Abstract
Systemic lupus erythematosus is an autoimmune disease characterized by multi-system involvement and autoantibody production. Abnormal T cell DNA methylation and type-I interferon play an important role in the pathogenesis of lupus. We performed a genome-wide DNA methylation study in two independent sets of lupus patients and matched healthy controls to characterize the DNA methylome in naïve CD4+ T cells in lupus. DNA methylation was quantified for over 485,000 methylation sites across the genome, and differentially methylated sites between lupus patients and controls were identified and then independently replicated. Gene expression analysis was also performed from the same cells to investigate the relationship between the DNA methylation changes observed and mRNA expression levels. We identified and replicated 86 differentially methylated CG sites between patients and controls in 47 genes, with the majority being hypomethylated. We observed significant hypomethylation in interferon-regulated genes in naïve T cells from lupus patients, including IFIT1, IFIT3, MX1, STAT1, IFI44L, USP18, TRIM22 and BST2, suggesting epigenetic transcriptional accessibility in these genetic loci. Indeed, the majority of the hypomethylated genes (21 out of 35 hypomethylated genes) are regulated by type I interferon. The hypomethylation in interferon-regulated genes was not related to lupus disease activity. Gene expression analysis showed overexpression of these genes in total but not naïve CD4+ T cells from lupus patients. Our data suggest epigenetic “poising” of interferon-regulated genes in lupus naïve CD4+ T cells, argue for a novel pathogenic implication for abnormal T cell DNA methylation in lupus, and suggest a mechanism for type-I interferon hyper-responsiveness in lupus T cells.
Keywords: Lupus, naïve CD4+ T cells, methylome, DNA methylation
1. Introduction
Systemic lupus erythematosus is a chronic autoimmune disease characterized by the production of antinuclear antibody and multiple organ involvement. The etiology of lupus is incompletely understood, but clear evidence suggests an important role for abnormal T cell DNA methylation in the pathogenesis of the disease [1]. Indeed, demethylated T cells are sufficient to cause a lupus-like disease in mouse models [2].
DNA methylation is an epigenetic mechanism that regulates gene expression by altering transcriptional accessibility of regulatory regions within gene sequences. This chemical modification of cytosine residues most commonly occurs in CG dinucleotides, and is mediated by DNA methyltransferase enzymes [3]. In general, methylation of CG dinucleotides in regulatory sequences induces gene silencing, while hypomethylation allows for transcriptional chromatin accessibility, and active gene expression when appropriate transcription factors are available [3]. DNA methylation induces chromatin inaccessibility by several mechanisms, including recruitment of histone deacetylases that remove acetyl groups from histone tails thereby increasing the charge attraction between DNA and histone proteins to generate more compact chromatin configuration that prevents access by the transcriptional machinery [4].
DNA methylation plays an important role in T cell differentiation. Indeed, the interferon gamma locus demethylates upon TH1 differentiation, and the interleukin (IL)-4,5 and 13 common locus control region demethylates upon TH2 differentiation, allowing for interferon gamma, and IL-4, IL-5, and IL-13 production in differentiated TH1 and TH2 cells, respectively [5]. In contrast, both loci are heavily methylated in naïve CD4+ T cells [5].
We have previously characterized DNA methylation changes in total CD4+ T cells from lupus patients and revealed wide-spread DNA methylation changes in patients compared to healthy controls [6]. Herein, we performed an extensive genome-wide DNA methylation study in naïve CD4+ T cells from lupus patients and controls, coupled with gene expression profiling from the same cells. We identified DNA methylation changes prior to T cell differentiation and activation in lupus and determined the effect of these methylation changes on gene expression.
2. Methods
2.1 Lupus patients and controls
We studied two independent sets of female lupus patients and controls, each consisting of 36 participants (18 lupus patients and 18 healthy controls). We designed our study to include a discovery cohort and a second independent cohort for replication. The discovery cohort was recruited from the Oklahoma Lupus Cohort at the Oklahoma Medical Research Foundation (OMRF), and the replication cohort was subsequently recruited from the University of Michigan rheumatology clinics. Patients and controls in both sets were matched for age (+/− 5 years) and ethnicity (Table I). Our study was approved by the institutional review boards at OMRF and the University of Michigan. All study participants singed a written informed consent prior to participation in the study.
Table I.
Demographic characteristics and disease activity as measured by Systemic Lupus Erythematosus Disease Activity Index (SLEDAI) scores in the lupus patients included in our study. Two independent cohorts of lupus patients and age-, sex- and ethnicity-matched controls were included in this study.
