of intensity depending on the species analysed. The weak CO interference (COI) observed in the tiger highlights the uniqueness of this taxon and its relevance in the study of the mechanisms controlling CO formation and resolution.
2. Results
(a). Immunolocalization of proteins implicated in the formation of crossovers suggests that phylogenetic relationships among mammalian groups are influencing recombination patterns
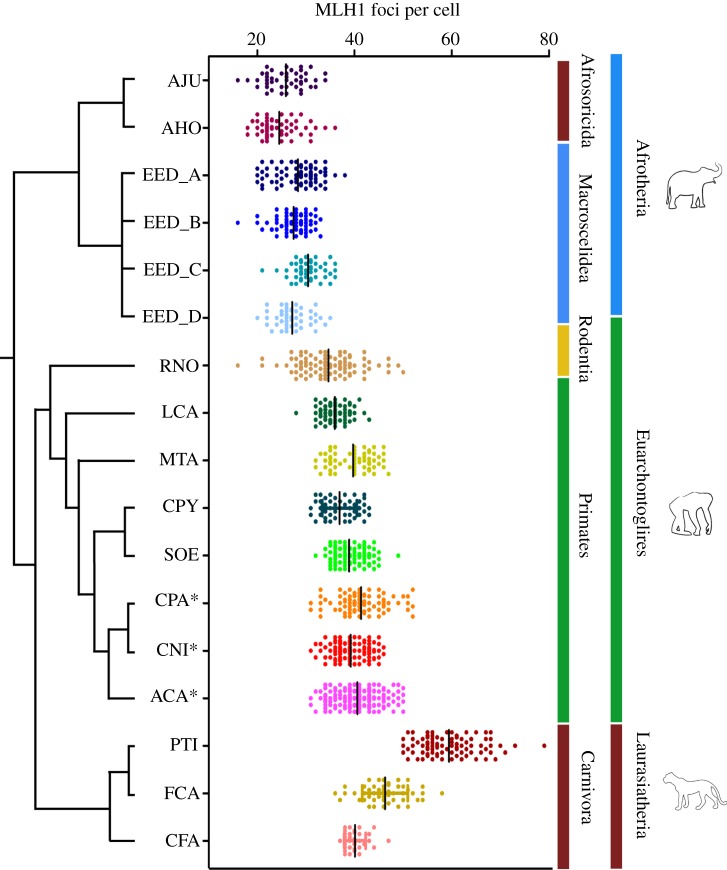
Meiotic recombination in 11 species was studied using immunolocalization against different meiotic proteins involved in CO formation. These were drawn from the Afrosoricida (Amblysomus julianae and Amblysomus hottentotus) and Macroscelidea (Elephantulus edwardii)—both falling within Afrotheria. The Rodentia species (Rattus norvegicus) and Primates species (Miopithecus talapoin, Callithrix pygmaea, Saguinus oedipus and Lemur catta) represented Euarchontoglires, and Canis familiaris, Felis catus and Panthera tigris (Order Carnivora) represented the Laurasiatheria (see electronic supplementary material, table S1). Published data from three Primates species, previously studied in our laboratory, were also included (Cebus paraguayanus, Cebus nigritus and Alouatta caraya) [16]. The observed numbers of crossover events detected by immunofluorescence (IF) of the recombination protein MLH1 (a marker of COs) at pachynema of the species analysed herein are presented in figure 1 and electronic supplementary material, tables S1 and S2. We have studied one specimen per species, with the exception of E. edwardii, where four animals were available for analysis. Between 33 and 165 cells per specimen were recorded (see electronic supplementary material, table S2). We detected variability in the mean number of autosomal MLH1 foci per cell; these range from 24.5 (± 4.1) in A. hottentotus to 59.4 (± 5.9) in P. tigris (figure 1; electronic supplementary material, table S2). With the exception of L. catta and C. familiaris, large standard deviations are associated with the mean values of MLH1 foci (between 3.1 and 6.2), reflecting cell-to-cell differences in MLH1 foci numbers for most of the species (see electronic supplementary material, table S2).
Figure 1.
The number of MLH1 foci observed per cell in specimens analysed and their phylogenetic relationships. A. julianae (AJU), A. hottentotus (AHO), E. edwardii (four individuals, EED_A to EED_D), R. norvegicus (RNO), L. catta (LCA), S. oedipus (SOE), C. pygmaea (CPY), M. talapoin (MTA), C. paraguayanus (CPA), C. nigritus (CNI), A. caraya (ACA), C. familiaris (CFA), F. catus (FCA) and P. tigris (PTI). Asterisk denotes data obtained from [16]. The phylogenetic branches are not proportional to DTs (phylogenetic tree redrawn from [17]). Taxonomy follows [18]. See electronic supplementary material, table S2 for information on standard deviation, standard error, 95% CI for the coefficient of mean and coefficient of variation values.
The afrotherians, thought to have diverged approximately 80.9 million years ago (Ma) [17], show the lowest numbers of MLH1 foci detected per cell. Amblysomus julianae and A. hottentotus (Afrosoricida, Chrysochloridae) share identical karyotypes (2n = 30 [19]) and these two species did not differ significantly in the mean number of COs per cell (25.9±4.1 for A. julianae and 24.5±4.1 for A. hottentotus). On the other hand, E. edwardii, a macroscelid representative with 2n = 26 [19,20], and thus a lower diploid number than the Amblysomus species, had a higher overall mean number of COs (28.3±3.8). These ranged from 27.2±3.4 (in EED_D) to 30.4±3.2 (in EED_C). The Afrosoricida is considered to have diverged approximately 68.2 Ma and the Macroscelidea approximately 49.1 Ma [17].
