Summary
Renal ciliopathies are a leading cause of kidney failure, but their exact etiology is poorly understood. NEK8/NPHP9 is a ciliary kinase associated with two renal ciliopathies in humans and mice, nephronophthisis (NPHP) and polycystic kidney disease. Here, we identify NEK8 as a key effector of the ATR-mediated replication stress response. Cells lacking NEK8 form spontaneous DNA double-strand breaks (DSBs) which further accumulate when replication forks stall, and they exhibit reduced fork rates, unscheduled origin firing, and increased replication fork collapse. NEK8 suppresses DSB formation by limiting cyclin A-associated CDK activity. Strikingly, a mutation in NEK8 that is associated with renal ciliopathies affects its genome maintenance functions. Moreover, kidneys of NEK8 mutant mice accumulate DNA damage, and loss of NEK8 or replication stress similarly disrupts renal cell architecture in a 3D-culture system. Thus, NEK8 is a critical component of the DNA damage response that links replication stress with cystic kidney disorders.
Keywords: NEK8, ATR, DNA replication, DNA damage response (DDR), polycystic kidney disease, DNA double-strand breaks (DSBs), replication fork, replication stress, nephronophthisis, renal ciliopathies, cyclin-dependent kinase (CDK), cyclin
Introduction
Ciliopathies are a recently defined class of genetic disorders that can present with a broad range of pathologies, including renal cysts, retinal degeneration, mental retardation, and polydactyly (Hildebrandt et al., 2011). These diseases are thought to result from dysfunction of the primary cilium, which plays essential roles in signaling pathways and organ development. However, the underlying mechanisms that link ciliar signaling defects to disease development remain largely unknown.
Intriguingly, some ciliopathies have been recently associated with defects in the DNA damage response (DDR). Nephronophthisis (NPHP) is a degenerative, recessive form of ciliopathy characterized by end-stage kidney disease, and many patients have extra-renal symptoms as well (Hildebrandt et al., 2011). Mutations in 15 different genes (NPHP1-NPHP15) are known to cause NPHP, two of which, CEP164 and ZNF423, have links to the DDR. The well-studied DDR protein, MRE11, has also been found to be mutated in two patients with another form of ciliopathy (Chaki et al., 2012; Sivasubramaniam et al., 2008). Furthermore, mutations in another DDR protein, the Fanconi-associated nuclease 1 (FAN1), are a cause of progressive kidney failure in patients with karyomegalic interstitial nephritis, a degenerative renal disease (Zhou et al., 2012). These findings raise the possibility that defective DDR signaling contributes to the development of progressive kidney diseases, including renal ciliopathies.
The cellular DDR is a complex process required for the maintenance of genome stability, and multiple DDR pathways act at different cell cycle stages and following different types of DNA damage (Ciccia and Elledge, 2010). The genome is particularly vulnerable during DNA replication, as replication forks can stall at DNA lesions, protein-DNA complexes or secondary DNA structures. Accordingly, stalled replication forks are rapidly recognized by pathways which act to stabilize the replication fork structure, arrest the cell cycle and promote replication fork restart (Branzei and Foiani, 2010; Paulsen and Cimprich, 2007). Many of these events are coordinated by the ATR (ATM and Rad3-related) protein kinase, which acts together with its downstream effector kinase CHK1 (Ciccia and Elledge, 2010; Cimprich and Cortez, 2008). Defects in this response to stalled forks, or persistent stalled forks, lead to replication fork collapse and the formation of DNA double-strand breaks (DSBs) (Branzei and Foiani, 2010).
One important function of the ATR-CHK1 signaling pathway is to regulate the firing of origins during S-phase through the inhibition of cyclin-dependent kinase (CDK) activity (Maya-Mendoza et al., 2007; Petermann et al., 2010; Shechter et al., 2004). This regulation is crucial to prevent DNA damage accumulation. ATR and CHK1 regulate CDK activity by inhibiting CDC25A, which activates CDK through dephosphorylation of threonine 14 (T14) and tyrosine 15 (Y15) (Sanchez et al., 1997; Sorensen and Syljuasen, 2012). The protein kinase WEE1 also acts in parallel to the ATR-CHK1 pathway to negatively regulate CDK activity through phosphorylation of CDK at tyrosine 15, and the inhibition of WEE1 leads to the accumulation of DNA damage during normal DNA replication (Beck et al., 2012; Dominguez-Kelly et al., 2011; Jones and Petermann, 2012; Sorensen and Syljuasen, 2012).
The NIMA-related kinase (NEK) family of serine/threonine kinases is emerging as important mediators of the cell cycle, and some NEK family kinases have also been linked to activation of the DDR (Fry et al., 2012; Moniz et al., 2011). One member of this family is NEK8/NPHP9, which has a broad tissue distribution. NEK8 was discovered in a positional cloning analysis of the juvenile cystic kidney (jck) mouse, a model for another ciliopathy, polycystic kidney disease (PKD) (Liu et al., 2002). Affected mice are homozygous for a missense mutation in the presumptive regulatory domain, which has homology to the nucleotide exchange factor RCC1 (regulator of chromatin condensation 1) (Hadjebi et al., 2008). Missense mutations in the RCC domain of Nek8 have also been identified in patents with NPHP and in the Lewis polycystic kidney rat model (McCooke et al., 2012; Otto et al., 2008). NEK8 localizes to primary cilia, and Nek8−/− mice exhibit asymmetry defects and impaired kidney development (Mahjoub et al., 2005; Manning et al., 2013).
Here we reveal an unexpected function for NEK8 in the maintenance of genomic stability and provide critical insights into the mechanism by which it acts. Loss of NEK8 leads to spontaneous DNA damage and a defect in the response of cells to replication stress. These events are a result of elevated CDK activity in the absence of NEK8, and indeed we find that NEK8 interacts with cyclin A-CDK2 complexes and regulates their protein levels. Consistent with effects on CDK activity, we show that NEK8 promotes replication fork progression, suppresses aberrant origin firing, and stabilizes stalled forks. In addition, NEK8 interacts physically and functionally with components of the ATR-mediated DDR. We also show that the disease-related jck mutant of NEK8 fails to interact with ATR pathway proteins and to rescue the genome maintenance defects associated with NEK8 knockdown. Furthermore, NEK8 knockdown or aphidicolin treatment reduces ciliary frequency and disrupts epithelial organization in 3D renal cell cultures. We propose that NEK8 is a critical effector of the replication stress response, and provide evidence that deregulation of this response may contribute to the pathogenesis of renal ciliopathies.
Results
Identification of NEK8 as a mediator of genome stability
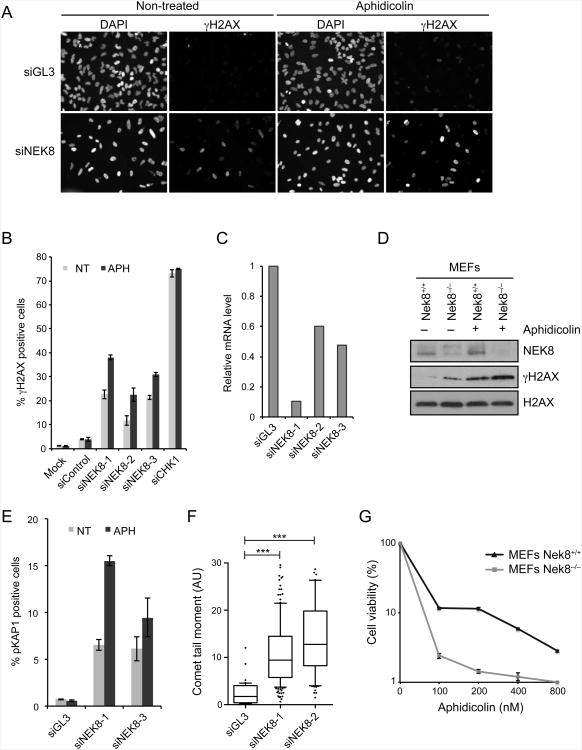
We identified NEK8 in an ongoing genome-wide siRNA screen designed to discover novel genes and pathways that prevent the accumulation of DNA damage in human cells exposed to replication stress. A complete report of this screen will be described elsewhere. Briefly, our strategy was to identify which genes, when knocked down, enhanced the phosphorylation of H2AX following treatment with a low dose of aphidicolin, a DNA polymerase inhibitor. γH2AX, the product of H2AX phosphorylation, is an early marker of DNA damage that we used in a related screen (Paulsen et al., 2009). Among those genes we found to cause aphidicolin-induced γH2AX, NEK8 emerged as a high-confidence hit. Although the knockdown of NEK8 induced H2AX phosphorylation in mock-treated cells, the addition of aphidicolin significantly enhanced this phenotype with three different siRNAs (Figures 1A-1C). Similar results were observed in Nek8−/− mouse embryo fibroblasts (MEFs) (Figure 1D) and upon the knockdown of NEK8 in several additional human cell lines (Figure 6B, Figure S1E), indicating that the effect of NEK8 is not limited to one cell type or even one species. NEK8 knockdown also led to aphidicolin-induced phosphorylation of KAP1 (KRAB-domain associated protein 1), a marker for DSBs (Figure 1E) (Ziv et al., 2006). Furthermore, we observed an increase in tail moment when neutral comet assays were performed (Figure 1F) after knockdown of NEK8, providing direct evidence for the formation of DSBs. Lastly, Nek8−/− MEFs displayed hypersensitivity to low doses of aphidicolin (Figure 1G). Taken together, these data demonstrate that NEK8 is required for genome maintenance, particularly under conditions of replication stress, and that in its absence DNA DSBs are formed.
Figure 1.
Loss of NEK8 leads to accumulation of DNA damage.
