Summary
Retinoic acid (RA) regulates many cellular behaviors during embryonic development and adult homeostasis. Like other morphogens, RA forms gradients through the use of localized sources and sinks, feedback, and interactions with other signals; this has been particularly well studied in the context of hindbrain segmentation in vertebrate embryos. Yet, as a small lipophilic molecule derived from a dietary source—vitamin A—RA differs markedly from better-studied polypeptide morphogens in its mechanisms of transport, signaling, and removal. Computational models suggest that the distinctive features of RA gradients make them particularly robust to large perturbations. Such features include combined positive and negative feedback effects via intracellular fatty acid binding proteins and RA-degrading enzymes. Here, we discuss how these features, together with feedback interactions among RA target genes, help enable RA to specify multiple, accurate pattern elements in the developing hindbrain, despite operating in an environment of high cellular and biochemical uncertainty and noise.
Introduction
A critical issue in developmental biology is how morphogen gradients are established and interpreted by cells. Most studies have focused either on cytoplasmic morphogens of syncytial embryos (e.g. Bicoid (Bcd) in Drosophila), or secreted proteins such as members of the TGFβ, FGF, EGF, Hedgehog (Hh), and Wnt families [1–5]. Non-polypeptide morphogens also exist, however, the best-studied example of which is retinoic acid (RA). RA influences the behaviors of numerous cell types and tissues during embryonic development, as well as adult stem cells (neuronal, pancreatic), cancers (leukemia) and regenerating organs (cardiomyocytes) [6–10]. RA derives from vitamin A, and its lipophilic nature and use of nuclear receptors make its movements within tissues and signal transduction properties distinctive among morphogens. Here we discuss how RA gradients, and the cellular responses to them, are established and maintained. We review literature suggesting that several unique features of RA signal regulation make it extraordinarily robust, yet precise, in defining patterning and sharp boundaries of target gene expression.
Robust RA gradient formation
Most morphogen gradients are thought to form through the action of localized sources of production (e.g. sites of synthesis or deposition) and localized or distributed sinks (e.g. uptake, degradation). Because morphogens act at a distance from their source of production, eliciting distinct cellular responses in a concentration-dependent manner [11], a traditional focus of both experimental and theoretical work on their roles in pattern formation has been on how steady-state gradients form, and how their shapes are controlled. More recently, it has also become clear that cells can respond to the temporal dynamics of morphogen signaling, i.e. they can sense the rate of change in morphogen concentration, and their responses to a morphogen can change over time [5,12–15].
Whereas early work on morphogen gradients treated cells as perfect detectors of invariant gradients, attention over the last decade has increasingly focused on the robustness of morphogen-mediated patterning [16,17]. This refers to the relative insensitivity of pattern formation to variability and uncertainty in the molecular processes underlying both morphogen gradient formation and readout. Such variability arises from a multitude of sources, including environmental factors (e.g. temperature and nutrition), individual genetic differences, and the intrinsically stochastic nature of biochemical processes (such as RNA and protein synthesis). Achieving robustness to such variation is also related to the problem of making morphogen gradients scale, i.e. making their steady-state shapes automatically expand or contract in response to variations in the size of the tissue field being patterned [18,19].
The collective magnitude of all of this variability is expected to be in the range where the need for robustness (or scaling) places severe constraints on the design of morphogen patterning systems. One of the most generic strategies for achieving robustness to variability in morphogen production is to exploit negative feedback, which indeed seems to be used in most morphogen systems [11]. For example, in gradients formed by TGFβ, FGF, EGF, Hh and Wnt family members, there can be negative feedback through: 1) regulation of morphogen synthesis, secretion, or transport; 2) regulation of morphogen-receptor interaction (e.g. through induction of secreted antagonists or sequestering components of the extracellular matrix [ECM]); 3) regulation of receptor expression; or 4) regulation of signal transduction events downstream of morphogens (Figure 1A). An especially wide variety of such strategies has been described in Decapentaplegic (Dpp) and Sonic hedgehog (Shh) signaling pathways [20–27].
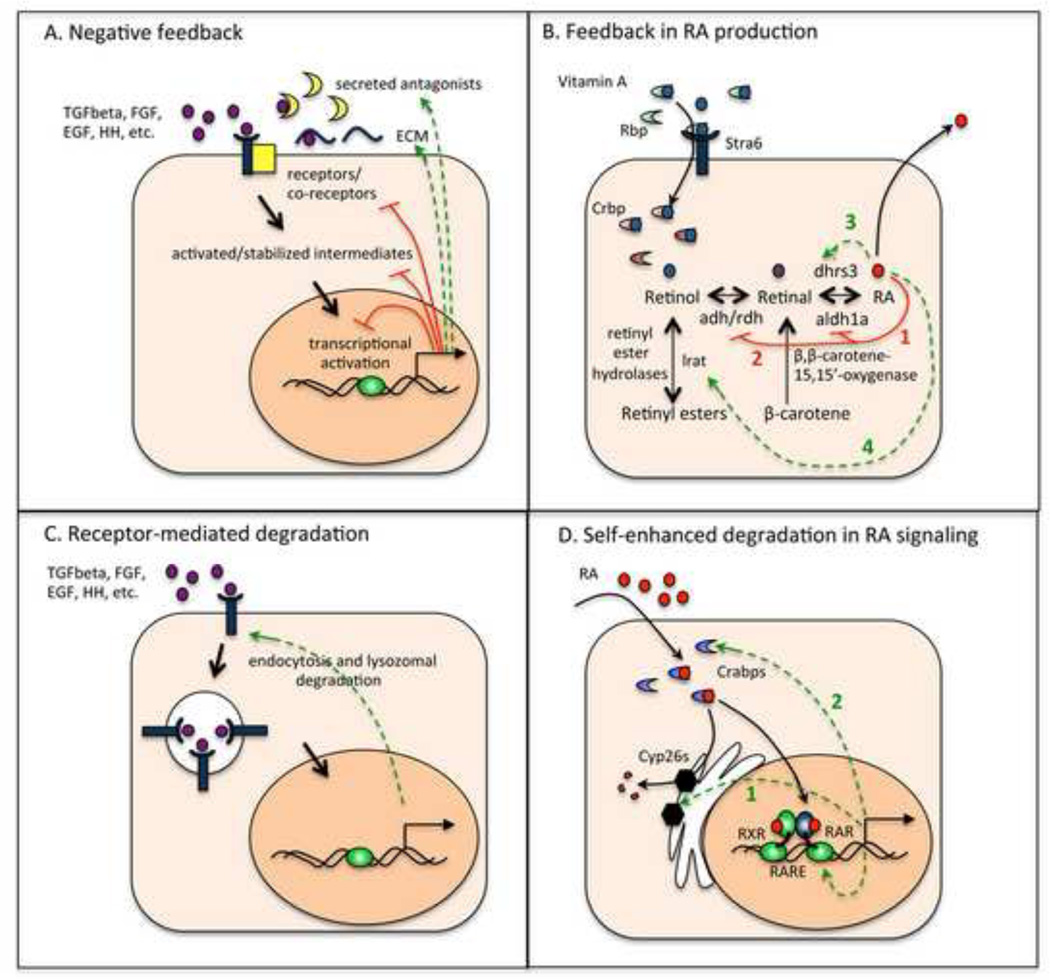
Figure 1.
