Abstract
Genetic variation patterns within and between species may change along geographic gradients and at different spatial scales. This was revealed by microsatellite data at 29 loci obtained from 119 accessions of three Oryza series Sativae species in Asia Pacific: Oryza nivara Sharma and Shastry, O. rufipogon Griff., and O. meridionalis Ng. Genetic similarities between O. nivara and O. rufipogon across their distribution are evident in the clustering and ordination results and in the large proportion of shared alleles between these taxa. However, local-level species separation is recognized by Bayesian clustering and neighbor-joining analyses. At the regional scale, the two species seem more differentiated in South Asia than in Southeast Asia as revealed by FST analysis. The presence of strong gene flow barriers in smaller spatial units is also suggested in the analysis of molecular variance (AMOVA) results where 64% of the genetic variation is contained among populations (as compared to 26% within populations and 10% among species). Oryza nivara (HE = 0.67) exhibits slightly lower diversity and greater population differentiation than O. rufipogon (HE = 0.70). Bayesian inference identified four, and at a finer structural level eight, genetically distinct population groups that correspond to geographic populations within the three taxa. Oryza meridionalis and the Nepalese O. nivara seemed diverged from all the population groups of the series, whereas the Australasian O. rufipogon appeared distinct from the rest of the species.
Keywords: Species divergence, SSR diversity, sympatric populations, wild Oryza
Introduction
The unwavering pursuit to fully understand the rice gene pool is reflected in the growing number of publications on Oryza rufipogon and O. nivara. These two closest relatives of Asian cultivated rice (O. sativa L.) are morphologically distinct (Ng et al. 1981; Uga et al. 2003; Banaticla-Hilario 2012), whereas genetic isolation is effected by differences in habitat, mating system, and flowering time. Their geographic distributions show overlap in tropical continental Asia with O. rufipogon extending southeastward to insular Southeast Asia and Australasia.
Oryza nivara is variously treated as a distinct species (Sharma and Shastry 1965; Ng et al. 1981; Lu 1999; Lu et al. 2001) or as an ecotype of O. rufipogon (Tateoka 1963; Oka 1988; Vaughan et al. 2003). This taxonomic ambivalence is also reflected in incongruencies between the results of different molecular data, where isozymes (Second 1985), random amplification of polymorphic DNAs (Ren et al. 2003), allozymes and restriction fragment length polymorphisms (Cai et al. 2004), transposon display markers (Kwon et al. 2006), tourist sequences (Iwamoto et al. 1999), miniature inverted-repeat transposable elements in amplified fragment length polymorphisms (Park et al. 2003), microsatellites (Ren et al. 2003), sequence tagged sites (Huang et al. 2012a), and various genes sequences (Zhu and Ge 2005; Zhou et al. 2008; Zheng and Ge 2010) did not detect divergence between O. nivara and O. rufipogon, whereas AFLPs (Aggarwal et al. 1999), microsatellites (Kuroda et al. 2007; Singh et al. 2013), combined sequences from chloroplast, mitochondrial and nuclear DNA (Duan et al. 2007), and single nucleotide polymorphisms (SNPs) (Xu et al. 2012) did indicate a separation at species level. In this study, the annual taxon is tentatively considered as a distinct species.
Genetic differentiation between O. nivara and O. rufipogon has been examined globally using populations sampled across the species' total geographic distribution (Zheng and Ge 2010; Huang et al. 2012a) and at a regional scale by comparing patterns in South and Southeast Asia (Lu et al. 2008). Local-scale studies were conducted by Kuroda et al. (2007) and Singh et al. (2013) using Lao and Indian populations, respectively. However, spatial patterns of intra- and interspecific differentiation remain unclear for these two taxa.
The closely related taxon, O. meridionalis, is a genetically distinct species (Xu et al. 2005; Kwon et al. 2006) that exhibits a similar life cycle, breeding habit, phenology, and habitat to that of O. nivara, but geographically overlaps only with the southern limit of O. rufipogon. Therefore, it seems worthwhile to compare the genetic differences between O. meridionalis and O. rufipogon with those between O. nivara and O. rufipogon.
A number of questions on the relationship between O. nivara and O. rufipogon remain: Are the observed genetic similarities/differences consistent along spatial gradients and across varying geographical units? Are locally sympatric populations of O. nivara and O. rufipogon more differentiated than the nonsympatric ones? How does geography influence the variations within and between the three Oryza species?
In an effort to answer these questions and uncover underlying spatial variation patterns, this study analyzes locally sympatric accession pairs (i.e., populations of different species collected from the same locality) of O. nivara and O. rufipogon from across South Asia and continental Southeast Asia and of O. meridionalis and O. rufipogon in Australasia (New Guinea and Australia) as well as O. rufipogon populations from insular Southeast Asia. These three taxa, along with cultivated rice compose Oryza series Sativae in the Asia-Pacific area.
For this study we use simple sequence repeat (SSR) markers to: (1) determine global-, regional-, and local-scale differentiation between O. nivara and O. rufipogon; (2) infer geographic population groups in Asia-Pacific Oryza series Sativae; and (3) assess genetic diversity at the population group and species level.
Material and Methods
Plant material
One hundred nineteen accessions from the International Rice Genebank (IRG) at the International Rice Research Institute (IRRI) in the Philippines were selected to represent sympatric populations of O. nivara and O. rufipogon and of O. meridionalis and O. rufipogon across their distribution range, as well as O. rufipogon populations that are nonsympatric to both annual species (Table S1). Due to limited availability and germination issues, only one accession from China was sampled. The same plant material was used in a previous phenotyping experiment (Banaticla-Hilario 2012) wherein some accessions were tentatively classified as intermediate forms (i.e., intermediate between two wild species or between O. sativa and a wild species) (Table S1). We included these intermediate forms in this study to determine their genetic affinity with the other Oryza series Sativae species in Asia. IRGC 81837, 89228, and 106152 displayed two different plant types within the accession and were thus represented as two separate subpopulations (N26A and N26B, R5A and R5B, and R29A and R29B, respectively). Six O. sativa accessions were also included for comparison (Table S1). The plant material was grown in the Genetic Resources Center screenhouse at IRRI, the Philippines. Leaf samples were harvested from five individual plants per accession.
Genomic DNA was extracted from fresh leaf samples by applying the modified CTAB (cetyl trimethyl ammonium bromide) extraction protocol (Fulton et al. 1995). The DNA samples were quantified using spectrophotometry (NanoDrop™ 1000 spectrophotometer, Thermo Fisher Scientific, Wilmington, DE) and gel densitometry (using Lambda DNA as a standard), and then normalized to 5 ng/μL concentration.
SSR genotyping
The markers used (Table 1) were from the panel of 30 standard SSR markers developed by the Generation Challenge Program for rice diversity analysis (http://gramene.org/markers/microsat/50_ssr.html). However, RM514 did not amplify well in most of the samples and was dropped from the analysis.
Table 1.
Basic information and overall diversity of the 29 SSR markers used in the study
| Marker | Chr | Oryza sativa varietal group (germplasm) | Motif | AT (°C) | AN | RAN | MAF | PIC |
|---|---|---|---|---|---|---|---|---|
| RM237 | 1 | indica (IR36) | (CT)18 | 55 | 23 | 17 | 0.1860 | 0.9056 |
| RM283 | 1 | indica (IR36) | (GA)18 | 61 | 20 | 13 | 0.2140 | 0.8869 |
| RM431 | 1 | japonica (Nipponbare) | (AG)16 | 55 | 19 | 11 | 0.1280 | 0.8925 |
| RM495 | 1 | japonica (Nipponbare) | (CTG)7 | 55 | 4 | 0 | 0.7480 | 0.3870 |
| RM154 | 2 | unknown | (GA)21 | 61 | 24 | 19 | 0.2480 | 0.8416 |
| RM452 | 2 | japonica (Nipponbare) | (GTC)9 | 61 | 9 | 5 | 0.7500 | 0.3985 |
| OSR13 | 3 | unknown | (GA)n | 53 | 19 | 12 | 0.2080 | 0.8783 |
| RM338 | 3 | indica (IR36) | (CTT)6 | 55 | 4 | 2 | 0.9280 | 0.1293 |
| RM124 | 4 | japonica (Nipponbare) | (TC)10 | 67 | 9 | 6 | 0.7100 | 0.4477 |
| RM161 | 5 | japonica (Nipponbare) | (AG)20 | 61 | 17 | 11 | 0.2570 | 0.8442 |
| RM413 | 5 | japonica (Nipponbare) | (AG)11 | 53 | 22 | 15 | 0.2580 | 0.8745 |
| RM507 | 5 | japonica (Nipponbare) | (AAGA)7 | 55 | 7 | 4 | 0.7320 | 0.3984 |
| RM133 | 6 | japonica (Nipponbare) | (CT)8 | 63 | 7 | 3 | 0.4470 | 0.6093 |
| RM162 | 6 | unknown | (AC)20 | 61 | 22 | 15 | 0.2790 | 0.8467 |
| RM118 | 7 | japonica (Nipponbare) | (GA)8 | 67 | 9 | 6 | 0.4000 | 0.6216 |
| RM125 | 7 | japonica (Nipponbare) | (GCT)8 | 63 | 13 | 6 | 0.2420 | 0.8362 |
| RM455 | 7 | japonica (Nipponbare) | (TTCT)5 | 57 | 3 | 1 | 0.8970 | 0.1691 |
| RM44 | 8 | indica (IR36) | (GA)16 | 53 | 18 | 7 | 0.1690 | 0.8897 |
| RM152 | 8 | unknown | (GGC)10 | 53 | 9 | 5 | 0.4450 | 0.6453 |
| RM408 | 8 | japonica (Nipponbare) | (CT)13 | 55 | 11 | 8 | 0.4150 | 0.6948 |
| RM447 | 8 | japonica (Nipponbare) | (CTT)8 | 55 | 15 | 8 | 0.1610 | 0.8723 |
| RM284 | 8 | indica (IR36) | (GA)8 | 55 | 10 | 4 | 0.5990 | 0.5447 |
| RM433 | 8 | japonica (Nipponbare) | (AG)13 | 53 | 18 | 11 | 0.2030 | 0.8591 |
| RM215 | 9 | indica (IR36) | (CT)16 | 55 | 21 | 13 | 0.1460 | 0.8904 |
| RM316 | 9 | indica (IR36) | (GT)8-(TG)9(TTTG)4(TG)4 | 55 | 32 | 28 | 0.1900 | 0.9048 |
| RM271 | 10 | indica (IR36) | (GA)15 | 55 | 23 | 14 | 0.1590 | 0.9135 |
| RM484 | 10 | japonica (Nipponbare) | (AT)9 | 55 | 8 | 3 | 0.3080 | 0.7381 |
| RM536 | 11 | japonica (Nipponbare) | (CT)16 | 55 | 12 | 7 | 0.3540 | 0.7064 |
| RM277 | 12 | indica (IR36) | (GA)11 | 55 | 9 | 5 | 0.3510 | 0.6941 |
| Mean | 14 | 9 | 0.3909 | 0.6934 |
Chr, chromosome number; AT, annealing temperature; AN, number of alleles; RAN, number of rare alleles (allele frequency ≤5%); MAF, frequency of major allele; PIC, polymorphism information content.
