Key Points
STAT3+ T cells are found not only in detected concomitant LGL-BMFs, but in cases in which an LGL expansion was not suspected.
Transformation via acquisition of a somatic mutation in T cells may be a mechanism of immune, mainly hypoplastic, bone marrow failure.
Abstract
Large granular lymphocyte leukemia (LGL) is often associated with immune cytopenias and can cooccur in the context of aplastic anemia (AA) and myelodysplastic syndromes (MDS). We took advantage of the recent description of signal transducer and activator of transcription 3 (STAT3) mutations in LGL clonal expansions to test, using sensitive methods, for the presence of these mutations in a large cohort of 367 MDS and 140 AA cases. STAT3 clones can be found not only in known LGL concomitant cases, but in a small proportion of unsuspected ones (7% AA and 2.5% MDS). In STAT3-mutated AA patients, an interesting trend toward better responses of immunosuppressive therapy and an association with the presence of human leukocyte antigen-DR15 were found. MDSs harboring a STAT3 mutant clone showed a lower degree of bone marrow cellularity and a higher frequency of developing chromosome 7 abnormalities. STAT3-mutant LGL clones may facilitate a persistently dysregulated autoimmune activation, responsible for the primary induction of bone marrow failure in a subset of AA and MDS patients.
Introduction
Large granular lymphocyte leukemia (LGL) is often associated with immune cytopenias and can cooccur in other bone marrow failure (BMF) disorders such as aplastic anemia (AA) and myelodysplastic syndromes (MDSs).1-3 Previously, using molecular analysis of the T-cell receptor variable β-chain (Vβ) repertoire in these diseases, we demonstrated oligoclonal skewing of the cytotoxic T-cell lymphocyte (CTL) spectrum, indicating expansions of CTLs clones that may recognize hematopoietic targets.4,5 However, the strongest evidence in support of the immune pathogenesis of these conditions stems from the success of immunosuppressive therapies (ISTs) in AA and also in some forms of MDS6,7; however, responses to immunosuppression are incomplete and the prediction of patients who will benefit from this treatment modality is not evident.
Several decades of clinical observations and laboratory experiments have suggested a pivotal role for the immune system in AA and MDS,8,9 but a key question remains: what drives autoimmunity in this context? Among the possible causative hypotheses are a breach of tolerance and/or an exacerbated immune response, and recent findings brought focus to a third possibility: immortalization of an autoimmune clone by acquisition of a somatic lesion. The impetus for investigation of this final theory came from our description of somatic mutations in the SH2 dimerization and activation domain of signal transducer and activator of transcription 3 (STAT3). These mutations were frequently observed in patients with T-cell LGL (T-LGL) and natural killer chronic lymphoproliferative disorders, suggesting that aberrant STAT3 signaling underlies the pathogenesis of clonal expansions of LGL.10,11
For the purpose of this study, we hypothesized that, similar to LGL, a disease in which we detected pathologic STAT3 mutations, a clonal CTL population can arise in the context of AA and MDS through acquisition of somatic STAT3 mutations indicating autoimmune autonomy. We propose that the presence of STAT3 mutations might be diagnostically useful and associated with clinical features in a subset of patients with acquired BMF.
Methods
Patients
Blood sample collection from patients at diagnosis was performed after informed consent, according to the protocols approved by the Institutional Review Board of Cleveland Clinic and by The Pennsylvania State University, The Moffit Cancer Center, and King´s College in accordance with the Declaration of Helsinki. The study population consisted of 367 patients with MDS and 140 with idiopathic AA, all of them diagnosed in adulthood, and no constitutional BMF patient was included.
We followed the diagnostic criteria of the World Health Organization classification of tumors of hematopoietic and lymphoid tissues.12 For the diagnosis of AA, only patients lacking dysplasia in the hematopoietic series were included, and only patients with normal or noninformative (no growth) cytogenetics were classified as having AA. Disease severity was defined by standard criteria.13 A diagnosis of hypocellular MDS was made based on the presence of dysplastic ASfeatures and the overall clinical presentation including the presence of cytopenias, the presence of blasts (<5% in the bone marrow or <2% in peripheral blood), and/or chromosomal abnormalities by metaphase cytogenetics and a decreased cellularity of the marrow of <20%.
Amplification refractory mutation system-polymerase chain reaction for STAT3 mutations
A tetra-primer amplification refractory mutation system (ARMS) assay on whole blood DNA was performed as previously described.11,14
Pyrosequencing
Polymerase chain reaction (PCR) primers and sequencing primers were designed using Pyrosequencing Assay Design software 1.0. PCR reactions were performed using HotstarTaq DNApolymerase (Qiagen) for 45 cycles. Aliquots of PCR products were verified to have single bands on agarose gels but were not gel extracted. PCR products were then sequenced and analyzed with a PyroMark Q24 1.0.10. using the standard protocol and reagents supplied by Qiagen—STAT3 pyro-forward: 5′ GGCTTAAGTCTTTTCCCCTTCGAG 3′; STAT3 pyro-reverse-biotin: 5′ TGCCTCCTCCTTGGGAATGT 3′; STAT3 pyro-sequencing: 5′ CCAGTCCGTGGAACC 3′.