(A-B) Induction of γH2AX upon NEK8 knockdown. HeLa cells were transfected with siRNAs targeting luciferase (siGL3) or NEK8 (siNEK8), treated with or without aphidicolin (APH, 400 nM) for 24 h, stained with γH2AX antibodies and DAPI, and analyzed by immunofluorescence. (A) Representative images of γH2AX and DAPI staining. (B) Quantification of percent γH2AX-positive cells. Each data point represents the mean of three experimental replicates. Error bars represent standard error of mean (SEM, n=3). (C) mRNA levels of NEK8 were assessed by RT-qPCR. (D) γH2AX levels in wild-type (Nek8+/+) and knockout (Nek8−/−) MEFs determined by western blotting. APH (400 nM) was used for 18 h. γtubulin was used as a loading control. (E) Quantification of percent pKAP1-positive cells. Each data point represents the mean of three experimental replicates. Error bars represent standard error of mean (SEM, n=3). (F) Quantification of tail moment from neutral comet assay in HeLa cells transfected with siGL3 or siNEK8. Box and whiskers indicate 25-75 and 10-90 percentiles, respectively. The lines represent the median values. Asterisks indicate the P-value of the statistical test (Mann-Whitney rank sum t-test, ***p < 0.0001). Data not included between the whiskers are plotted as outliers (dots). (G) Sensitivity of Nek8−/− MEFs to replication stress. Cells were treated with increasing doses of aphidicolin for 18 h. Surviving colonies were scored 12 days later. Each data point represents the mean of three experimental replicates. Error bars represent standard error of mean (SEM, n=3). In all panels, the data are from three independent experiments. See also Figure S1.
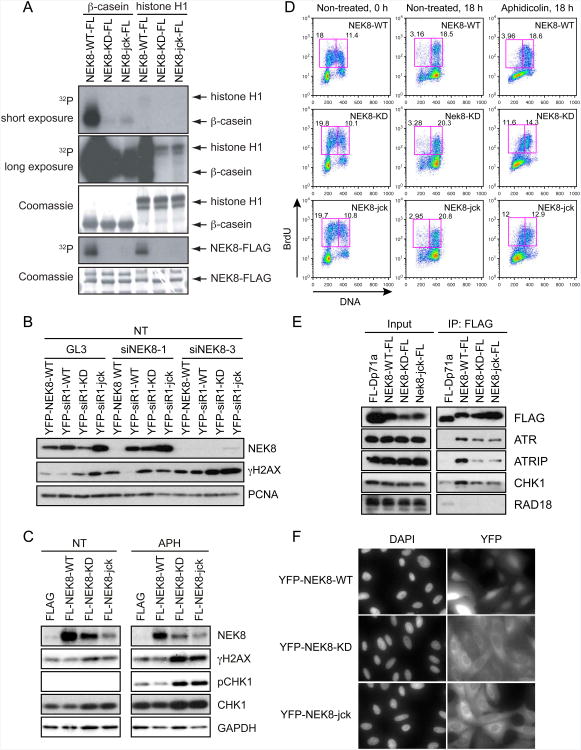
Figure 6.
Kinase-inactive NEK8 and the jck mutants are defective in genome maintenance functions.
(A) HEK293T cells were transfected with FLAG-tagged wild-type (NEK8-FL-WT), kinase-deficient (NEK8-FL-KD), or jck (NEK8-FL-jck) NEK8 constructs for 48 h. After cell lysis, FLAG-tagged proteins were isolated and in vitro kinase reactions were performed with β-casein or histone H1. (B) γH2AX complementation assay in U2OS cell lines stably expressing EYFP-tagged wild-type (YFP-NEK8-WT), siNEK8-1 -resistant wild-type (YFP-siR1-WT), kinase deficient (YFP-siR1-KD), or jck (YFP-siR1-jck) NEK8. Cells were transfected with the indicated siRNAs and lysates were analyzed by western blotting. (C) Overexpression of kinase-deficient and jck NEK8 mutants leads to DNA damage. HeLa cells were transfected with the indicated FLAG-tagged NEK8 constructs for 48 h, left untreated (NT) or aphidicolin (APH)-treated (400 nM) for 24 h, and then processed as described in (B). (D) Effects of overexpressing wild-type and mutant NEK8 on cell cycle progression before and after exposure to APH (400 nM) were determined as described in Figures 2C-2D. (E) Kinase-deficient and jck mutants of NEK8 show a decreased interaction with ATR, ATRIP and CHK1. Samples were processed as described in Figure 5A. (F) Subcellular localization of wild-type or mutant NEK8. U2OS cells stably expressing EYFP-tagged wild-type (YFP-NEK8-WT), kinase deficient (YFP-NEK8-KD), or jck (YFP-NEK8-jck) NEK8 were subjected to immunofluorescence. All of the results are from three independent experiments. See also Figure S6.
NEK8 is not required for cell cycle arrest after DNA damage
The accumulation of DNA damage in NEK8-deficient cells treated with aphidicolin could result from premature mitosis. Thus, we asked whether NEK8-deficient cells showed defects in their ability to arrest cell cycle progression at the G2/M transition following DNA damage. Unexpectedly, cells treated with siRNAs targeting NEK8, as well as Nek8−/− MEFs, displayed normal cell cycle arrest following treatment with ionizing radiation (IR) and ultraviolet (UV) radiation, suggesting the G2/M arrest response was intact (Figure S1A-S1C). We also found no significant difference in the kinase activity of CHK1 in wild-type and Nek8−/− MEFs (Figure S1D). Lastly, we observed no effect of NEK8 knockdown on CHK1 phosphorylation or on CDC25A degradation, two events needed for G2/M arrest (Figure S1E) (Sorensen and Syljuasen, 2012). Thus, we conclude that the observed effects of NEK8 loss are not due to a defect in CHK1 phosphorylation or an inability to arrest with DNA damage at the G2/M transition.
NEK8 prevents DNA damage from arising at stalled replication forks
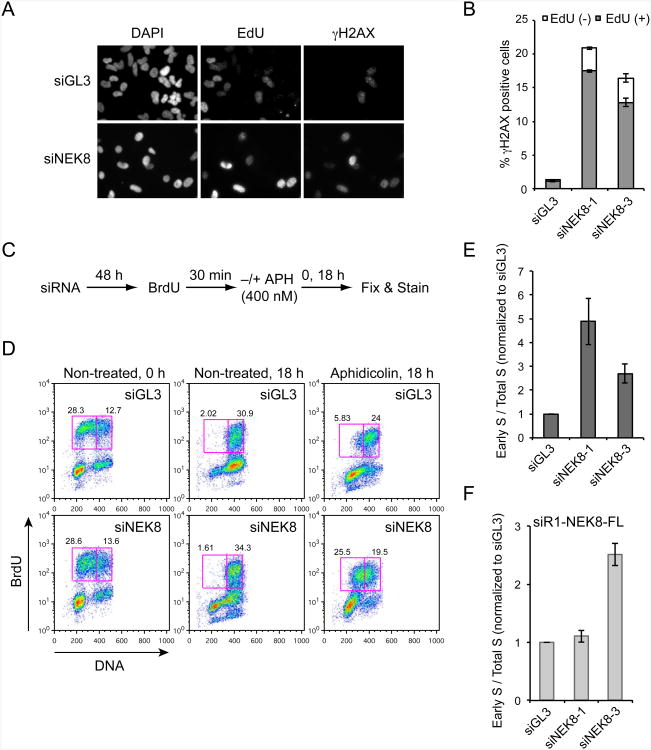
Another possible source of the DNA damage observed upon loss of NEK8 is from defects in the ability to process stalled replication forks. To test this hypothesis, we pulse-labeled replicating cells with EdU and then costained them for EdU and γH2AX. As shown in Figure 2A and 2B, H2AX phosphorylation induced by NEK8 knockdown appeared predominantly in EdU-positive cells, indicating that depletion of NEK8 results in the formation of replication-dependent DSBs.
Figure 2.
NEK8 knockdown causes replication-associated DNA damage and defects in S-phase progression.
(A-B) The γH2AX induced by NEK8 knockdown is enriched in S-phase cells. HeLa cells were transfected with siGL3 or siNEK8 for 72 h, pulse-labeled with EdU for 30 min, fixed and labeled for EdU and γH2AX, then analyzed by immunofluorescence. Representative images (A) and quantification (B) of EdU and γH2AX staining are shown. Error bars represent standard error of mean (SEM, n=3). (C) Schematic of replication progression assay. (D) Effects of NEK8 knockdown on cell cycle progression. The percentages of cells in early S-phase (left box) and late S/G2-phase (right box) are shown. (E) Data represent the ratio of early S-phase cells to total S-phase cells observed in the presence vs absence of aphidicolin for a given siRNA, and were normalized to the same number for siGL3. Error bars represent standard error of mean (SEM, n=3). (F) siRNA-resistant NEK8 rescues the defect in S-phase progression. HeLa cells stably expressing FLAG-tagged wild-type NEK8 resistant to siNEK8-1 (siR1-NEK8-FLAG) were transfected with siNEK8-1 or siNEK8-2 and analyzed as in (C-E). Error bars represent standard error of mean (SEM, n=3). All the results are from three independent experiments. See also Figure S2
Next, we asked whether NEK8 is needed for the efficient completion of DNA replication. To visualize S-phase progression, we pulse-labeled HeLa cells with BrdU and followed the labeled cells through S-phase (Figure 2C). NEK8-depleted cells were able to progress through S-phase nearly as well as control cells in the absence of aphidicolin (Figure 2D). Consistent with this finding, overall cell cycle progression was largely unaffected by the reduction of NEK8 levels in untreated HeLa cells and MEFs (Figure S2). Interestingly, however, loss of NEK8 dramatically affected S-phase progression in the presence of aphidicolin (Figure 2D-2E). 25 % of the cells treated cells remained in early S-phase after knockdown of NEK8, compared with less than 6 % of the cells in the control sample. Importantly, siRNA-resistant NEK8 rescued the effects of knocking down NEK8 (Figure 2F). Thus, NEK8 is important for efficient S-phase progression when cells are exposed to replication stress.
NEK8 regulates replication fork stability, fork speed and origin firing
The dramatic effects of aphidicolin on NEK8-deficient cells raised the possibility that NEK8 may play a role in the stabilization or restart of stalled replication forks. Indeed, we found that Nek8−/− MEFs and HeLa cells transfected with siRNAs targeting NEK8 displayed a profound defect in their ability to resume DNA synthesis upon release from aphidicolin (Figure S3).