Polymerase chain reaction (PCR) was conducted in 20 μL reaction volume composed of 5.92 μL sterilized ultrapure water, 2 μL each of 10× MgCl2 free buffer, 10 mmol/L deoxynucleotide triphosphates, and 25 mmol/L MgCl2 (iNtRON Biotechnology, Kyungki-Do, South Korea), 0.08 μL of 5U/μL i-Taq™ DNA polymerase (iNtRON Biotechnology), 1 μL of 1 μmol/L labeled M13 forward primer (IRDye 700 or 800, LI-COR Biosciences, Lincoln, NE), 1 μL of 1 μmol/L M13-tailed SSR forward primer (Invitrogen, Carlsbad, CA), 2 μL of 1 μmol/L SSR reverse primer (Invitrogen), and 4 μL of genomic DNA. The program started with denaturation at 95°C for 2 min, succeeded by 32 cycles of denaturation at 95°C (30 sec), annealing at 55°C (30 sec) and elongation at 72°C (50 sec), then followed by a 2 min final extension step at 72°C. The annealing temperature was adjusted to match the optimal value for each marker as indicated in Table 1.
PCR products were multiplexed by combining 2 μL each of IRDye 700- and IRDye 800-labeled samples, 5 μL sterilized nanopure water, and 5 μL loading dye. Gel electrophoresis was performed on the 4300 LI-COR DNA analyzer system. The LI-COR IRDye 50–350 bp size standard ladder was used to estimate allele size. The gels were analyzed and scored with the SAGA Generation 2 software (LI-COR, Biosciences, Lincoln, NE).
Data analyses
Genetic diversity measures were estimated for markers, populations, inferred population groups, and species. The number of alleles and rare alleles (frequency ≤5%), frequency of major allele (allele with the highest frequency), observed heterozygosity, unbiased estimate of gene diversity (Weir 1996), polymorphism information content (PIC), and inbreeding coefficient (FIS) were obtained using PowerMarker 3.25 (Liu and Muse 2005). Allelic richness values were determined with FSTAT (Goudet 2001).
A cluster analysis was conducted with PowerMarker 3.25 (Liu and Muse 2005). A neighbor-joining (NJ) tree based on C.S. Chord distance (Cavalli-Sforza and Edwards 1967) was constructed and bootstrapping with 1000 replicates was performed. The output trees were viewed and edited with MEGA 5.05 (Tamura et al. 2011).
Principal coordinate analysis (PCoA) of C.S. Chord distance matrices between species across distribution was performed with GenAlEx 6.4 (Peakall and Smouse 2006).
Genetically distinct populations were inferred by applying the nonspatial and spatially explicit Bayesian clustering algorithms of STRUCTURE 2.2 (Pritchard et al. 2000; Falush et al. 2003) and TESS 2.3.1 (Durand et al. 2009), respectively.
A STRUCTURE model allowing for admixture and assuming correlated allele frequencies was implemented. Cluster values ranging from K = 1 to K = 15 were tested with 20 independent runs for each K with a burn-in of 20,000 iterations and run length of 20,000 iterations per run. The 10 runs with the highest posterior probability ln P(D) values were selected from each K and their average ln P(D) were used in calculating the delta K (ΔK), a statistic based on the rate of changes in the likelihood distribution between successive K values (Evanno et al. 2005). The cluster value corresponding to the highest peak in the ΔK plot is considered as the appropriate cluster solution.
As the TESS program uses spatial prior information, the geographically underrepresented set of O. sativa accessions were excluded from the analysis. Misidentified accessions detected by NJ, PCoA, and STRUCTURE analysis were also removed from the runs. Prior to analysis, the “generate spatial coordinates” option of TESS was used to create individual sample coordinates based on accession coordinates. In the TESS runs, the conditional autoregressive (CAR) model of admixture (admixture parameter = 1.0, spatial interaction parameter = 0.6) was implemented with a linear trend surface. The maximal number of clusters was set to range from Kmax = 2 to Kmax = 10. Each Kmax was tested with 100 runs, and each run had 20,000 burn-in sweeps followed by another 30,000 sweeps. To determine the appropriate number of TESS clusters, the deviance information criterion (DIC) should be analyzed and the stability of the bar plots should be considered (Durand et al. 2009). From each Kmax, the 10 runs with the lowest DIC were selected and their mean DIC values were plotted against Kmax. The optimum cluster solution is the Kmax value that coincides with the plateau of the DIC curve.
For each K/Kmax (from K = 2 to K = 8), the 10 STRUCTURE runs with maximal ln (P[D]) values as well as the 10 TESS runs with the lowest DIC values were aligned and averaged using CLUMPP 1.1.2 (Jakobsson and Rosenberg 2007), employing the Greedy algorithm with 10,000 random permutations (except for K = 8 where the LargeKGreedy algorithm with 10,000 random permutations was used). The output files (bar graphs) were viewed and edited with the DISTRUCT software (Rosenberg 2004).
GenAlEx 6.4 (Peakall and Smouse 2006) was used in conducting an analysis of molecular variance (AMOVA) and in estimating pairwise FST values between inferred population groups.
Results
Overall microsatellite diversity in Asia-Pacific Oryza series Sativae
Across the 119 Oryza series Sativae populations from Asia Pacific, 417 alleles (62% of which are rare) were detected at the 29 SSR loci. The number of alleles per locus varied from three (RM455) to 32 (RM316), with an average of 14. The number of rare alleles ranged from zero (RM495) to 28 (RM316), with an average of nine. The most common allele in each locus had a mean frequency of 0.39 and varied from 0.13 (RM431) to 0.93 (RM338). The PIC values differed from 0.13 (RM338) to 0.91 (RM237 and RM271) and had a mean value of 0.69 (Table 1). Based on the mentioned parameters, the most diverse loci were RM154, RM271, RM237, and RM316, whereas the least diverse were RM338, RM455, and RM495.
Cluster analysis
The NJ tree (Fig. 1) shows the general tendency of accessions from the same species to cluster together. An admixed cluster composed of eight O. nivara, seven O. rufipogon, two O. sativa var. indica, and nine phenotypically intermediate accessions was also produced. However, the O. nivara and O. rufipogon clusters as well as the admixed cluster have very low bootstrap values (less than 50%) and only the O. meridionalis and Nepalese O. nivara clusters have strong bootstrap support (both with 99% bootstrap value). Oryza nivara and O. meridionalis join in a large cluster where the latter forms a distinct group that seems closer to the Southeast Asian populations of the former. The Nepalese O. nivara forms a separate branch within the admixed cluster. The aromatic and japonica populations of O. sativa group with several South Asian accessions of O. rufipogon, whereas the Australasian population joins a cluster composed of both South and Southeast Asian O. nivara. Three O. nivara populations from India (N21, N26A, and N26B) and an intermediate population from Sri Lanka (N37) separated from the annual species and form a cluster. Three O. nivara accessions (N39, N49, and N52) group with O. rufipogon while the intermediate populations are distributed among different clusters.
Figure 1.
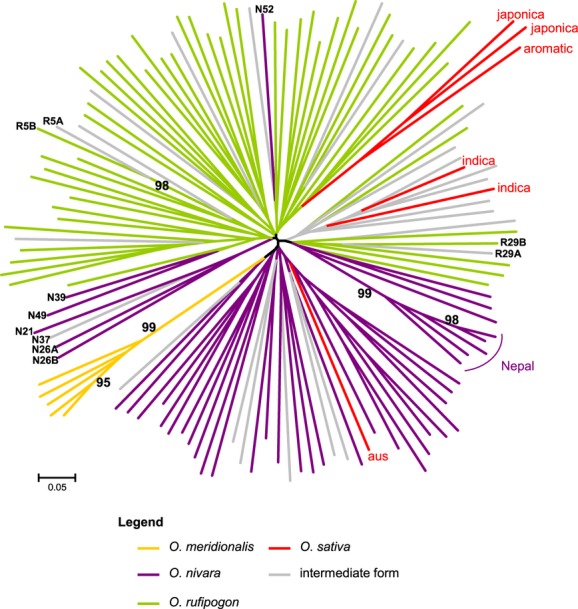
NJ tree of Oryza series Sativae populations based on C.S. Chord distance. Bootstrap values above 50% are displayed at the root of the supported cluster. The populations are colored according to their morphological classification.