Amplicon sequencing of STAT3
Locus-specific primers covering STAT3 exon 21 were designed for maximum 300-nucleotide-long PCR amplicons (Primer3; http://primer3.wi.mit.edu/). Sequence tails corresponding to the Illumina adapter sequences (5′-ACACTCTTTCCCTACACGACGCTCTTCCGATCT and 5′-AGACGTGTGCTCTTCCGATCT) were added to the 5′ end of the forward and reverse locus-specific primers, respectively (Sigma-Aldrich, St. Louis, MO). The PCR reaction was cycled in a DNA Engine Tetrad 2 (Bio-Rad Laboratories, Hercules, CA) or in a G-Storm GS4 (G-Storm; Somerton, Somerset, United Kingdom) thermal cycler. Purified sample pools were analyzed for amplification yield and performance on an Agilent 2100 Bioanalyzer (Agilent Technologies). Data were analyzed on Illumina MiSeq Reporter Software version 1.3 or an earlier version (Illumina). To increase specificity, cases were considered negative when the ratio between the known somatic variant and background variant errors was <1. All deep sequencing–confirmed cases had a ratio >50.
Flow cytometry and cell sorting
Blood mononuclear cells were isolated using a density gradient (Mediatech, Manassas, VA) and stained using CD8 phycoerythrin-Texas red (ECD) (5 μL), CD57 fluorescein isothiocyanate (10 μL), CD3 PC5 (5 μL), and one of the following anti–T-cell receptor PE conjugated antibodies (20 μL): Vβ1, Vβ2, Vβ3, Vβ4, Vβ5.1, Vβ5.2, Vβ5.3, Vβ7.1, Vβ7.2, Vβ8, Vβ9, Vβ11, Vβ12, Vβ13.1, Vβ13.2, Vβ13.6, Vβ14, Vβ16, Vβ17, Vβ18, Vβ20, Vβ21.3, Vβ22, or Vβ23 (all antibodies were from Beckman Coulter). Specific Vβ immunodominant expansions were then sorted on a Beckman Coulter Epics Altra or by magnetic beads (Miltenyi).
Sanger sequencing
Exons of selected genes (ie, TET2, IDH1, IDH2, DNMT3A, CBL, RUNX1, ASXL1, UTX [KDM6A], EZH2, NRAS, KRAS, TP53, JAK2, SF3B1, U2AF1, SRSF2, and SETBP1) were amplified and underwent direct genomic sequencing by standard techniques on a ABI 3730xl DNA analyzer (Applied Biosystems, Foster City, CA) as previously described.15-18
Statistical analysis
Comparisons of proportions and ranks of variables between groups were performed by the χ2 test, Fisher’s exact test, Student t test, or Mann-Whitney U test, as appropriate. We used the Kaplan-Meier and the Cox method to analyze overall survival and progression-free survival, with a 2-sided, P ≤ .05 considered significant. In Cox models, examination of log (−log) survival plots and partial residuals was performed to ensure the underlying assumption of proportional hazards was met.
Results
Screening, confirmation, and distribution of mutations
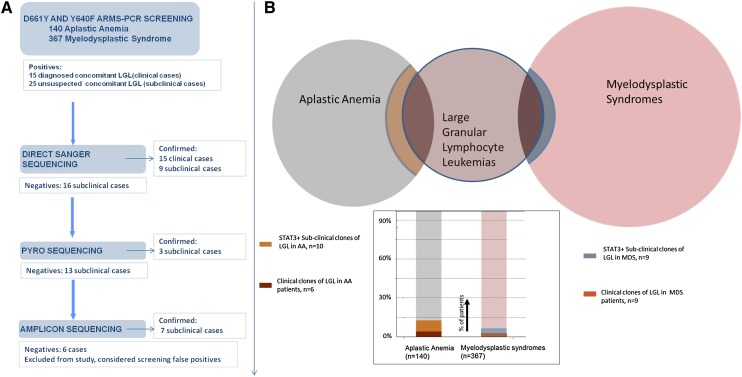
A large series of patients with acquired BMF syndromes, including 367 with MDS and 140 with AA, was screened for the presence of D661Y and Y640F mutations in STAT3 using a sensitive DNA tetra-primer ARMS assay. Patients with a positive testing result were further examined by direct sequencing and next-generation amplicon and pyrosequencing and were confirmed in 34 of 40 cases. STAT3 mutant clones subjected to deep sequencing were analyzed, and their “size” was judged by percentage of positive reads ranging from 0.3% to 13.2% (Table 1). For 6 of 40 positive cases, the results of the ARMS assay could not be confirmed; in 4 of these 6 cases, a variable number of variant reads was found, all of which remained under the set detection threshold as defined by the noise level. In the remaining 2 cases, no band was visible in an ARMS-PCR resequencing using 30 cycles, in contrast to the positive result produced by the original 40 amplification cycles. Consequently, the unconfirmed cases were excluded from further analysis.