Comparing feedback mechanisms in morphogen signaling. (A) A cell responding to a typical polypeptide morphogen (purple ovals). Green dashed arrows point to extracellular inhibitory factors induced by the signal. Red lines denote intracellular mechanisms of repression. (B) An RA-producing cell showing the biosynthetic pathway for RA. Retinyl esters and beta-carotene are important stores of RA precursor. Green dashed arrows point to components of the pathway induced by RA signaling. Red lines denote repression of adhs and aldh1as by RA. (C) Polypeptide morphogens typically induce their own receptors, which are subsequently degraded through receptor-mediated endocytosis. (D) An RA-responding cell showing signaling and catabolism. RA (red circles) enters cells and binds Crabps (blue crescents), which transport it either to RARs (blue and green ovals) in the nucleus (orange) for signaling or to Cyp26s (black hexagons) for degradation. Adhs, alcohol dehydrogenases; aldh1as, aldehyde dehydrogenase 1as.
In the case of RA morphogen gradients, the need for robustness to variation in morphogen production is exacerbated by the fact that RA is synthesized from a precursor, vitamin A or retinol, the levels of which are highly sensitive to dietary conditions. A first line of defense against this problem seems to be negative feedback control of the synthesis of RA from its precursors (Figure 1B) [6]. Indeed, it has been shown that RA downregulates, in a dose-dependent manner, the expression of Aldh1a2 (arrow 1 in Fig. 1B), the major aldehyde dehydrogenase required for conversion of retinaldehyde to RA in embryos [28], and recent evidence demonstrates that it also downregulates retinol dehydrogenase (RDH10, arrow 2), the enzyme that converts retinol to retinal [29]. Mutant screens in zebrafish for enhancers/suppressors of an RA-treated phenotype recently revealed that additional negative feedback occurs through upregulation of Dhrs3 (arrow 3), a dihyroreductase that catalyzes the conversion of all-trans RA to vitamin A [30]. Curiously, there also appears to be some positive feedback of RA on its own production, through upregulation of retinyl ester hydrolases, such as lecithin:retinol acyl transferase (Lrat, arrow 4), that produce Retinol [31]. Although the roles of such mixed positive and negative feedback are unknown, the existence of so many levels of feedback on RA synthesis suggests that tight regulation is very important.
Degradation and robustness in RA gradients
Quite independent of their ability to regulate RA synthesis, RA morphogen gradients also achieve extraordinary robustness through mechanisms that act downstream of synthesis, a fact we know thanks to a fortuitous situation in the zebrafish hindbrain. During early embryogenesis, the anterior-posterior (A-P) axis of the hindbrain is subdivided into seven rhombomeres (r1-7) by a posterior-to-anterior RA gradient. The shape and orientation of that gradient is so greatly determined by the location of sites of RA degradation, that the normal, endogenous, posteriorly-localized RA source can be eliminated and completely replaced with a uniform application of exogenous RA to the entire embryo. Remarkably, such embryos not only pattern the early hindbrain normally, they produce nearly the same pattern over a 20-fold range in applied exogenous RA concentration [32,33]].
Mathematical models have provided some insight into this phenomenon, identifying feedback regulation of morphogen degradation as one means for compensating for unreliable levels (or locations) of morphogen production. Such regulation exemplifies “self-enhanced degradation” [34] (or, more generally, self-enhanced decay), a type of negative feedback that has also been argued to make both Wnt and Hh gradients more robust to fluctuations in morphogen production [1–3,35] For polypeptide morphogens, self-enhanced degradation typically involves activity-dependent regulation of receptor expression or receptor-mediated endocytosis (Figure 1C). For example, both fly and vertebrate Hh proteins induce their receptor, Patched, and Shh in vertebrates also induces the matrix proteoglycan Glypican3 (GPC3), and these in turn regulate Hh endocytosis and degradation [36,37]. Effective feedback control may also occur through induction of proteases or other ECM components – as long as morphogen activity increases morphogen degradation, it will reduce sensitivity to perturbations in morphogen production. However, because this type of feedback necessarily makes gradients shallower with distance from the morphogen source, it tends to exacerbate the difficulty of forming sharp boundaries of target-gene expression [16], a problem we return to later.