Principal coordinate analysis
The first two principal coordinate axes reflect separate but partially overlapping clusters of O. nivara and O. rufipogon (Fig. 2A). Oryza meridionalis and Nepalese O. nivara accessions form distinct clusters isolated by axes 1 (21.83% proportion of variance) and 2 (19.10%). Oryza sativa accessions are distributed throughout the plot with aromatic and japonica populations joining O. rufipogon, Australasian, and one indica accession grouping with O. nivara, and the other indica accession in the middle of the O. nivara – O. rufipogon complex (Fig. 2A). The third and fourth principal coordinate axes do not separate O. nivara from O. rufipogon (Fig. 2B). Axis 3 (16.43%) isolates O. meridionalis, whereas axis 4 (15.52%) separates the Nepalese O. nivara from the rest of the taxa. The succeeding principal coordinate axes displayed uninformative clustering patterns.
Figure 2.
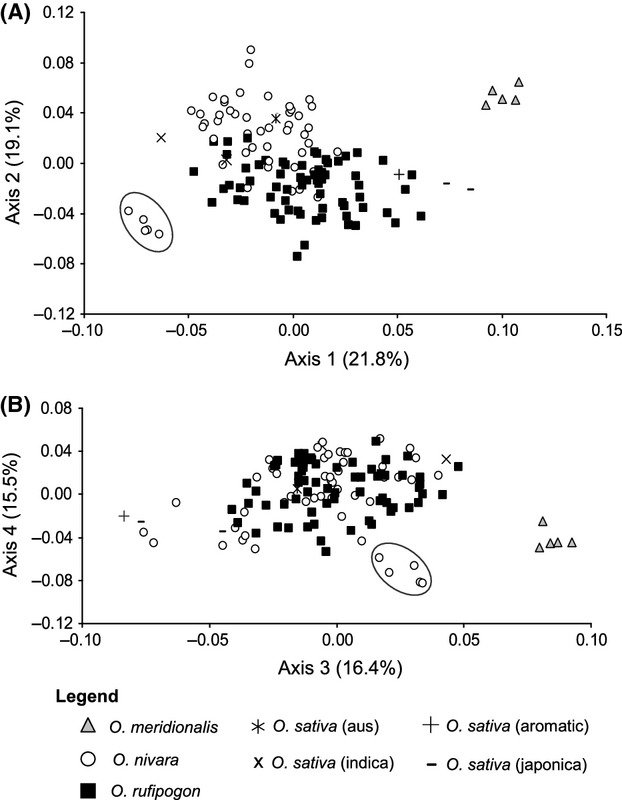
Principal coordinate plots revealing the genetic distinctiveness among Oryza rufipogon, O. meridionalis, O. nivara, and O. sativa accessions. Oryza nivara accessions from Nepal are encircled. (A) Axis 1 versus axis 2. (B) Axis 3 versus axis 4.
Bayesian clustering
TESS exhibits more consistent runs and produces more stable population clusters (with less fragmented members) than STRUCTURE as indicated in the average membership coefficients of each K (from K = 2 to K = 8) obtained by CLUMPP (Fig. S1). Across different K values in the STRUCTURE runs, populations N39 and N49 cluster with O. rufipogon, whereas R10, R43, and R50 are grouped with O. nivara. These apparently mislabelled populations also do not cluster with their supposed species groups in the NJ and PCA results and were excluded from the TESS runs.
The ΔK plot of the STRUCTURE runs displays distinct peaks at K = 2 (the highest value), K = 4, and K = 6 (Fig. 3A). However, K = 2 is rejected as an optimal cluster value as the cluster solution produced by STRUCTURE fails to distinguish O. meridionalis as a distinct population (Fig. S1). The relatively stable membership coefficient plots of both STRUCTURE and TESS runs at K = 4 (Fig. S1) suggest that four clusters optimally define the population structure of the data set. The four population groups depicted similarly by STRUCTURE and TESS are as follows: (C1) South Asian O. nivara; (C2) Southeast Asian O. nivara; (C3) continental and insular Asian O. rufipogon; and (C4) O. meridionalis and Australasian O. rufipogon (Figs. 4, 5). However, at K = 4, certain populations are occasionally swapped between different clusters across the 10 STRUCTURE runs (Fig. S2). Australasian O. rufipogon frequently joins O. meridionalis but also groups with either the rest of O. rufipogon or the japonica group of O. sativa in the other runs. The Nepalese and the Indian–Bangladeshi populations of O. nivara cluster together once, but in the rest of the runs, the former groups with either O. rufipogon or O. meridionalis while the latter joins either the Southeast Asian O. nivara or O. meridionalis (Fig. S2). The 10 TESS runs show more consistent population clustering at K = 4, with eight runs grouping the Australasian O. rufipogon with O. meridionalis and nine runs splitting O. nivara into South Asian and Southeast Asian populations (Fig. S3B).
Figure 3.
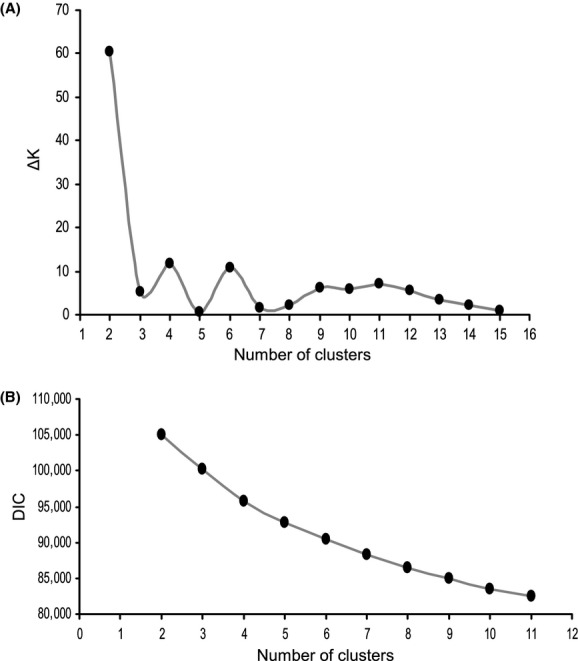
Criteria used in determining the appropriate cluster solution: (A) Delta K plot of STRUCTURE runs based on ln P(D) values; and (B) Plot of the mean DIC value of each TESS cluster solution. Plot lines were added to help visualize trends.
Figure 4.
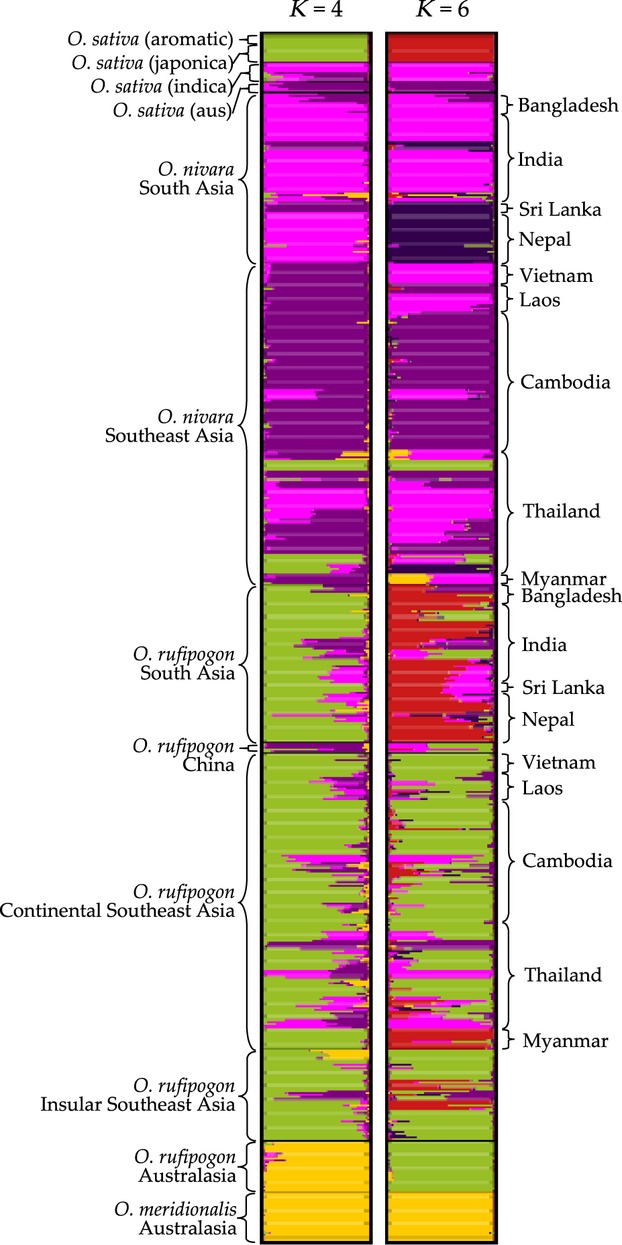
Population clusters of STRUCTURE at K = 4 and K = 6 (based on the modal clustering pattern). The predefined population assignments are on the left-hand side and the geographic origin of sympatric populations are on the right-hand side of the figure.
Figure 5.
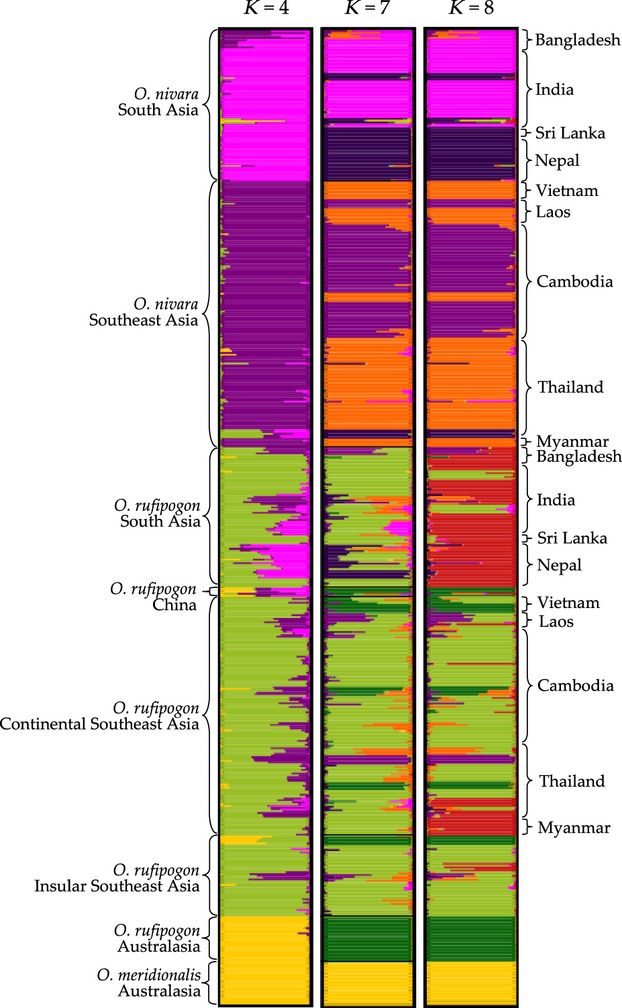
Population clusters of TESS at K = 4, K = 7, and K = 8 (based on the modal clustering pattern). The predefined population assignments are on the left-hand side and the geographic origin of sympatric populations are on the right-hand side of the figure.