Table 1.
Deep amplicon results from 13 subclinical cases
| Sample ID | Mutation in ARMS-PCR screening | Genomic coordinates | Deep sequencing somatic variant result | Percent of mutant variant reads | Percent of next “noise” variant read | Coverage | Diagnosis |
|---|---|---|---|---|---|---|---|
| UPN92 | D661Y | chrl7:40474420 | Positive | 1.8 | 0.002 | 2318 | MDS |
| UPN10 | D661Y | chrl7:40474420 | Positive | 13.2 | 0.002 | 5271 | MDS |
| UPN28 | DG61Y | chrl7:40474420 | Positive | 0.5 | 0.01 | 2585 | AA |
| UPN11 | D661Y | chrl7:40474419 | Positive | 5.4 | 0.01 | 7413 | MDS |
| UPN2 | D661Y | chrl7:40474420 | Positive | 0.9 | 0.002 | 17 200 | AA |
| UPN1 | Y640F | chrl7:40474482 | Positive | 1.7 | 0.003 | 9032 | AA |
| UPN3 | Y640F | chr17:40474482 | Positive | 0.3 | 0.0004 | 2352 | AA |
| UPN22 | Y640F | chrl7:40474482 | Negative | 0.00008 | 0.003 | 12 306 | MDS |
| UPN52 | D661Y | chrl7:40474420 | Negative | 0.6 | 1.3 | 14 680 | MDS |
| UPN57 | Y640F | chrl7:40474482 | Negative | 0 | 0 | 1335 | AA |
| UPN72 | Y640F | chrl7:404744S2 | Negative | 0 | 0.002 | 17 562 | AA |
| UPN60 | D661Y | chrl7:40474420 | Negative | 0.6 | 1.4 | 23 012 | AA |
| UPN32 | D661Y | chrl7:40474420 | Negative | 0.6 | 1.4 | 5620 | AA |
Six cases were considered ARMS-PCR false positives because either no variant reads were found or the number of reads was under the set detection threshold as defined by the noise level (multiple bases calls). The percentage of next “noise” variant reads corresponds to the base calls considered background errors (different from the reference or the known somatic mutated variant). Coverage indicates the numbers of reads generated in every case.
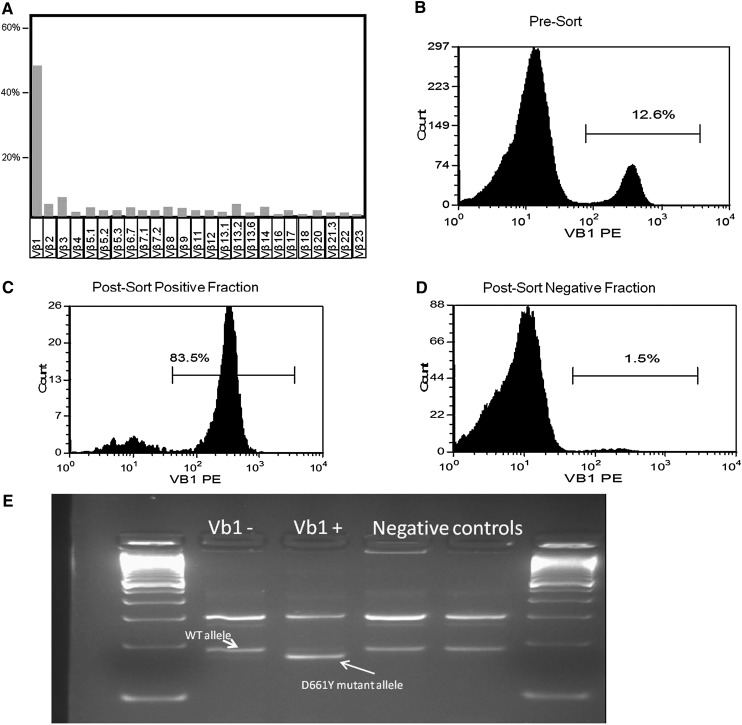
First, we identified 24 MDS patients with known concomitant LGL and screened them for the presence of STAT3 mutations: in 9 of 24 patients, STAT3 mutations were found. Based on this finding, we screened an additional cohort of 343 patients with MDS and no suspected concomitant LGL: we identified 9 additional patients with STAT3-mutated clones. In like manner, we moved on to search for STAT3 CTL clones in AA (N = 140). In total, we identified 10 (7%) AA patients with unsuspected LGL but STAT3-mutated clones, together with 6 mutated cases in 11 known AA-LGL patients. Those 7% of AA patients and 2.5% of MDS patients with subclinical STAT3 clones could now be added to the classic Venn diagram of overlapping acquired BMF syndromes (Figure 1). We then investigated whether the STAT3 mutations were present in the CTL compartment and could be found in the Vβ expanded population. In previous work, we showed in a T-LGL patient how the mutation was present exclusively in the CD3+ fraction.11 We herein chose 2 patients with MDS (both mutated for D661Y) and 1 AA patient with an Y640F mutation. DNA was extracted from CD3+CD8+CD57+ CTLs that were sorted, in addition, according to the immunodominant clone defined by expression of pathognomonic Vβ chain. In these 3 cases, the mutation was found in the sorted compartment using a direct sequencing method. Figure 2 illustrates 1 of these cases, showing the negative results in the negative fraction of the Vβ1-expanded population.