Among Primates, there was a good correlation among phylogenetic groups and the number of COs detected for each species (figure 1). Lemur catta (35.9±2.6 MLH1 foci per cell) belongs to the more basal group of Primates, the prosimians, which diverged from the last common ancestor of Catarrhini (Old World monkeys) and Platyrrhini (New World monkeys) approximately 75 Ma [21]. Moreover, dates for Platyrrhini diversification are younger (21.4 Ma) than for Catarrhini (29.6 Ma) [21]. In our study, M. talapoin (39.7±4.2 MLH1 foci per cell) is a representative of Catarrhini, whereas S. oedipus (38.9±3.1 MLH1 foci per cell), Callithrix pygmaea (36.9±3.5 MLH1 foci per cell), Cebus paraguayanus (41.3±4.9 MLH1 foci per cell), C. nigritus (39.2±3.3 MLH1 foci per cell) and Alouatta caraya (40.6±4.3 MLH1 foci per cell) are all grouped within Platyrrhini. Finally, the highest overall mean number of COs detected among mammals was in Carnivora (48.6 MLH1 foci per cell), represented herein by the dog (Canidae, 40.1±2.2 MLH1 foci per cell), cat (Felidae, 46.2±4.7 MLH1 foci per cell) and Sumatran tiger (Felidae, 59.4±5.9 MLH1 foci per cell). The MLH1 values obtained for the dog are in agreement with previous reports [22]. These two taxonomic groups (Felidae and Canidae) are considered to have diverged approximately 52.9 Ma [23].
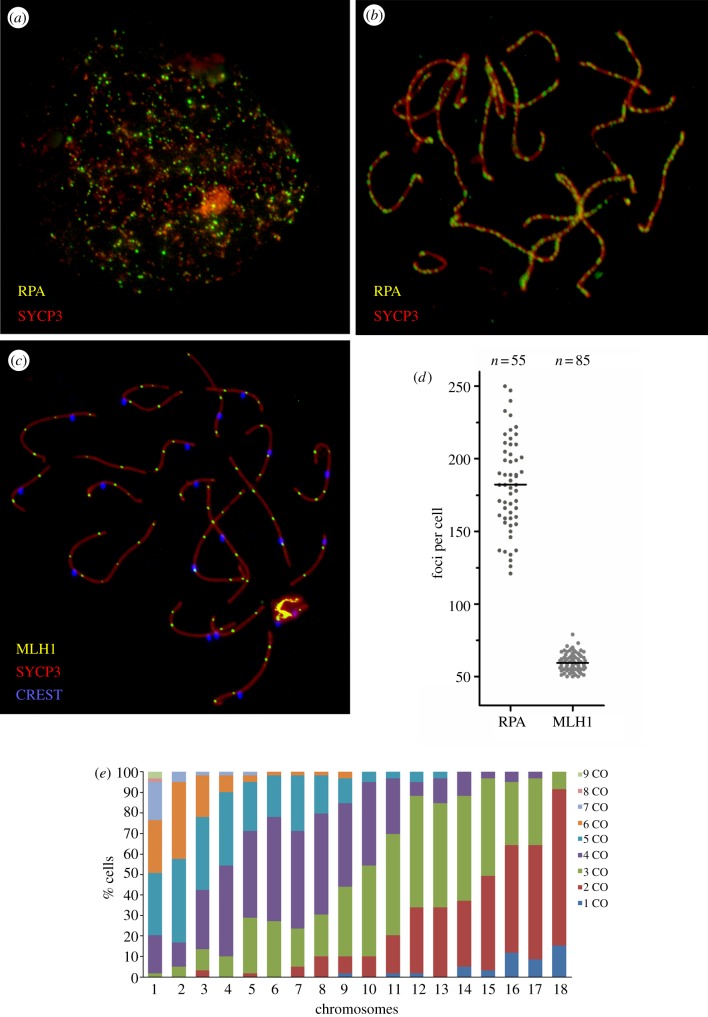
Owing to the high mean MLH1 scores observed in the tiger (59.4±5.9, figure 1), we investigated whether a similar pattern was reflected by the proteins implicated in the formation and repair of DSBs in the early stages of prophase I. Meiotic recombination is initiated by DSBs generated by the protein Spo11 [24]. The RPA protein associates with ssDNA following DSBs formation and subsequently accumulates at the DSBs sites [25]. Therefore, by analysing the number of RPA sites in early pachynema, we can determine the progression of DSBs. We observed the persistence of RPA foci in tiger spermatocytes from leptonema to pachynema (figure 2a,b). The analysis of early-pachytene spermatocytes in this species resulted in 182.2 RPA foci per cell (±31.8) with a range of 120–246 foci per cell (figure 2d). The ratio of RPA foci to COs (MLH1 foci) was approximately 3 : 1 in the tiger. This value was especially low when compared with what has been reported in human and mouse, where ratios range from 7 : 1 to 10 : 1 [5]. These results suggest that a higher proportion of DSBs are resolved as COs during early prophase I of the tiger, when compared with other mammalian species such as human and mouse.
Figure 2.
Immunolocalization of meiotic recombination events in tiger spermatocytes. (a) Representative spermatocyte at leptonema immunostained against SYCP3 (red) and RPA (green). (b) Representative spermatocyte at early pachynema immunostained against SYCP3 (red) and RPA (green). (c) Representative spermatocyte at pachynema showing triple immunostaining with SYCP3 (red), MLH1 (green) and the centromere with CREST (blue). (d) The numbers of early (RPA) and late (MLH1) recombination proteins detected in early pachynema are shown. Median numbers shown by horizontal bars. (e) Distribution (as a percentage of the cells analysed) of MLH1 foci (crossovers, COs) for tiger chromosomes.
(b). Chromosome-specific recombination patterns in tiger and elephant shrew show different levels of crossover interference
The comparative analysis of chromosome-specific recombination patterns entailed the selection of two species that were phylogenetically distant from each other, and which differed significantly with respect to the mean number of MLH1 foci per cell—E. edwardii (elephant shrew) and P. tigris (tiger). The former has the lowest (27.5±3.4) number of MLH1 foci per cell detected (see above) and, together with Xenarthra (i.e. Atlantogenata), is often thought to be basal in the eutherian ToL (figure 1; electronic supplementary material, table S2). The tiger on the other hand was characterized by the highest mean number of MLH1 foci per cell (59.4±5.9) in our analyses, and is representative of a more derived clade, the Laurasiatheria (figure 1; electronic supplementary material, table S1). The distribution of MLH1 foci along the autosomal synaptonemal complexes (SCs) at pachynema was analysed in both species. SCs were ranked on their relative lengths and the positions of their centromeres (SCs 1–12 for the elephant shrew and SCs 1–18 for the tiger; electronic supplementary material, tables S3 and S4). In the case of tiger spermatocytes, centromere position was established by IF using the antiserum CREST (figure 2c). For elephant shrew spermatocytes, we used an antibody against the histone H3 lysine 9 methylated (H3K9me), an epigenetic signal for constitutive heterochromatin [26]. This approach permitted the classification of elephant shrew chromosomes based on chromosomal length, centromere position and heterochromatin pattern (see electronic supplementary material, figure S1). The distribution of heterochromatic (hc) regions was consistent with previous cytogenetic studies [19], where chromosomes EED3, EED4, EED5 and EED7 comprised large interstitial hc blocks (see electronic supplementary material, figure S1). We did, however, detect small interstitial hc regions along the short and long arm of several chromosomes (EED1, EED2, EED4 and EED5) that had not previously been reported (see electronic supplementary material, figure S1).