How is self-enhanced degradation of RA achieved in the developing hindbrain? First, RA induces the expression of a cytochrome p450 enzyme (Cyp26a1) that specifically degrades RA [32] (Figure 1D, arrow 1). Second, RA induces expression of certain intracellular RA binding proteins (Crabps), one role of which is to promote the delivery of RA to Cyp26s [38](arrow 2 in Fig. 1D). Models that incorporate these two molecular features achieve considerably more robust gradients than models with either one alone [38]. Interestingly, Crabp2 can also mediate positive feedback in RA signaling, by promoting the delivery of RA to RA-receptors (Figure 2A). Modeling shows that this dual effect of Crabp2 can actually provide greater robustness, particularly in response to large perturbations [39]. Indeed, experiments in zebrafish have demonstrated that: 1) Cyp26a1 and Crabp2a are both induced by RA and 2) are both essential for the robustness of patterning to perturbations of the RA level [38].
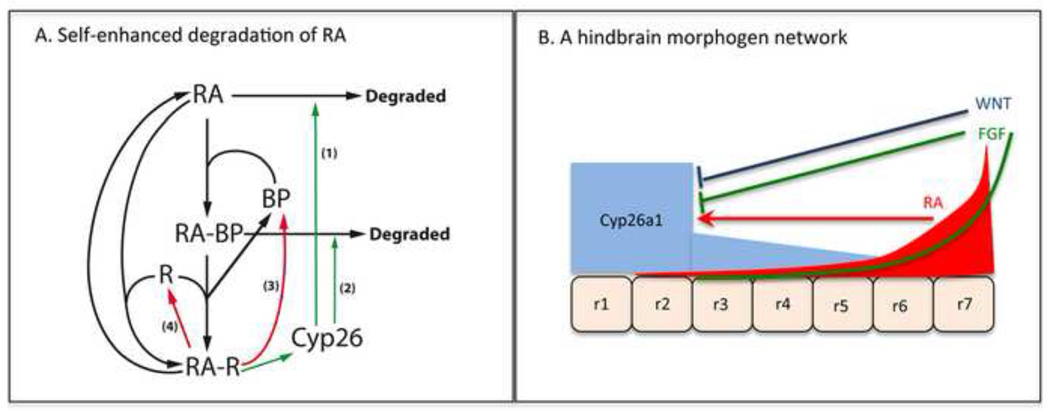
Figure 2.
Models for RA signaling during hindbrain development. (A) Arrow diagram illustrating different bound and unbound states of RA within a responding cell, and paths to degradation, incorporated in our computational models. (B) Morphogen model representing hindbrain rhombomeres (r1-7), anterior to the left. Cyp26a1 in blue, RA signaling in red, Fgf signaling in green, Wnt signaling in dark blue. In this model, Cyp26-mediated degradation is continuously under feedback and feedforward control from Wnt/Fgf and RA signaling, respectively, which shapes the RA gradient. This integrates time- and concentration-dependent effects of RA as its gradient grows, without increasing the rate of RA synthesis (adapted from [32]).
Robustness and interactions between RA and other morphogens
Even when the dynamics of morphogen gradient formation are sufficiently fast that they may be treated as steady-state systems, in many developing systems the sizes of the regions in which morphogens are produced and act undergo marked changes over time, due to tissue growth or cell movements. In such cases, in order for positional cues to maintain stable relative positions, it may be necessary for morphogen gradients to continually readjust themselves. In Drosophila imaginal discs, for example, there is evidence that, at least for a substantial fraction of larval life, the Dpp morphogen gradient both increases in amplitude and length-scale so as to compensate for disc growth [40]. Length-scale changes may reflect the presence of feedback mechanisms that involve diffusible “expander” molecules, such as Pentagone, whose concentration is thought to be a function of overall disc size [41].
In the hindbrain RA system there is also a gradual increase in size during patterning, due to cell movement during gastrulation, but the presence of an “expander” molecule has not been described. What has been described, however, is an interaction between the RA system and other morphogen systems. In particular, A-P patterning of the hindbrain is influenced by at least three “posteriorizing” signals, RA, Wnt and Fgf [42] (Figure 2B). All three are: 1) produced in the posterior mesoderm during gastrulation, and 2) induce posterior and suppress anterior expression of genes involved in rhombomere specification. Yet what would seem to be three independent morphogen gradient systems become intertwined at the level of RA gradient formation. This is because Fgf and Wnt inhibit the upregulation of Cyp26a1 by RA [32,42]. In this way, steadily increasing Fgf and Wnt expression may both drive the gastrulation movements that make the hindbrain field grow in size, and drive a compensatory expansion of the RA gradient, due to the resultant downregulation of Cyp26a1.
Although we can imagine how such compensatory mechanisms might broadly couple RA gradients to steady hindbrain elongation, the reality is that, as gastrulation proceeds, RA sources and sinks are much more dynamic and intricate. For example, not only is RA produced by the mesoderm that lies immediately posterior to the presumptive hindbrain (i.e. that will form somites), from which it diffuses anteriorly to pattern gene expression in the neural ectoderm (Figure 3A, upper panel), some RA is also synthesized and degraded in cranial mesoderm that lies on either side of the hindbrain. RA plays an essential role in A-P patterning of the mesoderm, which could, in turn secondarily influence hindbrain segmentation [43]. Moreover, within the cranial mesoderm, some of the targets of RA, such as Hoxa1 and Pbx1/2, are required for maintenance of expression of the RA biosynthetic enzyme Aldh1a2, i.e. mesodermal RA expression appears to be autocatalytic (Figure 3A, lower panel) [44]. Whether or not this dynamically changing nearby source of RA plays a role in the robustness of hindbrain patterning remains to be investigated.
