Abstract
Aims:
Our study aims were to determine the frequency of MODY mutations (HNF1A, HNF4A, glucokinase) in a diverse population of youth with diabetes and to assess how well clinical features identify youth with maturity-onset diabetes of the young (MODY).
Methods:
The SEARCH for Diabetes in Youth study is a US multicenter, population-based study of youth with diabetes diagnosed at age younger than 20 years. We sequenced genomic DNA for mutations in the HNF1A, HNF4A, and glucokinase genes in 586 participants enrolled in SEARCH between 2001 and 2006. Selection criteria included diabetes autoantibody negativity and fasting C-peptide levels of 0.8 ng/mL or greater.
Results:
We identified a mutation in one of three MODY genes in 47 participants, or 8.0% of the tested sample, for a prevalence of at least 1.2% in the pediatric diabetes population. Of these, only 3 had a clinical diagnosis of MODY, and the majority was treated with insulin. Compared with the MODY-negative group, MODY-positive participants had lower FCP levels (2.2 ± 1.4 vs 3.2 ± 2.1 ng/mL, P < .01) and fewer type 2 diabetes-like metabolic features. Parental history of diabetes did not significantly differ between the 2 groups.
Conclusions/Interpretation:
In this systematic study of MODY in a large pediatric US diabetes cohort, unselected by referral pattern or family history, MODY was usually misdiagnosed and incorrectly treated with insulin. Although many type 2 diabetes-like metabolic features were less common in the mutation-positive group, no single characteristic identified all patients with mutations. Clinicians should be alert to the possibility of MODY diagnosis, particularly in antibody-negative youth with diabetes.
Monogenic diabetes results from a single gene mutation. The most common types of monogenic diabetes are the autosomal dominant forms known as maturity-onset diabetes of the young (MODY). In patients with nonsyndromic diabetes, greater than 99% of MODY with a known genetic etiology results from mutations in hepatocyte nuclear factor (HNF)-1A (formerly MODY3), glucokinase (GCK) (MODY2), or HNF4A (MODY1) (1). Although mutations in other genes have been shown to cause MODY (2), they are very rare and genetic testing is not recommended unless other syndromic features are present (3). Thus, hereafter in this report, the general term MODY refers to HNF1A-, HNF4A-, and GCK-MODY. Studies have estimated that MODY accounts for less than 1%–2.4% of pediatric diabetes cases (4–6).
Patients with HNF1A- and HNF4A-MODY present with signs and symptoms of hyperglycemia, including polyuria, polydipsia, and nocturia. Patients with GCK-MODY are often diagnosed incidentally when mild hyperglycemia is found on routine blood glucose screening. The clinical diagnosis of MODY is usually based on the following criteria: autosomal inheritance of diabetes, insulin independence, and age at onset younger than 25 years (7). In recent years, researchers have attempted to identify inexpensive and widely available biomarkers that are sensitive and specific for identifying persons with MODY mutations. C-reactive protein (CRP) has exhibited the most promise for identifying HNF1A-MODY but not other MODY subtypes (8, 9).
The aims of this study were as follows: 1) to estimate the frequency of children with the 3 most common genetic etiologies of MODY in a diverse population of youth with diabetes, 2) to determine whether genetically diagnosed MODY patients were correctly clinically diagnosed and treated by their health care providers, and 3) to evaluate the clinical characteristics of genetically diagnosed MODY patients.
Materials and Methods
Study design
For this study of monogenic diabetes, a subset of SEARCH for Diabetes in Youth study participants who completed a research visit and had a fasting C-peptide (FCP) level of 0.8 ng/mL or greater and negative results for selected diabetes autoantibodies (DAAs) were tested for the 3 most common forms of MODY: HNF1A, HNF4A, and GCK. The rationale for selecting these 3 genes for testing is detailed in the online appendix.
Study population
SEARCH is a population-based study that ascertained cases of nongestational diabetes in youth ages less than 20 years of age in the United States with the goal of identifying all existing (prevalent) cases in 2001 and all newly diagnosed (incident) cases in subsequent calendar years. The SEARCH study was conducted in 6 clinical centers located in California, Colorado, Hawaii, Ohio, South Carolina, and Washington. Youth with diabetes mellitus were identified in 4 geographically defined populations in Ohio (8 counties encompassing and surrounding Cincinnati); Washington (5 counties encompassing and surrounding Seattle); South Carolina (4 counties); and Colorado (13 counties); among health plan enrollees in Hawaii (Hawaii Medical Service Association, MedQuest, Kaiser Permanente Hawaii), and California (Kaiser Permanente Southern California, excluding San Diego); and coordinated by the Colorado center, from health service beneficiary roles in several reservation-based American Indian populations. The study covers a population at risk of more than 5 million children, representing approximately 6.2% of the US population under the age of 20 years. A detailed description of the SEARCH study methods has been published elsewhere (10).
To identify all youth with diabetes, the centers established active surveillance systems based on networks of pediatric and adult endocrinologists, existing pediatric diabetes databases, hospitals, health plan databases, and other health care providers. Data sources included case reports from pediatric endocrinologists and linkage of clinical databases, which included information on prescriptions, inpatient, and outpatient encounters with an International Classification of Diseases, ninth revision, Clinical Modification, code for diabetes, and laboratory measures of glycosylated hemoglobin (A1c). Using a protocol that conformed to the Health Insurance Portability and Accountability Act of 1996, youth with diabetes were asked to complete a brief survey that included their age at diagnosis, race/ethnicity, and a limited health history. Diabetes type was based on the provider's diagnosis at the time of case ascertainment.