Figure 1.
Study flow diagram and frequency of confirmed STAT3-mutated patients. (A) Screening-confirmation pipeline. Several sequencing methods with increasing sensitivity were used to confirm the ARMS-PCR–positive cases. (B) Venn diagram showing the clinically detected overlapping of LGL with AA and MDS, and an additional percentage of STAT3+, clinically unsuspected concomitant LGL/BMF.
Figure 2.
D661Y STAT3 mutation in a hypocellular MDS case. (A-B) Immunodominant Vβ1 fraction before sorting; among the CD8 Vβ repertoire; and blood mononuclear cells. (C-D) Vβ1-positive and -negative fraction after blood mononuclear cell sorting. (E) Presence of the ARMS-PCR D661Y allele only in the Vβ1+ population.
Clinical correlates
The clinical characteristics of the 140 patients included in this study are summarized in supplemental Table 1. Clinically, AA patients with concomitant STAT3 mutant clones were characterized by a higher frequency of moderate severity disease (29% vs 21%) and a higher proportion of responders to first-line therapy (81% vs 61%), although no statistical significance was reached. We also found a significant correlation with the presence of human leukocyte antigen-DR15 (HLA-DR15) (75% vs 44%, P = .02; Table 2).
Table 2.
Comparison of clinical characteristics between AA patients according to the STAT3 mutation status
| Variable | AAs with STAT3 mutation, n = 16 | AAs without STAT3 mutation, n = 124 | P |
|---|---|---|---|
| Age, years (median, range) | 48 | 41 | .8 |
| Sex, % | |||
| Male | 66 | 52 | .3 |
| Female | 34 | 48 | |
| Moderate severity, % | 29 | 21 | .3 |
| Presence of PNH clone, % | 54 | 38 | .2 |
| HLA-DR15, % | 75 | 44 | .02 |
| Any response to first line, % | 81 | 61 | .1 |
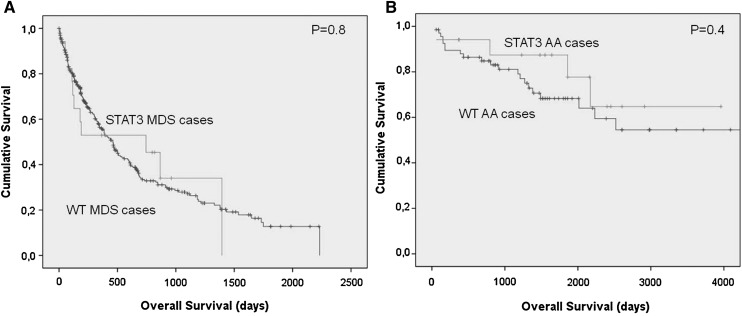
Of 367 patients with MDS, 64 had refractory cytopenia with unilineage dysplasia, 107 had refractory anemia with ring sideroblasts, 73 had refractory cytopenia with multilineage dysplasia, 61 had refractory anemia with excess of blasts type I, 75 had refractory anemia with excess of blasts type 2, and 13 had MDS del(5q). We also included 48 patients with myelodysplastic/myeloproliferative disease (MDS/MPN). None of the patients with MDS/MPN were mutated for STAT3 D661Y or Y640F. While heterogeneous, the group of MDS patients with STAT3 mutations was characterized by a lower bone marrow cellularity (43% vs 61%; P < .0.034) and lower neutrophil count. There was also a trend for a higher frequency of del7/7q and likelihood of having low or intermediate risk based on the International Prognosis Scoring System (Table 3). The clinical detection of an abnormal CTL population did correlate with the size of the mutated clone. Using the sensitivity of the Sanger technique vs deep sequencing as a surrogate marker for determining the clone size, none of the mutant cases confirmed only by amplicon sequencing could be detected as a CTL expansion during the clinical workup at presentation. However, no differences in the severity of the disease, depth of cytopenias, bone marrow cellularity, or development of karyotypic abnormalities could be found between clinical and subclinical cases (P > .150). No differences in survival were found when comparing AA and MDS patients with or without STAT3 mutation (Figure 3).
Table 3.