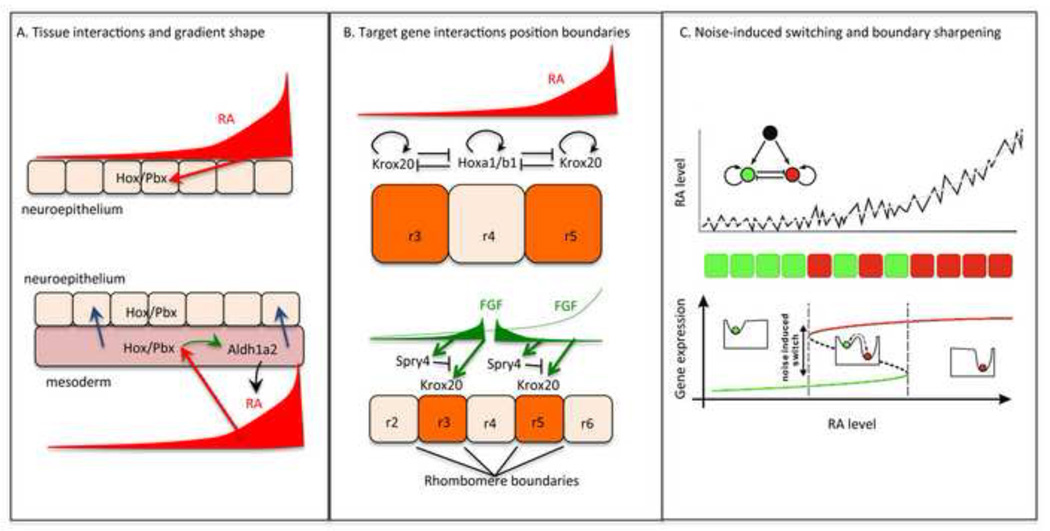
Figure 3.
Robustness, boundary sharpening and noise attenuation in RA signaling. (A) Modified morphogen model incorporating roles of adjacent cranial mesoderm on patterning of hindbrain rhombomeres, anterior to the left. The RA gradient (dark red), RA in the cranial mesoderm (light red), and putative influences on the hindbrain neuroepithelium (blue arrows). (B) Models showing RA gradient (red), cross-inhibition and auto-activation of Krox20 and Hoxa1/b1 at boundaries of r3-5 and influences of Fgfs signaling (green) - initially from a posterior source and later from r4 – together with Spry4-mediated negative feedback, on krox20 induction. (C) Schematic illustrating the concept of noise-induced switching at a rhombomere boundary. Fluctuations in RA levels, together with the gene regulatory network (upper panel) lead to fluctuations in target gene expression cell-by-cell (red and green squares) near a boundary, and noise in gene expression helps push cells into one stable state in this bistable region (e.g. from green to red).
The role of downstream gene regulatory networks
Typically, the ultimate functions of morphogen gradients are to specify distinct, spatial domains of gene expression and cell differentiation. From a mechanistic standpoint, generating even a single, sharp domain boundary from a shallow morphogen gradient is tricky enough, as it requires large changes in gene expression in response to small differences in morphogen signal; such strong amplification is also likely to amplify cell-to-cell variation (i.e. noise) in the morphogen response, thereby degrading boundary sharpness [39]. Remarkably, many morphogen gradient systems solve this problem not just at one threshold morphogen concentration, but at several, so that multiple sharp boundaries are specified. In this section, we consider the roles played by gene regulatory networks in providing solutions to the multiple sharp boundary problem, including those downstream of RA in the hindbrain. In the next section we will consider how cell-to-cell noise affects the sharpness of such boundaries, and how noise itself can be used to counteract these effects.
So far, the most detailed studies of how gene regulatory networks shape the outputs of morphogen gradients have come from studies of Bicoid in Drosophila and the Shh morphogen gradient system of the vertebrate neural tube. In both cases, morphogen signals feed into networks of cross-regulatory gene expression that are rich in mutual inhibition [13,23,45,46]. Such feedback is critical in creating “attractor states”, i.e. states of gene expression to which cells are driven from any of a large number of possible starting configurations. When either of two stable states is possible depending solely upon initial conditions, we refer to the situation as bistability (two stable attractors), but for complex networks the attractors can be more numerous, and can include dynamic behaviors, such as oscillations or defined trajectories (i.e. where gene expression changes continuously in a reliable manner). In the network downstream of Bicoid, where the number of mutually interacting genes is fairly large, and where additional positional information is provided by maternally-derived gradients, there are many potential attractors, both stable and dynamic [47,48]. In the Shh system that patterns the dorsal-ventral (D-V) axis of the neural tube, a more modest (so far) network of inhibitory gene regulatory interactions involving the genes Nkx2.2Olig2 and Pax6, creates three stable states [13,46], which correspond to three distinct domains of gene expression that emerge at different distances from the ventral Shh source.
The existence of attractor state networks downstream of morphogens has at least three important consequences. First, due to strong positive feedback created by the loops of mutual inhibition typically found in such systems, switching from one attractor to another often occurs with only very small changes in input (morphogen). Thus, such systems provide an explanation for how shallow gradient information is converted into spatially abrupt changes in cell fate (Figure 3B). Second, because a cell’s choice of attractor depends not just on morphogen concentration but on the dynamics over which that concentration changes, positioning of boundaries can be more robust than could be achieved given measurements of morphogen concentration alone [47,49]. Third, as discussed below, the effect of dynamics on attractor selection can make it possible to specify a greater number of spatial domains than there are attractor states.