We calculated the absolute length (in µm), the mean number of MLH1 foci/cell and the CO density (MLH1 µm−1) for each autosomal chromosome (see electronic supplementary material, tables S3 and S4) in each of the species analysed. The elephant shrew MLH1 values were lower than tiger (mean 2.3±0.8 on autosomal SCs per cell, range 4.2±1.0 in EED1 to 1.4±0.5 in EED12 versus mean 3.5±1.0 on autosomal SCs per cell, range 5.6±1.3 in PTI1 to 1.9±0.5 in PTI18; electronic supplementary material, tables S3 and S4). The differences between species were a consequence of CO density, which showed an almost twofold increase in the tiger (0.4±0.04 MLH1 foci µm−1) when compared with the elephant shrew (0.2±0.06 MLH1 foci µm−1). Despite these differences, a positive correlation exists between SC length and the mean number of MLH1 foci in both the elephant shrew (r = 0.947, p < 0.0001), and the tiger (r = 0.987, p < 0.0001; electronic supplementary material, figure S2). We found a negative correlation between RR and chromosome size (r = 0.916 in elephant shrew and r = 0.897 in tiger, p < 0.0001), suggesting that smaller chromosomes have higher RRs than do larger ones. These results corroborate previous observations in mammals that show larger chromosomes tend to accumulate greater numbers of COs, and that each chromosome generally presents at least one CO to ensure proper chromosomal segregation [27].
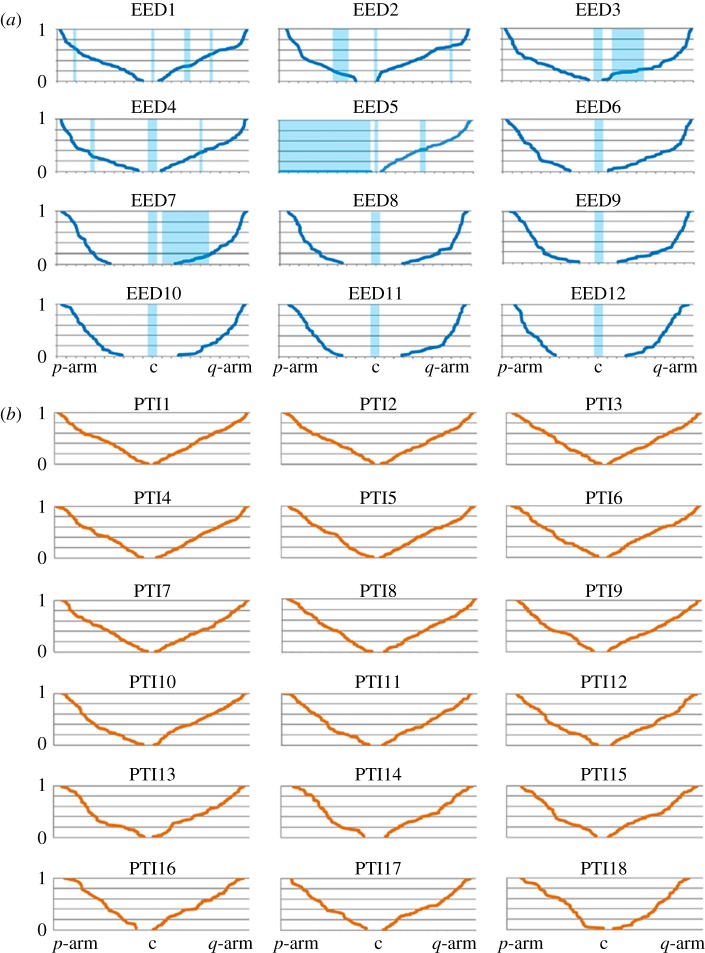
Once the total number of MLH1 foci per chromosome was determined for both species, we moved to analyse their relative positions along each SC (figure 3). The position of individual MLH1 foci was calculated for each chromosome using the centromere as reference point (i.e. from the centromere to the telomere in the p-arm, and from the centromere to the telomere in the q-arm; figure 3). Thus, for comparison among chromosomes and species, the MLH1 position was expressed as the relative position of each CO to the length of the chromosome (the length of each SC was divided into 10% intervals). The general pattern of MLH1 distribution on these chromosomes varies depending on the chromosome and the species studied. We observed that CO localization in the elephant shrew occurred preferentially at the ends of chromosomes (65–98% of the chromosomal length; figure 3a). Moreover, we detected suppression of recombination in the proximity of the centromere (the so-called centromere effect [1]) given that MLH1 foci were rare in the proximal 10–30% of each SC (figure 3a). It was also evident that interstitial (or terminal) hc blocks have an influence on the distribution of MLH1 foci, because these were rarely detected in these regions (figure 3a). In the case of the tiger, the autosomal recombination maps revealed unexpected results. First, we detected a high number of MLH1 foci per SC. Large bivalents such as P. tigris chromosome 1 (PTI1) showed a maximum of 9 MLH1 foci per SC (figure 2e). Second, the cumulative frequencies of MLH1 foci followed a linear distribution along the SC indicating a uniform distribution of COs (figure 3b). Finally, and in marked contrast to the elephant shrew, no centromere effect was noted for any chromosome except the smallest of the tiger complement, PTI18 (figure 3b).
Figure 3.