Youth with nonsecondary diabetes mellitus who responded to this survey were invited to a study visit (registered SEARCH cases with baseline in person visit, Figure 1). Written informed assent and/or consent were obtained from all study participants and from parents/guardians of participants younger than 18 year of age in accordance with the guidelines established by the local institutional review board. During the study visit, additional information was collected, including symptoms at presentation, family history of diabetes in first-degree relatives and grandparents, and medication use. Blood was drawn for measurement of the DAAs glutamic acid decarboxylase 65 (GAD65) and islet-antibody 2 (IA2), A1c, FCP, high-sensitivity CRP (hsCRP), and lipids; genomic DNA was extracted from peripheral leukocytes and stored for genetic studies. The visit occurred after an overnight fast, under conditions of metabolic stability, defined as no episode of diabetic ketoacidosis during the previous month and a fasting blood glucose of less than 300 mg/dL. All medicines as well as rapid-acting insulin administration were discontinued the night before the visit. A physical examination was performed to measure blood pressure, height, weight, and waist circumference and to assess for acanthosis nigricans. Body mass index (BMI) was calculated and converted to BMI Z-score using the standard Centers for Disease Control and Prevention approach. Insulin sensitivity index was estimated using the following equation: insulin sensitivity (IS) = exponential [4.64725 − 0.02032(waist, centimeters) − 0.09779(A1c, percentage) − 0.00235(triglycerides, milligrams per deciliter)] (11). The insulin sensitivity index was developed and validated in a subset of SEARCH participants and matched nondiabetic control individuals who had direct measurements of glucose disposal rate from euglycemic-hyperinsulinemic clamps (11). The major component of the formula explaining 60% of the variance in measured glucose disposal, regardless of provider-determined diabetes type, or case/control status, was waist circumference (11). We then established the range of IS for nondiabetic youth by applying the aforementioned equation to 2860 multiracial nondiabetic youth aged 12–20 years participating in the US National Health and Nutrition Examination Survey in 1999–2004 (12).
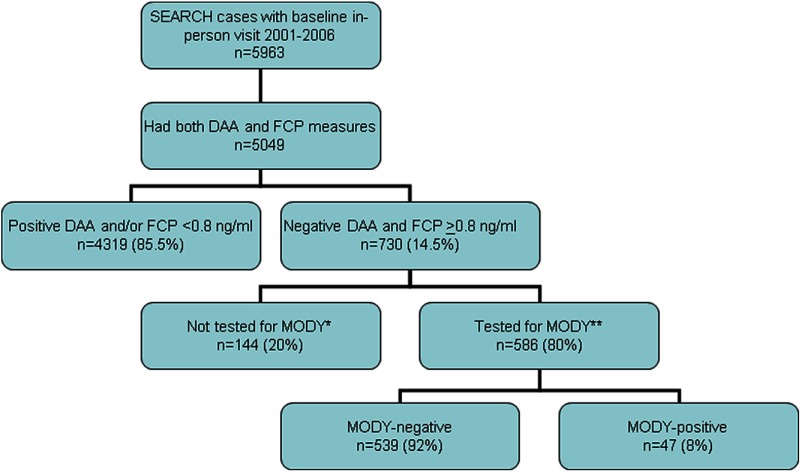
Figure 1.
Study sample, criteria used to select sample for screening for MODY, and MODY screening results. *, Reasons for not testing for HNF1A, HNF4A, or GCK included lack of consent for genetic testing, failed sequencing, and insufficient DNA sample; **, only 357 patients (61%) with A1c less than 75% were tested for GCK-MODY. MODY included HNF1A, HNF4A, and GCK. DAAs measured included glutamic acid decarboxylase 65 IA2.
Selection criteria for study sample
This report is based on DNA from 586 youth: 140 prevalent cases in 2001 and 446 incident cases diagnosed in 2002 through 2006 who participated in the SEARCH study visit and met eligibility criteria for genetic screening (see Figure 1). Eligibility criteria included the following: negativity for both the GAD65 and IA2 DAAs, FCP of 0.8 ng/mL or greater, stored DNA, and written consent for genetic testing. Additional selection criteria for GCK testing included A1c less than 7.5%, as in 303 UK patients with GCK mutations younger than 40 years of age, and no individual having an A1c greater than 7.5% (Hattersley, A. T., and S. Ellard, unpublished data).
Laboratory methods
GAD65 and IA2 autoantibodies were measured using a standardized radioligand-binding assay protocol and a common serum calibrator developed by the National Institute of Diabetes and Digestive and Kidney Diseases-sponsored standardization group (13). FCP levels were obtained during conditions of metabolic stability, at the same time as DAA, at a median of 13.0 (interquartile range 6.0–22.0) months after diagnosis, and were measured by a 2-site immunoenzymetric assay (Tosoh 1800; Tosoh Bioscience Inc). Measurements of serum lipids (cholesterol, triglycerides, low-density lipoprotein and high-density lipoprotein cholesterol) were performed using Roche reagent on a Roche Modular-P autoanalyzer (Roche Diagnostics). A1c was measured by a dedicated ion exchange HPLC instrument (TOSOH G7; Tosoh Bioscience). hsCRP was measured using Siemens reagent on a BN2 nepholometer autoanalyzer (Siemens Instruments). All analyses were performed at the Northwest Lipid Research Laboratory (University of Washington, Seattle, Washington).