At K = 6, the groups recognized by STRUCTURE are as follows (Fig. S2; Fig. 4): an O. meridionalis cluster (in 70% of the runs); two clusters in O. rufipogon (a South Asian group joined by the aromatic and japonica accessions of O. sativa and a Southeast Asian cluster in 40% of the runs); and three clusters in O. nivara (one cluster is predominantly Cambodian and groups with O. sativa aus, another cluster is mainly Nepalese, and the third cluster comprises the rest of O. nivara and is grouped with O. sativa indica, in 20% of the runs). Nevertheless, the output of the six-cluster solution of STRUCTURE (and even TESS) seems unstable because certain populations (particularly, the Australasian O. rufipogon and the non-Nepalese South Asian O. nivara) appear fragmented and/or are swapped between different clusters (Figs. S2, S3C).
The DIC plot of the TESS runs does not exhibit a well-defined plateau as the DIC values continuously decrease at higher Kmax (Fig. 3B). Across the 10 TESS runs, K = 8 shows the most consistent grouping of populations (Figs. S1, S3D). Moreover, higher Kmax values (K = 9 and K = 10), display less stable clustering and do not recognize additional distinct population clusters aside from the groups inferred at K = 8 (Fig. S3D and E). This indicates that the eight-cluster solution fits the lower population structure level of the data set. K = 4 and K = 7 also produce stable bar plots (Figs. S1, S3B and C) and will be discussed for comparison purposes. The clustering pattern of K = 4 was discussed previously with the STRUCTURE results. At K = 7, the inferred groups are as follows: (C1) Indian and Bangladeshi O. nivara; (C2) Cambodian O. nivara; (C3) continental and insular Asian O. rufipogon; (C4) O. meridionalis; (C5) Nepalese O. nivara; (C6) non-Cambodian O. nivara; and (C7) Australasian O. rufipogon (Fig. 5). At K = 8, the same population groups are recognized except for C3 (Asian O. rufipogon) that is split into the Southeast Asian O. rufipogon (C3 of K = 8) and South Asian O. rufipogon (C8 of K = 8) clusters (Fig. 5). The geographic subdivisions in O. nivara and O. rufipogon are illustrated in the distribution map of the eight population clusters (Fig. 6), where the local separation of the two species across their range is also depicted.
Figure 6.
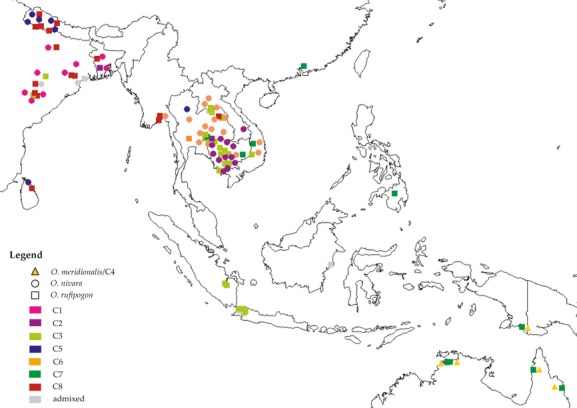
Distribution map of the eight population groups of Asia-Pacific Oryza series Sativae inferred by TESS. The geographic coordinates were slightly adjusted to allow better graphical representation of overlapping accessions.
Following Garris et al. (2005) and Agrama et al. (2010), a cut-off membership coefficient (Table S2) of ≥0.6 was imposed to assign each population to a cluster. The relationships between population clusters at K = 4, K = 7, and K = 8 are depicted by the PCoA plots in Figures S4, S5. The fourth inferred group (C4 – Australasian O. rufipogon and O. meridionalis) at K = 4 seems dubious as the genetic uniqueness of O. meridionalis detected by PCoA is not recognized in the said cluster solution (Figs. S4A, S5A). At K = 7 and K = 8, C1, C2, and C6 form the O. nivara cluster and C3, C7, and C8 (for K = 8) comprise the O. rufipogon group (Fig. S4B and C), whereas C4 (O. meridionalis) and C5 (Nepalese O. nivara) form distinct clusters (Figs. S4B and C, S5B and C). Principal coordinate axis 1 separates C7 (Australasian O. rufipogon) from the rest of O. rufipogon at K = 7 and K = 8 (Fig. S4B and C), whereas axis 4 separates the South Asian clusters (C1, C5, and C8) from the Southeast Asian groups (C2, C3, and C6) at K = 8 (Fig. S5C).
Genetic diversity of species and population groups
The annual species and population groups (O. meridionalis and O. nivara) exhibit higher FIS values (0.94–0.98) than the perennial taxa (0.85–0.87) (Table 2). Among the three species, O. rufipogon contains the largest genetic variation as it displays the highest values in all diversity parameters. In contrast, O. meridionalis has the lowest values, rendering it the least diverse species and population group.
Table 2.
Microsatellite diversity of species and population groups in Asia-Pacific Oryza series Sativae (based on 29 SSR loci)
| Taxon (TESS cluster number) | S | AL | RAL | RS | HO | HE | PIC | FIS |
|---|---|---|---|---|---|---|---|---|
| Species | ||||||||
| O. nivara | 229 | 9.83 | 5.03 | 7.47 | 0.02 | 0.67 | 0.65 | 0.97 |
| O. rufipogon | 226 | 11.79 | 6.72 | 8.43 | 0.09 | 0.70 | 0.68 | 0.87 |
| O. meridionalis (C4) | 24 | 2.24 | 0.21 | 2.24 | 0.01 | 0.24 | 0.23 | 0.98 |
| Population clusters | ||||||||
| Indian and Bangladeshi O. nivara (C1) | 46 | 4.28 | 0.86 | 4.06 | 0.02 | 0.52 | 0.48 | 0.97 |
| Nepalese O. nivara (C5) | 35 | 3.59 | 0.76 | 3.47 | 0.02 | 0.39 | 0.37 | 0.94 |
| Cambodian O. nivara (C2) | 72 | 6.55 | 2.31 | 5.70 | 0.03 | 0.59 | 0.56 | 0.95 |
| Non-Cambodian O. nivara (C6) | 73 | 6.48 | 1.59 | 5.88 | 0.02 | 0.61 | 0.59 | 0.97 |
| South Asian O. rufipogon (C8) | 69 | 7.97 | 3.31 | 6.85 | 0.10 | 0.64 | 0.62 | 0.86 |
| Southeast Asian O. rufipogon (C3) | 122 | 9.48 | 5.07 | 7.36 | 0.10 | 0.66 | 0.64 | 0.85 |
| Australasian O. rufipogon (C7) | 44 | 5.38 | 1.41 | 5.04 | 0.07 | 0.59 | 0.56 | 0.88 |
S, number of samples; AL, mean number of alleles per locus; RAL, mean number of rare alleles per locus; RS, allelic richness; HO, observed heterozygosity; HE, gene diversity (unbiased estimate); PIC, polymorphism information content; FIS, inbreeding coefficient.
Based on allelic richness and gene diversity, C3 (Southeast Asian O. rufipogon) is the most diverse among the population groups, followed by C8 (South Asian O. rufipogon) (Table 2). The genetic variation in Southeast Asian O. nivara clusters C2 and C6 is comparable to that of C7 (Australasian O. rufipogon) and greater than those of South Asian O. nivara clusters C1 and C5. Next to C4 (O. meridionalis), C5 (Nepalese O. nivara) shows the least diversity among the population groups. Heterozygosity is greater in the O. rufipogon clusters C3, C7, and C8 (0.07–0.1) than in the rest of the population groups (0.01–0.03). Clusters with the highest proportion of rare alleles are C3 (53.5%), C8 (41.6%), and C2 (35.3%) (Table 2).
Unique and shared alleles
Forty-seven alleles are common to the three species (Fig. 7 and Table S3). Oryza meridionalis shares two alleles with O. nivara and five alleles with O. rufipogon. In stark contrast, O. nivara and O. rufipogon share 192 alleles making up more than half of the total alleles detected in the annual (68.6%) and perennial (56.1%) taxa. Of the 192 alleles, 14 are exclusively present in Southeast Asian populations (C2, C3, and C6), only one allele is endemic to South Asian populations (C1 and C8), whereas the remaining 177 alleles are not restricted to regionally sympatric populations (Table S3).
Figure 7.
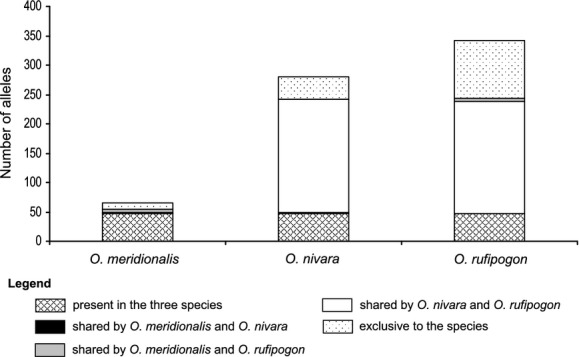
The proportion of private and shared alleles in Oryza meridionalis, O. nivara, and O. rufipogon in Asia Pacific.