Chromosome-specific recombination maps for the elephant shrew and the tiger. Cumulative frequency plots for each of the (a) elephant shrew and (b) tiger autosomal SC. The x-axis represents the length of the SC from the centromere (c) to the telomere of each arm (p-arm and q-arm), whereas the y-axis shows the cumulative frequency of MLH1 foci (from 0 to 1). Each chromosome is divided into 10% intervals based on SC length. The areas shaded blue in (a) represent the constitutive hc regions detected by H3K9me signals.
Given the unusual distribution of COs observed in the tiger, we investigated whether a relaxation of COI could account for our observations. COI dictates that adjacent COs on the same chromosome tend to occur at sites that are further apart than expected if they were randomly distributed. In this context, the gamma distribution is a useful model for estimating the presence and the strength of COI [28]—this being reflected by the interference parameter ν (see electronic supplementary material for details). The higher ν, the more even the distribution of COs and the greater the influence of COI (i.e. ν = 1 is an indication of absence of COI [28]). We observed that the frequency of interfocus distances fits a gamma distribution in both species as suggested by the low p-values (see electronic supplementary material, tables S3 and S4). When estimating the interference parameter ν, we noted that the elephant shrew chromosomes displayed variable ν values; larger bivalents (such as EED1, EED3, EED4 and EED6) had values of ν = 2.9 (p = 0.005; electronic supplementary material, table S3), whereas ν increased (ν = 20.0, p = 0.000) for smaller chromosomes (i.e. EED7–EED12), indicating the influence of a strong COI effect (see electronic supplementary material, table S3). By contrast, however, tiger chromosomes showed very low ν values for all bivalents (ν = 2.9, p = 0.000) independent of SC size (see electronic supplementary material, table S4), indicating a weak interference effect among COs corroborating the homogeneous distribution of MLH1 foci observed in the cumulative frequency plots (figure 3b). These values are especially low when compared with MLH1 interfocus distances described for the mouse (ν = 13.7 [28]).
(c). Analysis of the factors affecting recombination rates among mammals
Our analysis of recombinational events (reflected by the number and relative positions of MLH1 foci) among different species suggests a strong phylogenetic component to the patterns observed in mammals (figure 1). We observed that specimens of the same species (i.e. E. edwardii) did not differ significantly in the mean number of autosomal MLH1 foci per cell (Kruskal–Wallis, p > 0.05; figure 1). The same pattern was detected among specimens of the same phylogroup (taxon)—that is, there were no statistical differences in the mean number of autosomal MLH1 foci per cell among species of the Chrysochloridae (A. julianae and A. hottentotus) or within Primates (L. catta, S. oedipus, M. talapoin, C. pygmaea, C. paraguayanus, C. nigritus and A. caraya; Kruskal–Wallis, p > 0.05). Moreover, we found statistically significant differences among all five major phylogroups (Macroscelidea, Afrosoricida, Rodentia, Primates and Carnivora) when considering the number of MLH foci per cell (Kruskal–Wallis, p < 0.0001). The most basal of the mammalian clades examined by us (Afrotheria) was characterized by lower mean numbers of MLH1 foci per cell (25.1±4.1 for Amblysomus and 28.29±3.8 for Elephantulus), followed by Rodentia (34.6±6.1), Primates (39.3±4.1) and Carnivora (51.7±9.5). We therefore proceeded with the analysis of several variables that could potentially influence RRs. These included diploid number, chromosomal morphology, divergence times (DTs) and phylogenetic relationships among species. The data obtained from the analysis of MLH1 foci were combined with recombination data in Marsupialia, Carnivora, Primates, Eulipotyphla, Perissodactyla and Cetartiodactyla available in the literature (see electronic supplementary material, table S1). We anticipated that this would better inform our understanding of RR variation across the mammalian ToL.
(d). Diploid numbers and chromosomal morphology
In order to explain the differences in RRs found among species, the effect of chromosomal number variation on recombination was investigated. Some reports argue that one crossover per bivalent is sufficient to ensure correct segregation during meiosis [3], whereas others are of the view that the more common situation in mammals is one crossover per chromosome arm [3,29,30]. Our data seem to be more consistent with the second view given that we detected a stronger correlation between the mean number of MLH1 foci per cell and number of autosomal chromosome arms (Spearman r = 0.655, p = 0.0025) than when considering haploid chromosome number in isolation (Spearman r = 0.541, p = 0.0167; electronic supplementary material, figure S3). With respect to chromosomal morphology, however, we found that species with a high proportion of acrocentric chromosomes (in our study LCA and CFA) showed the lowest standard deviation in number of MLH1 foci per cell (2.6 and 2.2, respectively; Kruskal–Wallis, p = 0.083) when compared with those with metacentric chromosomes (see electronic supplementary material, table S2). These results suggest that chromosomal morphology similarly has an influence on the distribution of COs. Species with karyotypes comprising high numbers of acrocentric chromosomes showed lower cell-to-cell differences in recombination foci numbers (lower coefficients of variation, CV; electronic supplementary material, table S2; Kruskal–Wallis, p = 0.083) than those whose karyotypes comprised a mix of chromosomal morphologies (metacentric, submetacentric and acrocentric).
(e). Divergence times and phylogenetic relationships
We performed further analyses to explore the effects that RR and phylogenetic distance may have had on the variation in recombination detected among the species studied. By combining our experimental data with published information on RRs in different mammals (see electronic supplementary material, table S1), we were able to identify factors that affect variability in RRs. In order to best estimate how the RR varied among species, autosomal genetic length (cM) and genomic length (Mb) were correlated for each taxon included in our analysis (see electronic supplementary material, table S1).