Human leukocyte antigen (HLA) class II genotyping for HLA DRB1, DQA1, and DQB1 was performed in the laboratories of Drs L. K. Gaur (Genomic Research Laboratory, Puget Sound Blood Center, Seattle Washington) and H. Erlich (Roche Molecular Systems). Oligonucleotide probes corresponding to known polymorphic sequence motifs in HLA DRB1, DQA1, and DQB1 loci were immobilized on nylon membranes. The polymorphic second exons of DQA1, DQB1, and DRB1 were amplified using labeled primers (biotinylated), denatured, and hybridized to the immobilized probe array. Preliminary genotype data were imported into Sequence Compilation and Rearrangement Evaluation software for final genotype determination (14, 15). HLA genotypes were categorized as follows: susceptible, DR3/4 (4 genotypes), DR4/4 (4 genotypes), DR4/8 (4 genotypes), DR4/1 (1 genotype), DR 4/13 (1 genotype), DR 3/3 (1 genotype), DR 3/9 (1 genotype), DR 4/9 (2 genotypes), and DR 9/9 (1 genotype); neutral, DR4**/X*, DR3/X (where X = other non-high risk genotype), DR3/4-non-DQB1*0302, and DR4/DR4-DQB1*0301; and protective, DR-0403, DR2-DQB1*0602, DR7-DQB1*0303, and DR14-DQB1*0503, as recommended by the type 1 Diabetes Genetic Consortium, with modifications for the multiethnic population (14).
Mutational analysis
Genetic testing for HNF1A, HNF4A, and GCK was performed by the Department of Molecular Genetics, Royal Devon and Exeter National Health Service Foundation Trust (United Kingdom) as follows: the coding exons and conserved splice sites of HNF1A (n = 586), HNF4A (n = 569), and GCK (n = 357) were amplified by PCR. Primers and reaction conditions are available upon request. Sequence-specific primers for each amplicon were tagged with 5′ M13 tails to allow sequencing to be performed with a universal M13 primer. Single-strand sequencing was carried out using standard methods on an ABI 3730 (Applied Biosystems). Sequences were compared with the published reference sequences (accession numbers NM_000545.5, NM_000457.3, and NM_000162.3) using Mutation Surveyor version 3.24 (SoftGenetics). Any changes in the sequence were checked against published polymorphisms and mutations, the Exeter laboratory database, which includes GCK, HNF1A, and HNF4A sequence data for more than 1000 individuals. Novel missense mutations or in-frame deletions were classified as highly likely to be pathogenic if they affect an amino acid conserved through evolution.
Statistical analysis
Summary statistics on clinical and laboratory measures were calculated for the MODY+ and the MODY− groups. Means and SDs were calculated for continuous measures and percentages for categorical measures. Differences for categorical and continuous measures between the MODY+ and the MODY− groups were evaluated using nonparametric Fisher's exact and Wilcoxon rank-sum tests, respectively, due to the relatively small sample size of the MODY+ group. Statistical comparisons between different MODY subtypes were not performed due to the low numbers of participants in each group. All analyses were computed using SAS version 9.2 (SAS Institute), and significance was determined using a 2-sided significance level of P = .05.
Results
Frequency of MODY variants in the study sample
Among 586 youth, we identified 49 variants in the HNF1A, HNF4A, or GCK genes in 47 participants that were either proven mutations (n = 37) or novel highly likely pathogenic mutations (n = 12). The specific mutations are listed in Supplemental Table 1, published on The Endocrine Society's Journals Online web site at http://jcem.endojournals.org, and include 27 HNF1A mutations, 8 HNF4A mutations, and 14 GCK mutations. One participant had 2 different HNF1A mutations and one had mutations in both HNF1A and HNF4A; the latter participant's data are subsequently included with the HNF1A mutation carriers. Thus, the frequency of mutation carriers was 4.4% for HNF1A, 1.2% for HNF4A, and 2.4% for GCK. The overall frequency of having one of the 3 most common forms of MODY among DAA-negative participants with preserved FCP was 8.0%.
Clinical diagnosis
Only 6% of MODY+ individuals (3 of 47) were correctly identified as MODY by their provider clinically: 2 with HNF1A and 1 with HNF4A. Most MODY+ participants were diagnosed by the provider as type 1 diabetes (36%) or type 2 diabetes (T2D; 51%).
Demographic/clinical/biochemical characteristics
Most MODY+ participants were of minority ethnicity (64%), largely African American (20%) and Hispanic (31%), as well as Asian/Pacific Islander (11%) (Table). Frequency of minority ethnicity in MODY+ did not significantly differ from the MODY− group (64% vs 67%, P = ns), suggesting that it was reflective of the underlying racial/ethnic distribution of the group tested for MODY but did differ from the larger SEARCH population (64% vs 31%, P < .01).
A comparison of classical MODY diagnostic criteria (age of diagnosis < 25 years, not insulin dependent, and a dominant family history) between MODY+ and MODY− groups showed these were not good discriminating factors. MODY+ participants were slightly younger at diagnosis (11.5 ± 3.9 vs 13.3 ± 3.1 years, P < .01) and had a lower FCP (2.2 ± 1.4 vs 3.2 ± 2.1 ng/mL, P < .01) (Table 1), but these measures did not differentiate well on an individual basis. Parental history of diabetes did not differ between the 2 groups (50% vs 51%, P = 1.00). Susceptible HLA genotype differed significantly between MODY+ and MODY− groups (2% vs 12%, P < .05). MODY+ (vs MODY−) participants demonstrated a significantly lesser degree of T2D-like features, including a lower BMI Z-score (1.2 ± 1.0 vs 1.8 ± 1.0, P < .01), lower prevalence of acanthosis nigricans (40% vs 61%, P < .01), a smaller waist circumference (84.9 ± 19.2 vs 103.0 ± 25.1 cm, P < .01), lower CRP levels (0.15 ± 0.22 vs 0.45 ± 1.01 mg/dL, P < .01), and a higher IS index (8.6 ± 4.0 vs 5.4 ± 3.4, P < .01). Of note, the differences in hsCRP and C-peptide between the 2 groups were no longer significant after adjusting for waist circumference and BMI Z-score, suggesting that these differences are not inherent to the 2 groups but are primarily due to adiposity differences (Table 1). The MODY+ and MODY− groups did not have any significant differences in symptoms at clinical presentation, including weight loss (44% vs 49%), polyuria, polydipsia, or nocturia (82% vs 86%), gastrointestinal symptoms (59% vs 65%), or diabetic ketoacidosis within 6 months of diagnosis (23% vs 25%, all comparisons MODY+ vs MODY−). Other clinical and biochemical characteristics that were similar between the MODY+ and MODY− groups included A1c, birth weight, hypertension, hypertriglyceridemia, and dyslipidemia (see Table 1).