Sixteen of the 52 alleles (31%) shared by O. meridionalis and O. rufipogon are detected in at least one of the five locally sympatric population pairs, whereas the remaining 36 alleles (69%) are shared by nonsympatric populations (Fig. S6). Among the 239 shared alleles of O. nivara and O. rufipogon, 98 (41%) are found in locally sympatric populations and 141 are found in nonsympatric populations (Fig. S6).
Oryza rufipogon has the largest proportion of unique alleles (98 alleles, 28.7%), followed by O. meridionalis (11 alleles, 16.9%) and O. nivara (39 alleles, 13.9%) (Table S3; Fig. 7). The most highly discriminating markers for O. meridionalis are RM124, RM316, and RM413, as they distinguish all accessions of the Australasian species from the rest of Asia-Pacific Oryza series Sativae populations (Table S3). RM44, RM431, RM118, and RM161 discriminate 12.5–20.8% of O. meridionalis populations, whereas RM237 and RM433 distinguish less than 5% of the taxon. Certain alleles of RM154, RM413, RM44, RM433, and RM495 are found exclusively in all geographic populations of O. rufipogon, but in limited frequencies ranging from 0.007 to 0.432. RM118 differentiates 47% of Australasian O. rufipogon, and at least one allele from each of the 26 loci (RM277, RM455, and RM536 are not included) discriminates a small proportion (allele frequencies ranging from 0.004 to 0.205) of one or two O. rufipogon population group/s. No allele is present throughout the distribution range of O. nivara. The 39 unique alleles from 20 markers discriminate at least one of the four O. nivara population groups in frequencies ranging from 0.007 to 0.304 (Table S3).
Genetic differentiation
Based on the AMOVA results, genetic variation in Asia-Pacific Oryza series Sativae (excluding O. sativa) resides mainly among accessions (explaining 64% of the total variance) and to a lower degree within accessions (26%) as well as among the three species (10%) (Table 3). Significant and moderate differentiation can be observed between accessions (ΦPT = 0.74) and between species (ΦRT = 0.1), respectively (both at P < 0.001 level).
Table 3.
Analysis of molecular variance among species, among populations, and within populations of Asia-Pacific Oryza series Sativae accessions based on 29 SSR markers
| Source | Degree of freedom | Sum of squares | Mean sum of squares | Estimated variance | Percentage of variance | P-value |
|---|---|---|---|---|---|---|
| Among species | 2 | 1449.896 | 724.948 | 4.408 | 10 | 0.001 |
| Among populations | 100 | 13,853.252 | 138.533 | 27.102 | 64 | 0.001 |
| Within populations | 382 | 4205.283 | 11.009 | 11.009 | 26 | 0.001 |
| Total | 484 | 19,508.431 | 42.518 | 100 |
The P-values are based on 999 permutations.
The population clusters identified by TESS at K = 8 display different degrees of differentiation (Fig. 8) with pairwise FST values ranging from 0.08 (between South [C8] and Southeast Asian [C3] O. rufipogon) to 0.58 (between O. meridionalis [C4] and Nepalese O. nivara [C5]). Oryza meridionalis is clearly the most distinct population group. The mean pairwise FST value between O. nivara clusters (0.23) is greater than between O. rufipogon clusters (0.13) and even between clusters of O. nivara and O. rufipogon (0.19) suggesting deep genetic divisions within the annual species. Oryza nivara from Nepal (C5) is clearly differentiated from the rest of the clusters. The Australasian cluster C7 seems the most distinct among the O. rufipogon population groups. The pairwise FST values are lower between the Southeast Asian clusters of O. nivara (C2 and C6) and O. rufipogon (C3) than between the South Asian clusters of the two species.
Figure 8.
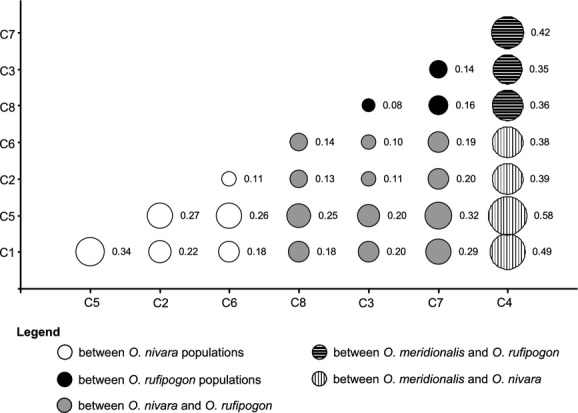
Pairwise FST of the eight population groups of Oryza series Sativae from Asia Pacific. All values are significant at P < 0.001 level based on 999 permutations. C1, Indian and Bangladeshi O. nivara; C5, Nepalese O. nivara; C2, Cambodian O. nivara; C6, non-Cambodian Southeast Asian O. nivara; C8, South Asian O. rufipogon; C3, Southeast Asian O. rufipogon; C7, Australasian O. rufipogon; C4, O. meridionalis. Circle size is proportional to its corresponding FST value (indicated at its right-hand side).
Discussion
Global overlapping and local differentiation
NJ (Fig. 1) and ordination (Fig. 2) methods reveal a lack of clear-cut genetic division between O. nivara and O. rufipogon across their distribution range, concurring with results of previous molecular studies (Second 1985; Barbier 1989; Iwamoto et al. 1999; Park et al. 2003; Ren et al. 2003; Cai et al. 2004; Zhu and Ge 2005; Zhou et al. 2008; Zheng and Ge 2010; Huang et al. 2012a). None of the markers used in this study can discriminate the majority of either O. nivara or O. rufipogon accessions from the rest of the series. The relatively large extent of allele sharing between nonsympatric populations from different geographic regions (Table S3 and Fig. S6) renders it more probable that most of the similarities can be traced to common ancestry, although gene flow cannot be ruled out as an explanation for the genetic overlap of the two species (Zhou et al. 2008; Zheng and Ge 2010). However, genetic separation of O. nivara and O. rufipogon was detected by Bayesian clustering methods at the highest population structure level (K = 2), even earlier than the recognition of O. meridionalis as a distinct group (at K = 3).
Despite the genetic overlap, species separation is apparent at a local scale. Oryza nivara and O. rufipogon populations from the same locality cluster apart from each other (except N43 and R43) in the NJ tree. Spatially explicit Bayesian clustering differentiated the two species in sympatric population pairs throughout their distribution range (Fig. 5). Kuroda et al. (2007) have reported species separation of O. nivara and O. rufipogon populations in Vientiane, Laos. Likewise, Singh et al. (2013) observed species divergence in local populations found within the Indo-Gangetic Plains of India. Therefore, contrary to the claim of Zheng and Ge (2010), molecular divergence is not completely absent between the two species and exists locally in sympatric populations indicative of adequately strong barriers to gene flow (e.g., differences in phenology and mating system) operating over smaller spatial units. It is evident from the AMOVA results that gene flow is more restricted between populations than between species (Table 3). Indeed, crossability studies suggest that reproductive barriers between O. nivara and O. rufipogon as well as between O. meridionalis and O. rufipogon tend to intensify under sympatric conditions (Banaticla-Hilario et al. 2013).
The failure of molecular data to clearly separate the two taxa led some scientists to treat O. nivara as an ecotype of O. rufipogon (Ren et al. 2003; Zhu and Ge 2005; Zheng and Ge 2010). However, we might have a different situation at hand. Recently, some authors postulated the acceptance of the “genic view” of speciation (Wu 2001; Lexer and Widmer 2008). In this view, species reproductive barriers are somewhat permeable to gene flow, and speciation can be triggered by expression of relatively few genes that affect differential adaptation and reproductive isolation. These “speciation genes” remain diverged while neutral loci are more freely exchanged between species (Wu 2001; Feder and Nosil 2010; Rieseberg and Blackman 2010; Nosil and Schluter 2011; Southcott and Ostevik 2011). Similar to the case of O. nivara and O. rufipogon, adaptive divergence in the face of massive allele sharing but resulting in reproductive isolation has been observed in closely related, recently diverged, and geographically overlapping species of Howea (Savolainen et al. 2006), Silene (Bratteler et al. 2006), Lupinus (Drummond and Hamilton 2007), Helianthus (Yatabe et al. 2007), and Pitcairnia (Palma-Silva et al. 2011), where species discrimination appears to involve only a few loci/genes. Whether O. nivara and O. rufipogon are “genic” species remains to be seen as their speciation genes still await ascertainment. A good starting point is the work of Grillo et al. (2009), where quantitative trait loci (QTLs) with moderate to large effect on flowering time as well as QTLs with small to moderate effect on floral and panicle traits associated with the mating system of O. nivara were identified. The same authors also implicated the role of directional selection in the fixation of majority of the QTL alleles of O. nivara. The loci/genes used in earlier studies that confirmed species separation could also hold clues to the identity of their supposed speciation genes. Kuroda et al. (2007) and Singh et al. (2013) used a total of 30 SSR markers that differentiated local populations of O. nivara and O. rufipogon. Likewise, species divergence was evident in the results of Duan et al. (2007) based on sequences of the chloroplast trnL intron and trnL-trnF spacer, the mitochondrial nad1 intron 2, and the nuclear internal transcribed spacer, and in those of Xu et al. (2012) based on 6.5 million SNPs.
Regional divergence
The population groups of O. nivara and O. rufipogon in South Asia exhibit lower diversity (Table 2) and higher intra- and interspecific differentiation (Fig. 8) than their Southeast Asian counterparts. Moreover, only one allele is exclusively shared by O. nivara and O. rufipogon in South Asia, whereas 14 alleles are endemic and common between the two species in Southeast Asia. This evidence indicates stronger gene flow barriers in the South Asian region. Such a geographic pattern conforms to the morphological variations reported by Banaticla-Hilario (2012) but contradicts an earlier SSR experiment that reported greater species differentiation in Southeast Asia (Lu et al. 2008).