The DTs used [17] were approximately 82 Ma (interval 67.9–97.2) for Marsupialia, approximately 80 Ma (74.4–96.5) for Afrotheria, approximately 77.3 Ma (70.7–85.8) for Eulipotyphla, approximately 71.5 Ma (64.3–78.4) for Primates, approximately 69.0 Ma (64.1–74.8) for Rodentia, approximately 65.4 Ma (62.3–68.5) for Cetartiodactyla, approximately 56.8 Ma (55.1–61.0) for Perissodactyla and, finally, Carnivora at approximately 54.7 Ma (47.4–60.6). While there is some disagreement with respect to several of the more specific mammalian nodes [17,31–33], there is broad consensus among recent studies regarding these estimates. Based on differences in RR data, we were able to distinguish three distinct groups: (i) Marsupialia (mean RR = 0.20) and Afrotheria (mean RR = 0.28); (ii) Eulipotyphla (mean RR = 0.44), Rodentia (mean RR = 0.51) and Primates (mean RR = 0.61); and (iii) Carnivora (mean RR = 0.98), Perissodactyla (mean RR = 1.02) and Cetartiodactyla (mean RR = 1.19). Our results showed a strong correlation (Spearman r = 0.808, p < 0.0001) between RR and DT, suggesting that deeper lineages in the mammalian ToL, such as Marsupialia and Afrotheria, have lower RRs than do species that have diverged more recently, such as Carnivora, Perissodactyla and Catartiodactyla (see electronic supplementary material, figure S4). These observations were formally corroborated by a phylogenetic analysis which suggested that a directional Brownian model, with a low ancestral value of recombination (about 0.2 cM Mb−1) that increases with time, best fits the evolution of RR in mammals (table 1). This is consistent with the low values of recombination observed for Metatheria, the closest ancestor to eutherian mammals [17]. The other relevant evolutionary parameters κ, δ and λ are related to different rescalings of the tree: κ is a measure of punctuated evolution, δ shows if evolution of recombination slows down or accelerates with time and λ describes how well the phylogeny explains the observed pattern of recombination [34]. According to the ‘best-fit’ model for our data (model B inferring the parameters κ and δ, referenced as B(κ,δ) in table 1), the evolution of recombination in mammals is well described by the phylogeny (λ = 1) and RR appears to be strongly punctuated (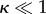 ). Furthermore, highest values occur at recent nodes (δ ≃ 3), suggesting a pattern of species- or genera-dependent adaptation. These results are consistent with our analysis of 1351 trees obtained after analysing the posterior distributions of the studied phylogeny [17] in which all analyses converged on the same model; that is, B(κ,δ) (table 1). Moreover, the results were robust even when considering different topologies of the deep branches of the tree; the ‘best-fit’ model obtained for a phylogeny based on mitochondrial genomes was similarly determined to be B(κ,δ) (see electronic supplementary material).
). Furthermore, highest values occur at recent nodes (δ ≃ 3), suggesting a pattern of species- or genera-dependent adaptation. These results are consistent with our analysis of 1351 trees obtained after analysing the posterior distributions of the studied phylogeny [17] in which all analyses converged on the same model; that is, B(κ,δ) (table 1). Moreover, the results were robust even when considering different topologies of the deep branches of the tree; the ‘best-fit’ model obtained for a phylogeny based on mitochondrial genomes was similarly determined to be B(κ,δ) (see electronic supplementary material).
Table 1.
Likelihood and ML values of parameters for the evolutionary models tested following the phylogeny provided by Meredith et al. [17]. n.a., not applied.
| model | log(L) | AIC | p-value versus A | p-value versus B | α | β | var | κ | δ | λ |
|---|---|---|---|---|---|---|---|---|---|---|
| A | −7.14 | −18.29 | 1.000 | n.a. | 0.43 | 0 | 1.232 | 1 | 1.0 | 1.0 |
| B | −6.89 | −19.77 | 0.472 | 1.000 | 0.98 | −1.82 | 1.196 | 1 | 1.0 | 1.0 |
| A(κ) | −3.21 | −12.43 | 0.005 | n.a. | 0.30 | 0 | 0.038 | 0 | 1.0 | 1.0 |
| A(δ) | −5.46 | −16.91 | 0.066 | n.a. | 0.47 | 0 | 5.167 | 1 | 3.0 | 1.0 |
| A(λ) | −5.53 | −17.05 | 0.072 | n.a. | 0.45 | 0 | 0.679 | 1 | 1.0 | 0.8 |
| B(κ) | −1.00 | −9.99 | 0.002 | 0 | −0.07 | 0.13 | 0.030 | 0 | 1.0 | 1.0 |
| B(δ) | −5.39 | −18.78 | 0.173 | 0.084 | 0.54 | −2.51 | 5.128 | 1 | 3.0 | 1.0 |
| B(λ) | −5.27 | −18.55 | 0.154 | 0.072 | 0.90 | −1.49 | 0.668 | 1 | 0.8 | 1.0 |
| A(κ,δ) | −0.22 | −8.43 | 0.001 | n.a. | 0.25 | 0 | 0.001 | 0 | 3.0 | 1.0 |
| A(κ,λ) | −2.84 | −13.69 | 0.014 | n.a. | 0.31 | 0 | 0.029 | 0 | 1.0 | 0.9 |
| A(δ,λ) | −5.08 | −18.16 | 0.127 | n.a. | 0.48 | 0 | 2.551 | 1 | 2.5 | 0.9 |
| B(κ,δ) | 4.45 | −1.00 | 0 | 0 | 0.21 | 0 | 0 | 0 | 3.0 | 1.0 |
| B(κ,λ) | 0.94 | −8.11 | 0.001 | 0 | −0.07 | 0.13 | 0.013 | 0 | 1.0 | 0.5 |
| B(δ,λ) | −5.00 | −19.99 | 0.231 | 0.151 | −0.59 | −1.60 | 1.878 | 1 | 2.2 | 0.8 |
| A(κ,δ,λ) | −0.22 | −10.43 | 0.003 | n.a. | 0.25 | 0 | 0.001 | 0 | 3.0 | 1.0 |
| B(κ,δ,λ) | 4.49 | −3.01 | 0 | 0 | 0.22 | 0 | 0 | 0 | 3.0 | 0.9 |
Moreover, body size (log-scaled) appears to be strongly and positively correlated with RR (r ≃ 0.65). This is intriguing given the inverse relation between body size and effective population size. However, the correlation is not significant after the phylogeny is taken into account (p ≃ 0.8), suggesting that it could be due to the similar evolution of the two traits. In fact, both traits fit a model of punctuated directional evolution—their values increase and accelerate with time, resulting in an apparent correlation. A similar relationship exists for metabolism and temperature—both of which are related to body size, for which fewer data exist—while generation time is not correlated with recombination (see electronic supplementary material).