Table 1.
Demographic, Clinical, and Biochemical Characteristics and Pharmacological Treatment at the Time of the SEARCH Study Visit of Youth Screened for MODY by Genetically Defined MODY Status and MODY Type
| Total Sample Screened (n = 586) |
Positive for MODY by Genetic Testing (n = 47) |
||||
|---|---|---|---|---|---|
| MODY− (n = 539) | MODY+ (n = 47) | HNF1A-MODY (n = 26) | HNF4A-MODY (n = 7) | GCK-MODY (n = 14) | |
| Demographic characteristics | |||||
| Male gender | 41% | 38% | 27% | 50% | 54% |
| Race/ethnicity | |||||
| Non-Hispanic white | 33% | 36% | 31% | 50% | 38% |
| African American | 34% | 20% | 23% | 0% | 23% |
| Hispanic | 23% | 31% | 35% | 17% | 31% |
| Asian/Pacific Islander | 9% | 11% | 8% | 33% | 8% |
| Other/unknown | 1% | 2% | 4% | 0% | 0% |
| Clinical/biochemical characteristics ascertained at the time of the SEARCH study visit | |||||
| Age at diagnosis, y | 13.3 ± 3.1a | 11.5 ± 3.9 | 12.2 ± 3.0 | 11.4 ± 5.2 | 10.2 ± 4.7 |
| Age at the research visit, y | 15.2 ± 3.2 | 13.6 ± 3.4 | 13.7 ± 2.9 | 15.8 ± 3.8 | 12.3 ± 3.7 |
| Diabetes duration, y | 1.6 ± 1.8 | 1.3 ± 1.7 | 1.2 ± 1.1 | 2.1 ± 3.1 | 1.1 ± 1.8 |
| Parental history of diabetes, % | 51% | 50% | 52% | 71% | 30% |
| A1c, % | 7.5 ± 2.2 | 6.8 ± 1.5 | 6.9 ± 1.7 | 7.3 ± 1.4 | 6.4 ± 0.4 |
| Weight, kg | 88.2 ± 31.4a | 65.5 ± 29.6 | 70.4 ± 27.8 | 69.0 ± 29.1 | 53.9 ± 32.5 |
| Height, m | 1.64 ± 0.14a | 1.57 ± 0.18 | 1.60 ± 0.17 | 1.64 ± 0.15 | 1.48 ± 0.18 |
| BMI, kg/m2 | 32.4 ± 10.3a | 25.4 ± 7.5 | 26.7 ± 6.8 | 25.2 ± 8.0 | 22.8 ± 8.4 |
| BMI Z-score | 1.8 ± 1.0a | 1.2 ± 1.0 | 1.5 ± 0.9 | 0.8 ± 1.4 | 0.9 ± 1.1 |
| Waist circumference, cm | 103.0 ± 25.1a | 84.9 ± 19.2 | 89.9 ± 18.6 | 78.8 ± 12.4 | 76.6 ± 20.5 |
| Fasting C-peptide, ng/mL | 3.2 ± 2.1a | 2.2 ± 1.4 | 2.3 ± 1.2 | 2.1 ± 1.4 | 2.1 ± 1.8 |
| Fasting C-peptide, ng/mL, adjusted by zBMI (±SE) | 3.2 ± 0.1 | 2.8 ± 0.3 | 2.1 ± 0.2 | 2.4 ± 0.5 | 2.4 ± 0.3 |
| hsCRP, mg/dLb | 0.45 ± 1.01a | 0.15 ± 0.22 | 0.09 ± 0.11 | 0.07 ± 0.10 | 0.26 ± 0.32 |
| hsCRP, mg/dL, adjusted by zBMI (±SE)b | 0.43 ± 0.05 | 0.24 ± .19 | 0.06 ± 0.05c | 0.09 ± 0.11 | 0.29 ± 0.07 |
| IS index | 5.4 ± 3.4a | 8.6 ± 4.0 | 7.3 ± 3.6 | 9.8 ± 2.8 | 10.7 ± 4.2 |
| IS index, adjusted by zBMI, gender, and age at the time of the SEARCH study visit (±SE) | 5.6 ± 0.1a | 6.5 ± 0.3 | 8.5 ± 0.4 | 8.9 ± 0.8 | 9.2 ± 0.5 |
| Birth weight, kg | 3.3 ± 0.7 | 3.1 ± 0.9 | 3.1 ± 1.0 | 3.2 ± 0.3 | 3.2 ± 0.5 |
| Hypertension, % | 17% | 9% | 8% | 33% | 0% |
| Elevated triglycerides, % | 30% | 20% | 27% | 17% | 8% |
| Dyslipidemia, % | 73% | 60% | 69% | 67% | 38% |
| Acanthosis nigricans, % | 61%a | 40% | 50% | 33% | 23% |
| Susceptible HLA, %c | 12%c | 2% | 4% | 0% | 0% |
| Pharmacological treatment at the time of the SEARCH study visit | |||||
| Treated with insulin, % | 61% | 51% | 58% | 67% | 27% |
| Insulin dose, U/kg · dd | 0.57 ± 0.40 | 0.47 ± 0.30 | 0.49 ± 0.33 | 0.61 ± 0.20 | 0.14 ± 0.03 |
| Treated with OHA, % | 61% | 51% | 65% | 50% | 23% |
| Sulfonylurea, % | 5% | 5% | 8%e | 0%e | 0% |
| Metformin, % | 51% | 41% | 48% | 50% | 23% |
| Other/unknown OHA, % | 14% | 13% | 18% | 0% | 0% |
| No pharmacological treatment, % | 10% | 19% | 8% | 0% | 55%e |
Abbreviation: OHA, oral hypoglycemic agent. Data are percentage or mean ± SD (or ±SE for adjusted measures). Hypertension is the systolic or diastolic blood pressure greater than the 95th percentile for age, sex, and height (32); elevated triglycerides are fasting triglycerides greater than 150 mg/dL; dyslipidemia is triglycerides greater than 150 mg/dL, low-density lipoprotein greater than 100 mg/dL, high-density lipoprotein less than 35 mg/dL, or taking lipid-lowering medication; and susceptible HLA is HLA types predisposing to type 1 diabetes, as defined in Materials and Methods. Some HNF1A-MODY and HNF4A-MODY patients were treated with both insulin and an oral glucose-lowering agent.