| Cohort 1 |
Cohort 2 |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Patients |
Controls |
Patients |
Controls |
||||||||||||||
| Number | Age | Sex | Ethnicity | SLEDAI | Number | Age | Sex | Ethnicity | Number | Age | Sex | Ethnicity | SLEDAI | Number | Age | Sex | Ethnicity |
| 1 | 33 | Female | EA | 6 | C1 | 35 | Female | EA | 19 | 22 | Female | AA | 6 | C19 | 25 | Female | AA |
| 2 | 58 | Female | EA | 0 | C2 | 53 | Female | EA | 20 | 51 | Female | EA | 3 | C20 | 47 | Female | EA |
| 3 | 33 | Female | EA | 4 | C3 | 30 | Female | EA | 21 | 58 | Female | EA | 6 | C21 | 55 | Female | EA |
| 4 | 50 | Female | AA | 4 | C4 | 52 | Female | AA | 22 | 54 | Female | EA | 10 | C22 | 50 | Female | EA |
| 5 | 36 | Female | EA | 0 | C5 | 41 | Female | EA | 23 | 25 | Female | EA | 4 | C23 | 24 | Female | EA |
| 6 | 25 | Female | EA | 2 | C6 | 26 | Female | EA | 24 | 18 | Female | EA | 4 | C24 | 23 | Female | EA |
| 7 | 58 | Female | EA | 2 | C7 | 58 | Female | EA | 25 | 36 | Female | EA | 2 | C25 | 36 | Female | EA |
| 8 | 35 | Female | AA | 0 | C8 | 35 | Female | AA | 26 | 40 | Female | EA | 2 | C26 | 45 | Female | EA |
| 9 | 30 | Female | EA | 2 | C9 | 27 | Female | EA | 27 | 34 | Female | AA | 4 | C27 | 32 | Female | AA |
| 10 | 30 | Female | AA | 4 | C10 | 32 | Female | AA | 28 | 41 | Female | EA | 6 | C28 | 38 | Female | EA |
| 11 | 40 | Female | EA | 4 | C11 | 40 | Female | EA | 29 | 63 | Female | EA | 0 | C29 | 58 | Female | EA |
| 12 | 57 | Female | AA | 2 | C12 | 57 | Female | AA | 30 | 38 | Female | EA | 12 | C30 | 36 | Female | EA |
| 13 | 41 | Female | EA | 4 | C13 | 45 | Female | EA | 31 | 39 | Female | EA | 2 | C31 | 41 | Female | EA |
| 14 | 29 | Female | EA | 2 | C14 | 34 | Female | EA | 32 | 53 | Female | EA | 2 | C32 | 53 | Female | EA |
| 15 | 25 | Female | AA | 0 | C15 | 27 | Female | AA | 33 | 24 | Female | EA | 4 | C33 | 26 | Female | EA |
| 16 | 26 | Female | Asian | 4 | C16 | 29 | Female | Asian | 34 | 33 | Female | AA | 4 | C34 | 34 | Female | AA |
| 17 | 66 | Female | EA | 0 | C17 | 64 | Female | EA | 35 | 40 | Female | EA | 6 | C35 | 40 | Female | EA |
| 18 | 34 | Female | EA | 2 | C18 | 30 | Female | EA | 36 | 34 | Female | AA | 2 | C36 | 38 | Female | AA |
SLEDAI, Systemic Lupus Erythematosus Disease Activity Index; EA, European-American; AA, African-American.
2.2 Naïve CD4+ T cell isolation and purity
Peripheral blood mononuclear cells (PBMCs) were isolated from fresh blood samples obtained from patients and controls using density gradient centrifugation (Amersham Biosciences, Uppsala, Sweden). Naïve CD4+ T cells were separated from PBMCs using the Naive CD4+ T Cell Isolation Kit II (Miltenyi Biotec, Cambridge, MA) which allows for the indirect isolation of untouched naïve CD4+ T cells. Naïve CD4+ T cell purity was confirmed by flow cytometry using fluorochrome-conjugated antibodies against CD4 and CD45RA; we have consistently achieved cell purity of >95% (Supplementary Figure 1). DNA was isolated using the DNeasy Kit (Qiagen, Valencia, CA) for subsequent use in the DNA methylation studies. An aliquot of naïve CD4+ T cells from a subset of the samples was stored in TRIzol® reagent at −80C and later used for RNA extraction and mRNA expression experiments.
2.3 DNA methylation studies and array validation
Genome-wide DNA methylation in naïve CD4+ T cells from lupus patients and controls included in the discovery and the replication cohorts was assessed using the Illumina Infinium HumanMethylation450 BeadChip array. This array allows for the interrogation of over 485,000 methylation sites within the entire genome. This array covers 99% of RefSeq genes, with an average of 17 CG sites per gene across the promoter region, 5'-UTR, first exon, gene body, and 3'-UTR. It also covers 96% of CG islands. Non CpG methylated sites recently identified in human stem cells are also covered as well as microRNA promoter regions. Validation of the array data was performed using bisulfite DNA sequencing in known hypermethylated and hypomethylated regions as previously described [6].
2.4 Gene expression studies
RNA extraction was performed using a combination of TRIzol (Invitrogen, Carlsbad, CA) and RNeasy kits (Qiagen, Valencia, CA) as previously described [7]. RNA concentration was determined with a NanoDrop 1000 Spectrophotometer (Thermo Scientific, Wilmington, DE) and then qualitatively assessed for degradation with 28:18S ribosomal RNA, using a capillary gel electrophoresis system (Agilent 2100 Bioanalyzer, Agilent, Wilmington, DE). Gene expression profiling was performed in naïve CD4+ T cells from lupus patients and controls using the HumanHT-12 v4 Expression BeadChip array (Illumina).
2.5 Statistical and bioinformatics analysis
DNA methylation analysis was performed using GenomeStudio methylation analysis package (Illumina) as previously described [6]. The average level of DNA methylation (β) on each CG site was first compared between lupus patients and controls in the discovery cohort. Differentially methylated CG sites were identified in the discovery cohort and were defined as CG site with an average difference in DNA methylation level of at least 1.2-fold, and differential methylation score of ≥ 33 (P≤ 0.001) after adjusting for multiple testing using the Benjamini and Hochberg false discovery rate of 5%. Differential methylation score is defined as 10*sgn(βRisk-βProtective)*log10P. CG sites assessed with probes with a genetic variant located within 10bp of the 3’ end of the probe were excluded from the analysis. CG sites that met the aforementioned criteria for being differentially methylated between patients and controls in the discovery cohort were assessed in the replication cohort. For these CG sites to be considered established differentially methylated sites and included in further analysis, we required an FDR-adjusted differential methylation score of at least 13 (P≤ 0.05) in the replication set. Microarray expression data were normalized and gene expression differences between patients and controls were detected using a 2-tailed t-test adjusted for multiple testing using Bonferroni correction. Gene ontology, network, and pathway analysis was performed using Ingenuity Pathway Analysis (Redwood City, CA) and the IRIDESCENT algorithm [8].