Evidence and modeling suggest that all of these phenomena are important in A-P patterning of the vertebrate hindbrain by RA. For example, the RA target genes hoxb1a and krox20 are self-activating and mutually-repressing, creating three stable states: hoxb1a-on/krox20-off, which is adopted by the territory that will become rhombomere 4 (r4); krox20-on/hoxb1a-off, which occurs both in r3 and r5; and a both-off state. Activation of hoxb1a expression occurs first, in a diffuse pattern that simply reflects RA levels. Later, induction of krox20 becomes possible, probably reflecting the fact that it is a more indirect target of RA [50]. Recent modeling [51] indicates that, given appropriate assumptions about relative strengths of mutual inhibition and self-activation of hoxb1a and krox20, and their relative sensitivity to RA, three spatial outcomes can result.
Posterior to r4, where RA concentration is highest and the input to both genes approaches maximal values, krox20—the stronger inhibitor—dominates, and krox20-on becomes the only available steady state. Anterior to r4, where RA concentration is lowest and therefore cells have not previously expressed hoxb1a, the greater sensitivity of krox20 to RA, as well as its stronger autoactivation, leads to the adoption of a stable krox20 state as well. In r4 itself, however, hoxb1a is induced more rapidly than krox20, so that when krox20 is induced hoxb1a levels are high enough to block its expression and cells fall into the hoxb1 attractor i.e. the hoxb1a-on/krox20-off state. In this manner, three sharply-defined domains of gene expression are generated (Figure 3B).
In this model, the attractor landscape, and thus the locations where cell fates switch from krox20 to hoxb1a and back again, emerges out of the strength and history of the mutually inhibitory interactions between these genes. One would thus expect that any perturbation that gives an “advantage” to either gene—by hastening or strengthening its expression or autoactivation—would result in predictable movements of gene expression boundaries. Recent work on the effects of Fgf signaling during hindbrain patterning supports this view. Early during the patterning of r4, its cells begin to express Fgf [52–54]. Laballete et al., [55] observed that knocking down Sprouty4, which normally inhibits Fgf signaling, leads to a marked expansion of r3 and r5 at the expense of r4. This is accompanied by an earlier than normal onset of krox20 expression, which they showed is a direct Fgf target. This result agrees with model predictions, which state that a greater constitutive input to krox20 should enable it to compete more effectively with hoxb1a and thus be expressed in a wider territory. Interestingly, because Fgf is produced by r4, yet promotes the ability of krox20 to drive cells toward an r3 or r5 fate (Figure 3B), the Fgf effect may be seen as a form of stabilizing negative feedback on the width of r4, i.e. it should help make that width more robust to the parameters of hoxb1a/krox20 mutual inhibition.
Overcoming noise and boundary sharpening
When we refer to morphogen gradient robustness, we typically mean the relative insensitivity of the boundary positions established by morphogen gradients to perturbations that affect all cells equally. Yet morphogen gradients must also deal with cell-to-cell variability, which creates problems of a different type: to the extent that such variability makes cells misread the morphogen concentration in their immediate vicinity, gene expression boundaries should take on a ragged, or salt-and-pepper, appearance.
Indeed, many morphogen gradient systems display such rough boundaries, at least early on during patterning, but often the situation corrects itself. In the zebrafish hindbrain, for example, rhombomere boundaries start out rough and sharpen over a few hours – 9–11 hours postfertilization [56]. This is accomplished only in part by cell rearrangement [56]. The rest is due to cells switching patterns of gene expression to better reflect their true positional environment. Indeed, individual cells of forming rhombomeres have been shown to remain distinctly plastic—able to up- or down-regulate Hox expression after initiation—for some time [57,58].
How does such plasticity help cells figure out their true locations, if the signals they receive remain corrupted by a constant level of noise? It turns out that the same gene regulatory networks that create multiple attractor states create an opportunity to implement a strategy known as “noise-induced switching”, which can help cells improve their positional choices. Noise-induced switching depends upon hysteresis, the property that switching from one attractor state to another will occur at one morphogen threshold, while switching back in the other direction will occur at a distinctly different threshold. If the thresholds are well-separated enough, and the noise is large enough to drive most cells across one or the other threshold at least a few times, cells will tend to settle into steady states that much better reflect the average morphogen concentration than if they had to rely upon a single noisy measurement. Stochastic modeling has shown how the interaction of noise with the hysteresis produced by the mutual inhibition between hoxb1a and krox20 is sufficient to explain much of the spontaneous sharpening of rhombomere boundaries during hindbrain patterning (Figure 3C) [51].
What makes noise-induced switching such a counter-intuitive strategy is that it requires noise to work, i.e. it takes uncertainty to overcome uncertainty. Although several instances have been described in which noise-induced switching influences the behaviors of isolated cells undergoing differentiation [59], its role in improving the precision of patterning boundaries has not generally been appreciated. Given that the gene regulatory networks downstream of many morphogen systems are distinctly hysteretic [46,47], it seems likely that this strategy is quite broadly exploited in vivo.
Conclusions and Perspectives
In this review we have highlighted how RA signaling is regulated—in ways that are sometimes distinct from, and sometimes similar to, those employed by polypeptide morphogens—so that it can form gradients that are surprisingly robust, precise, and capable of inducing multiple sharp boundaries of target gene expression. Mechanisms that enhance robustness include: 1) tight feedback regulation of RA synthesis, 2) multiple paths of self-enhanced degradation, and 3) interactions between RA and other morphogens. Mechanisms that enhance the precision of boundary formation include: 1) target gene regulatory networks that drive cells toward distinct attractor states and 2) a surprising beneficial role for noise in facilitating the switching of target gene expression between bistable states, enabling individual cells to choose their fates more accurately.