P < 0.01; MODY− compared with all MODY+ combined.
Data were available on a subset of participants (334 MODY−, 27 MODY+, 14 HNF1A-MODY, 3 HNF4A-MODY, and 10 GCK-MODY).
P < .05.
Insulin dosage information available for 261 of 586 youth in the cohort.
Appropriate clinical treatment for each MODY subtype.
Pharmacological treatment
Participants with genetically identified MODY were seldom on an appropriate treatment, with only 11 (24%) being on sulfonylureas or no pharmacological treatment (Table 1). Most participants with HNF1A and HNF4A mutations were treated with insulin (58% and 67%, respectively) and/or metformin (48% and 50%, respectively). Of the 3 MODY+ participants identified as MODY by their provider, 2 were treated appropriately, one with a sulfonylurea and the other with diet alone. None of the HNF4A-MODY participants were treated with a sulfonylurea, including one who was diagnosed by the provider with MODY and being treated with insulin and metformin. Nearly half of the GCK-MODY participants were treated with insulin (27%) and/or an oral glucose-lowering agent (23%). The treatment regimens of the MODY+ group as a whole were similar to that of the MODY− group.
Discussion
In this report of systematic screening for the 3 most common MODY subtypes in SEARCH participants who were DAA negative with a fasting C-peptide of 0.8 ng/mL or greater, the frequency of a genetic diagnosis of HNF1A-, HNF4A-, or GCK-MODY was 8.0%. Applied to the larger sample of youth who participated in a SEARCH study visit and had negative DAA and preserved FCP (n = 730), this results in an estimated prevalence of these 3 MODY subtypes of at least 1.2% in this US population-based study of pediatric diabetes. Applying this proportion to the SEARCH prevalence estimate of 182 diabetes cases per 100 000 for youth younger than 20 years of age in 2001 (16), the prevalence of monogenic diabetes in the US pediatric population is approximately 2.1 per 100 000. This is likely to be a minimum prevalence because exonic deletions and noncoding regions outside the conserved splice sites were not studied. Furthermore, there may be other MODY cases who were not screened because they were DAA positive or had a FCP less than 0.8 ng/ml.
We tested for GCK-MODY only in those participants with A1c less than 7.5, based on prior experience in a European cohort that was predominantly normal weight; it is possible that we may have missed some cases of glucokinase-MODY by excluding those with an A1c of 7.5% or greater because some youth in our diverse cohort may have had both GCK-MODY plus obesity-associated insulin resistance, resulting in more severe hyperglycemia. In addition, we assumed that youth who participated in a study visit were similar in clinical characteristics to those who did not (50% of the study population). Nevertheless, this study provides the best prevalence estimate to date of MODY in the US pediatric population. Several European studies have estimated that MODY accounts for anywhere from less than 1% to 2.4% of pediatric diabetes cases (4–6), but these estimates are largely based on clinical, rather than genetic, diagnoses. In addition, these studies were performed in studies that were largely Caucasians. Our results, which are consistent with the European estimates, represent the first systematic study of MODY mutations in a large pediatric diabetes cohort, unselected by referral pattern or clinical features such as parental history of diabetes.
Negative DAA and preserved β-cell function were used as screening criteria because previous studies have shown that the vast majority of individuals confirmed to have MODY in a population-based screening would fall into that category (17). To our knowledge, no patients below the age of 20 years have been reported with the common subtypes of MODY and undetectable C-peptide levels (18). Although there have been reports of positive DAA in patients with MODY (6), in a large robustly conducted study of more than 500 patients with MODY, only 1% of subjects had positive antibodies (17). Based on our current understanding of the pathophysiology of MODY, autoimmunity is not likely to play a major role, so few cases will be missed by this screening approach.