3. Results
We identified and then validated DNA methylation changes in naïve CD4+ T cells in lupus using two independent sets of lupus patients and age-, sex-, and ethnicity-matched controls (Table I). We identified and replicated 86 CG sites that are differentially methylated in naïve CD4+ T cells from lupus patients. Sixty sites were hypomethylated and 26 were hypermethylated in patients compared to controls (Supplementary Table I). A total of 47 differentially methylated unique genes were identified in naïve CD4+ T cells from lupus patients, with the majority (35 genes, 75%) being hypomethylated. Bisulfite DNA sequencing was used to validate the DNA methylation array results in a number of known hypermethylated and hypomethylated loci in the genome as described previously [6] in all the samples included in the discovery set (cohort 1). The correlation r2 value between the array data and bisulfite sequencing data was 0.864.
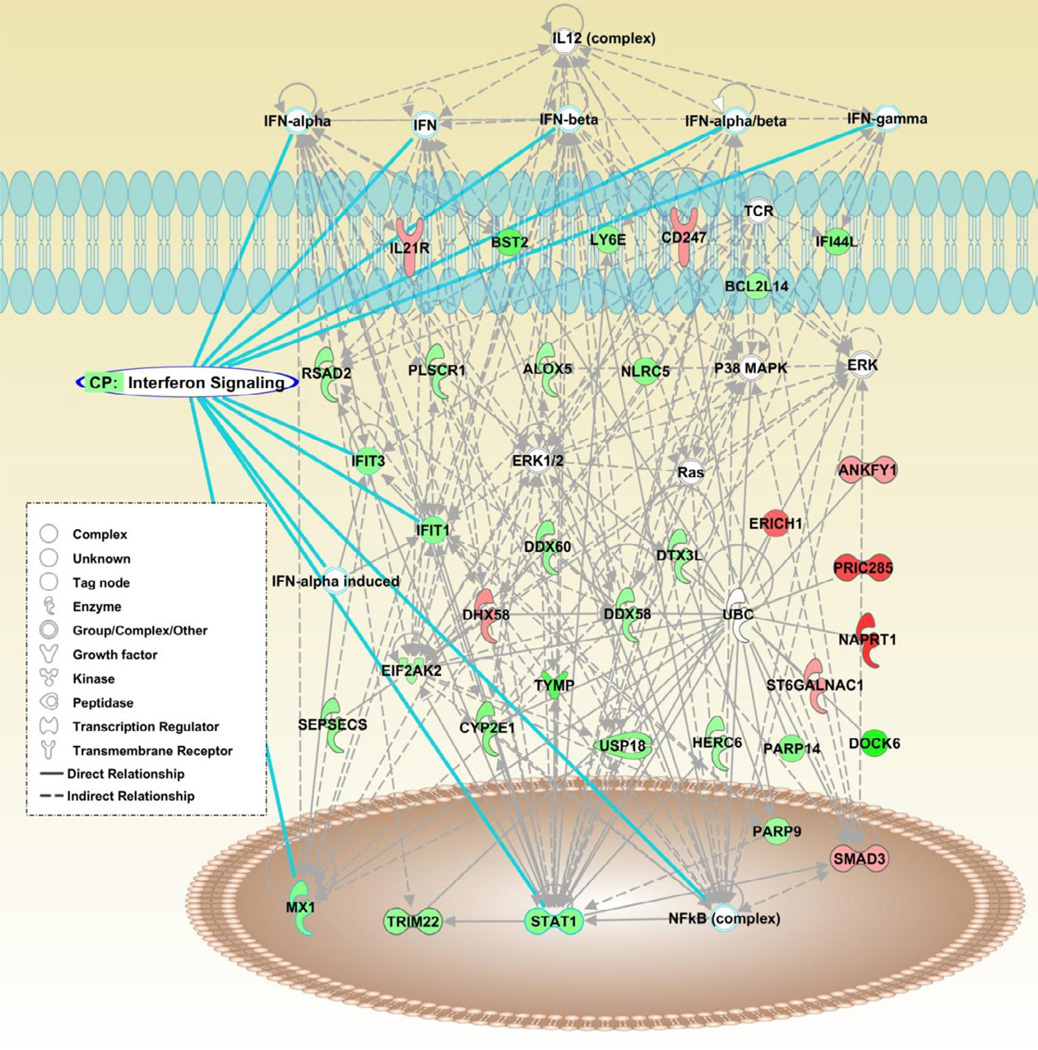
Canonical pathway analysis identified interferon-signaling as the most significant pathway unifying the differentially methylated genes in naïve CD4+ T cells from lupus patients (P=8.99×10−7). Network analysis revealed a number of type-I interferon-regulated genes among the differentially methylated loci detected, and most of these genes were hypomethylated in naïve CD4+ T cells from lupus patients (Figure 1). Indeed, the majority of the hypomethylated genes (21 out of 35 hypomethylated genes) are interferon-regulated. Significantly hypomethylated genes regulated by type I interferon include IFIT1, IFIT3, MX1, STAT1, IFI44L, USP18, TRIM22 and BST2 among others (Table II). An analysis of transcription factor binding site enrichment within the promoters of the differentially methylated genes identified canonical binding sites for numerous interferon regulatory factors (Table III).
Figure 1.