Acknowledgements
This work was supported by the Center for Complex Biological Systems and the National Institutes of Health P50GM076516 (AL, QN, TS), R01DE13828 (TS), R01GM067247 (AL, QN), and the National Science Foundation DMS-0917492 (QN) and DMS-1161621 (QN).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
- 1.Lander AD. Morpheus unbound: reimagining the morphogen gradient. Cell. 2007;128:245–256. doi: 10.1016/j.cell.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 2.Wartlick O, Kicheva A, González-Gaitán M. Morphogen gradient formation. Cold Spring Harb Perspect Biol. 2009;1:a001255. doi: 10.1101/cshperspect.a001255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meinhardt H. Models for the generation and interpretation of gradients. Cold Spring Harb Perspect Biol. 2009;1:a001362. doi: 10.1101/cshperspect.a001362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wolpert L. Positional information and patterning revisited. J Theor Biol. 2011;269:359–365. doi: 10.1016/j.jtbi.2010.10.034. [DOI] [PubMed] [Google Scholar]
- 5.Nahmad M, Lander AD. Spatiotemporal mechanisms of morphogen gradient interpretation. Curr Opin Genet Dev. 2011;21:726–731. doi: 10.1016/j.gde.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.White RJ, Schilling TF. How degrading: Cyp26s in hindbrain development. Dev Dyn. 2008;237:2775–2790. doi: 10.1002/dvdy.21695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duester G. Retinoic acid synthesis and signaling during early organogenesis. Cell. 2008;134:921–931. doi: 10.1016/j.cell.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Niederreither K, Dollé P. Retinoic acid in development: towards an integrated view. Nat Rev Genet. 2008;9:541–553. doi: 10.1038/nrg2340. [DOI] [PubMed] [Google Scholar]
- 9.Tang XH, Gudas LJ. Retinoids, retinoic acid receptors, and cancer. Annu Rev Pathol. 2011;6:345–364. doi: 10.1146/annurev-pathol-011110-130303. [DOI] [PubMed] [Google Scholar]
- 10.Rhinn M, Dollé P. Retinoic acid signalling during development. Development. 2012;139:843–858. doi: 10.1242/dev.065938. [DOI] [PubMed] [Google Scholar]
- 11.Rogers KW, Schier AF. Morphogen gradients: from generation to interpretation. Annu Rev Cell Dev Biol. 2011;27:377–407. doi: 10.1146/annurev-cellbio-092910-154148. [DOI] [PubMed] [Google Scholar]
- 12.Dessaud E, Yang LL, Hill K, Cox B, Ulloa F, Ribeiro A, Mynett A, Novitch BG, Briscoe J. Interpretation of the sonic hedgehog morphogen gradient by a temporal adaptation mechanism. Nature. 2007;450:717–720. doi: 10.1038/nature06347. [DOI] [PubMed] [Google Scholar]
- 13.Dessaud E, Ribes V, Balaskas N, Yang LL, Pierani A, Kicheva A, Novitch BG, Briscoe J, Sasai N. Dynamic assignment and maintenance of positional identity in the ventral neural tube by the morphogen sonic hedgehog. PLoS Biol. 2010;8:e1000382. doi: 10.1371/journal.pbio.1000382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sjödal M, Edlund T, Gunhaga L. Time of exposure to BMP signals plays a key role in the specification of the olfactory and lens placodes ex vivo. Dev Cell. 2007;13:141–149. doi: 10.1016/j.devcel.2007.04.020. [DOI] [PubMed] [Google Scholar]
- 15.Nahmad M, Stathopoulos A. Dynamic interpretation of hedgehog signaling in the Drosophila wing disc. PLoS Biol. 2009;7:e1000202. doi: 10.1371/journal.pbio.1000202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lander AD, Lo WC, Nie Q, Wan FY. The measure of success: constraints, objectives, and tradeoffs in morphogen-mediated patterning. Cold Spring Harb Perspect Biol. 2009;1:a002022. doi: 10.1101/cshperspect.a002022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barkai N, Shilo BZ. Robust generation and decoding of morphogen gradients. Cold Spring Harb Perspect Biol. 2009;1:a001990. doi: 10.1101/cshperspect.a001990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ben-Zvi D, Barkai N. Scaling of morphogen gradients by an expansionrepression integral feedback control. Proc Natl Acad Sci U S A. 2010;107:6924–6929. doi: 10.1073/pnas.0912734107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ben-Zvi D, Shilo BZ, Barkai N. Scaling of morphogen gradients. Curr Opin Genet Dev. 2011;21:704–710. doi: 10.1016/j.gde.2011.07.011. [DOI] [PubMed] [Google Scholar]
- 20.Affolter M, Basler K. The Decapentaplegic morphogen gradient: from pattern formation to growth regulation. Nat Rev Genet. 2007;8:663–674. doi: 10.1038/nrg2166. [DOI] [PubMed] [Google Scholar]