Comparison of clinical characteristics between MDS patients according to the STAT3 mutation status
| Variable | MDS with STAT3 mutation, n = 18 | MDS without STAT3 mutation, n = 349 | P |
|---|---|---|---|
| Age, years (median, range) | 69 (27-70) | 66 (56-72) | .07 |
| Sex, % | |||
| Male | 59 | 56 | .6 |
| Female | 41 | 44 | |
| White blood cell count, ×109/L (median, IQR) | 5.1 (2.1-13.5) | 11.9 (2.7-25.7) | .04 |
| Absolute neutrophil count, ×109/L (median, IQR) | 1.23 | 4.41 (1-6.8) | ≤.001 |
| Hemoglobin, g/dL (mean ± SD) | 9.3 ± 1.9 | 9.9 ± 1.3 | .180 |
| Mean corpuscular volume, fL (median, IQR) | 95 (85-99) | 94 (86-104.8) | .875 |
| Platelets, ×109/L (median, IQR) | 139 (22-171) | 126 (24-186) | .743 |
| BM cellularity, % (median, IQR) | 43 (22-61) | 61 (45-90) | .034 |
| Abnormal cytogenetics, % | 52 | 51 | .9 |
| Patients with chromosome 7 abnormality, % | 32 | 9 | .09 |
| IPSS* | |||
| Low/intermediate-1 | 44 | 66 | .074 |
| Intermediate-2/high | 56 | 34 |
Boldface indicates significant P values.
BM, bone marrow; IPSS, International Prognostic Scoring System; IQR, interquartile range; SD, standard deviation.
Forty-eight patients with MDS/MPN disease were excluded from this subanalysis.
Figure 3.
Differences in survival outcomes in acquired BMF patients according to the presence of a STAT3 mutated clone. (A-B) No difference is seen in survival when comparing AA and MDS patients with or without the STAT3-mutated CTL clone.
We then expanded the study of concomitant mutations by traditional methods to 10 STAT3-mutated MDS cases for screening of mutations of 17 well-known genes in myeloid cancers (ie, TET2, IDH1, IDH2, DNMT3A, CBL, RUNX1, ASXL1, UTX [KDM6A], EZH2, NRAS, KRAS, TP53, JAK2, SF3B1, U2AF1, SRSF2, and SETBP1). Of note, the most frequent concomitant lesions were found in NRAS (50% of cases) (supplemental Figure 1).
Discussion
The clinical relationship between acquired, immune-mediated BMF syndromes and CTL “LGL-like” clonal expansions has been known since the description of this latter entity2,19 and has inspired various theories of immune-mediated suppression of hematopoiesis for several decades. Because of the nonaberrant phenotype of the LGL expansion and the evidence that patients with T-LGL leukemia may display a small, unnoticed, population of circulating LGLs,20,21 it is believed that this association may be underappreciated, similar to the fact that LGL may be underestimated in relation to rheumatoid arthritis.
In this study, we took advantage of the recent description of STAT3 mutations in classical LGL clonal lymphoproliferations to test whether clonal transformation through acquisition of somatic mutations (ie, somatic STAT3 mutations in autoimmune T cells) could be also be present in AA and MDS patients.4,5 Our analysis shows that somatic STAT3 clones can be found not only in AA and MDS patient with a known concomitant LGL, but also in a proportion of unsuspected cases. Additionally, the issue of whether STAT3 mutations reside within Vβ expansions is of essential importance. We show here how cells from a MDS patient harboring the D661Y STAT3 mutation were sorted by Vβ1 (TRBV9) into positive and negative fractions. ARMS-PCR results from both fractions clearly showed that the mutation is present in the Vβ1+ population. This issue was also addressed previously, in which the mutation in a T-LGL patient was present exclusively in the CD3+ fraction of LGL patients.11
Similar to the discovery of somatic phosphatidylinositol glycan class A (PIG-A) mutations as a clonal marker in paroxysmal nocturnal hemoglobinuria (PNH) and subsequently in other immune-mediated marrow failure syndrome clones,22 detection of STAT3 mutations in CTLs of idiopathic AA and MDS opens interesting concepts as to the pathogenesis of the autoimmune process (immune-mediated production cytopenia) in these diseases. However, although the presence of the PNH clone reflects hematopoietic cells’ secondary escape to the immune damage, the STAT3 LGL clone may be postulated to represent the cause for persistence of autoimmune clones and thereby of immune attack itself responsible for the primary induction of BMF. In MDS, the presence of STAT3 clones may point toward the possibility that, to a degree, cytopenias are related to a CTL-mediated process and may be potentially amenable to IST.