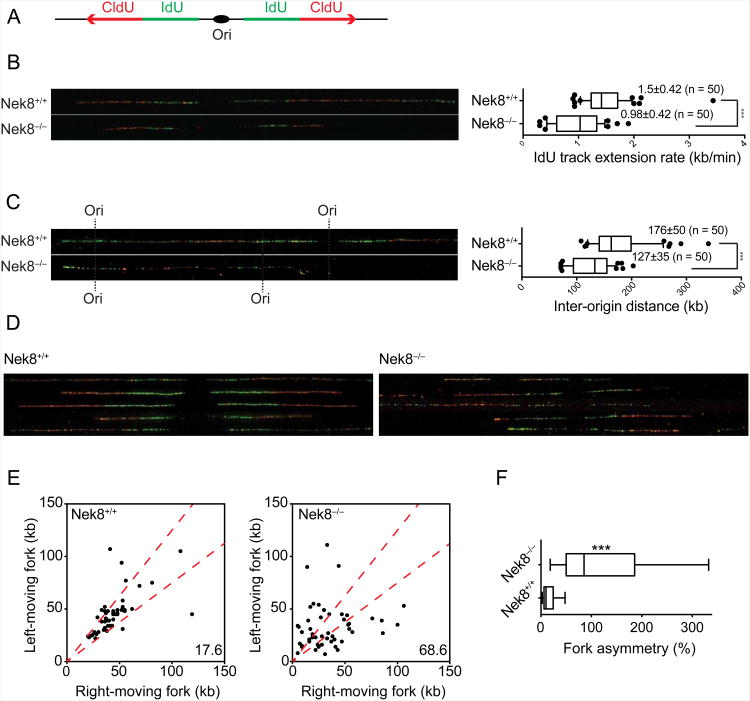
To gain further insight into the mechanisms leading to defects in DNA replication in NEK8-deficient cells, we used DNA combing to examine replication fork dynamics (Bianco et al., 2012; Michalet et al., 1997). We analyzed individual DNA fibers from untreated, asynchronously growing primary wild-type and Nek8−/− MEFs after sequential pulse-labeling of cells with IdU and CldU (Figure 3A). Interestingly, we found that IdU tracks were significantly shorter in Nek8−/− MEFs (Figure 3B). This observation suggests that replication forks move at a slower speed in the absence of NEK8 (0.98 kb/min) versus in its presence (1.5 kb/min). We also compared the inter-origin distance in wild-type and Nek8−/− MEFs by measuring the center-to-center distances between two adjacent initiation sites during the IdU pulse. The median inter-origin distance in Nek8−/− MEFs was significantly decreased (127 kb) relative to that observed in wild-type cells (176 kb), suggesting that Nek8−/− MEFs exhibit an increase in origin firing (Figure 3C). Finally, we asked whether the slower replication fork speed observed in Nek8−/− MEFs could be due to increased fork stalling and collapse by comparing the lengths of the left and right replication tracks around the same origin, a measure of the progression of sister replication forks (Figure 3D-3F). In wild-type cells, sister forks progressed bi-directionally at similar rates and generated symmetrical replication tracts. However, Nek8−/− MEFs exhibited a significant increase in fork asymmetry relative to wild-type cells. Thus, NEK8 is required to suppress origin firing, promote normal replication fork speed, and assure continued replication fork progression. Defects in any or all of these processes could contribute to the increased DNA damage observed in NEK8-depleted cells.
Figure 3.
NEK8 regulates origin firing, replication fork speed, and fork stability.
(A) Schematic of DNA combing. Primary MEFs were sequentially labeled with IdU (green) and CldU (red) for 30 min each. Individual DNA fibers were stretched and then visualized by immunofluorescence. Representative images of DNA fibers are presented. (B) Graph depicts the distribution of IdU track lengths (right panel). (C) Inter-origin distances were measured as the distance between two adjacent initiation sites during IdU pulse and median values are indicated in kb (right panel). (D) Representative images of DNA fibers show the fork asymmetry in Nek8−/− MEFs. (E) Scatter plot shows the length of left- and right-moving sister forks during CldU pulse. The central area marked with red lines indicates sister forks with less than a 25% length difference and the percentage of outliers is shown. (F) Fork asymmetry between left and right forks was scored as the ratio of the longest distance to the shortest covered during CldU labeling for each sister replication fork. Approximately 50 fibers were analyzed in each condition. Box and whiskers in all graphs indicate 25-75 and 10-90 percentiles, respectively. The lines represent the median values. Asterisks indicate the P-value of the statistical test (Mann-Whitney rank sum t-test, ***p < 0.0001). All the data are from three independent experiments. See also Figure S3.
NEK8 regulates cyclin A-associated CDK activity to suppress DSBs
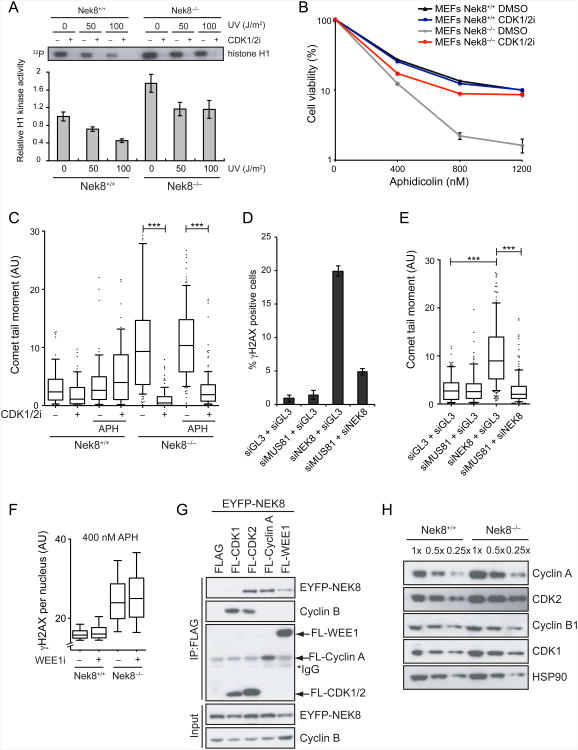
The results from DNA combing suggest that NEK8 plays an important role in coordinating replication fork dynamics and origin firing, and S-phase cyclin-CDK complexes have been previously implicated in regulating these processes. In fact deregulation of CDK activity through cyclin E or cyclin A overexpression, or inhibition of WEE1, results in DNA damage and genome instability (Bartkova et al., 2005; Beck et al., 2010; Dominguez-Kelly et al., 2011; Jones et al., 2012; Tane and Chibazakura, 2009). Thus, we reasoned that NEK8 might regulate the CDK activities needed to fire origins in S-phase. As cyclin A regulates origin firing in mammalian cells (Katsuno et al., 2009; Nakanishi et al., 2010; Sorensen and Syljuasen, 2012), we first carried out an in vitro kinase assay on cyclin A-bound CDKs isolated from wild-type and Nek8−/− MEFs to test our hypothesis. Surprisingly, Nek8−/− MEFs displayed a reproducible increase in cyclin A-associated CDK activity relative to wild-type MEFs (Figure 4A), while cyclin B-associated CDK activity was unaffected (Figure S4A). More importantly, cyclin A-associated CDK activity was only slightly reduced in Nek8−/− MEFs following DNA damage treatment, in contrast with the effect of DNA damage on wild-type MEFs. This finding suggests that NEK8 suppresses cyclin A-associated CDK activity and is also needed to downregulate this activity following DNA damage.
Figure 4.
NEK8 suppresses cyclin A-associated CDK activity to prevent DSB formation.
(A) Nek8−/− MEFs have higher levels of cyclin A-associated CDK activity. Immortalized MEFs were UV-irradiated as indicated and allowed to recover for 1 h, then cyclin A was immunoprecipitated and used in an in vitro kinase assay with histone H1 as a substrate. The CDK1/2 inhibitor (CDK1/2i) was used to demonstrate specificity. An autoradiograph of 32P-labeled histone H1 is shown. The kinase activity was quantified by densitometry. Each data point represents the mean of three experimental replicates, and error bars represent standard error of mean (SEM, n=3) (B) MEFs were treated with indicated concentrations of aphidicolin and either DMSO or CDK1/2i (200 nM) for 24 h. Surviving clones were scored 9 days later. Each data point represents the mean of three experimental replicates and cell viability was analyzed as in Figure 1G. (C) A neutral comet assay was performed in MEFs treated with or without APH (400 nM) and/or CDK1/2i (200 nM) for 18 h. Boxes and whiskers represent the 25-75 and 10-90 percentile, respectively. Horizontal lines mark the medians. Dots represent the data not included between the whiskers. Differences between distributions were assessed with the Mann-Whitney rank sum test (***p < 0.0001). (D & E) Knockdown of MUS81 rescues DNA damage induced by NEK8 loss. HeLa cells were transfected with the indicated siRNAs for 72 h then γH2AX level was assessed by immunofluorescence (D). Each data point represents the mean of three experimental replicates. Error bars represent standard error of mean (SEM, n=3). The neutral comet assay was performed and analyzed (E) as described in (C). (F) NEK8 and WEE1 act in independent pathways to suppress the accumulation of DNA damage. γH2AX levels were analyzed by immunofluorescence in MEFs treated with APH (400 nM) and/or WEE1 inhibitor (WEE1 i, 10 μM) for 18 h. The mean γH2AX intensity for each cell is plotted in box and whisker format with the same statistical analysis as in (C). (G) NEK8 interacts with cyclin A-CDK2 and WEE1. EYFP-tagged NEK8 was co-expressed with FLAG-tagged CDK1, CDK2, cyclin A or WEE1 in HEK293T cells. The cells were lysed 48 h later, and FLAG-tagged proteins and associated proteins were immunoprecipitated using M2 beads, followed by western blotting as indicated. (H) NEK8 regulates cyclin A and CDK2 protein levels. Extracts from Nek8+/+ or Nek8−/− MEFs were collected and cyclin A, cyclin B, CDK1 or CDK2 levels were analyzed by western blotting. All the results are from three independent experiments. AU, arbitrary units. See also Figure S4.