Network analysis identifies the interferon pathway among differentially methylated genes in naïve CD4+ T cells from lupus patients. Genes hypomethylated in lupus patients in this network are in green, and hypermethylated genes are in red. Genes and molecules that are brought into the network by the analysis software due to statistically enriched relationships with differentially methylated genes are in colorless shapes. Analysis was performed using ingenuity software.
Table II.
Differentially methylated genes in naïve CD4+ T cells between lupus patients and controls that are regulated by type-I interferon.
| Cohort 1 |
Cohort 2 |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene | CG site ID | Methylation fraction cases |
Methylation fraction controls |
Differential methylation score |
Case/control ratio |
Methylation fraction cases |
Methylation fraction controls |
Differential methyltaion score |
Case/control ratio |
| BST2 | cg20092122 | 0.125 | 0.253 | −198.143 | 0.495 | 0.124 | 0.231 | −143.391 | 0.535 |
| cg16363586 | 0.334 | 0.436 | −66.219 | 0.766 | 0.293 | 0.419 | −115.114 | 0.699 | |
| cg01329005 | 0.209 | 0.344 | −161.483 | 0.607 | 0.211 | 0.322 | −107.917 | 0.654 | |
| cg09993699 | 0.216 | 0.311 | −74.424 | 0.694 | 0.210 | 0.307 | −81.454 | 0.684 | |
| cg01254505 | 0.083 | 0.162 | −101.579 | 0.511 | 0.085 | 0.155 | −78.928 | 0.550 | |
| cg12090003 | 0.090 | 0.158 | −70.596 | 0.569 | 0.089 | 0.151 | −61.905 | 0.587 | |
| cg11558551 | 0.073 | 0.125 | −46.832 | 0.579 | 0.077 | 0.117 | −27.406 | 0.653 | |
| CYP2E1 | cg19469447 | 0.224 | 0.298 | −39.366 | 0.753 | 0.195 | 0.287 | −75.182 | 0.682 |
| cg11445109 | 0.157 | 0.249 | −89.576 | 0.628 | 0.136 | 0.207 | −59.610 | 0.655 | |
| cg05194426 | 0.386 | 0.469 | −37.092 | 0.824 | 0.359 | 0.432 | −30.861 | 0.831 | |
| cg27214960 | 0.182 | 0.248 | −36.638 | 0.734 | 0.161 | 0.216 | −28.197 | 0.749 | |
| DDX58 | cg14286514 | 0.427 | 0.558 | −112.776 | 0.765 | 0.458 | 0.558 | −63.246 | 0.820 |
| DDX60 | cg05883128 | 0.215 | 0.295 | −49.264 | 0.731 | 0.192 | 0.303 | −112.274 | 0.635 |
| DHX58 | cg00647820 | 0.274 | 0.183 | 76.314 | 1.497 | 0.305 | 0.213 | 70.713 | 1.428 |
| DTX3L | cg08122652 | 0.647 | 0.824 | −338.273 | 0.785 | 0.587 | 0.796 | −338.604 | 0.737 |
| cg22930808 | 0.668 | 0.841 | −338.273 | 0.794 | 0.588 | 0.831 | −338.604 | 0.708 | |
| EIF2AK2 | cg14126601 | 0.541 | 0.658 | −97.114 | 0.821 | 0.496 | 0.650 | −174.378 | 0.763 |
| cg16795804 | 0.348 | 0.433 | −41.450 | 0.804 | 0.333 | 0.427 | −56.593 | 0.781 | |
| EPSTI1 | cg03763873 | 0.179 | 0.276 | −88.342 | 0.650 | 0.150 | 0.300 | −242.248 | 0.501 |
| HERC6 | cg14212360 | 0.617 | 0.767 | −213.059 | 0.804 | 0.582 | 0.741 | −219.492 | 0.786 |
| IFI44L | cg05696877 | 0.380 | 0.607 | −338.273 | 0.626 | 0.377 | 0.598 | −338.604 | 0.632 |
| cg03607951 | 0.594 | 0.759 | −250.414 | 0.783 | 0.552 | 0.760 | −338.604 | 0.727 | |
| cg06872964 | 0.453 | 0.611 | −178.132 | 0.741 | 0.410 | 0.601 | −270.808 | 0.682 | |
| cg17980508 | 0.575 | 0.697 | −114.657 | 0.824 | 0.521 | 0.693 | −237.606 | 0.751 | |
| cg00855901 | 0.371 | 0.517 | −147.359 | 0.717 | 0.337 | 0.506 | −207.807 | 0.666 | |
| IFIT1 | cg05552874 | 0.687 | 0.834 | −261.613 | 0.824 | 0.629 | 0.836 | −338.604 | 0.752 |
| cg26974214 | 0.237 | 0.349 | −100.191 | 0.679 | 0.235 | 0.354 | −114.714 | 0.664 | |
| IFIT3 | cg06188083 | 0.327 | 0.487 | −185.861 | 0.671 | 0.311 | 0.478 | −207.565 | 0.650 |
| IL21R | cg00050618 | 0.316 | 0.237 | 45.801 | 1.336 | 0.357 | 0.252 | 83.514 | 1.413 |
| LY6E | cg14392283 | 0.630 | 0.891 | −338.273 | 0.707 | 0.548 | 0.871 | −338.604 | 0.629 |
| cg12110437 | 0.315 | 0.439 | −106.719 | 0.717 | 0.283 | 0.427 | −155.888 | 0.663 | |
| MX1 | cg21549285 | 0.481 | 0.728 | −338.273 | 0.661 | 0.419 | 0.736 | −338.604 | 0.569 |