3. Discussion
(a). The influence of the phylogeny in the variation of recombination rates across the mammalian tree of life
We have shown that there is a phylogenetic dimension to RRs among mammals. Recombination can vary among individuals of the same species (i.e. E. edwardii), but these differences are not statistically significant. This pattern is also observed in species belonging to the same phylogroup (i.e. within Macroscelidea—two species—or within Primates—seven species), suggesting that recombination can evolve (the so-called ‘evolvability of recombination rates’ [35]), but within certain limits imposed by the intrinsic genetic characteristics and population dynamics of each taxon. Moreover, RRs seem to be specific to each major taxonomic group. The same clustering (i.e. a negative correlation found between RRs and DTs; Spearman r = 0.808, p < 0.0001) was observed when data from genetic linkage analyses conducted in Marsupialia, Carnivora, Perissodactyla and Cetartiodactyla are included. The reasons for this are not obvious. At first glance, a plausible hypothesis is that closely related species share similar recombination patterns on average when broader (i.e. genome-wide) scales are considered. It has been argued that rates of recombination might vary considerably between species when comparing high-resolution (kb) recombination maps [12,13], but these differences disappear at a broader scale (Mb; revised in [14]). Moreover, recent studies have suggested the existence of a phylogenetic effect in RRs by indicating that closely related species tend to have similar average rates of recombination [15,16,36]. This explains an earlier report [34] of a phylogenetic signal in the distribution of genomic RRs in the genetic maps of domesticated species, and why the evolution of recombination in mammals is well described by DT estimates. Put differently, based on Meredith et al.'s [17] phylogenetic time tree of mammalian families, species representative of basal lineages have lower RRs than those that are more derived. In this framework, the rate of recombination appears to be directional and strongly punctuated. Moreover, it occurs mostly at times of cladal divergence, suggesting selection is a driving evolutionary force. Alternatively, there may be a positive correlation between recombination and substitution rate since in this case longer branches would have higher RRs. Importantly, however, a strongly positive correlation between recombination and body mass/metabolism was detected, as has been observed in previous studies [37] (i.e. a negative correlation between body mass/metabolism and substitution rate). Consequently, a correlation between both recombination and substitution rate should be negative or absent. Our data support the first explanation since we found that basal mammals had low RRs, and that recombination increased in more recent mammalian lineages, suggesting that selective forces are behind the lineage-dependent increment in RRs.
We can, however, only speculate which selective forces are instrumental in driving lineage-dependent RRs. Simulations to determine the conditions under which high RRs can be achieved have bolstered the dearth of available empirical data. In general, conditions associated with the process of domestication, such as adaptation to new environments, strong directional selection at multiple loci and a reduction in population size, are thought to affect RRs [38]. However, whether additional factors influence or ameliorate the process cannot be discounted at this stage.
(b). Mechanistic factors affect crossover distribution along chromosomes
The mechanisms by which cells regulate the spatial distribution of COs along chromosomes are poorly understood. That said, the total number and distribution of COs along a specific chromosome seems to be dependent on chromosome size and morphology. Larger chromosomes tend to accumulate larger numbers of COs, and each chromosome arm generally presents at least one CO [3]. Our finding of a positive correlation between SC length and the mean number of MLH1 foci in the elephant shrew and tiger supports this view (see electronic supplementary material, figure S2). However, we also show that this is certainly not universal and that chromosomal morphology (acrocentric versus metacentric) may also affect CO distribution. We observed that cell-to-cell differences in numbers of recombination sites (reflected by the standard deviation of the mean; electronic supplementary material, table S2) were especially low in species with karyotypes comprising a high number of acrocentric chromosomes (dog—all acrocentric chromosomes; lemur—85% acrocentric chromosomes). The same pattern is found in mouse and Chinese muntjac, both of which have strictly acrocentric karyotypes, where standard deviations were similarly low (± 2.0–2.7) [15,39]. On the other hand, species with karyotypes comprising metacentric and submetacentric chromosomes show high cell-to-cell variability in the mean number of MLH1 foci (see electronic supplementary material, table S1), and this was particularly pronounced in the rat (±6.2) and tiger (±5.9). This suggests that chromosomal morphology plays a mechanistic role in the distribution of COs, probably owing to forces that are associated with chromosome pairing and synapses during early meiotic prophase [40].
Consistent with the suggestion that chromosomal morphology is affecting recombination, we observed that autosomal chromosomal arm number in a karyotype shows the highest correlation with the average number of COs in mammals. It has been documented that an ‘obligatory chiasma’ occurs to ensure the proper disjunction of homologous chromosomes [3]. The alternative—non-disjunction—generally leads to zygotic death or aneuploidy [41], and consequently selection will negatively impact where less than one CO per chromosomal pair prevails. By contrast, there is an alternative view based on early studies [29,30] that documents a strong correlation between the number of chiasmata and the haploid number of chromosome arms. In this respect, our results appear to be more consistent with the general requirement for at least one crossover per chromosome arm, rather than the classical expectation of one crossover per chromosome.
(c). Low levels of crossover interference in the tiger: an exception to the rule in mammals?
A particularly surprising finding of this investigation was the low level of crossover interference found in the tiger. This species presented a higher number of MLH1 foci per cell (59.4±5.9) than what is expected based on both diploid number (2n = 38) and the number of autosomal chromosomal arms in its karyotype. The cat (a closely related species with the same diploid number, 2n = 38) similarly showed high RRs (figure 1; electronic supplementary material, table S2) [42], suggesting that this feature might be characteristic of the Felidae. On the other hand, and probably as a result of the high RRs observed, chromosome-specific recombination maps of the tiger show that MLH1 foci are uniformly distributed along SCs (figure 3b). As a result, low values for ν were detected in all chromosomes reflecting the effects of weak COI in this species (see electronic supplementary material, table S4).
Although COI has been described in most of the organisms that have been studied (S. cerevisiae, Neurospora crassa, C. elegans, Zea mays, Drosophila melanogaster, Danio rerio [9]; and, importantly for the present investigation, mammalian species such as mice, humans, dogs and the Chinese muntjac [9,15,22]), the tiger has the lowest levels of COI, suggesting that interference is a complex process that requires multiple controls. Moreover, it is noteworthy that although values obtained for early pachynema RPA foci in tiger (a marker of DSBs formation) were slightly elevated, but within the same range (182.2±31.8; figure 2) as those of mice (144.9±30.9; see [5]), the ratio of DBSs/COs increased remarkably (3 : 1) when compared with human and mouse (from 7 : 1 to 10 : 1). Consequently, the high CO number detected in the tiger is probably not directly related to the initial number of DSBs that are formed in early stages of meiosis. Rather, additional (yet undescribed) mechanisms might act when resolving DSBs into COs in later stages of prophase I, and these would determine the final balance of COs. These data suggest that the Felidae will be particularly useful for the study of the mechanisms controlling crossover formation, distribution and resolution.