To test whether the elevated CDK activity was an underlying cause for the genome maintenance defects observed in Nek8−/− MEFs, we treated cells with a CDK1/2 inhibitor (CDK1/2i) and monitored γH2AX, DSB formation and cell survival in the absence and the presence of aphidicolin. Aphidicolin did not strongly increase the number of DSBs observed under the conditions monitored although γH2AX was increased, suggesting forks stalled by aphidicolin have not yet collapsed. Remarkably, however, inhibition of CDK activity rescued the sensitivity of Nek8−/− MEFs to replication stress, and it also significantly reduced the accumulation of γH2AX and DSBs in these cells (Figure 4B-4C, Figure S4B). These effects were observed using concentrations of CDK1/2i that did not significantly alter cell cycle progression (Figure S4C), suggesting that the effect is not due to preventing entry into S-phase. Based on these findings, we hypothesize that elevated cyclin A-associated CDK activity resulting from the absence of NEK8 leads to aberrant replication fork dynamics and DNA damage accumulation.
The DSBs that form upon inhibition of WEE1 depend on the activity of MUS81, a structure-specific endonuclease that cleaves stalled replication forks (Beck et al., 2012; Dominguez-Kelly et al., 2011; Hanada et al., 2007). Thus, we asked whether MUS81 activity could account for the DNA damage observed upon down-regulation of NEK8. Strikingly, we found that co-depletion of MUS81 and NEK8 reduced the γH2AX signal observed after knockdown of NEK8 alone (Figure 4D, Figure S4D), and it also led to a reduction in DSB formation as assessed by neutral comet assay (Figure 4E). These observations indicate that MUS81 processes stalled replication forks into DSBs when NEK8 is absent, and taken together with previous findings support the idea that regulation of cyclin A-CDK activity by NEK8 is important for suppressing DSB formation.
To determine if NEK8 acts with WEE1 to suppress DSB formation, we exposed wild-type and Nek8−/− MEFs to an inhibitor of WEE1 and measured the intensity of γH2AX (as opposed to the percentages of γH2AX positive cells) to detect differences in the amount of DNA damage per cell. We found that the WEE1 inhibitor increased the mean γH2AX intensity, consistent with previous observations (Beck et al., 2010; Dominguez-Kelly et al., 2011), while the loss of NEK8 had a modest effect (Figure S4E). These observations suggest that NEK8 is acting in a pathway distinct from that of WEE1, and that WEE1 has functions not shared by NEK8 that help suppress DNA damage formation in unstressed cells. In contrast, when cells were stressed with low levels of aphidicolin, the Nek8−/− MEFs exhibited high levels of γH2AX relative to wild-type MEFs and the addition of the WEE1 inhibitor had a small effect (Figure 4F). These observations indicate that NEK8 plays the primary role in preventing DSB formation after replication stress. Furthermore, they suggest that NEK8 and WEE1 act in distinct ways or at distinct times to regulate S-phase CDK activity and to suppress the accumulation of DNA damage.
To gain insight into the mechanism by which NEK8 controls cyclin A-associated CDK activity, we asked whether NEK8 physically interacts with CDKs, cyclins, or WEE1. We found that EYFP-tagged NEK8 coimmunoprecipitated with FLAG-tagged cyclin A and CDK2, and also weakly with WEE1 (Figure 4G). The interaction between NEK8 and cyclin A was confirmed in a pulldown experiment using bacterially expressed GST-cyclin A and further mapped to the C-terminal half of cyclin A (Figure S4F). Interestingly, GST-CDK2 did not interact with NEK8 in this format, suggesting the CDK2-NEK8 interaction we observed via coimmunoprecipitation might be indirect and through cyclin A-NEK8. Importantly, we also found that Nek8−/− MEFs reproducibly contained significantly elevated levels of cyclin A and CDK2 (Figure 4H). Moreover, inhibitory phosphorylation of CDK1/2 was still observed in Nek8−/− MEFs, consistent with the idea that WEE1 is still active in the absence of NEK8 (Figure S4G). Taken together, these results demonstrate that NEK8 regulates CDK function in a manner distinct from that of WEE1, and they raise the possibility that NEK8 acts via control of cyclin A.
NEK8 interacts with ATR and the ATR-CHK1 pathway and travels with the replication fork
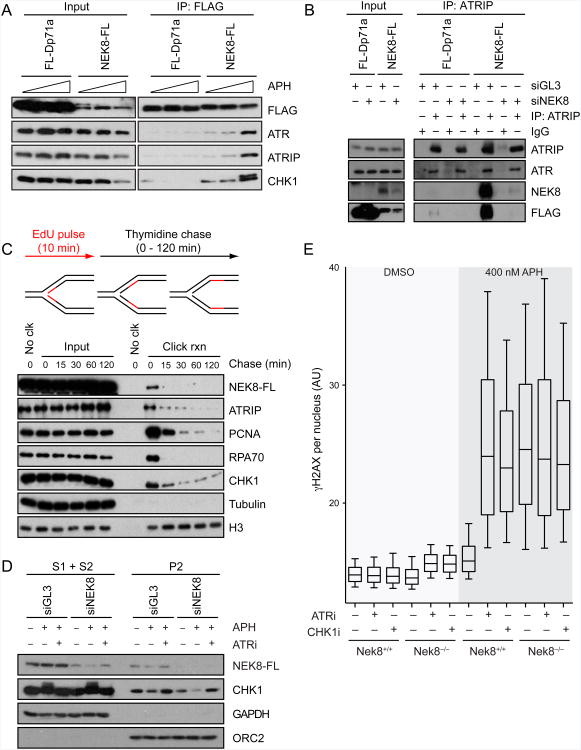
S-phase CDK activity and replication fork dynamics are regulated by the ATR-CHK1 pathway, raising the possibility that NEK8 and ATR are part of a common signaling pathway (Cimprich and Cortez, 2008; Sorensen and Syljuasen, 2012). Therefore, we asked whether we could detect components of the ATR signaling pathway in immunoprecipitates from cells stably expressing FLAG-tagged NEK8. We found that NEK8 interacts with ATR, the ATR-interacting protein ATRIP, and CHK1, while interaction with RAD18, another DNA repair protein, was not detected (Figure 5A, Figure 6E and Figure S5B). Importantly, these interactions were significantly enhanced after treatment of cells with aphidicolin (Figure 5A). We could also detect stably expressed NEK8-FLAG when endogenous ATRIP was immunoprecipitated (Figure 5B). Moreover, we found that bacterially expressed and purified GST-tagged NEK8 pulled down FLAG-tagged ATRIP (Figure S5C, left panel). These results demonstrate that NEK8 interacts with ATR and other checkpoint proteins, and the effect of aphidicolin further suggests that the interactions may be regulated by the DDR.
Figure 5.
NEK8 physically and functionally interacts with the ATR-CHK1 pathway.
(A) NEK8 interacts with ATR-ATRIP and CHK1 proteins and aphidicolin promotes the interactions. HEK293T cells stably expressing FL-Dp71a as a FLAG control or wild-type NEK8-FL were mock-treated or treated with APH (400 nM or 5 μM) for 18 h. Cell lysates (input) and FLAG immunoprecipitates were analyzed by western blotting with the indicated antibodies. (B) Protein complexes immunoprecipitated with IgG or ATRIP antibodies from HEK293T cells stably expressing FL-Dp71a or wild-type NEK8-FL were analyzed by western blotting using the indicated antibodies. (C) NEK8 is enriched at the replication fork. HEK293T cells stably expressing wild-type NEK8 were pulsed with EdU for 10 min then chased with thymidine for the time shown. The nascent DNA-protein complexes were purified by iPOND procedures and proteins were analyzed by western blotting. (D) ATR-dependent dissociation of NEK8 from chromatin. HEK293T cells stably expressing NEK8-WT-FL were transfected with siGL3 or siNEK8 for 48 h and mock-treated or treated with APH (5 μM) and/or ATR inhibitor (ATRi, 1 μM) for 6 h. Cells were harvested and fractionated as described in the Extended Experimental Procedures and proteins were analyzed by western blotting. Cytoplasmic fraction (S1) and soluble nuclear fraction (S2) were pooled. P2 is the chromatin fraction. (E) NEK8 and ATR-CHK1 act together to prevent DNA damage formation. γH2AX intensity was analyzed by immunofluorescence in wild-type and Nek8−/− MEFs treated with or without APH (400 nM), ATRi (1 μM), and/or CHK1i (30 nM) for 18 h. Boxes and whiskers represent the 25-75 and 10-90 percentile, respectively. Horizontal lines mark the medians. All the data are from at least three independent experiments. AU, arbitrary units. See also Figure S5.
Next, we asked whether NEK8 might be acting at the replication fork by monitoring its interaction with nascent DNA using iPOND (isolation of proteins on nascent DNA) (Sirbu et al., 2011). We found that stably expressed NEK8-FLAG associated with EdU-labeled DNA in untreated cells and was chased off the EdU-labeled DNA with thymidine (Figure 5C). Thus, NEK8 travels with the replication fork in undamaged cells along with RPA70, ATRIP, CHK1 and other replisome components.
We also asked if ATR or aphidicolin might affect the interaction of NEK8 with chromatin by monitoring the chromatin binding of stably expressed FLAG-tagged NEK8. Surprisingly, we found that NEK8 was released from chromatin following aphidicolin treatment. Moreover, this release was suppressed by a specific ATR inhibitor (Figure 5D). Interestingly, CHK1 also dissociates from chromatin in an ATR-dependent manner after DNA damage, and this dissociation is important for a functional checkpoint (Smits et al., 2006). Taken together, these observations suggest that NEK8 acts with ATR at the replication fork and that it is regulated in an ATR-dependent manner under conditions of replication stress.
To further explore the relationship between ATR-CHK1 and NEK8, we tested the effects of specific inhibitors of ATR and CHK1 in wild-type and Nek8−/− MEFs as we did with the WEE1 inhibitor. In untreated wild-type MEFs, there was little effect on the mean intensity of γH2AX when ATR or CHK1 was inhibited and the signal in these samples was similar to that observed in untreated Nek8−/− MEFs. Importantly, however, the inhibition of ATR or CHK1 led to further increases in γH2AX intensity in Nek8−/− MEFs (Figure 5E). In contrast, after aphidicolin treatment, the addition of ATR or CHK1 inhibitors to wild-type MEFs, as well as NEK8 loss, dramatically increased the γH2AX intensity. However, the inhibition of ATR or CHK1 did not lead to a further increase in the γH2AX levels in Nek8−/− MEFs in these conditions. These findings are consistent with the idea that NEK8 acts to control origin firing and suggests that ATR-CHK1 and NEK8 can compensate for the lack of the other in the absence of stress. They also indicate that when large numbers of forks are stalled, as with aphidicolin, both pathways are critical. Taken together with the chromatin binding and interaction data, they suggest that ATR-CHK1 may regulate NEK8 to prevent DNA damage accumulation at stalled forks.