When a patient with diabetes first presents for medical attention, the clinician must assess diabetes type to institute appropriate management. Classic MODY criteria of age at diagnosis younger than 25 years, an autosomal dominant family history for diabetes, and preserved β-cell function were established before T2D was diagnosed in children and are best for discriminating MODY from type 1 diabetes (7). Although a strong autosomal dominant mode of inheritance of diabetes may provide information to distinguish between type 1 diabetes and MODY, this study demonstrates that family history is not very sensitive, with only half of MODY+ participants reporting a parental history of diabetes. Family history was not useful in differentiating MODY+ from MODY− participants, nor was it useful in differentiating them from clinically diagnosed T2D participants (data not shown). We are likely to have underestimated the true frequency of positive family history of diabetes because we used self-report as the method of assessment, and parental diabetes may be undiagnosed if they have not been tested. However, this is not likely to be significantly different between groups or from that occurring in clinical practice.
With a rising prevalence of obesity in children (19), T2D is becoming increasingly more common, with approximately 12% of teenagers with newly diagnosed diabetes in the United States now classified as having T2D (20). In our study, the exclusion by DAA and FCP measurements meant that our patients predominantly had nonautoimmune diabetes; hence, the major differential diagnosis was with T2D. Although many T2D-like characteristics were significantly more common in MODY− compared with MODY+ patients, none of these were discriminatory at the individual level. For example, more than half of the MODY+ participants had at least one metabolic abnormality associated with T2D and 64% were of minority ethnicity. MODY has been studied in some non-Caucasian populations (21, 22), but most genetic testing has been performed in homogeneous European Caucasian populations (1, 4, 5, 23–25). A study of UK children with clinical diagnoses other than type 1 diabetes found that MODY is present in UK Asians, but the number of Asian families referred for genetic testing was lower than expected, either because these families declined testing or because they were assumed to have T2D (26).
In our study, nonadjusted hsCRP levels were lower in the MODY+ group as a whole; however, using less than 0.075 mg/dL as a cutoff for genetic screening (8) would have missed 41% of MODY patients (11 of 27), including 5 of 14 HNF1A-MODY patients. In addition, the observed significant in differences in hsCRP and FCP between the 2 groups disappeared after adjustments were made for adiposity differences, suggesting that such differences are not inherent to being MODY positive or negative. Thus, this study questions whether the classical clinical criteria or the currently available biomarkers are sufficiently specific and sensitive to reliably identify candidates for genetic testing.
The difficulty of identifying appropriate patients to screen for MODY mutations is illustrated by the very low rate of correct clinical MODY diagnoses in the SEARCH participants. Only 3 of the 47 MODY+ participants had a clinical diagnosis of MODY. An important consequence of misdiagnosis is incorrect clinical treatment. Mutations in the transcription factors HNF1A and HNF4A result in a reduced insulin secretory response to glucose but show marked sensitivity to low-dose sulfonylureas (27–29). This therapy is a marked departure from the exogenous insulin requirement seen in most youth with diabetes. Furthermore, patients with GCK mutations have altered glucose sensing by the β-cell, resulting in a mild defect in insulin secretion (30). These patients do not require any pharmacological treatment and do not develop diabetes complications (2, 31). Another consequence of misdiagnosis is the missed opportunity for genetic counseling and early diagnosis in siblings and other relatives including the patients' offspring who have a 50% risk for developing the same condition.
The major strengths of this study are the large size, diversity of the tested cohort, and the systematic approach to genetic testing using laboratory rather than clinical criteria. Limitations include the fact that selection criteria for genetic testing were based on 2 DAAs only and FCP levels obtained at a baseline visit on participants with variable diabetes duration. We did not screen participants who were DAA positive and/or who had low FCP; therefore, our approach could potentially have underestimated the prevalence of MODY. In addition, potential causal mutations outside coding exons or heterozygous deletions of coding exons or regulatory sequences would be missed by the genetic tests performed, also contributing to the possible underestimation of the prevalence of MODY. Finally, in most cases for novel mutations identified by this study, the pathogenicity was determined by assessment of protein conservation across species, rather than by tracking cosegregation of mutations with disease within a family because family testing was not performed.
In conclusion, MODY due to the 3 most common genetic etiologies is not uncommon in the pediatric diabetes population, with an estimated prevalence of at least 1.2%, the diagnosis is often missed, and MODY patients are often inappropriately treated. Among a population enriched for MODY+ participants (nonautoimmune, with preserved β-cell function), clinical and biochemical characteristics, including a family history, do not clearly identify those with monogenic diabetes. A correct diagnosis of MODY has clinical implications for the patients and potentially for family members. Misdiagnosis and inappropriate treatment may have a significant impact on quality of life, long-term health outcomes, and cost associated with unnecessary treatment with insulin. Thus, a MODY diagnosis should be considered in pediatric patients who lack evidence of autoimmunity and have preserved β-cell function. Further studies are needed to examine the relative costs and benefits of genetic testing for MODY in youth with nonautoimmune diabetes.
Supplementary Material
Acknowledgments
The SEARCH for Diabetes in Youth Study is indebted to the many youth and their families, and their health care providers, whose participation made this study possible. We thank Andrew Parrish, Marcus Mitchell, and Kevin Colclough for technical assistance with the molecular genetic testing.
The following authors made contributions to this study: L.K.G. made substantial contributions to the study conception and design, was involved with the analysis and interpretation of the data, drafted the article, gave final approval of the version to be published, and takes responsibility for the contents of the manuscript; C.P., S.E., and A.T.H. made substantial contributions to the study conception and design, were involved with the acquisition of data and the analysis and interpretation of the data, revised the article critically for important intellectual content, and gave final approval of the version to be published; and D.D., C.D., L.M.D., C.J.G., G.I., J.M.L., S.M.M., E.M.-D., B.L.R., A.K.S., and D.E.W. were involved with the acquisition of the data and analysis and the interpretation of data, revised the article critically for important intellectual content, and gave final approval of the version to be published.