| cg22862003 | 0.632 | 0.766 | −168.583 | 0.825 | 0.572 | 0.764 | −338.604 | 0.749 | |
| cg26312951 | 0.262 | 0.350 | −53.435 | 0.749 | 0.242 | 0.359 | −109.111 | 0.674 | |
| cg08924203 | 0.142 | 0.211 | −50.293 | 0.674 | 0.143 | 0.225 | −75.550 | 0.637 | |
| NLRC5 | cg07839457 | 0.225 | 0.341 | −111.160 | 0.660 | 0.245 | 0.330 | −54.351 | 0.744 |
| PARP9 | cg08122652 | 0.647 | 0.824 | −338.273 | 0.785 | 0.587 | 0.796 | −338.604 | 0.737 |
| cg22930808 | 0.668 | 0.841 | −338.273 | 0.794 | 0.588 | 0.831 | −338.604 | 0.708 | |
| PLSCR1 | cg06981309 | 0.546 | 0.722 | −260.559 | 0.757 | 0.482 | 0.697 | −338.604 | 0.691 |
| RSAD2 | cg10959651 | 0.185 | 0.253 | −37.604 | 0.734 | 0.186 | 0.250 | −36.625 | 0.744 |
| STAT1 | cg14951497 | 0.360 | 0.522 | −187.309 | 0.689 | 0.372 | 0.489 | −89.425 | 0.761 |
| cg00676801 | 0.313 | 0.463 | −163.414 | 0.676 | 0.339 | 0.440 | −67.217 | 0.770 | |
| TRIM22 | cg26724018 | 0.303 | 0.409 | −76.230 | 0.741 | 0.322 | 0.431 | −79.525 | 0.749 |
| TYMP | cg16367976 | 0.041 | 0.088 | −51.879 | 0.462 | 0.033 | 0.092 | −91.502 | 0.360 |
| USP18 | cg14293575 | 0.355 | 0.555 | −298.906 | 0.640 | 0.339 | 0.534 | −286.306 | 0.634 |
Differential methylation score=10*sgn(βRisk-βProtective)*log10P. β= methylation fraction.
Table III.
Transcription factor analysis showing clear enrichment of interferon-regulated transcription factor binding sites in the promoter regions of differentially methylated genes in naïve CD4+ T cells from lupus patients
| Transcription factor |
P value of overlap |
Target differentially methylated genes |
|---|---|---|
| NKX2-3 | 5.19E-16 | DDX58,DDX60,DHX58,EIF2AK2,LY6E,PARP14,PARP9,PLSCR1,PRIC285,STAT1,TRIM22,TYMP,USP18 |
| IRF7 | 3.18E-15 | DDX58,DHX58,IFI44L,IFIT1,IFIT3,MX1,PLSCR1,RSAD2,STAT1,TRIM22,USP18 |
| STAT1 | 4.15E-12 | CYP2E1,EIF2AK2,HERC6,IFIT1,LY6E,RSAD2,SMAD3,STAT1,TRIM22,TYMP,USP18 |
| TRIM24 | 4.54E-11 | BST2,DDX58,DDX60,DHX58,HERC6,IFIT3,STAT1,USP18 |
| BRCA1 | 4.92E-10 | DDX58,FHIT,IFIT1,IFIT3,MX1,PLSCR1,SMAD3,STAT1 |
| IRF1 | 1.09E-09 | EIF2AK2,IFI44L,IFIT1,IFIT3,MX1,RSAD2,STAT1,TRIM22 |
| IRF3 | 8.59E-09 | DDX58,DHX58,IFIT1,IFIT3,MX1,RSAD2,USP18 |
| STAT3 | 1.66E-05 | EIF2AK2,IFIT1,IFIT3,MX1,PLSCR1,STAT1,TRIM22 |
| STAT2 | 4.88E-05 | IFIT1,IFIT3,MX1 |
| IRF5 | 1.88E-04 | IFIT1,PLSCR1,RSAD2 |
| NFATC2 | 2.85E-04 | MX1,PRIC285,RSAD2,STAT1 |
| GFI1 | 8.95E-04 | EIF2AK2,SMAD3,STAT1 |
| SATB1 | 1.60E-03 | EPSTI1,PRIC285,TRIM22 |
| IRF9 | 1.89E-03 | IFIT3,STAT1 |
| SP1 | 3.31E-03 | ALOX5,EIF2AK2,IL21R,SMAD3,STAT1 |
| SMARCB1 | 5.31E-03 | CYP2E1,EIF2AK2,MX1 |
| HDAC3 | 5.33E-03 | ALOX5,USP18 |
| IRF2 | 7.40E-03 | EIF2AK2,USP18 |
| TBL1XR1 | 1.33E-02 | USP18 |
| ISGF3 | 1.77E-02 | RSAD2 |
| BRD7 | 1.77E-02 | TRIM22 |
| TBL1X | 1.77E-02 | USP18 |
| IRF8 | 1.97E-02 | IFI44L,STAT1 |
| Stat1-Stat2 | 2.20E-02 | EIF2AK2 |
| ZNF148 | 2.64E-02 | STAT1 |
| COPS5 | 2.86E-02 | CD247 |
| ESR1 | 3.47E-02 | ODF3B,SMAD3,STAT1 |
| IKZF3 | 3.50E-02 | ALOX5 |
| NR1D1 | 3.72E-02 | STAT1 |
| ELF1 | 5.00E-02 | CD247 |
To determine the functional consequences of the methylation changes we observed in naïve CD4+ T cells from lupus patients, we performed gene expression analysis using RNA extracted from the same naïve CD4+ T cells isolated from a subset of the study participants. Interestingly, none of the hypomethylated interferon-related genes were overexpressed in naïve CD4+ T cells from lupus patients (Supplementary Table II). In contrast, analysis of gene expression profiles for total CD4+ T cells from lupus patients and healthy controls (Lauwerys et al.; GEO accession: GSE4588) showed that most of these interferon-regulated genes are significantly overexpressed in total CD4+ T cells in lupus (Supplementary Table III).