4. Conclusion
Our results show how the evolution of recombination in mammals is influenced by phylogeny. RRs appear to be directional and occur mostly at times of cladal divergence, suggesting selection is a driving evolutionary force. Moreover, the construction of chromosome-specific recombination maps for distantly related taxa shows that crossover interference is especially weak in species with high RRs, suggesting control by additional mechanisms in mammals. These results indicate that recombination landscapes reflect a trade-off between the selective forces that affect the DNA sequence itself (determined by population genetics and the evolutionary history of each taxon), the position and distribution of COs along the chromosomes, and the mechanistic forces that control how the DNA is packaged into chromosomes during meiosis.
5. Material and methods
Testicular samples from the following species were analysed: E. edwardii, A. julianae, A. hottentotus, M. talapoin, S. oedipus, L. catta, C. pygmaea, R. norvegicus, C. familiaris, F. catus and P. tigris. See electronic supplementary material for details on sample processing, IF, estimates of RRs and phylogenetic inferences.
Acknowledgements
We thank I. Eduardo for insightful discussions during the development of the work, and W. Murphy and M. Springer for making their raw data available to us.
Funding statement
A.R-H.'s laboratory is funded by Ministerio de Ciencia e Innovacion (MICINN-Spain, CGL2010 20170) and the Barcelona Zoological Gardens (BSM, Zoo Barcelona). T.J.R.'s research is supported by a grant from the South African National Research Foundation. M.O.-B. is funded by Instituto de Salud Carlos III (CP07/0258 and PI08/1185).
References
- 1.Farré M, Micheletti D, Ruiz-Herrera A. 2013. Recombination rates and chromosomal reorganizations between human and chimpanzee genomes: a new twist in the chromosomal speciation theory. Mol. Biol. Evol. 30, 853–864 (doi:10.1093/molbev/mss272) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bishop DK, Zickler D. 2004. Early decision; meiotic crossover interference prior to stable strand exchange and synapsis. Cell 117, 9–15 (doi:10.1016/S0092-8674(04)00297-1) [DOI] [PubMed] [Google Scholar]
- 3.Kauppi L, Jeffreys AJ, Keeney S. 2004. Where the crossovers are: recombination distributions in mammals. Nat. Rev. Genet. 5, 413–424 (doi:10.1038/nrg1346) [DOI] [PubMed] [Google Scholar]
- 4.Youds JL, et al. 2010. RTEL-1 enforces meiotic crossover interference and homeostasis. Science 327, 1254–1258 (doi:10.1126/science.1183112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cole F, Kauppi L, Lange J, Roig I, Wang R, Keeney S, Jasin M. 2012. Homeostatic control of recombination is implemented progressively in mouse meiosis. Nat. Cell Biol. 14, 424–430 (doi:10.1038/ncb2451) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hollingsworth NM, Brill SJ. 2004. The Mus81 solution to resolution: generating meiotic crossovers without Holliday junctions. Genes Dev. 18, 117–125 (doi:10.1101/gad.1165904) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Phadnis N, Hyppa RW, Smith GR. 2011. New and old ways to control meiotic recombination. Trends Genet. 27, 411–421 (doi:10.1016/j.tig.2011.06.007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hillers KJ, Villeneuve AM. 2003. Chromosome-wide control of meiotic crossing over in C. elegans. Curr. Biol. 13, 1641–1647 (doi:10.1016/j.cub.2003.08.026) [DOI] [PubMed] [Google Scholar]
- 9.Berchowitz LE, Copenhaver GP. 2010. Genetic interference: don't stand so close to me. Curr. Genomics 11, 91–102 (doi:10.2174/138920210790886835) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holloway JK, Booth J, Edelmann W, McGowan CH, Cohen PE. 2008. MUS81 generates a subset of MLH1-MLH3-independent crossovers in mammalian meiosis. PLoS Genet. 4, e1000186 (doi:10.1371/journal.pgen.1000186) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Woods LM, Hodges CA, Baart E, Baker SM, Liskay M, Hunt PA. 1999. Chromosomal influence on meiotic spindle assembly: abnormal meiosis I in female Mlh1 mutant mice. J. Cell Biol. 145, 1395–1406 (doi:10.1083/jcb.145.7.1395) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ptak SE, Hinds DA, Koehler K, Nickel B, Patil N, Ballinger DG, Przeworski M, Frazer KA, Pääbo S. 2005. Fine-scale recombination patterns differ between chimpanzees and humans. Nat. Genet. 37, 429–434 (doi:10.1038/ng1529) [DOI] [PubMed] [Google Scholar]
- 13.Paigen K, Petkov P. 2010. Mammalian recombination hot spots: properties, control and evolution. Nat. Rev. Genet. 11, 221–233 (doi:10.1038/nrg2712) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smukowski CS, Noor MA. 2011. Recombination rate variation in closely related species. Heredity 107, 496–508 (doi:10.1038/hdy.2011.44) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dumont BL, Payseur BA. 2011. Evolution of the genomic recombination rate in murid rodents. Genetics 187, 643–657 (doi:10.1534/genetics.110.123851) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garcia-Cruz R, Pacheco S, Brieño MA, Steinberg ER, Mudry MD, Ruiz-Herrera A, Garcia-Caldés M. 2011. A comparative study of the recombination pattern in three species of Platyrrhini monkeys (primates). Chromosoma 120, 521–530 (doi:10.1007/s00412-011-0329-6) [DOI] [PubMed] [Google Scholar]
- 17.Meredith RW, et al. 2011. Impacts of the cretaceous terrestrial revolution and KPg extinction on mammal diversification. Science 334, 521–524 (doi:10.1126/science.1211028) [DOI] [PubMed] [Google Scholar]