The optimal four-cluster solution of STRUCTURE recognizes the South Asian and Southeast Asian populations of O. nivara as two genetically distinct groups (Figs. 4, 5). Oryza nivara is confined to areas with a pronounced dry season and its occurrence has not been reported in the more humid, western part of Myanmar (Vaughan et al. 2008) where the regional boundary of tropical continental Asia lies. This geoclimatic factor probably restricts gene flow between the South and Southeast Asian populations of O. nivara. The vicariance displayed by O. nivara is also evident from phenotype data (Banaticla-Hilario 2012) and could be a plausible explanation of regional differences in the extent of genetic differentiation between O. nivara and O. rufipogon.
Australasian populations are distinct
The results confirmed the genetic disparities of O. meridionalis from the rest of the series (Figs. 1, 2, 8) and of the Australasian populations of O. rufipogon from the rest of the perennial species (Figs. 4, 5, 8). The latter disagrees with the findings of Huang et al. (2012a) where O. rufipogon exhibited two genetic groups (i.e., China-centered Ruf-I and South Asia-centered Ruf-II) displaying a clinal variation pattern. This China–South Asia division cannot be confirmed in the present analysis as only one accession from China was sampled.
Nevertheless, the pattern obtained in this study corresponds to the previously reported morphological (Banaticla-Hilario 2012) and genetic (Waters et al. 2012) divergence of Australasian O. rufipogon. Genetic and morphological differentiation between continental and insular populations has been reported in many other plant species (e.g., Howcroft and Davidson 1973; Rivera-Ocasio et al. 2006; Fievet et al. 2007; Fedorenko et al. 2009). Geographic isolation probably restrained gene exchange between these Australasian taxa and the other population groups and may have caused genetic bottlenecks as indicated by the poor genetic diversity of O. meridionalis (Table 2). However, a larger number of accessions and multiple individuals from the accessions should be analyzed to confirm this conclusion. Oryza rufipogon populations in Australasia have been reported to flourish vegetatively and produce less seeds in their natural habitats (Vaughan et al. 2003, 2008), which could also be a reason for their low genetic diversity.
The geographic separation and low diversity of O. meridionalis and Australasian O. rufipogon predispose them to inbreeding depression and subsequent genetic deterioration as observed in other island populations (Frankham 1997, 1998; Fedorenko et al. 2009). Therefore, these population groups should be carefully examined and considered in reviewing and designing management practices for their protection and preservation.
Oryza nivara in Nepal: a discrete genetic entity
The genetic distinctiveness of O. nivara populations in Nepal is comparable to that of O. meridionalis as explicitly shown in the NJ (Fig. 1), ordination (Fig. 2), and FST (Fig. 8) results. However, the Nepalese O. nivara group seems distinguishable at lower population structure levels. Bayesian methods detect this group at K = 5 following the recognition of the South–Southeast Asia split in O. nivara at K = 4 (Figs. 4, 5). The uniqueness of O. meridionalis is evident also at higher population structure levels (K = 3).
Low diversity and genetic isolation from the rest of the species expose the Nepalese O. nivara to inbreeding depression and genetic erosion. More in-depth studies are needed not just to confirm the unique genetic identity of these regional populations but also to further establish variation patterns that will aid in formulating in and ex situ conservation strategies.
Associations with O. sativa
The reported consanguinity of O. rufipogon with O. sativa var. japonica and of O. nivara with O. sativa var. indica (Cheng et al. 2003; Yamanaka et al. 2003; Ohtsubo et al. 2004; Xu et al. 2007, 2012) is evident from this study (Figs. 1, 2, 4). At the uppermost hierarchical level of population structure (K = 2), the japonica and aromatic varietal groups join up with O. rufipogon, whereas the indica and aus groups do so with O. nivara (Fig. S1).
Geographic clustering patterns are further displayed by the cultivated varieties at lower structural levels. Starting at K = 4, aus consistently groups with Cambodian O. nivara (Fig. 4), whereas starting at K = 7, indica clusters with O. nivara from Thailand. This is analogous to the clustering patterns revealed by 6.5 million SNPs where indica and aus appeared similar to different populations of O. nivara (Xu et al. 2012). However, the limited number of cultivated and Chinese wild rice populations analyzed in this study limits the validity of the clustering patterns obtained. Phylogenetic analyses based on ∼8 million SNPs indicated that japonica is genetically closer to O. rufipogon from China than to any other O. rufipogon populations in Asia (Huang et al. 2012b). Phylogeographic results (Londo et al. 2006) agree with the genetic association of indica with wild rice in Thailand (Fig. 4), but are in discordance with the observed merging of aromatic and japonica with South Asian O. rufipogon at K = 6 (Fig. 4).
It is worth mentioning that of the six populations morphologically classified as weedy forms (i.e., intermediate between O. sativa and either O. nivara or O. rufipogon) (Banaticla-Hilario 2012), one was detected by STRUCTURE (at K = 6) as a genetic admixture of O. nivara and O. rufipogon, whereas the other populations were included in the O. nivara–indica group. Caution should be taken when interpreting SSR diversity patterns as the presence of interaction between cultivated and wild taxa could be masked by the genetic similarities within Oryza series Sativae. Vaughan et al. (2008) warned that some genebank accessions of the Asian wild rice might have introgressed with cultivated rice, as most of these accessions were collected from disturbed habitats.
Conclusions
This research imparts a more detailed account of the genetic variation patterns in O. nivara and O. rufipogon, less so in the geographically restricted O. meridionalis. The recognition of local differentiation in the midst of global similarities reconciles the conflicting results of prior studies (Second 1985; Barbier 1989; Aggarwal et al. 1999; Iwamoto et al. 1999; Park et al. 2003; Ren et al. 2003; Cai et al. 2004; Zhu and Ge 2005; Duan et al. 2007; Kuroda et al. 2007; Takahashi et al. 2008; Zhou et al. 2008; Zheng and Ge 2010; Xu et al. 2012; Singh et al. 2013). Furthermore, regional differences in the strength of interspecific gene flow have been detected indicating that the extent of genetic differentiation between O. nivara and O. rufipogon varies at different geographic scales.
The revealed geographic partitions within species as well as the inferred population groupings within the series can be considered in assessing the genetic representativeness of genebank collections and in selecting plant materials for in and ex situ conservation and research purposes. Especially, the uniqueness and vulnerability to genetic degradation of O. meridionalis, O. nivara in Nepal, and O. rufipogon in Australasia call for immediate conservation measures. Furthermore, the vast amount of genetic variation detected among populations justifies the maintenance of a large collection of Asian wild rice germplasm.
Acknowledgments
This study was supported by the T.T. Chang Genetic Resources Center (TTC-GRC) in IRRI and the Biosystematics Group of Wageningen University. The authors would like to thank: the Genomic Diversity Laboratory team and the Wild Rice Nursery support staff of TTC-GRC for their help in DNA extraction and SSR genotyping; Sheila Mae Mercado, Elizabeth Naredo, and M. J. M. Smulders for their advice on SSR analyses; and Marc Sm. Sosef for his help in editing the manuscript.
Conflict of Interest
None declared.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Figure S1. Cluster solutions produced by STRUCTURE and TESS from K = 2 to K = 8. The average membership coefficient of 10 runs from each K is shown. The predefined populations are represented as follows: 1, Oryza sativa (aromatic); 2, O. sativa (japonica); 3, O. sativa (indica); 4, O. sativa (aus); 5, O. nivara from South Asia; 6, O. nivara from Southeast Asia; 7, O. rufipogon from South Asia; 8, O. rufipogon from China; 9, O. rufipogon from continental Southeast Asia; 10, O. rufipogon from insular Southeast Asia; 11, O. rufipogon from Australasia; 12, O. meridionalis from Australasia.
Figure S2. Membership coefficients of 10 aligned STRUCTURE runs at K = 4 and K = 6. The predefined populations are represented as follows: 1, Oryza sativa (aromatic); 2, O. sativa (japonica); 3, O. sativa (indica); 4, O. sativa (aus); 5, O. nivara (from South Asia); 6, O. nivara (from Southeast Asia); 7, O. rufipogon (from South Asia); 8, O. rufipogon (from China); 9, O. rufipogon (from continental Southeast Asia); 10, O. rufipogon (from insular Southeast Asia); 11, O. rufipogon (from Australasia); 12, O. meridionalis (from Australasia).
Figure S3. (A) Membership coefficients of the 10 aligned TESS runs at K = 2 and K = 3. The predefined populations are represented as follows: 1, South Asian Oryza nivara; 2, Southeast Asian O. nivara; 3, South Asian O. rufipogon; 4, Chinese O. rufipogon; 5, continental Southeast Asian O. rufipogon; 6, insular Southeast Asian O. rufipogon; 7, Australasian O. rufipogon; and 8, O. meridionalis. (B) Membership coefficients of the 10 aligned TESS runs at K = 4 and K = 5. The predefined populations are represented as follows: 1, South Asian O. nivara; 2, Southeast Asian O. nivara; 3, South Asian O. rufipogon; 4, Chinese O. rufipogon; 5, continental Southeast Asian O. rufipogon; 6, insular Southeast Asian O. rufipogon; 7, Australasian O. rufipogon; and 8, O. meridionalis. (C) Membership coefficients of the 10 aligned TESS runs at K = 6 and K = 7. The predefined populations are represented as follows: 1, South Asian O. nivara; 2, Southeast Asian O. nivara; 3, South Asian O. rufipogon; 4, Chinese O. rufipogon; 5, continental Southeast Asian O. rufipogon; 6, insular Southeast Asian O. rufipogon; 7, Australasian O. rufipogon; and 8, O. meridionalis. (D) Membership coefficients of the 10 aligned TESS runs at K = 8 and K = 9. The predefined populations are represented as follows: 1, South Asian O. nivara; 2, Southeast Asian O. nivara; 3, South Asian O. rufipogon; 4, Chinese O. rufipogon; 5, continental Southeast Asian O. rufipogon; 6, insular Southeast Asian O. rufipogon; 7, Australasian O. rufipogon; and 8, O. meridionalis. (E) Membership coefficients of the 10 aligned TESS runs at K = 10. The predefined populations are represented as follows: 1, South Asian O. nivara; 2, Southeast Asian O. nivara; 3, South Asian O. rufipogon; 4, Chinese O. rufipogon; 5, continental Southeast Asian O. rufipogon; 6, insular Southeast Asian O. rufipogon; 7, Australasian O. rufipogon; and 8, O. meridionalis.