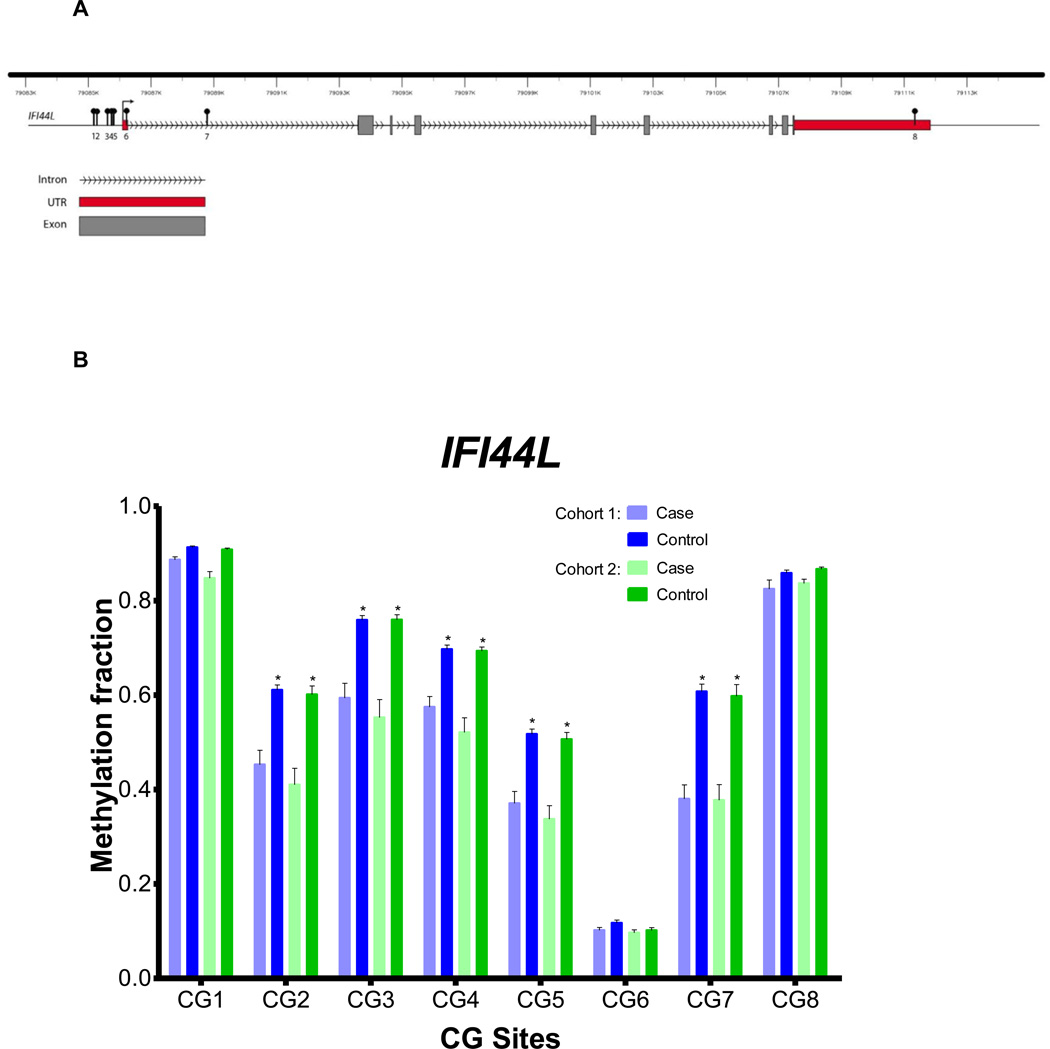
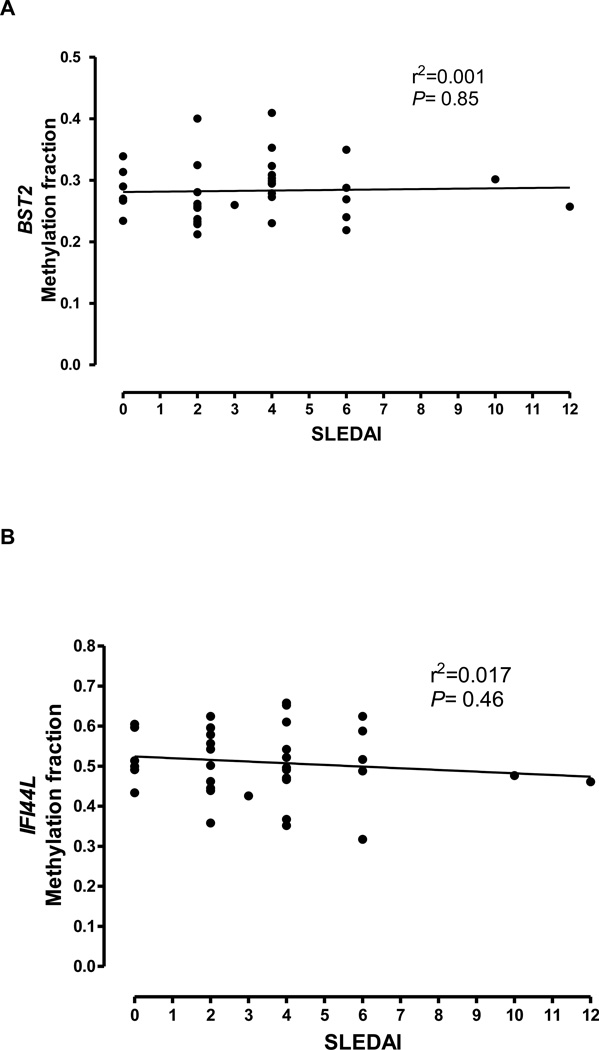
Hypomethylation of some of these same interferon-regulated genes, such as BST2 and IFI44L, was also previously observed in total CD4+ lupus T cells using a different methylation platform [6]. We show consistent hypomethylation across multiple CG sites in the promoter regions of both genes in naïve CD4+ T cells (Figure 2, 3), without evidence of overexpression. In contrast, BST2 and IFI44L are ~2 times and 18 times overexpressed in total CD4+ T cells from lupus patients (P= 6.25×10−5 and 5.94×10−8respectively). Indeed, hypomethylation of BST2 and IFI44L in naïve CD4+ T cells does not correlate with disease activity as measured by SLEDAI scores in lupus patients (r2= 0.001 and 0.017, and P= 0.85 and 0.46, respectively), suggesting that the hypomethylation observed in naïve CD4+ T cells in lupus is not influenced by disease activity (Figure 4).