STAT3 mutations can be used as a sensitive clonal marker. We used ultrasensitive techniques and found an additional 7% of AA patients carrying an unsuspected STAT3-mutated LGL clone. The presence of an acquired molecular lesion in these clones might help to distinguish truly transformed populations from reactive immune responses that could appear in the context of these BMF. In addition, improvement in identifying true, concomitant malignant clones could help us to define concrete clinical associations; we show here an interesting trend for better therapeutic responses in those patients harboring the mutation. This trend needs to be investigated in a larger series of mutated patients. The association of STAT3 mutant CTLs with the presence of HLA-DR15 is in further agreement with the higher proportion of responders in this group.23
As in acquired AA, several cytokine abnormalities have been described in MDS that suggest an underlying immune process.24-26 Additionally, skewed T-cell populations are also observed in some MDS cases, the reduction of which parallels the response to IST.5,27,28 In our cohort, we found an additional 9 patients with subclinical STAT3 mutant CTL clones in addition to the 9 patients with known overt LGL harboring the same mutations; in total, 5% of our MDS cohort presented with STAT3 mutant CTL expansions and were characterized by profound neutropenia, a lower degree of bone marrow cellularity, and a higher frequency of developing chromosome 7 abnormalities that often evolve in the context of immune-mediated AA.29 In this series, 5 of 18 STAT3 mutant MDS cases presented with del7. In addition, we expanded our molecular characterization by Sanger sequencing 10 STAT3-mutated MDS cases. Of note, we found a high proportion of concomitant NRAS mutations. Ras oncogenes mutations are one of the most common gain-of-function mutations found in human cancers,30 and they have been described in 15% to 20% of acute myeloid leukemia, 10% to 50% of MDS, and 11% to 27% of MDS/MPN cases.31-33 Although the number of patients studied is small, whether cells escaping autoimmunity may acquire a survival advantage through NRAS anomalies emerges as an interesting hypothesis to explore.
We acknowledge that the described screening-confirmation method confers both strength and weakness to our study. The ARMS system is rapid and reliable.14 The use of 2 reactions with internal controls ensures that false-negative results are not obtained, yet a certain percentage of false-positive amplicons has been reported.34,35 To not miss cases found by the high analytical sensitivity of ARMS-PCR, we used 3 different methods for confirmation. We are aware that because only 2 (ie, most frequent) canonical STAT3 mutations were tested, CTL clones with other mutations may remain undetected. In fact, the less common mutations of STAT3 and the recently described lesions in STAT5b could increase the detection rate.10,11,36 During high-throughput sequencing data generation or analysis, errors can occur related to, among others, the fidelity of the polymerase used, biases for high GC content areas, or asynchronous sequencing. These potential background errors could be considered actual sequence variants in the downstream analysis.37 We tried to avoid those errors by a strict threshold for positive cases, acknowledging that we give priority to specificity in this study, limiting the potential false-positive cases.
In summary, our study shows that STAT3 mutant CTL clones can be found in a small proportion of patients with acquired BMF syndromes, including AA and hypocellular MDS. Immortalization/transformation via acquisition of a promoting somatic mutation in CTLs may be a mechanism not restricted to immune-mediated BMF; it might be involved in classical autoimmune diseases such as rheumatoid arthritis. Sensitive detection methods may facilitate the answer to this fascinating theory.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health grants R01 CA98472/CA/NCI NIH HHS (to T.P.L.) and 2K24HL077522/HL/NHLBI NIH HHS (to J.P.M.), a grant from Fundacion CajaMadrid (to A.J.), and grants from the Academy of Finland, the Finnish Cancer Societies, the Sigrid Juselius Foundation, the National Clinical Graduate School, and the Signe and Ane Gyllenberg Foundation.
Footnotes
The online version of this article contains a data supplement.
There is an Inside Blood commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked ‘‘advertisement’’ in accordance with 18 USC section 1734.
Authorship
Contribution: J.P.M., H.M., M.J.C., and A.J. were responsible for overall design, data collection, data analysis, data interpretation, statistical analysis, manuscript preparation, writing, and completion and final approval of manuscript; and H.R., I.G.-S., T.O., K.M., B.P., A.K., M.A., H.D.H., N.H., F.L., S.L., D.Z., P.E., A.E.L., A.T., C.N., A.W.-F., G.J.M., A.F.L., S.M., and T.P.L. gathered data and edited and approved the final manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jaroslaw P. Maciejewski, Taussig Cancer Institute/R40, 9500 Euclid Ave, Cleveland, OH 44195; e-mail: maciejj@ccf.org.