The jck mutant of NEK8 exhibits reduced kinase activity
To better understand the links between NEK8's function in the replication stress response and kidney diseases and to explore the role of its kinase activity in its functions, we generated a kinase-inactive mutant of NEK8 as well as the jck form found in the mouse model for PKD. The jck mutant has a single amino acid change (G442V) within the RCC1 domain (Figure S5A) (Liu et al., 2002). To confirm the impact of the kinase-inactivating mutation on NEK8 activity and to explore the regulation of this activity, we first established conditions for an in vitro kinase assay using transiently expressed epitope-tagged NEK8 protein immunoprecipitated using EYFP or FLAG antibodies (Figure 6A and Figure S6A-S6B). We found that wild-type NEK8 could strongly phosphorylate β-casein, and to a lesser extent histone H1. Autophosphorylation of NEK8 was also observed. As expected, the kinase-defective mutant was unable to phosphorylate itself or the β-casein and histone H1 substrates. Several other mutations in the kinase domain also abrogated NEK8's in vitro kinase activity (Figure S6C). Surprisingly, the jck mutant also showed reduced kinase activity compared with wild-type NEK8 (Figure 6A and Figure S6A-S6B). This is in contrast to the results of (Zalli et al., 2012) who find no difference in the activities of wild-type NEK8 and the jck mutant. This discrepancy is likely due to their placement of the epitope tag, which significantly reduces maximal kinase activity (Figure S6D). Also consistent with the effect of the jck mutation on kinase activity, we demonstrate that a different mutation (H425Y) identified in a human patient with NPHP and which lies in the same RCC1 repeat as the jck mutation, reduced NEK8 kinase activity (Figure S6C) (Otto et al., 2008). Lastly, we examined the kinase activity of NEK8 after isolation from UV-damaged cells, but we detected no change in activity under the conditions used. Taken together, these findings strongly suggest that disease-causing mutations in the second repeat of NEK8's RCC1 domain can affect its catalytic activity.
The kinase activity and RCC1 domain of NEK8 are critical for its functions during replication
We next asked whether either the kinase-inactive NEK8 or the jck mutant had defects in NEK8's genome maintenance functions, by knocking down NEK8 in cell lines stably expressing siRNA-resistant forms of wild-type, kinase-deficient, or jck NEK8. Strikingly, wild-type NEK8, but not the other mutants, suppressed the H2AX phosphorylation observed after knockdown of NEK8 (Figure 6B). H2AX and CHK1 phosphorylation were also increased in cells with endogenous NEK8 when either the kinase-deficient or jck mutants were expressed, and this was enhanced further when aphidicolin was present (Figure 6C). Furthermore, overexpression of these mutants exacerbated the effects of aphidicolin on S-phase progression (Figure 6D). Thus, both loss-of-function and overexpression experiments indicate that the kinase activity of NEK8 as well as its RCC1 domain are required to prevent the accumulation of DNA damage in S-phase cells.
Next, we assessed interactions of the kinase-inactive and jck mutants of NEK8 with DDR proteins. Intriguingly, we found that immunoprecipitates of both NEK8 mutants contained less ATR, ATRIP, and CHK1 than immunoprecipitates of wild-type NEK8, suggesting that kinase inactive NEK8 and the jck mutant interacted poorly with these DDR proteins (Figure 6E and Figure S5B). Similar results were observed in pull-down experiments with GST-tagged versions of wild-type, kinase-inactive, or jck NEK8 and FLAG-ATRIP (Figure S5C, left panel). Interestingly, however, all three forms of GST-NEK8 interacted with FLAG-NEK8 equally. This finding suggests that NEK8 is a dimer or higher order oligomer (Figure S5C, right panel), but it also indicates the failure of ATR and ATRIP to interact with the NEK8 mutant proteins is not due to gross misfolding of these proteins.
Lastly, we determined the subcellular localization of stably expressed EYFP-tagged NEK8 proteins. The localization of wild-type NEK8 was predominantly nuclear, consistent with its potential nuclear function in the response to DNA damage, and it was not altered by DNA damage (Figure 6F and data not shown). Surprisingly, however, the localization of both kinase-deficient and jck NEK8 mutants was predominantly cytoplasmic. To test if defective nuclear localization of the NEK8 mutants might account for their inability to execute genome maintenance functions, we added an SV40 nuclear localization signal to the C-terminus of each protein and generated cell lines stably expressing these proteins. Nuclear targeting did not restore the kinase activity of either mutant, its ability to rescue H2AX phosphorylation or its interaction with ATR, ATRIP and CHK1 (Figure S6E-S6G, data not shown). Therefore, relocalization of NEK8 mutants to the nucleus is not sufficient to functionally compensate for wild-type NEK8. These data indicate that the kinase activity of NEK8 and its RCC1 domain are critical for preventing the accumulation of DNA damage as well as NEK8's interaction with checkpoint proteins. Furthermore, the inability of the jck mutants to rescue replication defects suggests that replication stress could be a contributing factor in the etiology of cystic kidney disease.
NEK8 knockdown and replicative stress reduce ciliary frequency and renal cell epithelial organization in 3D spheroid assays
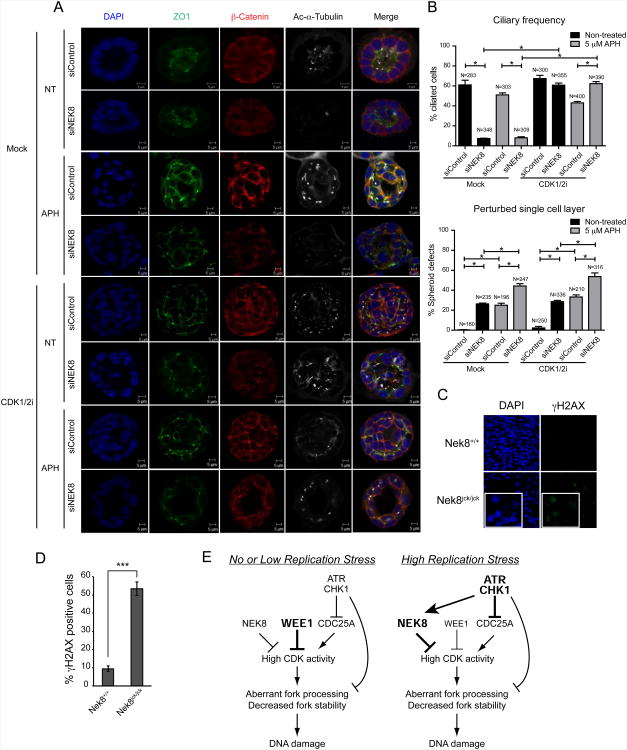
Loss-of-function mutations in NEK8 result in renal fibrosis and cyst formation. To more directly address the potential relationship between NEK8, replication stress and kidney disease, we tested the effect of knocking down NEK8 in a murine collecting duct system we have previously used to model nephronophthisis (Sang et al., 2011). First, we validated the siRNA knockdown of NEK8 by RT-qPCR and confirmed that this knockdown led to increased phosphorylation of H2AX in mouse IMCD3 (inner medullary collecting duct 3) cells (data not shown). Next, IMCD3 cells were transfected with control or NEK8-targeting siRNA in conditions promoting spheroid growth. Immunostaining the spheroids for cilia, tight junctions and adherens junctions revealed no gross changes of polarity upon NEK8 knockdown compared to controls. However, ciliation was significantly affected by NEK8 depletion (7 % siNEK8 vs 61 % siControl), and frequently we observed a perturbed single cell layer at the spheroid equator (26 % siNEK8 vs 0 % siControl) (Figure 7A-B, Figure S7). To analyze the effect of replication stress on this system, spheroids were incubated with aphidicolin (5 μM, 6 h or 400 nM, 18 h) with or without NEK8 knockdown. Decreased ciliary frequencies were observed after 18 h of aphidicolin treatment (33 % siControl). The observed modest effect of aphidicolin on ciliary frequency at the 6 h time point despite the higher dose of aphidicolin might suggest that aphidicolin-induced cilia loss is secondary to S-phase prolongation. However, the perturbed single cell layer phenotype was present at both concentrations of aphidicolin (25 % at 5 μM APH, 6 h, 53 % at 400 nM APH, 18 h), and was further exacerbated by NEK8 knockdown at the 6 h time point (44 % at 5 μM APH, 6 h), relating this phenotype to replicative stress.
Figure 7.
NEK8 knockdown and replication stress perturb ciliogenesis and morphology in 3D spheroid culture, and kidneys from jck mice accumulate DNA damage in vivo.
(A-B) Loss of cilia and induction of perturbed single cell layer upon NEK8 knockdown and/or APH and/or CDK1/2i treatment. (A) Immunostaining of spheroids for cilia (acetylated tubulin, white), tight junctions (ZO1, green) and adherens junctions (β-catenin, red) with DAPI counterstaining (blue) shows loss of cilia and perturbed single cell layer in cells treated with siNEK8, APH (5 μM, 6 h), and/or CDK1/2i (1 μM, 18 h). (B) Quantification of ciliary frequency (* p < 0.006) and perturbed single layer events (* p < 0.01) in spheroids show significant differences between control spheroids and spheroids depleted for NEK8 with an additive effect of treatment with APH (5 μM for 6 h) and a rescue of ciliary frequency upon 18 h treatment with CDK1/2i (1 μM). Number of events scored is indicated in bar graphs (N). Error bars represent SEM (n = 4). (C-D) DNA damage accumulation in homozygous jck mouse kidneys. (C) Representative images of γH2AX and DAPI staining in kidney sections from wild-type and homozygous jck mice at 3 weeks of age are shown; insets indicate a higher magnification. (D) Quantification of percent γH2AX-positive cells in (C). Error bars represent standard error of mean (SEM, n=3). Data are from three independent experiments. (E) Model highlighting the role of NEK8 in preventing DNA damage accumulation through regulation of S-phase CDK activity. See text for details. See also Figure S7.