Figure 2.
BST2 gene DNA methylation in naïve CD4+ T cells from lupus patients and controls. A) The BST2 gene showing the location of the CG sites evaluated in this study. B) DNA methylation fractions across the CG sites evaluated in the BST2 gene in the two independent cohorts of lupus patients and controls.
Figure 3.
IFI44L gene DNA methylation in naïve CD4+ T cells from lupus patients and controls. A) The IFI44L gene showing the location of the CG sites evaluated in this study. B) DNA methylation fractions across the CG sites evaluated in the IFI44L gene in the two independent cohorts of lupus patients and controls.
Figure 4.
Correlation between the DNA methylation in naïve CD4+ T cells and lupus disease activity in BST2 and IFI44L (panels A and B, respectively). Lupus disease activity is measured using SLEDAI (x-axis) is plotted against the average DNA methylation level on all CG sites evaluated in BST2 and IFI44L (y-axis).
4. Discussion
We performed a genome-wide DNA methylation study coupled with a gene expression profiling experiment in naïve CD4+ T cells from lupus patients and controls. DNA methylation levels were quantified in over 485,000 methylation sites in naïve CD4+ cells across the entire genome in two independent sets of lupus patients and age-, sex-, and ethnicity-matched controls. Differentially methylated CG sites in lupus were discovered and then replicated, and the effect of the methylation changes detected upon gene expression was examined using a subset of the same sample. The majority of the differentially methylated genes in CD4+ T cells from lupus patients were hypomethylated, consistent with a methylation defect previously described in lupus T cells [1, 6].
We found significant enrichment of interferon-regulated genes among hypomethylated loci in naïve CD4+ T cells from lupus patients. An analysis of canonical pathways, biological interaction networks, and transcription factor binding site enrichment highlighted the interferon signaling pathway in the genes differentially methylated in lupus. Indeed, type-I interferon is strongly emphasized among differentially methylated genes in agreement with the interferon expression signature described in lupus PBMCs [9, 10]. However, we show that despite being hypomethylated, none of these interferon-regulated genes were overexpressed in naïve CD4+ T cells in contrast to total CD4+ T cells in lupus patients.
We propose that the lack of active expression of these hypomethylated genes may be explained by the naïve status of these CD4+ T cells. Active gene expression requires both chromatin accessibility and appropriate transcription factors, and these transcription factors would be lacking in naïve T cells. Thus, these same interferon-related genes are overexpressed in total CD4+ T cells from lupus patients, but not in naïve CD4+ T cells. We suggest that the presence of hypomethylated interferon-regulated genes makes the naïve T cells from lupus patients epigenetically “poised” to produce a rapid type-I interferon response. Indeed, PBMCs from at least a subset of lupus patients have been previously shown to have increased sensitivity to IFN-α [11]. The mechanism of this increased sensitivity is currently unknown, and our data provide a novel possible explanation and adds to the knowledge of the role of type-I interferon in the pathogenesis of lupus.
Among the hypomethylated genes in naïve CD4+ T cells from lupus patients, we found significant hypomethylation in BST2, an interferon-regulated gene involved in preventing the release of viral particles from infected cells [12]. We have previously reported hypomethylation and overexpression of BST2 in total CD4+ T cells from lupus patients [6]. More recently, we have also reported similar hypomethylation in BST2 in T cells from healthy individuals that carry the lupus-risk variant in methyl-CpG-binding protein 2 (MECP2) compared to individuals with the protective genetic variants in this locus [13]. MECP2 is a confirmed susceptibility locus for lupus and encodes for a transcription regulator for the expression of methylation-sensitive genes.
Two of the hypermethylated genes that we identified in naïve CD4+ T cells in lupus (CD247 and IL21R), are known lupus genetic susceptibility loci [14–16]. CD247 encodes for the TCR-zeta chain which is downregulated in lupus T cells [17]. Apoptosis-related genes, including BCL2L14, BCL2L15, and SMAD3 were also among the differentially methylated genes detected (Supplementary Table I).