- 21.Clarke DC, Liu X. Decoding the quantitative nature of TGF-beta/Smad signaling. Trends Cell Biol. 2008;18:430–442. doi: 10.1016/j.tcb.2008.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bénazet JD, Zeller R. Vertebrate limb development: moving from classical morphogen gradients to an integrated 4-dimensional patterning system. Cold Spring Harb Perspect Biol. 2009;1:a001339. doi: 10.1101/cshperspect.a001339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ribes V, Briscoe J. Establishing and interpreting graded Sonic Hedgehog signaling during vertebrate neural tube patterning: the role of negative feedback. Cold Spring Harb Perspect Biol. 2009;1:a002014. doi: 10.1101/cshperspect.a002014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pourquié O. Vertebrate segmentation: from cyclic gene networks to scoliosis. Cell. 2011;145:650–663. doi: 10.1016/j.cell.2011.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wojcinski A, Nakato H, Soula C, Glise B. DSulfatase-1 fine-tunes Hedgehog patterning activity through a novel regulatory feedback loop. Dev Biol. 2011;358:168–180. doi: 10.1016/j.ydbio.2011.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Allen BL, Song JY, Izzi L, Althaus IW, Kang JS, Charron F, Krauss RS, McMahon AP. Overlapping roles and collective requirement for the coreceptors GAS1, CDO, and BOC, in SHH pathway function. Dev Cell. 2011;20:775–787. doi: 10.1016/j.devcel.2011.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Félix MA, Barkoulas M. Robustness and flexibility in nematode vulva development. Trends Genet. 2012;28:185–195. doi: 10.1016/j.tig.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 28.Niederreither K, McCaffery P, Dräger UC, Chambon P, Dollé P. Restricted expression and retinoic acid-induced downregulation of the retinaldehyde dehydrogenase type 2 (RALDH-2) gene during mouse development. Mech Dev. 1997;62:67–78. doi: 10.1016/s0925-4773(96)00653-3. [DOI] [PubMed] [Google Scholar]
- 29.Strate I, Min TH, Iliev D, Pera EM. Retinol dehydrogenase 10 is a feedback regulator of retinoic acid signalling during axis formation and patterning of the central nervous system. Development. 2009;136:461–472. doi: 10.1242/dev.024901. [DOI] [PubMed] [Google Scholar]
- 30. Feng L, Hernandez RE, Waxman JS, Yelon D, Moens CB. Dhrs3a regulates retinoic acid biosynthesis through a feedback inhibition mechanism. Dev Biol. 2010;338:1–14. doi: 10.1016/j.ydbio.2009.10.029. This paper describes a dihydroreductase that converts all-trans RA to vitamin A, which is RA-inducible and provides additional negative feedback which may influence signal robustness.
- 31.Kim YK, Wassef L, Hamberger L, Piantedosi R, Palczewski K, Blaner WS, Quadro L. Retinyl ester formation by lecithin: retinol acyltransferase is a key regulator of retinoid homeostasis in mouse embryogenesis. J Biol Chem. 2008;283:5611–5621. doi: 10.1074/jbc.M708885200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.White RJ, Nie Q, Lander AD, Schilling TF. Complex regulation of cyp26a1 creates a robust retinoic acid gradient in the zebrafish embryo. PLoS Biol. 2007;5:e304. doi: 10.1371/journal.pbio.0050304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hernandez RE, Putzke AP, Myers JP, Margaretha L, Moens CB. Cyp26 enzymes generate the retinoic acid response pattern necessary for hindbrain development. Development. 2007;134:177–187. doi: 10.1242/dev.02706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eldar A, Shilo BZ, Barkai N. Elucidating mechanisms underlying robustness of morphogen gradients. Curr Opin Genet Dev. 2004;14:435–439. doi: 10.1016/j.gde.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 35.Eldar A, Rosin D, Shilo BZ, Barkai N. Self-enhanced ligand degradation underlies robustness of morphogen gradients. Dev Cell. 2003;5:635–646. doi: 10.1016/s1534-5807(03)00292-2. [DOI] [PubMed] [Google Scholar]
- 36.Irons DJ, Wojcinski A, Glise B, Monk NA. Robustness of positional specification by the Hedgehog morphogen gradient. Dev Biol. 2010;342:180–193. doi: 10.1016/j.ydbio.2010.03.022. [DOI] [PubMed] [Google Scholar]
- 37.Capurro MI, Shi W, Filmus J. LRP1 mediates the Shh-induced endocytosis of the GPC3-Shh complex. J Cell Sci. 2012 doi: 10.1242/jcs.098889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cai AQ, Radtke K, Linville A, Lander AD, Nie Q, Schilling TF. Cellular retinoic acid-binding proteins are essential for hindbrain patterning and signal robustness in zebrafish. Development. 2012;139:2150–2155. doi: 10.1242/dev.077065. This work combines mathematical modeling and genetic studies in zebrafish to examine robustness of the RA morphogen gradient. A model incorporating Crabps was generated, and its main predictions were validated experimentally.
- 39. Wang L, Xin J, Nie Q. A critical quantity for noise attenuation in feedback systems. PLoS Comput Biol. 2010;6:e1000764. doi: 10.1371/journal.pcbi.1000764. This paper shows that Signed Activation Time (SAT) is a key feature that can enable noise reduction in a regulatory system. Since SAT is straightforward to obtain, it is a convenient computational tool for studies of noise propagation in gene regulatory networks.
- 40. Wartlick O, Mumcu P, Kicheva A, Bittig T, Seum C, Jülicher F, González-Gaitán M. Dynamics of Dpp signaling and proliferation control. Science. 2011;331:1154–1159. doi: 10.1126/science.1200037. This paper demonstrates that the morphogen gradient of Dpp in the Drosophila wing disc increases in amplitude and length-scale, which can help compensate for disc growth during larval development.