To investigate the effect of CDK inhibition on our system, we exposed the spheroids to 1 μM CDK1/2 inhibitor for the last 18 h of growth and scored for cilia and perturbed cell layer. In cells transfected with siControl, no difference was observed upon addition of CDK1/2i. However, the profound ciliary defect observed in cells depleted of NEK8 was entirely rescued by exposure to CDK1/2i both in the presence and absence of APH (Figures 7A-7B, Figure S7). These data strongly support a physiological relationship between NEK8 depletion and CDK activation. The perturbed cell layer, however, was not affected by CDK1/2i, presumably because that process is not reversible in the last 18 h of spheroid growth. Therefore, we suggest that replication stress (caused by aphidicolin, NEK8 loss or CDK activation) may etiologically contribute to the architectural renal collecting duct disorganization associated with nephronophthisis.
DNA damage signaling is activated in cystic jck homozygous kidneys
Finally, we asked whether DNA damage accumulates in the early states of kidney disease in the jck mouse. The kidneys of three-week-old jck homozygotes exhibit some signs of cystic disease but their mass is still comparable to the kidneys of wild-type mice. Thus, we examined γH2AX immunostaining in kidney sections from wild-type and homozygous jck mice at this stage of development (Manning et al., 2013; Smith et al., 2006). Importantly, the γH2AX signal was markedly increased in kidney sections from the jck/jck mice compared to those from wild-type mice (Figure 7C-7D), strongly suggesting that these kidneys have high basal levels of DNA damage. Therefore, DNA damage accumulation resulting from the jck mutation is observed in animal models with cystic kidney disease, as well as in cell culture.
Discussion
An unexpected link between renal cilia function and the DNA damage response was recently established when families with renal ciliopathies were found to carry mutations in genes associated with impaired DDR signaling (Chaki et al 2012). However, the underlying mechanism was not addressed, nor are most of the other NPHP genes directly connected to the DDR as yet. In this study, we not only identify a new function for the NPHP-associated protein NEK8 in preventing replication-associated DNA damage, but we also implicate replication stress in the pathogenesis of renal ciliopathies.
Prior to this work, there was no known role for NEK8 in the DDR, and in fact its cellular functions were poorly understood. We demonstrate that NEK8 is a critical regulator of replication fork progression that prevents fork collapse and DSB accumulation through the regulation of CDK activity. Several findings support this model. First, cells lacking NEK8 accumulate DSBs in S-phase and exhibit a defect in S-phase progression, particularly when challenged with inhibitors of replication. Second, NEK8-deficient cells are unable to recover after release from a replication block, and they exhibit abnormal origin firing and decreased fork stability. Third, NEK8 suppresses S-phase CDK activity, and CDK1/2 inhibitors can rescue the DNA damage and aphidicolin sensitivity arising from loss of NEK8. Lastly, NEK8 and ATR act together to suppress the accumulation of DNA damage. Importantly, our data also suggest there is a role for replication stress in the pathogenesis of certain renal ciliopathies. A disease-causing mutation in NEK8 (jck) that produces cystic kidneys leads to defects in the cellular response to replication stress, and this NEK8 mutant fails to interact with ATR-pathway proteins. Moreover, the kidneys of mice with the jck mutation have elevated DDR signaling, and loss of NEK8 function, or replication stress alone, perturbs ciliation and spheroid architecture in a similar and distinctive way. Taken together, these findings indicate that NEK8 plays a crucial function at stalled replication forks through its regulation of CDK activity, and that the absence of NEK8 and other DDR proteins may contribute to renal ciliopathies by raising the basal level of replication stress in cells.
NEK8 Regulates Cyclin A-CDK2 Activity and Levels To Modulate DNA Replication Dynamics and Maintain Genomic Integrity
Our studies establish an unanticipated but critical role for NEK8 in regulating replication fork dynamics and origin firing. Given the discussed effects of CDK activity on the regulation of origin firing, it seems likely that at least some of NEK8's effects are due to the regulation of S-phase CDK activity by NEK8. One possibility is that the increased CDK activity caused by NEK8 loss leads to increased origin firing and fork stalling by depleting nucleotide pools and key replication factors and/or by causing frequent collisions with transcriptional machinery. Excessive origin firing would also prevent the use of dormant origins in the rescue of stalled replication forks, further affecting fork stability and recovery, particularly in the presence of aphidicolin. Cleavage of stalled forks by MUS81, the activity of which is elevated when CDK activity is elevated, would then lead to fork collapse and DSB formation. Although we cannot exclude the possibility that NEK8 has additional functions that prevent DNA damage accumulation, this model is supported by the ability of CDK inhibitors and MUS81 depletion to rescue the DNA damage induced by NEK8 loss, as well as the effect of CDK inhibitors on the survival of Nek8−/− MEFs treated with aphidicolin.
Our findings also provide insight into how NEK8 regulates CDK activity. A crucial regulator of S-phase CDK activity is WEE1, and previous studies have shown that phenotypes similar to those we observe upon loss of NEK8 arise from deregulation of WEE1 in unperturbed cells (Beck et al., 2010; Dominguez-Kelly et al., 2011). However, we find that NEK8 regulates CDK activity in a manner distinct from that of WEE1. Nek8−/− MEFs have significantly elevated levels of cyclin A and CDK2. Furthermore, NEK8 can interact with cyclin A and CDK2, raising the possibility that its effect on their levels is direct. We therefore suggest that NEK8's effects on cyclin A and CDK2 levels are responsible for its effects on CDK activity and genome stability. Further experiments will be required to fully elucidate the mechanism by which this occurs.
NEK8 Acts with the ATR-CHK1 Pathway to Prevent the Accumulation of DNA Damage
Previous work has shown that ATR-CHK1 signaling also downregulates CDK activity during S-phase to reduce origin firing (Maya-Mendoza et al., 2007; Petermann et al., 2010; Shechter et al., 2004), and our data are consistent with the idea that NEK8 acts with the ATR-CHK1 signaling pathway to prevent DNA damage accumulation. Indeed, the effects of NEK8 phenocopy those of ATR and CHK1 loss in several ways. Cells lacking NEK8, ATR or CHK1 activity exhibit spontaneous DNA damage, have similar defects in replication fork dynamics, are sensitive to replication stress, and have profound defects in the recovery from aphidicolin treatment ((Jones and Petermann, 2012) and results herein). However, experiments with ATR and CHK1 inhibitors suggest that ATR-CHK1 and NEK8 have overlapping but non-redundant functions during normal replication. One possibility is that ATR and CHK1 can stabilize or restart spontaneously stalled forks resulting from the increased origin firing in NEK8's absence, while in ATR or CHK1's absence there are still sufficient origins available to rescue forks that collapse due to the lack of ATR.
Importantly, however, experiments with ATR and CHK1 inhibitors also suggest that ATR may regulate NEK8 under conditions of replication stress. This hypothesis is supported by the physical interactions and localization of NEK8. NEK8 interacts with the ATR-ATRIP complex as well as CHK1, and these interactions are enhanced by aphidicolin treatment. Additionally, the dynamic association of NEK8 with chromatin following replication stress is regulated by ATR kinase activity. Intriguingly, this behavior of NEK8 is similar to that of CHK1, suggesting both proteins are downstream effectors of ATR with functions important for maintaining replication fork stability (Smits et al., 2006).
In summary, our data on the cellular functions and regulation of NEK8 suggest that it is an important S-phase CDK modulator that can be regulated by the ATR-CHK1 pathway under conditions of replication stress. Furthermore, both the distinct roles of ATR-CHK1 and NEK8 in unperturbed cells as well as the convergence of their functions following replicative stress may contribute to the maintenance of genome integrity (Figure 7E).
NEK8 Links Genome Stability to Renal Ciliopathies
The first links between the DDR and chronic kidney disease were established when patients with NPHP-like disease or progressive renal failure were found with mutations in DDR proteins (Chaki et al., 2012; Zhou et al., 2012). Our findings extend this connection by demonstrating a role for NEK8 in the DDR, and they also provide mechanistic insight into the molecular basis of the link between the DDR and renal ciliopathies. Indeed, several of our findings strongly suggest that the phenotypes associated with some cystic kidney diseases result from replication stress, either induced by exogenous insults or as a result of defects in the replication stress response. First, we show that NEK8 mutations which cause disease in the jck mouse model lead to defects in the replication stress response and in the interactions of NEK8 with ATR, ATRIP and CHK1. Second, we demonstrate that kidneys from jck homozygous mice have elevated levels of γH2AX at an early stage of disease progression, suggesting the DDR is activated and DNA damage is accumulating in these kidneys. Third, in direct support of the idea that replication stress itself may promote cystic kidney disease, we show that knockdown of NEK8, which causes replication stress, or treatment with aphidicolin alone, reduces ciliary frequency and disturbs epithelial organization in 3D renal cell cultures. Intriguingly, we also find that DNA damage accumulation and aphidicolin sensitivity in Nek8−/− MEFs, as well as the ciliary defect observed after NEK8 knockdown in a 3D culture system, can be suppressed with an inhibitor of this activity. This is a particularly intriguing observation because disease progression in the jck mouse can be significantly delayed with the CDK inhibitor roscovitine (Bukanov et al., 2006), and it therefore ties disease progression to the DNA damage phenotype. Thus, our findings provide a mechanistic basis for the therapeutic effect of roscovitine, and reveal a new direction for further research into renal ciliopathies.
Our results are consistent with the idea that DDR defects may account for the distinct phenotypes of dysplasia and degeneration observed in several tissues of patients with NPHP or related ciliopathies, but they also extend these findings in a significant way. It has been speculated that a defect in the DDR together with replication stress may underlie disease pathogenesis, causing dysplasia and defects in morphogenesis in early development when cells are rapidly proliferating and degenerative phenotypes when tissues are maintained (Chaki et al., 2012). Although a defect in the DDR may still contribute to disease progression, our studies suggest that replication stress itself may be sufficient to give rise to some of the phenotypes observed. In the case of NEK8, it would thus be the high levels of replication stress brought about by changes in replication fork dynamics that induce the phenotypic changes associated with disease progression.