5. Conclusions
We identified and replicated DNA methylation changes in naïve CD4+ T cells from lupus patients for the first time. These data indicate that abnormal DNA methylation exists in lupus T cells even before activation and differentiation. Therefore, our findings emphasize the role of DNA methylation defect in the pathogenesis of the disease. Notably, we propose a model whereby interferon-regulated genes are epigenetically poised to respond to type-I interferon upon T cell activation and provide evidence for an epigenetic architecture favoring, and providing an explanation for, hyper-responsiveness to type-I interferon in lupus T cells.
Supplementary Material
Research Highlights.
The DNA methylome of naïve CD4+ T cells in lupus patients has been characterized
We identified and replicated 86 CG sites that are differentially methylated in naïve CD4+ T cells from lupus patients
Interferon-regulated genes are significantly hypomethylated in naïve CD4+ T cells from lupus patients
Naïve CD4+ T cells are epigenetically poised to respond to type-1 interferon in lupus patients
Acknowledgements
This work was made possible by support from the Lupus Research Institute and the NIH grant number R01AI097134 from the National Institute of Allergy and Infectious Diseases.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Financial conflict of interest: None of the authors has any financial conflict of interest to report.
References
- 1.Zhang Y, Zhao M, Sawalha AH, Richardson B, Lu Q. Impaired DNA methylation and its mechanisms in CD4(+)T cells of systemic lupus erythematosus. J Autoimmun. 2013 doi: 10.1016/j.jaut.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 2.Richardson B, Sawalha AH, Ray D, Yung R. Murine models of lupus induced by hypomethylated T cells (DNA hypomethylation and lupus…) Methods Mol Biol. 2012;900:169–180. doi: 10.1007/978-1-60761-720-4_8. [DOI] [PubMed] [Google Scholar]
- 3.Jeffries MA, Sawalha AH. Epigenetics in systemic lupus erythematosus: leading the way for specific therapeutic agents. Int J Clin Rheumtol. 2011;6:423–439. doi: 10.2217/ijr.11.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jones PL, Veenstra GJ, Wade PA, Vermaak D, Kass SU, Landsberger N, et al. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 5.Sawalha AH. Epigenetics and T-cell immunity. Autoimmunity. 2008;41:245–252. doi: 10.1080/08916930802024145. [DOI] [PubMed] [Google Scholar]
- 6.Jeffries MA, Dozmorov M, Tang Y, Merrill JT, Wren JD, Sawalha AH. Genome-wide DNA methylation patterns in CD4+ T cells from patients with systemic lupus erythematosus. Epigenetics. 2011;6:593–601. doi: 10.4161/epi.6.5.15374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Webb R, Wren JD, Jeffries M, Kelly JA, Kaufman KM, Tang Y, et al. Variants within MECP2, a key transcription regulator, are associated with increased susceptibility to lupus and differential gene expression in patients with systemic lupus erythematosus. Arthritis Rheum. 2009;60:1076–1084. doi: 10.1002/art.24360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wren JD, Garner HR. Shared relationship analysis: ranking set cohesion and commonalities within a literature-derived relationship network. Bioinformatics (Oxford, England) 2004;20:191–198. doi: 10.1093/bioinformatics/btg390. [DOI] [PubMed] [Google Scholar]
- 9.Elkon KB, Stone VV. Type I interferon and systemic lupus erythematosus. J Interferon Cytokine Res. 2011;31:803–812. doi: 10.1089/jir.2011.0045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Niewold TB. Interferon alpha as a primary pathogenic factor in human lupus. J Interferon Cytokine Res. 2011;31:887–892. doi: 10.1089/jir.2011.0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kariuki SN, Kirou KA, MacDermott EJ, Barillas-Arias L, Crow MK, Niewold TB. Cutting edge: autoimmune disease risk variant of STAT4 confers increased sensitivity to IFNalpha in lupus patients in vivo. J Immunol. 2009;182:34–38. doi: 10.4049/jimmunol.182.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuhl BD, Sloan RD, Donahue DA, Bar-Magen T, Liang C, Wainberg MA. Tetherin restricts direct cell-to-cell infection of HIV-1. Retrovirology. 2010;7:115. doi: 10.1186/1742-4690-7-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koelsch KA, Webb R, Jeffries M, Dozmorov MG, Frank MB, Guthridge JM, et al. Functional characterization of the MECP2/IRAK1 lupus risk haplotype in human T cells and a human MECP2 transgenic mouse. J Autoimmun. 2012 doi: 10.1016/j.jaut.2012.12.012. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gorman CL, Russell AI, Zhang Z, Cunninghame Graham D, Cope AP, Vyse TJ. Polymorphisms in the CD3Z gene influence TCRzeta expression in systemic lupus erythematosus patients and healthy controls. J Immunol. 2008;180:1060–1070. doi: 10.4049/jimmunol.180.2.1060. [DOI] [PubMed] [Google Scholar]
- 15.Li R, Yang W, Zhang J, Hirankarn N, Pan HF, Mok CC, et al. Association of CD247 with systemic lupus erythematosus in Asian populations. Lupus. 2012;21:75–83. doi: 10.1177/0961203311422724. [DOI] [PubMed] [Google Scholar]
- 16.Webb R, Merrill JT, Kelly JA, Sestak A, Kaufman KM, Langefeld CD, et al. A polymorphism within IL21R confers risk for systemic lupus erythematosus. Arthritis Rheum. 2009;60:2402–2407. doi: 10.1002/art.24658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nambiar MP, Enyedy EJ, Fisher CU, Krishnan S, Warke VG, Gilliland WR, et al. Abnormal expression of various molecular forms and distribution of T cell receptor zeta chain in patients with systemic lupus erythematosus. Arthritis Rheum. 2002;46:163–1674. doi: 10.1002/1529-0131(200201)46:1<163::AID-ART10065>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.