- 41.Vuilleumier R, Springhorn A, Patterson L, Koidl S, Hammerschmidt M, Affolter M, Pyrowolakis G. Control of Dpp morphogen signalling by a secreted feedback regulator. Nat Cell Biol. 2010;12:611–617. doi: 10.1038/ncb2064. [DOI] [PubMed] [Google Scholar]
- 42.Kudoh T, Wilson SW, Dawid IB. Distinct roles for, Fgf, Wnt and retinoic acid in posteriorizing the neural ectoderm. Development. 2002;129:4335–4346. doi: 10.1242/dev.129.18.4335. [DOI] [PubMed] [Google Scholar]
- 43.Bothe I, Tenin G, Oseni A, Dietrich S. Dynamic control of head mesoderm patterning. Development. 2011;138:2807–2821. doi: 10.1242/dev.062737. [DOI] [PubMed] [Google Scholar]
- 44. Vitobello A, Ferretti E, Lampe X, Vilain N, Ducret S, Ori M, Spetz JF, Selleri L, Rijli FM. Hox and Pbx factors control retinoic acid synthesis during hindbrain segmentation. Dev Cell. 2011;20:469–482. doi: 10.1016/j.devcel.2011.03.011. This paper demonstrates that some RA targets within the cranial mesoderm (i.e. Hoxa1 and Pbx1/2), maintain Aldh1a2 expression, which may then influence hindbrain patterning and signal robustness.
- 45.Grimm O, Coppey M, Wieschaus E. Modelling the Bicoid gradient. Development. 2010;137:2253–2264. doi: 10.1242/dev.032409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Balaskas N, Ribeiro A, Panovska J, Dessaud E, Sasai N, Page KM, Briscoe J, Ribes V. Gene regulatory logic for reading the Sonic Hedgehog signaling gradient in the vertebrate neural tube. Cell. 2012;148:273–284. doi: 10.1016/j.cell.2011.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gursky VV, Panok L, Myasnikova EM, Manu, Samsonova MG, Reinitz J, Samsonov AM. Mechanisms of gap gene expression canalization in the Drosophila blastoderm. BMC Syst Biol. 2011;5:118. doi: 10.1186/1752-0509-5-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim MS, Kim JR, Kim D, Lander AD, Cho KH. Spatiotemporal network motif reveals the biological traits of developmental gene regulatory networks in Drosophila melanogaster. BMC Syst Biol. 2012;6:31. doi: 10.1186/1752-0509-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Manu, Surkova S, Spirov AV, Gursky VV, Janssens H, Kim AR, Radulescu O, Vanario-Alonso CE, Sharp DH, Samsonova M, et al. Canalization of gene expression in the Drosophila blastoderm by gap gene cross regulation. PLoS Biol. 2009;7:e1000049. doi: 10.1371/journal.pbio.1000049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chomette D, Frain M, Cereghini S, Charnay P, Ghislain J. Krox20 hindbrain cisregulatory landscape: interplay between multiple long-range initiation and autoregulatory elements. Development. 2006;133:1253–1262. doi: 10.1242/dev.02289. [DOI] [PubMed] [Google Scholar]
- 51. Zhang L, Radtke K, Zheng L, Cai AQ, Schilling TF, Nie Q. Noise drives sharpening of gene expression boundaries in the zebrafish hindbrain. 2012:8. doi: 10.1038/msb.2012.45. This work combines mathematical modeling and gene expression studies in zebrafish to examine how sharp boundaries between rhombomeres can form in the face of noise in the RA morphogen gradient and in its downstream targets, hoxb1 and krox20. In part, because these two genes cross-inhibit one another and auto-activate themselves, noisy expression can actually facilitate boundary sharpening.
- 52.Maves L, Jackman W, Kimmel CB. FGF3 and FGF8 mediate a rhombomere 4 signaling activity in the zebrafish hindbrain. Development. 2002;129:3825–3837. doi: 10.1242/dev.129.16.3825. [DOI] [PubMed] [Google Scholar]
- 53.Wiellette EL, Sive H. vhnf1 and Fgf signals synergize to specify rhombomere identity in the zebrafish hindbrain. Development. 2003;130:3821–3829. doi: 10.1242/dev.00572. [DOI] [PubMed] [Google Scholar]
- 54.Hernandez RE, Rikhof HA, Bachmann R, Moens CB. vhnf1 integrates global RA patterning and local FGF signals to direct posterior hindbrain development in zebrafish. Development. 2004;131:4511–4520. doi: 10.1242/dev.01297. [DOI] [PubMed] [Google Scholar]
- 55. Labalette C, Bouchoucha YX, Wassef MA, Gongal PA, Le Men J, Becker T, Gilardi-Hebenstreit P, Charnay P. Hindbrain patterning requires fine-tuning of early krox20 transcription by Sprouty 4. Development. 2011;138:317–326. doi: 10.1242/dev.057299. This paper demonstrates that initiation and maintenance of krox20 expression occur through separate enhancer elements, and that Fgf promotes initiation of expression but also modulates it through negative feedback. This work reveals robustness mechanisms downstream of RA and Fgf in the hindbrain.
- 56.Cooke JE, Moens CB. Boundary formation in the hindbrain: Eph only it were simple. Trends Neurosci. 2002;25:260–267. doi: 10.1016/s0166-2236(02)02134-3. [DOI] [PubMed] [Google Scholar]
- 57.Trainor P, Krumlauf R. Plasticity in mouse neural crest cells reveals a new patterning role for cranial mesoderm. Nat Cell Biol. 2000;2:96–102. doi: 10.1038/35000051. [DOI] [PubMed] [Google Scholar]
- 58.Schilling TF, Prince V, Ingham PW. Plasticity in zebrafish hox expression in the hindbrain and cranial neural crest. Dev Biol. 2001;231:201–216. doi: 10.1006/dbio.2000.9997. [DOI] [PubMed] [Google Scholar]
- 59. Kuchina A, Espinar L, Garcia-Ojalvo J, Süel GM. Reversible and noisy progression towards a commitment point enables adaptable and reliable cellular decision-making. PLoS Comput Biol. 2011;7:e1002273. doi: 10.1371/journal.pcbi.1002273. This paper provides an example in which noise-induced switching helps facilitate the differentiation of isolated cells.