At this point, we can only speculate on the molecular mechanism by which the DNA damage accumulating in renal epithelial cells affects renal cell architecture but several intriguing possibilities arise. Because short treatments with aphidicolin induce perturbations in the spheroid assay yet have only a modest effect on ciliary formation, we hypothesize that replication stress ultimately impinges directly on pathways regulating the events that allow 3D-cell architecture to be established or maintained. One possibility is that the perturbed cell layer in the renal spheroid assays may reflect enhanced cell migration. Along these lines, depletion of NEK8 has been described to result in increased migration (Simpson et al., 2008). Another possibility is that replication stress may directly induce changes in the actin cytoskeleton and defects in morphogenesis, as reported in yeast (Enserink et al., 2006). Thus, it seems plausible that some of the characteristic architectural changes observed in patients with renal ciliopathies could be linked to these changes. Clearly, further studies will be needed to fully dissect the relationship between DNA damage accumulation and renal ciliopathies.
NEK8 Kinase Activity, Replication Stress and Kidney Disease
Our data also establish that the effects of NEK8 on replication fork dynamics require its kinase activity, and they raise an important point about the link between NEK8 kinase activity and the jck mutation found in the mouse model for PKD. We find that NEK8 kinase-inactive and jck mutants have similar phenotypes, consistent with our finding that the jck mutant exhibits reduced kinase activity. Although kinase inactivation might be considered a loss-of-function mutation, it appears that in the case of cyst formation in the jck mouse, and likely in patients, the situation is more complex. Indeed, Nek8−/− mice do not develop the same phenotypes as those with the jck mutation (Manning et al., 2013; Smith et al., 2006). Moreover, the Nek8−/jck compound heterozygotes develop less severe cystic kidney disease than the jck homozygotes. This suggests that the jck allele, which has reduced kinase activity, is actually a gain-of-function allele in the context of renal cyst formation. Consistent with such an idea, we find expression of the kinase-inactive or jck mutant leads to dominant negative effects (Figure 6B-6D). This finding is interesting and akin to the more severe phenotypes observed in the kinase-inactive ATM mouse relative to that seen in the Atm−/− mice (Daniel et al., 2012; Yamamoto et al., 2012).
Summary
In conclusion, our study establishes that NEK8 function is critical for a proper response to stalled replication forks and that replication stress, due to disruption of NEK8 or by other means, may directly contribute to the phenotypes observed in renal ciliopathies. The functions of NEK8 also raise the possibility that it could play an important role in cancer cells. Indeed, NEK8 is overexpressed in some cancer cell lines, and it will be interesting to explore the significance of this finding and whether this contributes to an ability to tolerate replication stress (Bowers and Boylan, 2004; Camilleri-Broet et al., 2007). One could also imagine that NEK8 might have tumor suppressive functions and that NEK8 downregulation could contribute to the genome instability observed in many cell types. The identification of NEK8 targets and NEK8 inhibitors will help address many of these interesting hypotheses.
Experimental Procedures
Cell Culture, Antibodies and Reagents
HeLa, U2OS, and HEK293T cells were cultured in DMEM (GIBCO) supplemented with 10 % FBS, 2 mM L-Glutamine, and penicillin/streptomycin in 5 % CO2, at 37 °C. IMCD3 cells were grown in DMEM:F12 (1:1) medium with 10 % FCS, 2 mM L-Glutamine, and penicillin/streptomycin. Primary Nek8+/+ and Nek8−/− and SV40 large T-antigen transformed MEFs were maintained in DMEM with 15 % FBS, 2 mM L-Glutamine, and penicillin/streptomycin. Sources of antibodies, reagents, and plasmids used in this study are presented in the Extended Experimental Procedures.
DNA Combing, Replication Fork Recovery and Restart Assays
DNA combing was performed in primary Nek8+/+ and Nek8−/− MEFs and processed as previously described. For the recovery and restart assay, siRNA-transfected HeLa cells were sequentially labeled with IdU and CldU and treated with or without aphidicolin. Cell were then fixed and stained with BrdU antibodies specific for IdU and CldU. Details can be found in the Extended Experimental Procedures.
Isolation of Proteins on Nascent DNA (iPOND) and Chromatin Fractionation
Detailed protocols for iPOND and chromatin fractionation have been reported (Sirbu et al., 2011; Smits et al., 2006). Some modifications in this study are described in the Extended Experimental Procedures.
Co-immunoprecipitation, GST Pull-down, and In Vitro Kinase Assays
FLAG-tagged proteins expressed in HEK293T cells were isolated with M2 agarose beads (Sigma). Anti-ATRIP and Protein G sepharose beads (Sigma) was used for immunoprecipitation. Bacterially expressed GST-fusion proteins were isolated by glutathione agarose beads (Sigma). Detailed conditions for cell lysis, protein purification and in vitro kinase assay are described in the Extended Experimental Procedures.
Immunofluorescence and Flow Cytometry
All experiments were performed as previously described (Beck et al., 2012; Paulsen et al., 2009). Some exceptions are described in the Extended Experimental Procedures.
Statistical Analysis
Statistical analyses represent the mean of at least three independent experiments. Error bars represent standard error of mean (SEM, n=3). P-values were calculated using a two-tailed Student's t test, if not specified.
Supplementary Material
Highlights.
NEK8 acts with ATR at the replication fork to control fork dynamics and origin firing
NEK8 suppresses DNA damage by regulating S-phase CDK activity
A ciliopathy-causing NEK8 mutant is defective in the replication stress response
Replication stress or loss of NEK8 perturbs ciliation and 3D renal cell architecture
Acknowledgments
We thank members of the Cimprich lab (especially Robert Driscoll) and Peter Jackson for a careful reading of this manuscript and helpful discussions. This work was supported by a Stanford Graduate Fellowship to HJCC, by a Department of Defense (Breast Cancer Research Program) predoctoral fellowship (W81XWH-09-1-0026) and NIH training grant (R90 DK071499) to JRL, an NIH training grant (T32 CA09151) and ACS postdoctoral fellowship to ACK, an EMBO long-term fellowship to JBV, a Cancer Research UK and an ERC Advanced Investigator Grant (RecMitMei) to SJB, and by NIH grants ES016486 and ES016867 to KAC. SJB is a recipient of a Royal Society Wolfson Research Merit Award, and KAC is an Ellison Medical Foundation Senior Scholar and Glenn Foundation recipient. GGS and RHG were supported by grants from the EU FP7/2009 Consortium “SYSCILIA” (241955) and the Dutch Kidney Foundation Kouncil Consortium Grant CP11.
Footnotes
Supplemental Information: Supplemental Information includes Extended Experimental Procedures and seven figures.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bartkova J, Horejsi Z, Koed K, Kramer A, Tort F, Zieger K, Guldberg P, Sehested M, Nesland JM, Lukas C, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–870. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- Beck H, Nahse V, Larsen MS, Groth P, Clancy T, Lees M, Jorgensen M, Helleday T, Syljuasen RG, Sorensen CS. Regulators of cyclin-dependent kinases are crucial for maintaining genome integrity in S phase. J Cell Biol. 2010;188:629–638. doi: 10.1083/jcb.200905059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck H, Nahse-Kumpf V, Larsen MS, O'Hanlon KA, Patzke S, Holmberg C, Mejlvang J, Groth A, Nielsen O, Syljuasen RG, et al. Cyclin-dependent kinase suppression by WEE1 kinase protects the genome through control of replication initiation and nucleotide consumption. Mol Cell Biol. 2012;32:4226–4236. doi: 10.1128/MCB.00412-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco JN, Poli J, Saksouk J, Bacal J, Silva MJ, Yoshida K, Lin YL, Tourriere H, Lengronne A, Pasero P. Analysis of DNA replication profile in budding yeast and mammalian cells using DNA combing. Methods. 2012;57:149–157. doi: 10.1016/j.ymeth.2012.04.007. [DOI] [PubMed] [Google Scholar]
- Bowers AJ, Boylan JF. Nek8, a NIMA family kinase member, is overexpressed in primary human breast tumors. Gene. 2004;328:135–142. doi: 10.1016/j.gene.2003.12.002. [DOI] [PubMed] [Google Scholar]
- Branzei D, Foiani M. Maintaining genome stability at the replication fork. Nature Reviews Molecular Cell Biology. 2010;11:208–219. doi: 10.1038/nrm2852. [DOI] [PubMed] [Google Scholar]
- Bukanov NO, Smith LA, Klinger KW, Ledbetter SR, Ibraghimov-Beskrovnaya O. Long-lasting arrest of murine polycystic kidney disease with CDK inhibitor roscovitine. Nature. 2006;444:949–952. doi: 10.1038/nature05348. [DOI] [PubMed] [Google Scholar]
- Camilleri-Broet S, Cremer I, Marmey B, Comperat E, Viguie F, Audouin J, Rio MC, Fridman WH, Sautes-Fridman C, Regnier CH. TRAF4 overexpression is a common characteristic of human carcinomas. Oncogene. 2007;26:142–147. doi: 10.1038/sj.onc.1209762. [DOI] [PubMed] [Google Scholar]