Figure S4. Placement of TESS clusters in the plot of the first two principal coordinate axes (axis 1 on x and axis 2 on y). (A) at K = 4, C1, South Asian Oryza nivara; C2, Southeast Asian O. nivara; C3, continental Asian and insular Southeast Asian O. rufipogon; and C4, O. meridionalis and Australasian O.rufipogon. (B) at K = 7, C1, Indian and Bangladeshi O. nivara; C2, Cambodian O. nivara; C3, continental Asian and insular Southeast Asian O. rufipogon; C4, O. meridionalis; C5, Nepalese O. nivara; C6, non-Cambodian O. nivara; and C7, Australasian O. rufipogon. (C) at K = 8, C1, Indian and Bangladeshi O. nivara; C2, Cambodian O. nivara; C3, Southeast Asian O. rufipogon; C4, O. meridionalis; C5, Nepalese O. nivara; C6, non-Cambodian O. nivara; C7, Australasian O. rufipogon; and C8, South Asian O. rufipogon. Admixed populations are those with less than 0.6 membership coefficient.
Figure S5. Placement of TESS clusters in the plot of the third and fourth principal coordinate axes (axis 3 on x and axis 4 on y). (A) at K = 4, C1, South Asian Oryza nivara; C2, Southeast Asian O. nivara; C3, continental Asian and insular Southeast Asian O. rufipogon; and C4, O. meridionalis and Australasian O. rufipogon. (B) at K = 7, C1, Indian and Bangladeshi O. nivara; C2, Cambodian O. nivara; C3, continental Asian and insular Southeast Asian O. rufipogon; C4, O. meridionalis; C5, Nepalese O. nivara; C6, Non-Cambodian O. nivara; and C7, Australasian O. rufipogon. (C) at K = 8, C1, Indian and Bangladeshi O. nivara; C2, Cambodian O. nivara; C3, Southeast Asian O. rufipogon; C4, O. meridionalis; C5, Nepalese O. nivara; C6, Non-Cambodian O. nivara; C7, Australasian O. rufipogon; and C8, South Asian O. rufipogon. Admixed populations are those with less than 0.6 membership coefficient.
Figure S6. Proportion of shared alleles detected in sympatric and nonsympatric population pairs of Oryza meridionalis and O. rufipogon (MR) and of O. nivara and O. rufipogon (NR).
Table S1. Genetic diversity exhibited by the 125 Oryza series Sativae populations across 29 microsatellite loci.
Table S2. TESS cluster membership of each population group at K = 8 (averaged over 10 runs).
Table S3. Allele sizes and frequencies of 29 SSR markers in Asia Pacific Oryza series Sativae. Alleles that can distinguish all or certain population groups within a species are highlighted. Highly discriminating alleles are boldfaced.
References
- Aggarwal RK, Brar DS, Nandi S, Huang N, Khush GS. Phylogenetic relationships among Oryza species revealed by AFLP markers. Theor. Appl. Genet. 1999;98:1320–1328. [Google Scholar]
- Agrama H, Yan W, Jia M, Fjellstrom R, McClung A. Genetic structure associated with diversity and geographic distribution in the USDA rice world collection. Nat. Sci. 2010;2:247–291. [Google Scholar]
- Banaticla-Hilario MCN. An ecogeographic analysis of Oryza series Sativae in Asia and the Pacific. the Netherlands: Wageningen University; 2012. Ph.D. thesis. [Google Scholar]
- Banaticla-Hilario MCN, McNally KL, Sackville Hamilton RG, Van den Berg NR. Crossability patterns within and among Oryza series Sativae species from Asia and Australia. Genet. Resour. Crop Evol. 2013 doi: 10.1007/s10722-013-9965-410.1007/s10722-013-9965-4. [Google Scholar]
- Barbier P. Genetic variation and ecotypic variation in the wild rice species Oryza rufipogon. I. Population differentiation in life-history traits and isozymic loci. Jpn. J. Genet. 1989;64:259–271. [Google Scholar]
- Bratteler M, Baltisberger M, Widmer A. QTL analysis of intraspecific differences between two Silene vulgaris ecotypes. Ann. Bot. 2006;98:411–419. doi: 10.1093/aob/mcl113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai HW, Wang XK, Morishima H. Comparison of population genetic structures of common wild rice (Oryza rufipogon Griff.), as revealed by analyses of quantitative traits, allozymes, and RFLPs. Heredity. 2004;92:409–417. doi: 10.1038/sj.hdy.6800435. [DOI] [PubMed] [Google Scholar]
- Cavalli-Sforza LL, Edwards AWF. Phylogenetic analysis models and estimation procedures. Am. J. Hum. Genet. 1967;19:233–257. [PMC free article] [PubMed] [Google Scholar]
- Cheng C, Motohashi R, Tsuchimoto S, Fukuta Y, Ohtsubo H, Ohtsubo E. Polyphyletic origin of cultivated rice: based on the interspersion pattern of SINEs. Mol. Biol. Evol. 2003;20:67–75. doi: 10.1093/molbev/msg004. [DOI] [PubMed] [Google Scholar]
- Drummond CS, Hamilton MB. Hierarchical components of genetic variation at a species boundary: population structure in two sympatric varieties of Lupinus microcarpus (Leguminosae) Mol. Ecol. 2007;16:753–769. doi: 10.1111/j.1365-294X.2006.03186.x. [DOI] [PubMed] [Google Scholar]
- Duan S, Lu B, Li Z, Tong J, Kong J, Yao W, et al. Phylogenetic analysis of AA-genome Oryza species (Poaceae) based on chloroplast, mitochondrial, and nuclear DNA sequences. Biochem. Genet. 2007;45:113–129. doi: 10.1007/s10528-006-9062-x. [DOI] [PubMed] [Google Scholar]
- Durand E, Chen C, Francois O. 2009. p. 30. TESS version 2.3 - reference manual. [DOI] [PubMed]
- Evanno G, Regnaut S, Goudet J. Detecting the name of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feder JL, Nosil P. The efficacy of divergence hitchhiking in generating genomic islands during ecological speciation. Evolution. 2010;64:1729–1747. doi: 10.1111/j.1558-5646.2010.00943.x. [DOI] [PubMed] [Google Scholar]
- Fedorenko OM, Gritskikh MV, Malysheva IE, Nikolaevskaya TS. Genetic diversity of insular natural populations of Festuca pratensis Huds.: RAPD analysis. Russ. J. Genet. 2009;45:1134–1138. [PubMed] [Google Scholar]
- Fievet V, Touzet P, Arnaud J-F, Cuguen J. Spatial analysis of nuclear and cytoplasmic DNA diversity in wild sea beet (Beta vulgaris ssp. maritima) populations: do marine currents shape the genetic structure? Mol. Ecol. 2007;16:1847–1864. doi: 10.1111/j.1365-294X.2006.03208.x. [DOI] [PubMed] [Google Scholar]
- Frankham R. Do island populations have less genetic variation than mainland populations? Heredity. 1997;78:311–327. doi: 10.1038/hdy.1997.46. [DOI] [PubMed] [Google Scholar]
- Frankham R. Inbreeding and extinction: island populations. Conserv. Biol. 1998;12:665–675. [Google Scholar]
- Fulton T, Chunwongse J, Tanksley S. Microprep protocol for extraction of DNA from tomato and other herbaceous plants. Plant Mol. Biol. Rep. 1995;13:207–209. [Google Scholar]
- Garris AJ, Tai TH, Coburn J, Kresovich S, McCouch S. Genetic structure and diversity in Oryza sativa L. Genetics. 2005;169:1631–1638. doi: 10.1534/genetics.104.035642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goudet J. 2001. FSTAT, a program to estimate and test gene diversities and fixation indices (version 2.9.3)
- Grillo MA, Li C, Fowlkes AM, Briggeman TM, Zhou A, Schemske DW, et al. Genetic architecture for the adaptive origin of annual wild rice, Oryza nivara. Evolution. 2009;63:870–883. doi: 10.1111/j.1558-5646.2008.00602.x. [DOI] [PubMed] [Google Scholar]
- Howcroft N, Davidson J. 1973. An international provenance trial of Pinus merkusii in Papua New Guinea. Tropical Forestry Research Note, Papua New Guinea. 1973. SR. 15, 27 P. 7.