The contents of this paper are solely the responsibility of the authors and do not necessarily represent the official views of the Centers for Disease Control and Prevention or the National Institute of Diabetes and Digestive and Kidney Diseases.
The SEARCH for Diabetes in Youth is supported by the Centers for Disease Control and Prevention (PAs 00097, DP-05-069, and DP-10-001) and supported by the National Institute of Diabetes and Digestive and Kidney Diseases. Site contract numbers included the following: Kaiser Permanente Southern California (U48/CCU919219, U01 DP000246, and U18DP002714); University of Colorado Denver (U48/CCU819241-3, U01 DP000247, and U18DP000247-06A1); Kuakini Medical Center (U58CCU919256 and U01 DP000245); Children's Hospital Medical Center (Cincinnati) (U48/CCU519239, U01 DP000248, and 1U18DP002709); University of North Carolina at Chapel Hill (U48/CCU419249, U01 DP000254, and U18DP002708-01), University of Washington School of Medicine (U58/CCU019235-4, U01 DP000244, and U18DP002710-01); and Wake Forest University School of Medicine (U48/CCU919219, U01 DP000250, and 200-2010-35171).
We also acknowledge the involvement of General Clinical Research Centers at the South Carolina Clinical, Translational Research Institute, at the Medical University of South Carolina [National Institutes of Health (NIH)/National Center for Research Resources Grant UL1RR029882]; the Children's Hospital and Regional Medical Center (Grant M01RR00037); the Colorado Pediatric General Clinical Research Center (Grant M01 RR00069) and the Barbara Davis Center at the University of Colorado at Denver (DERC NIH Grant P30 DK57516); and the Institutional Clinical and Translational Science Award), NIH/National Center for Research Resources at the University of Cincinnati (Grant 1UL1RR026314-01). The Monogenic Diabetes ancillary study was supported by the Juvenile Diabetes Research Foundation (Grant JDRF 9-2007-1700). The staff at the University of Exeter were supported by the European Union funding from FP7 Integrated Project CEED3 and Madam Curie initial funding network Biology of Liver and Pancreas Development (BOLD) and the Peninsula National Institute for Health Research Clinical Research Facility.
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- A1c
- glycosylated hemoglobin
- BMI
- body mass index
- CRP
- C-reactive protein
- DAA
- diabetes autoantibodies
- FCP
- fasting C-peptide
- GAD65
- glutamic acid decarboxylase 65
- GCK
- glucokinase
- HLA
- human leukocyte antigen
- HNF
- hepatocyte nuclear factor
- hsCRP
- high-sensitivity CRP
- IA2
- islet-antibody 2
- IS
- insulin sensitivity
- MODY
- maturity-onset diabetes of the young
- T2D
- type 2 diabetes.
References
- 1. Shields B, Hicks S, Shepherd M, Colclough K, Hattersley A, Ellard S. Maturity-onset diabetes of the young (MODY): how many cases are we missing? Diabetologia. 2010;53:2504–2508 [DOI] [PubMed] [Google Scholar]
- 2. Murphy R, Ellard S, Hattersley AT. Clinical implications of a molecular genetic classification of monogenic β-cell diabetes. Nat Clin Pract Endocrinol Metab. 2008;4:200–213 [DOI] [PubMed] [Google Scholar]
- 3. Ellard S, Bellanné-Chantelot C, Hattersley A. Best practice guidelines for the molecular genetic diagnosis of maturity-onset diabetes of the young. Diabetologia. 2008;51:546–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Galler A, Stange T, Müller G, et al. Incidence of childhood diabetes in children aged less than 15 years and its clinical and metabolic characteristics at the time of diagnosis: data from the childhood diabetes registry of Saxony, Germany. Horm Res Paediatr. 2010;74:285–291 [DOI] [PubMed] [Google Scholar]
- 5. Neu A, Feldhahn L, Ehehalt S, Hub R, Ranke MB, on behalf of the Diary Group Baden-Württemberg Type 2 diabetes mellitus in children and adolescents is still a rare disease in Germany: a population-based assessment of the prevalence of type 2 diabetes and MODY in patients aged 0–20 years. Pediatr Diabetes. 2009;10:468–473 [DOI] [PubMed] [Google Scholar]
- 6. Schober E, Rami B, Grabert M, et al. Phenotypical aspects of maturity-onset diabetes of the young (MODY diabetes) in comparison with type 2 diabetes mellitus (T2DM) in children and adolescents: experience from a large multicentre database. Diabet Med. 2009;26:466–473 [DOI] [PubMed] [Google Scholar]
- 7. Tattersall RB. Mild familial diabetes with dominant inheritance. Q J Med. 1974;43:339–357 [PubMed] [Google Scholar]
- 8. McDonald TJ, Shields BM, Lawry J, et al. High-sensitivity CRP discriminates HNF1a-MODY from other subtypes of diabetes. Diabetes Care. 2011;34:1860–1862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Owen KR, Thanabalasingham G, James TJ, et al. Assessment of high-sensitivity c-reactive protein levels as diagnostic discriminator of maturity-onset diabetes of the young due to HNF1a mutations. Diabetes Care. 2010;33:1919–1924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. SEARCH Study Group Search for Diabetes in Youth: a multicenter study of the prevalence, incidence and classification of diabetes mellitus in youth. Control Clin Trials. 2004;25:458–471 [DOI] [PubMed] [Google Scholar]