References
- 1.Loughran TP., Jr Clonal diseases of large granular lymphocytes. Blood. 1993;82(1):1–14. [PubMed] [Google Scholar]
- 2.Gastl G, Niederwieser D, Marth C, et al. Human large granular lymphocytes and their relationship to natural killer cell activity in various disease states. Blood. 1984;64(1):288–295. [PubMed] [Google Scholar]
- 3.Saunthararajah Y, Molldrem JL, Rivera M, et al. Coincident myelodysplastic syndrome and T-cell large granular lymphocytic disease: clinical and pathophysiological features. Br J Haematol. 2001;112(1):195–200. doi: 10.1046/j.1365-2141.2001.02561.x. [DOI] [PubMed] [Google Scholar]
- 4.Risitano AM, Maciejewski JP, Green S, Plasilova M, Zeng W, Young NS. In-vivo dominant immune responses in aplastic anaemia: molecular tracking of putatively pathogenetic T-cell clones by TCR beta-CDR3 sequencing. Lancet. 2004;364(9431):355–364. doi: 10.1016/S0140-6736(04)16724-X. [DOI] [PubMed] [Google Scholar]
- 5.Wlodarski MW, Gondek LP, Nearman ZP, Plasilova M, Kalaycio M, Hsi ED, Maciejewski JP. Molecular strategies for detection and quantitation of clonal cytotoxic T-cell responses in aplastic anemia and myelodysplastic syndrome. Blood. 2006;108(8):2632–2641. doi: 10.1182/blood-2005-09-3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frickhofen N, Kaltwasser JP, Schrezenmeier H, et al. The German Aplastic Anemia Study Group. Treatment of aplastic anemia with antilymphocyte globulin and methylprednisolone with or without cyclosporine. N Engl J Med. 1991;324(19):1297–1304. doi: 10.1056/NEJM199105093241901. [DOI] [PubMed] [Google Scholar]
- 7.Sloand EM, Wu CO, Greenberg P, Young N, Barrett J. Factors affecting response and survival in patients with myelodysplasia treated with immunosuppressive therapy. J Clin Oncol. 2008;26(15):2505–2511. doi: 10.1200/JCO.2007.11.9214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zeng W, Maciejewski JP, Chen G, Young NS. Limited heterogeneity of T cell receptor BV usage in aplastic anemia. J Clin Invest. 2001;108(5):765–773. doi: 10.1172/JCI12687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maciejewski J, Selleri C, Anderson S, Young NS. Fas antigen expression on CD34+ human marrow cells is induced by interferon gamma and tumor necrosis factor alpha and potentiates cytokine-mediated hematopoietic suppression in vitro. Blood. 1995;85(11):3183–3190. [PubMed] [Google Scholar]
- 10.Koskela HL, Eldfors S, Ellonen P, et al. Somatic STAT3 mutations in large granular lymphocytic leukemia. N Engl J Med. 2012;366(20):1905–1913. doi: 10.1056/NEJMoa1114885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jerez A, Clemente MJ, Makishima H, et al. STAT3 mutations unify the pathogenesis of chronic lymphoproliferative disorders of NK cells and T-cell large granular lymphocyte leukemia. Blood. 2012;120(15):3048–3057. doi: 10.1182/blood-2012-06-435297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Swerdlow SH. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Lyon, France: International Agency for Research on Cancer; 2008. [Google Scholar]
- 13.Marsh JC, Ball SE, Cavenagh J, et al. British Committee for Standards in Haematology. Guidelines for the diagnosis and management of aplastic anaemia. Br J Haematol. 2009;147(1):43–70. doi: 10.1111/j.1365-2141.2009.07842.x. [DOI] [PubMed] [Google Scholar]
- 14.Newton CR, Graham A, Heptinstall LE, et al. Analysis of any point mutation in DNA. The amplification refractory mutation system (ARMS). Nucleic Acids Res. 1989;17(7):2503–2516. doi: 10.1093/nar/17.7.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dunbar AJ, Gondek LP, O’Keefe CL, et al. 250K single nucleotide polymorphism array karyotyping identifies acquired uniparental disomy and homozygous mutations, including novel missense substitutions of c-Cbl, in myeloid malignancies. Cancer Res. 2008;68(24):10349–10357. doi: 10.1158/0008-5472.CAN-08-2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jankowska AM, Szpurka H, Tiu RV, et al. Loss of heterozygosity 4q24 and TET2 mutations associated with myelodysplastic/myeloproliferative neoplasms. Blood. 2009;113(25):6403–6410. doi: 10.1182/blood-2009-02-205690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Makishima H, Jankowska AM, McDevitt MA, et al. CBL, CBLB, TET2, ASXL1, and IDH1/2 mutations and additional chromosomal aberrations constitute molecular events in chronic myelogenous leukemia. Blood. 2011;117(21):e198–e206. doi: 10.1182/blood-2010-06-292433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gómez-Seguí I, Makishima H, Jerez A, et al. Novel recurrent mutations in the RAS-like GTP-binding gene RIT1 in myeloid malignancies [published online ahead of print June 14, 2013]. Leukemia. doi: 10.1038/leu.2013.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Go RS, Tefferi A, Li CY, Lust JA, Phyliky RL. Lymphoproliferative disease of granular T lymphocytes presenting as aplastic anemia. Blood. 2000;96(10):3644–3646. [PubMed] [Google Scholar]