- Huang PU, Molina J, Flowers JM, Rubinstein S, Jackson SA, Purugganan MD, et al. Phylogeography of Asian wild rice, Oryza rufipogon: a genome-wide view. Mol. Ecol. 2012a;21:4593–4604. doi: 10.1111/j.1365-294X.2012.05625.x. [DOI] [PubMed] [Google Scholar]
- Huang X, Kurata N, Wei X, Wang Z-X, Wang A, Zhao Q, et al. A map of rice genome variation reveals the origin of cultivated rice. Nature. 2012b;490:497–501. doi: 10.1038/nature11532. http://dx.doi.org/10.1038/nature11532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamoto M, Nagashima H, Nagamine T, Higo H, Higo K. A tourist element in the flanking region of the catalase gene CatA reveals evolutionary relationships among Oryza species with various genome types. Mol. Genet. Genomics. 1999;262:493–500. doi: 10.1007/s004380051110. [DOI] [PubMed] [Google Scholar]
- Jakobsson M, Rosenberg NA. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics. 2007;23:1801–1806. doi: 10.1093/bioinformatics/btm233. [DOI] [PubMed] [Google Scholar]
- Kuroda Y, Sato Y-I, Bounphanousay C, Kono Y, Tanaka K. Genetic structure of three Oryza AA genome species (O. rufipogon, O. nivara and O. sativa) as assessed by SSR analysis on the Vientiane Plain of Laos. Conserv. Genet. 2007;8:149–158. [Google Scholar]
- Kwon S-J, Lee JK, Hong S-W, Park Y-J, McNally KL, Kim N-S. Genetic diversity and phylogenetic relationship in AA Oryza species as revealed by Rim2/Hipa CACTA transposon display. Genes Genet. Syst. 2006;81:93–101. doi: 10.1266/ggs.81.93. [DOI] [PubMed] [Google Scholar]
- Lexer C, Widmer A. The genic view of plant speciation: recent progress and emerging questions. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2008;363:3023–3036. doi: 10.1098/rstb.2008.0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Muse SV. PowerMarker: integrated analysis environment for genetic marker data. Bioinformatics. 2005;21:2128–2129. doi: 10.1093/bioinformatics/bti282. [DOI] [PubMed] [Google Scholar]
- Londo JP, Chiang YC, Hung KH, Chiang TY, Schaal BA. Phylogeography of Asian wild rice, Oryza rufipogon, reveals multiple independent domestications of cultivated rice, Oryza sativa. Proc. Natl. Acad. Sci. USA. 2006;103:9578–9583. doi: 10.1073/pnas.0603152103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B-R. Taxonomy of the genus Oryza (Poaceae): historical perspective and current status. Int. Rice Res. Notes. 1999;24:4–8. [Google Scholar]
- Lu B-R, Ge S, Sang T, Chen J-K, Hong D-Y. The current taxonomy and perplexity of the genus Oryza (Poaceae) Acta Phytotaxon. Sin. 2001;39:373–388. [Google Scholar]
- Lu J-Z, Zhang X-L, Wang H-G, Yuan X-P, Xu Q, Wang Y-P, et al. SSR analysis on diversity of AA genome Oryza species in the Southeast and South Asia. Rice Sci. 2008;15:289–294. [Google Scholar]
- Ng NQ, Chang TT, Williams JT, Hawkes JG. Morphological studies of Asian rice and its related wild species and the recognition of a new Australian taxon. Biol. J. Linn. Soc. 1981;16:303–313. [Google Scholar]
- Nosil P, Schluter D. The genes underlying the process of speciation. Trends Ecol. Evol. 2011;26:160–167. doi: 10.1016/j.tree.2011.01.001. [DOI] [PubMed] [Google Scholar]
- Ohtsubo H, Cheng C, Ohsawa I, Tsuchimoto S, Ohtsubo E. Rice retroposon p-SINE1 and origin of cultivated rice. Breed. Sci. 2004;54:1–11. [Google Scholar]
- Oka HI. Origin of cultivated rice. Tokyo: Japan Science Society Press; 1988. [Google Scholar]
- Palma-Silva C, Wendt T, Pinheiro F, Barbara T, Fay MF, Cozzolino S, et al. Sympatric bromeliad species (Pitcairnia spp.) facilitate tests of mechanisms involved in species cohesion and reproductive isolation in Neotropical inselbergs. Mol. Ecol. 2011;20:3185–3201. doi: 10.1111/j.1365-294X.2011.05143.x. [DOI] [PubMed] [Google Scholar]
- Park KC, Kim NH, Cho YS, Kang KH, Lee JK, Kim NS. Genetic variations of AA genome Oryza species measured by MITE-AFLP. Theor. Appl. Genet. 2003;107:203–209. doi: 10.1007/s00122-003-1252-x. [DOI] [PubMed] [Google Scholar]
- Peakall ROD, Smouse PE. Genalex 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes. 2006;6:288–295. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren F, Lu B-R, Li S, Huang J, Zhu Y. A comparative study of genetic relationships among the AA-genome Oryza species using RAPD and SSR markers. Theor. Appl. Genet. 2003;108:113–120. doi: 10.1007/s00122-003-1414-x. [DOI] [PubMed] [Google Scholar]
- Rieseberg LH, Blackman BK. Speciation genes in plants. Ann. Bot. 2010;106:439–455. doi: 10.1093/aob/mcq126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Ocasio E, Aide TM, McMillan WO. The influence of spatial scale on the genetic structure of a widespread tropical wetland tree Pterocarpus officinalis (Fabaceae) Conserv. Genet. 2006;7:251–266. [Google Scholar]
- Rosenberg NA. Distruct: a program for the graphical display of population structure. Mol. Ecol. Notes. 2004;4:137–138. [Google Scholar]
- Savolainen V, Anstett M-C, Lexer C, Hutton I, Clarkson JJ, Norup MV, et al. Sympatric speciation in palms on an oceanic island. Nature. 2006;441:210–213. doi: 10.1038/nature04566. [DOI] [PubMed] [Google Scholar]
- Second G. Evolutionary relationships in the Sativa group of Oryza based on isozyme data. Genet. Sel. Evol. 1985;17:89–114. doi: 10.1186/1297-9686-17-1-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma SD, Shastry SVS. Taxonomic studies in the genus Oryza L. III. O. rufipogon Griff. sensu stricto and O. nivara Sharma et Shastry nom. nov. Indian J. Genet. Plant Breed. 1965;25:157–167. [Google Scholar]
- Singh A, Singh B, Panda K, Rai VP, Singh AK, Singh SP, et al. Wild rices of Eastern Indo-Gangetic plains of India constitute two sub-populations harbouring rich genetic diversity. Plant Omics J. 2013;6:121–127. [Google Scholar]
- Southcott L, Ostevik KL. Bromeliad population genetics reveals species cohesion against the odds. Mol. Ecol. 2011;20:3081–3083. doi: 10.1111/j.1365-294x.2011.05174.x. [DOI] [PubMed] [Google Scholar]
- Takahashi H, Sato Y-i, Nakamura I. Evolutionary analysis of two plastid DNA sequences in cultivated and wild species of Oryza. Breed. Sci. 2008;58:225–223. [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tateoka T. Taxonomic studies of Oryza. III. Key to the species and their enumeration. Bot. Mag. Tokyo. 1963;76:165–173. [Google Scholar]
- Uga Y, Fukuta Y, Ohsawa R, Fujimura T. Variations of floral traits in Asian cultivated rice (Oryza sativa L.) and its wild relatives (O. rufipogon Griff.) Breed. Sci. 2003;53:345–352. [Google Scholar]
- Vaughan DA, Morishima H, Kadowaki K. Diversity in the Oryza genus. Curr. Opin. Plant Biol. 2003;6:139–146. doi: 10.1016/s1369-5266(03)00009-8. [DOI] [PubMed] [Google Scholar]
- Vaughan DA, Lu B-R, Tomooka N. The evolving story of rice evolution. Plant Sci. 2008;174:394–408. [Google Scholar]
- Waters DLE, Nock CJ, Ishikawa R, Rice N, Henry RJ. Chloroplast genome sequence confirms distinctness of Australian and Asian wild rice. Ecol. Evol. 2012;2:211–217. doi: 10.1002/ece3.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir BS. Genetic data analysis II. Sunderland, MA: Sinauer Associates; 1996. [Google Scholar]
- Wu C-I. The genic view of the process of speciation. J. Evol. Biol. 2001;14:851–865. [Google Scholar]
- Xu J-H, Kurata N, Akimoto M, Ohtsubo H, Ohtsubo E. Identification and characterization of Australian wild rice strains of Oryza meridionalis and Oryza rufipogon by SINE insertion polymorphism. Genes Genet. Syst. 2005;80:129–134. doi: 10.1266/ggs.80.129. [DOI] [PubMed] [Google Scholar]
- Xu J-H, Cheng C, Tsuchimoto S, Ohtsubo H, Ohtsubo E. Phylogenetic analysis of Oryza rufipogon strains and their relations to Oryza sativa strains by insertion polymorphism of rice SINEs. Genes Genet. Syst. 2007;82:217–229. doi: 10.1266/ggs.82.217. [DOI] [PubMed] [Google Scholar]
- Xu X, Liu X, Ge S, Jensen JD, Hu F, Li X, et al. Resequencing 50 accessions of cultivated and wild rice yields markers for identifying agronomically important genes. Nat. Biotechnol. 2012;30:105–111. doi: 10.1038/nbt.2050. [DOI] [PubMed] [Google Scholar]
- Yamanaka S, Nakamura I, Nakai H, Sato Y-I. Dual origin of the cultivated rice based on molecular markers of newly collected annual and perennial strains of wild rice species, Oryza nivara and O. rufipogon. Genet. Resour. Crop Evol. 2003;50:529–538. [Google Scholar]
- Yatabe Y, Kane NC, Scotti-Saintagne C, Rieseberg LH. Rampant gene exchange across a strong reproductive barrier between the annual sunflowers, Helianthus annuus and H. petiolaris. Genetics. 2007;175:1883–1893. doi: 10.1534/genetics.106.064469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Ge S. Ecological divergence in the presence of gene flow in two closely related Oryza species (Oryza rufipogon and O. nivara. Mol. Ecol. 2010;19:2439–2454. doi: 10.1111/j.1365-294x.2010.04674.x. [DOI] [PubMed] [Google Scholar]
- Zhou H-F, Zheng X-M, Wei R-X, Second G, Vaughan DA, Ge S. Contrasting population genetic structure and gene flow between Oryza rufipogon and Oryza nivara. Theor. Appl. Genet. 2008;117:1181–1189. doi: 10.1007/s00122-008-0855-7. [DOI] [PubMed] [Google Scholar]
- Zhu Q, Ge S. Phylogenetic relationships among A-genome species of the genus Oryza revealed by intron sequences of four nuclear genes. New Phytol. 2005;167:249–265. doi: 10.1111/j.1469-8137.2005.01406.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