- 18.Asher RJ, Helgen KM. 2010. Nomenclature and placental mammal phylogeny. BMC Evol. Biol. 10, 102 (doi:10.1186/1471-2148-10-102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gilbert C, Maree S, Robinson TJ. 2008. Chromosomal evolution and distribution of telomeric repeats in golden moles (Chrysochloridae, Mammalia). Cytogenet. Genome Res. 121, 110–119 (doi:10.1159/000125836) [DOI] [PubMed] [Google Scholar]
- 20.Robinson TJ, Fu B, Ferguson-Smith MA, Yang F. 2004. Cross-species chromosome painting in the golden mole and elephant-shrew: support for the mammalian clades Afrotheria and Afroinsectiphillia but not Afroinsectivora. Proc. R. Soc. Lond. B 271, 1477–1484 (doi:10.1098/rspb.2004.2754) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Steiper ME, Young NM. 2009. Primates (Primates). In The timetree of life (eds Hedges SB, Kumar S.), pp. 482–486 Oxford, UK: Oxford University Press [Google Scholar]
- 22.Basheva EA, Bidau CJ, Borodin PM. 2008. General pattern of meiotic recombination in male dogs estimated by MLH1 and RAD51 immunolocalization. Chromosome Res. 16, 709–719 (doi:10.1007/s10577-008-1221-y) [DOI] [PubMed] [Google Scholar]
- 23.Eizirik E, Murphy WJ. 2009. Carnivores (Carnivora). In The timetree of life (eds Hedges SB, Kumar S.), pp. 504–507 Oxford, UK: Oxford University Press [Google Scholar]
- 24.Keeney S, Giroux CN, Kleckner N. 1997. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88, 375–384 (doi:10.1016/S0092-8674(00)81876-0) [DOI] [PubMed] [Google Scholar]
- 25.Moens PB, Marcon E, Shore JS, Kochakpour N, Spyropoulos B. 2007. Initiation and resolution of interhomolog connections: crossover and non-crossover sites along mouse synaptonemal complexes . J. Cell Sci. 120, 1017–1027 (doi:10.1242/jcs.03394) [DOI] [PubMed] [Google Scholar]
- 26.Hublitz P, Albert M, Peters A. 2009. Mechanisms of transcriptional repression by histone lysine methylation. Int. J. Dev. Biol. 10, 335–354 (doi:10.1387/ijdb.082717ph) [DOI] [PubMed] [Google Scholar]
- 27.Sun F, Oliver-Bonet M, Liehr T, Starke H, Turek P, Navarro J, Benet J. 2004. Human male recombination maps for individual chromosomes. Am. J. Hum. Genet. 74, 521–531 (doi:10.1086/382138) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Boer E, Stam P, Dietrich AJ, Pastink A, Heyting C. 2006. Two levels of interference in mouse meiotic recombination. Proc. Natl Acad. Sci. USA 103, 9607–9612 (doi:10.1073/pnas.0600418103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dutrillaux B. 1986. Role of chromosomes in evolution: a new interpretation. Ann. Genet. 29, 69–75 [PubMed] [Google Scholar]
- 30.Pardo-Manuel de Villena F, Sapienza C. 2001. Recombination is proportional to the number of chromosome arms in mammals. Mamm. Genome 12, 318–322 (doi:10.1007/s003350020005) [DOI] [PubMed] [Google Scholar]
- 31.Dos Reis M, Inoue J, Hasegawa M, Asher RJ, Donoghue PC, Yang Z. 2012. Phylogenomic datasets provide both precision and accuracy in estimating the timescale of placental mammal phylogeny. Proc. R. Soc. B 279, 3491–3500 (doi:10.1098/rspb.2012.0683) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nishihara H, Maruyama S, Okada N. 2009. Retroposon analysis and recent geological data suggest near-simultaneous divergence of the three superorders of mammals. Proc. Natl Acad. Sci. USA 106, 5235–5240 (doi:10.1073/pnas.0809297106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hallstrom BM, Janke A. 2010. Mammalian evolution may not be strictly bifurcating. Mol. Biol. Evol. 27, 2804–2816 (doi:10.1093/molbev/msq166) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pagel M. 1999. Inferring the historical patterns of biological evolution. Nature 401, 877–884 (doi:10.1038/44766) [DOI] [PubMed] [Google Scholar]
- 35.Butlin RK. 2005. Recombination and speciation. Mol. Ecol. 14, 2621–3265 (doi:10.1111/j.1365-294X.2005.02617.x) [DOI] [PubMed] [Google Scholar]
- 36.Dumont BL, Payseur BA. 2008. Evolution of the genomic rate of recombination in mammals. Evolution 62, 276–294 (doi:10.1111/j.1558-5646.2007.00278.x) [DOI] [PubMed] [Google Scholar]
- 37.Sayres MA, Venditti C, Pagel M, Makova KD. 2011. Do variations in substitution rates and male mutation bias correlate with life-history traits? A study of 32 mammalian genomes. Evolution 65, 2800–2815 (doi:10.1111/j.1558-5646.2011.01337.x) [DOI] [PubMed] [Google Scholar]
- 38.Ross-Ibarra J. 2004. The evolution of recombination under domestication: a test of two hypotheses. Am. Nat. 163, 105–112 (doi:10.1086/380606) [DOI] [PubMed] [Google Scholar]
- 39.Yang Q, Zhang D, Leng M, Yang L, Zhong L, Cooke HJ, Shi Q. 2012. Synapsis and meiotic recombination in male Chinese muntjac (Muntiacus reevesi). PLoS ONE 4, e19255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schertan H, Weich S, Schwegler H, Heyting C, Härle M, Cremer T. 1996. Centromere and telomere movements during early meiotic prophase of mouse and man are associated with the onset of chromosome pairing. J. Cell Biol. 134, 1109–1125 (doi:10.1083/jcb.134.5.1109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hassold T, Hall H, Hunt P. 2007. The origin of human aneuploidy: where we have been, where we are going. Hum. Mol. Genet. 15, R203–R208 (doi:10.1093/hmg/ddm243) [DOI] [PubMed] [Google Scholar]
- 42.Borodin PM, Karamysheva TV, Rubtsov NB. 2008. Immunofluorescent analysis of meiotic recombination and interference in the domestic cat. Tsitologiia 50, 62–66 [PubMed] [Google Scholar]