- 11. Dabelea D, D'Agostino R, Mason C, et al. Development, validation and use of an insulin sensitivity score in youths with diabetes: the SEARCH for Diabetes in Youth study. Diabetologia. 2011;54:78–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Centers for Disease Control and Prevention National Health and Nutrition Examination Survey. http://www.cdc.gov/nchs/nhanes/nhanes_questionnaires.htm Accessed February 27, 2006
- 13. Bonifacio E, Yu L, Williams AK, et al. Harmonization of glutamic acid decarboxylase and islet antigen-2 autoantibody assays for National Institute of Diabetes and Digestive and Kidney Diseases consortia. J Clin Endocrinol Metab. 2010;95:3360–3367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Erlich H, Valdes AM, Noble J, et al. HLA DR-DQ haplotypes and genotypes and type 1 diabetes risk. Diabetes. 2008;57:1084–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Maldonado M, Hampe CS, Gaur LK, et al. Ketosis-prone diabetes: dissection of a heterogeneous syndrome using an immunogenetic and β-cell functional classification, prospective analysis, and clinical outcomes. J Clin Endocrinol Metab. 2003;88:5090–5098 [DOI] [PubMed] [Google Scholar]
- 16. Liese A, D'Agostino RJ, Hamman R, et al. The burden of diabetes mellitus among US youth: prevalence estimates from the Search for Diabetes in Youth study. Pediatrics. 2006;118:1510–1518 [DOI] [PubMed] [Google Scholar]
- 17. McDonald TJ, Colclough K, Brown R, et al. Islet autoantibodies can discriminate maturity-onset diabetes of the young (MODY) from type 1 diabetes. Diabet Med. 2011;28:1028–1033 [DOI] [PubMed] [Google Scholar]
- 18. Besser REJ, Shepherd MH, McDonald TJ, et al. Urinary c-peptide creatinine ratio is a practical outpatient tool for identifying hepatocyte nuclear factor 1-α/hepatocyte nuclear factor 4-α maturity-onset diabetes of the young from long-duration type 1 diabetes. Diabetes Care. 2011;34:286–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ogden CL, Flegal KM, Carroll MD, Johnson CL. Prevalence and trends in overweight among US children and adolescents, 1999–2000. JAMA. 2002;288:1728–1732 [DOI] [PubMed] [Google Scholar]
- 20. Dabelea D, Bell R, D'Agostino RJ, et al. Incidence of diabetes in youth in the United States. JAMA. 2007;297:2716–2724 [DOI] [PubMed] [Google Scholar]
- 21. Yamada S, Nishigori H, Onda H, et al. Identification of mutations in the hepatocyte nuclear factor (HNF)-1α gene in Japanese subjects with IDDM. Diabetes. 1997;46:1643–1647 [DOI] [PubMed] [Google Scholar]
- 22. Radha V, Ek J, Anuradha S, Hansen T, Pedersen O, Mohan V. Identification of novel variants in the hepatocyte nuclear factor-1α gene in South Indian patients with maturity onset diabetes of the young. J Clin Endocrinol Metab. 2009;94:1959–1965 [DOI] [PubMed] [Google Scholar]
- 23. Bjørkhaug L, Sagen JV, Thorsby P, Søvik O, Molven A, Njølstad PR. Hepatocyte nuclear factor-1α gene mutations and diabetes in Norway. J Clin Endocrinol Metab. 2003;88:920–931 [DOI] [PubMed] [Google Scholar]
- 24. Frayling TM, Bulamn MP, Ellard S, et al. Mutations in the hepatocyte nuclear factor-1α gene are a common cause of maturity-onset diabetes of the young in the U.K. Diabetes. 1997;46:720–725 [DOI] [PubMed] [Google Scholar]
- 25. Vaxillaire M, Rouard M, Yamagata K, et al. Identification of nine novel mutations in the hepatocyte nuclear factor 1α are associated with maturity-onset diabetes of the young (MODY3). Hum Mol Genet. 1997;6:583–586 [DOI] [PubMed] [Google Scholar]
- 26. Porter JR, Rangasami JJ, Ellard S, et al. Asian MODY: are we missing an important diagnosis? Diabet Med. 2006;23:1257–1260 [DOI] [PubMed] [Google Scholar]
- 27. Pearson ER, Pruhova S, Tack CJ, et al. Molecular genetics and phenotypic characteristics of MODY caused by hepatocyte nuclear factor 4α mutations in a large European collection. Diabetologia. 2005;48:878–885 [DOI] [PubMed] [Google Scholar]
- 28. Pearson ER, Starkey BJ, Powell RJ, Gribble FM, Clark PM, Hattersley AT. Genetic cause of hyperglycaemia and response to treatment in diabetes. Lancet. 2003;362:1275–1281 [DOI] [PubMed] [Google Scholar]
- 29. Shepherd M, Shields B, Ellard S, Rubio-Cabezas O, Hattersley AT. A genetic diagnosis of HNF1A diabetes alters treatment and improves glycaemic control in the majority of insulin-treated patients. Diabet Med. 2009;26:437–441 [DOI] [PubMed] [Google Scholar]
- 30. Froguel P, Zouali H, Vionnet N, et al. Familial hyperglycemia due to mutations in glucokinase—definition of a subtype of diabetes mellitus. N Engl J Med. 1993;328:697–702 [DOI] [PubMed] [Google Scholar]
- 31. Hattersley A, Bruining J, Shield J, Njolstad P, Donaghue K. ISPAD Clinical Practice Consensus Guidelines 2006–2007. The diagnosis and management of monogenic diabetes in children. Pediatr Diabetes. 2006;7:352–360 [DOI] [PubMed] [Google Scholar]
- 32. Rodriguez BL, Dabelea D, Liese AD, et al. Prevalence and correlates of elevated blood pressure in youth with diabetes mellitus: the Search for Diabetes in Youth study. J Pediatr. 2010;157:245–251 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.