- Chaki M, Airik R, Ghosh AK, Giles RH, Chen R, Slaats GG, Wang H, Hurd TW, Zhou W, Cluckey A, et al. Exome capture reveals ZNF423 and CEP164 mutations, linking renal ciliopathies to DNA damage response signaling. Cell. 2012;150:533–548. doi: 10.1016/j.cell.2012.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nature Reviews Molecular Cell Ciology. 2008;9:616–627. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel JA, Pellegrini M, Lee BS, Guo Z, Filsuf D, Belkina NV, You Z, Paull TT, Sleckman BP, Feigenbaum L, et al. Loss of ATM kinase activity leads to embryonic lethality in mice. J Cell Biol. 2012;198:295–304. doi: 10.1083/jcb.201204035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Kelly R, Martin Y, Koundrioukoff S, Tanenbaum ME, Smits VA, Medema RH, Debatisse M, Freire R. Wee1 controls genomic stability during replication by regulating the Mus81-Eme1 endonuclease. J Cell Biol. 2011;194:567–579. doi: 10.1083/jcb.201101047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enserink JM, Smolka MB, Zhou H, Kolodner RD. Checkpoint proteins control morphogenetic events during DNA replication stress in Saccharomyces cerevisiae. J Cell Biol. 2006;175:729–741. doi: 10.1083/jcb.200605080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry AM, O'Regan L, Sabir SR, Bayliss R. Cell cycle regulation by the NEK family of protein kinases. J Cell Sci. 2012;125:4423–4433. doi: 10.1242/jcs.111195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjebi O, Casas-Terradellas E, Garcia-Gonzalo FR, Rosa JL. The RCC1 superfamily: from genes, to function, to disease. Biochim Biophys Acta. 2008;1783:1467–1479. doi: 10.1016/j.bbamcr.2008.03.015. [DOI] [PubMed] [Google Scholar]
- Hanada K, Budzowska M, Davies SL, van Drunen E, Onizawa H, Beverloo HB, Maas A, Essers J, Hickson ID, Kanaar R. The structure-specific endonuclease Mus81 contributes to replication restart by generating double-strand DNA breaks. Nat Struct Mol Biol. 2007;14:1096–1104. doi: 10.1038/nsmb1313. [DOI] [PubMed] [Google Scholar]
- Hildebrandt F, Benzing T, Katsanis N. Ciliopathies. N Engl J Med. 2011;364:1533–1543. doi: 10.1056/NEJMra1010172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones RM, Mortusewicz O, Afzal I, Lorvellec M, Garcia P, Helleday T, Petermann E. Increased replication initiation and conflicts with transcription underlie Cyclin E-induced replication stress. Oncogene. 2012 doi: 10.1038/onc.2012.387. [DOI] [PubMed] [Google Scholar]
- Jones RM, Petermann E. Replication fork dynamics and the DNA damage response. Biochem J. 2012;443:13–26. doi: 10.1042/BJ20112100. [DOI] [PubMed] [Google Scholar]
- Katsuno Y, Suzuki A, Sugimura K, Okumura K, Zineldeen DH, Shimada M, Niida H, Mizuno T, Hanaoka F, Nakanishi M. Cyclin A-Cdk1 regulates the origin firing program in mammalian cells. Proc Natl Acad Sci U S A. 2009;106:3184–3189. doi: 10.1073/pnas.0809350106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Lu W, Obara T, Kuida S, Lehoczky J, Dewar K, Drummond IA, Beier DR. A defect in a novel Nek-family kinase causes cystic kidney disease in the mouse and in zebrafish. Development. 2002;129:5839–5846. doi: 10.1242/dev.00173. [DOI] [PubMed] [Google Scholar]
- Mahjoub MR, Trapp ML, Quarmby LM. NIMA-related kinases defective in murine models of polycystic kidney diseases localize to primary cilia and centrosomes. J Am Soc Nephrol. 2005;16:3485–3489. doi: 10.1681/ASN.2005080824. [DOI] [PubMed] [Google Scholar]
- Manning DK, Sergeev M, van Heesbeen RG, Wong MD, Oh JH, Liu Y, Henkelman RM, Drummond I, Shah JV, Beier DR. Loss of the ciliary kinase nek8 causes left-right asymmetry defects. J Am Soc Nephrol. 2013;24:100–112. doi: 10.1681/ASN.2012050490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maya-Mendoza A, Petermann E, Gillespie DA, Caldecott KW, Jackson DA. Chk1 regulates the density of active replication origins during the vertebrate S phase. Embo J. 2007;26:2719–2731. doi: 10.1038/sj.emboj.7601714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCooke JK, Appels R, Barrero RA, Ding A, Ozimek-Kulik JE, Bellgard MI, Morahan G, Phillips JK. A novel mutation causing nephronophthisis in the Lewis polycystic kidney rat localises to a conserved RCC1 domain in Nek8. BMC Genomics. 2012;13:393. doi: 10.1186/1471-2164-13-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalet X, Ekong R, Fougerousse F, Rousseaux S, Schurra C, Hornigold N, van Slegtenhorst M, Wolfe J, Povey S, Beckmann JS, et al. Dynamic molecular combing: stretching the whole human genome for high-resolution studies. Science. 1997;277:1518–1523. doi: 10.1126/science.277.5331.1518. [DOI] [PubMed] [Google Scholar]
- Moniz L, Dutt P, Haider N, Stambolic V. Nek family of kinases in cell cycle, checkpoint control and cancer. Cell Div. 2011;6:18. doi: 10.1186/1747-1028-6-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi M, Katsuno Y, Niida H, Murakami H, Shimada M. Chk1-cyclin A/Cdk1 axis regulates origin firing programs in mammals. Chromosome Res. 2010;18:103–113. doi: 10.1007/s10577-009-9086-2. [DOI] [PubMed] [Google Scholar]
- Otto EA, Trapp ML, Schultheiss UT, Helou J, Quarmby LM, Hildebrandt F. NEK8 mutations affect ciliary and centrosomal localization and may cause nephronophthisis. J Am Soc Nephrol. 2008;19:587–592. doi: 10.1681/ASN.2007040490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen RD, Cimprich KA. The ATR pathway: fine-tuning the fork. DNA Repair. 2007;6:953–966. doi: 10.1016/j.dnarep.2007.02.015. [DOI] [PubMed] [Google Scholar]
- Paulsen RD, Soni DV, Wollman R, Hahn AT, Yee MC, Guan A, Hesley JA, Miller SC, Cromwell EF, Solow-Cordero DE, et al. A genome-wide siRNA screen reveals diverse cellular processes and pathways that mediate genome stability. Mol Cell. 2009;35:228–239. doi: 10.1016/j.molcel.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petermann E, Woodcock M, Helleday T. Chk1 promotes replication fork progression by controlling replication initiation. Proc Natl Acad Sci U S A. 2010;107:16090–16095. doi: 10.1073/pnas.1005031107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez Y, Wong C, Thoma RS, Richman R, Wu Z, Piwnica-Worms H, Elledge SJ. Conservation of the Chk1 checkpoint pathway in mammals: linkage of DNA damage to Cdk regulation through Cdc25. Science. 1997;277:1497–1501. doi: 10.1126/science.277.5331.1497. [DOI] [PubMed] [Google Scholar]
- Sang L, Miller JJ, Corbit KC, Giles RH, Brauer MJ, Otto EA, Baye LM, Wen X, Scales SJ, Kwong M, et al. Mapping the NPHP-JBTS-MKS protein network reveals ciliopathy disease genes and pathways. Cell. 2011;145:513–528. doi: 10.1016/j.cell.2011.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shechter D, Costanzo V, Gautier J. ATR and ATM regulate the timing of DNA replication origin firing. Nat Cell Biol. 2004;6:648–655. doi: 10.1038/ncb1145. [DOI] [PubMed] [Google Scholar]
- Simpson KJ, Selfors LM, Bui J, Reynolds A, Leake D, Khvorova A, Brugge JS. Identification of genes that regulate epithelial cell migration using an siRNA screening approach. Nat Cell Biol. 2008;10:1027–1038. doi: 10.1038/ncb1762. [DOI] [PubMed] [Google Scholar]
- Sirbu BM, Couch FB, Feigerle JT, Bhaskara S, Hiebert SW, Cortez D. Analysis of protein dynamics at active, stalled, and collapsed replication forks. Genes Dev. 2011;25:1320–1327. doi: 10.1101/gad.2053211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivasubramaniam S, Sun X, Pan YR, Wang S, Lee EY. Cep164 is a mediator protein required for the maintenance of genomic stability through modulation of MDC1, RPA, and CHK1. Genes Dev. 2008;22:587–600. doi: 10.1101/gad.1627708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith LA, Bukanov NO, Husson H, Russo RJ, Barry TC, Taylor AL, Beier DR, Ibraghimov-Beskrovnaya O. Development of polycystic kidney disease in juvenile cystic kidney mice: insights into pathogenesis, ciliary abnormalities, and common features with human disease. J Am Soc Nephrol. 2006;17:2821–2831. doi: 10.1681/ASN.2006020136. [DOI] [PubMed] [Google Scholar]
- Smits VA, Reaper PM, Jackson SP. Rapid PIKK-dependent release of Chk1 from chromatin promotes the DNA-damage checkpoint response. Curr Biol. 2006;16:150–159. doi: 10.1016/j.cub.2005.11.066. [DOI] [PubMed] [Google Scholar]
- Sorensen CS, Syljuasen RG. Safeguarding genome integrity: the checkpoint kinases ATR, CHK1 and WEE1 restrain CDK activity during normal DNA replication. Nucleic Acids Res. 2012;40:477–486. doi: 10.1093/nar/gkr697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tane S, Chibazakura T. Cyclin A overexpression induces chromosomal double-strand breaks in mammalian cells. Cell Cycle. 2009;8:3900–3903. doi: 10.4161/cc.8.23.10071. [DOI] [PubMed] [Google Scholar]
- Yamamoto K, Wang Y, Jiang W, Liu X, Dubois RL, Lin CS, Ludwig T, Bakkenist CJ, Zha S. Kinase-dead ATM protein causes genomic instability and early embryonic lethality in mice. J Cell Biol. 2012;198:305–313. doi: 10.1083/jcb.201204098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalli D, Bayliss R, Fry AM. The Nek8 protein kinase, mutated in the human cystic kidney disease nephronophthisis, is both activated and degraded during ciliogenesis. Hum Mol Genet. 2012;21:1155–1171. doi: 10.1093/hmg/ddr544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Otto EA, Cluckey A, Airik R, Hurd TW, Chaki M, Diaz K, Lach FP, Bennett GR, Gee HY, et al. FAN1 mutations cause karyomegalic interstitial nephritis, linking chronic kidney failure to defective DNA damage repair. Nat Genet. 2012;44:910–915. doi: 10.1038/ng.2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv Y, Bielopolski D, Galanty Y, Lukas C, Taya Y, Schultz DC, Lukas J, Bekker-Jensen S, Bartek J, Shiloh Y. Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nat Cell Biol. 2006;8:870–876. doi: 10.1038/ncb1446. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.