- 20.Evans HL, Burks E, Viswanatha D, Larson RS. Utility of immunohistochemistry in bone marrow evaluation of T-lineage large granular lymphocyte leukemia. Hum Pathol. 2000;31(10):1266–1273. doi: 10.1053/hupa.2000.19298. [DOI] [PubMed] [Google Scholar]
- 21.Semenzato G, Zambello R, Starkebaum G, Oshimi K, Loughran TP., Jr The lymphoproliferative disease of granular lymphocytes: updated criteria for diagnosis. Blood. 1997;89(1):256–260. [PubMed] [Google Scholar]
- 22.Bessler M, Mason PJ, Hillmen P, et al. Paroxysmal nocturnal haemoglobinuria (PNH) is caused by somatic mutations in the PIG-A gene. EMBO J. 1994;13(1):110–117. doi: 10.1002/j.1460-2075.1994.tb06240.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saunthararajah Y, Nakamura R, Nam JM, et al. HLA-DR15 (DR2) is overrepresented in myelodysplastic syndrome and aplastic anemia and predicts a response to immunosuppression in myelodysplastic syndrome. Blood. 2002;100(5):1570–1574. [PubMed] [Google Scholar]
- 24.Shetty V, Mundle S, Alvi S, et al. Measurement of apoptosis, proliferation and three cytokines in 46 patients with myelodysplastic syndromes. Leuk Res. 1996;20(11-12):891–900. doi: 10.1016/s0145-2126(96)00008-2. [DOI] [PubMed] [Google Scholar]
- 25.Kitagawa M, Saito I, Kuwata T, et al. Overexpression of tumor necrosis factor (TNF)-alpha and interferon (IFN)-gamma by bone marrow cells from patients with myelodysplastic syndromes. Leukemia. 1997;11(12):2049–2054. doi: 10.1038/sj.leu.2400844. [DOI] [PubMed] [Google Scholar]
- 26.Kornblau SM, McCue D, Singh N, Chen W, Estrov Z, Coombes KR. Recurrent expression signatures of cytokines and chemokines are present and are independently prognostic in acute myelogenous leukemia and myelodysplasia. Blood. 2010;116(20):4251–4261. doi: 10.1182/blood-2010-01-262071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kochenderfer JN, Kobayashi S, Wieder ED, Su C, Molldrem JJ. Loss of T-lymphocyte clonal dominance in patients with myelodysplastic syndrome responsive to immunosuppression. Blood. 2002;100(10):3639–3645. doi: 10.1182/blood-2002-01-0155. [DOI] [PubMed] [Google Scholar]
- 28.Nishimura M, Yamada T, Andoh T, et al. Granulocyte colony-stimulating factor (G-CSF) dependent hematopoiesis with monosomy 7 in a patient with severe aplastic anemia after ATG/CsA/G-CSF combined therapy. Int J Hematol. 1998;68(2):203–211. doi: 10.1016/s0925-5710(98)00047-4. [DOI] [PubMed] [Google Scholar]
- 29.Jerez A, Sugimoto Y, Makishima H, et al. Loss of heterozygosity in 7q myeloid disorders: clinical associations and genomic pathogenesis. Blood. 2012;119(25):6109–6117. doi: 10.1182/blood-2011-12-397620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bos JL. ras oncogenes in human cancer: a review. Cancer Res. 1989;49(17):4682–4689. [PubMed] [Google Scholar]
- 31.Kadia TM, Kantarjian H, Kornblau S, et al. Clinical and proteomic characterization of acute myeloid leukemia with mutated RAS. Cancer. 2012;118(22):5550–5559. doi: 10.1002/cncr.27596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muramatsu H, Makishima H, Maciejewski JP. Chronic myelomonocytic leukemia and atypical chronic myeloid leukemia: novel pathogenetic lesions. Semin Oncol. 2012;39(1):67–73. doi: 10.1053/j.seminoncol.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jankowska AM, Makishima H, Tiu RV, et al. Mutational spectrum analysis of chronic myelomonocytic leukemia includes genes associated with epigenetic regulation: UTX, EZH2, and DNMT3A. Blood. 2011;118(14):3932–3941. doi: 10.1182/blood-2010-10-311019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ayyadevara S, Thaden JJ, Shmookler Reis RJ. Discrimination of primer 3′-nucleotide mismatch by taq DNA polymerase during polymerase chain reaction. Anal Biochem. 2000;284(1):11–18. doi: 10.1006/abio.2000.4635. [DOI] [PubMed] [Google Scholar]
- 35.Latorra D, Campbell K, Wolter A, Hurley JM. Enhanced allele-specific PCR discrimination in SNP genotyping using 3′ locked nucleic acid (LNA) primers. Hum Mutat. 2003;22(1):79–85. doi: 10.1002/humu.10228. [DOI] [PubMed] [Google Scholar]
- 36.Rajala HL, Eldfors S, Kuusanmaki H, et al. Discovery of somatic STAT5b mutations in large granular lymphocytic leukemia. Blood. 2013;121(22):4541–4550. doi: 10.1182/blood-2012-12-474577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen K, Wallis JW, McLellan MD, et al. BreakDancer: an algorithm for high-resolution mapping of genomic structural variation. Nat Methods. 2009;6(9):677–681. doi: 10.1038/nmeth.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.