Abstract
Residues Tyr59, Gly78, Ser79, Met103, Gln107, Ile136 and Glu137 in human arsenic (+3 oxidation state) methyltransferase (hAS3MT) were deduced to form a potential hydrogen bond network around S-adenosylmethionine (SAM) from the sequence alignment between Cyanidioschyzon merolae arsenite S-adenosylmethyltransferase (CmArsM) and hAS3MT. Herein, seven mutants Y59A, G78A, S79A, M103A, Q107A, I136A and E137A were obtained. Their catalytic activities and conformations were characterized and models were built. Y59A and G78A were completely inactive. Only 7.0%, 10.6% and 13.8% inorganic arsenic (iAs) was transformed to monomethylated arsenicals (MMA) when M103A, Q107A and I136A were used as the enzyme. The Vmax (the maximal velocity of the reaction) values of M103A, Q107A, I136A and E137A were decreased to 8%, 22%, 15% and 50% of that of WT-hAS3MT, respectively. The KM(SAM) (the Michaelis constant for SAM) values of mutants M103A, I136A and E137A were 15.7, 8.9 and 5.1 fold higher than that of WT-hAS3MT, respectively, indicating that their affinities for SAM were weakened. The altered microenvironment of SAM and the reduced capacity of binding arsenic deduced from KM(As) (the Michaelis constant for iAs) value probably synergetically reduced the catalytic activity of Q107A. The catalytic activity of S79A was higher than that of WT despite of the higher KM(SAM), suggesting that Ser79 did not impact the catalytic activity of hAS3MT. In short, residues Tyr59 and Gly78 significantly influenced the catalytic activity of hAS3MT as well as Met103, Ile136 and Glu137 because they were closely associated with SAM-binding, while residue Gln107 did not affect SAM-binding regardless of affecting the catalytic activity of hAS3MT. Modeling and our experimental results suggest that the adenine ring of SAM is sandwiched between Ile136 and Met103, the amide group of SAM is hydrogen bonded to Gly78 in hAS3MT and SAM is bonded to Tyr59 with van der Waals, cation-π and hydrogen bonding contacts.
Introduction
S-adenosylmethionine (SAM), which is a conjugate of nucleotide adenosine and amino acid methionine, is an essential metabolic intermediate in every organism having several SAM-dependent enzymes [1]-[4]. One of the important biological roles of SAM as a cofactor is the transfer of its methyl group to different substrates, such as lipids, proteins, DNA and other small molecules (inorganic arsenic, chloride, bromide) [5]–[8]. All these methylation processes catalyzed by SAM-dependent methyltransferases significantly affect various essential biological processes including biosynthesis, protein repair, signal transduction, metabolism and detoxification [9], [10]. The methylation of arsenic catalyzed by arsenic (+3 oxidation state) methyltransferase (AS3MT) is considered to be the major metabolism pathway of arsenic [11], [12]. Besides AS3MT, the arsenic methylation process needs tris(2-carboxyethyl)-phosphine hydrochloride (TCEP), glutathione (GSH) or coupled reducing systems as the reductant and SAM as the methyl donor [12]–[16]. Mechanisms of arsenic methylation have been studied for many years [16], [17], but those of the methyl transfer from SAM to As atom have not been studied. Discovery of SAM and As-binding sites is a prerequisite for understanding the methylation of arsenic via hAS3MT.
Active and As-binding sites in AS3MT have been studied in earlier studies. Residues Cys157 and Cys207 in recombinant mouse AS3MT, Cys156 in rat AS3MT, Cys156 and Cys206 in human AS3MT (hAS3MT) and Cys72, Cys174 and Cys224 in Cyanidioschyzon merolae arsenite S-adenosylmethyltransferase (CmArsM) have been verified to be the As-binding sites and in the active sites of mouse, rat, human and cyanidioschyzon merolae AS3MT, respectively [18]–[21]. However, most of the SAM-binding sites in hAS3MT have been deduced from the sequence alignment between diverse species AS3MT [2], [11], [22], [23]. Only the function of the acidic residues of hAS3MT in the predicted SAM-binding motifs has been studied [24].
The residues in CmArsM, which form a potential hydrogen bond network around SAM deduced from the crystal structure of CmArsM-SAM, are Y70, G91, C92, D115, M116, Q120, I151, E152 and C174 [25]. The corresponding residues in hAS3MT are Y59, G78, S79, D102, M103, Q107, I136, E137 and C156. Gly78 and Ser79 belong to the SAM-binding motif I 74-ILDLGSGSG-82, which is inferred from the sequence alignment of various SAM-dependent methyltransferases [11], [23]. The functions of C156 and D102 have been studied [20], [24]. However, the functions of other residues have not been demonstrated. In order to determine whether residues Y59, G78, S79, M103, Q107, I136 and E137 bind to SAM, herein we designed seven mutants Y59A, G78A, S79A, M103A, Q107A, I136A and E137A by site-directed mutagenesis. The catalytic activities of the mutants were determined by high performance liquid chromatography-inductively coupled plasma-mass spectrometry (HPLC-ICP-MS), the mutant conformations were characterized by circular dichroism (CD) and attenuated total reflection Fourier transform infrared spectrometry (ATR-FTIR), and the models of hAS3MT-SAM were built by modeller9v8 utilizing the most updated protein template CmArsM with a sequence most similar to that of hAS3MT. We conclude that Tyr59 and Gly78 markedly influence the SAM-binding and catalytic activity of hAS3MT as well as Met103, Gln107, Ile136 and Glu137. The capacities of binding to SAM for mutants M103A, I136A and E137A are weaker than that of WT. Ser79 exerts no effect on the catalytic activity of hAS3MT. The KM value of SAM (KM(SAM)) for S79A suggests that Ser79 also affects the SAM binding slightly. Modeling of WT-hAS3MT with SAM shows that the adenine ring of SAM is sandwiched between Ile136 on one side and Met103 on the other side, the hydrogen bond is formed between the amide group of SAM and Gly78 rather than Ser79 estimated from the activity of the G78A and S79A. Tyr59 plays important roles in distinguishing and binding SAM, it might bond to the S+-CH3 of SAM with van der Waals contacts, the electron-rich π system of Tyr59 forms cation-π interaction with sulfonium S+-CH3 of SAM as well as the hydrogen bond formed between Tyr59 and the carboxyl group of SAM.
Materials and Methods
Caution: Arsenical compounds are human carcinogens and should be handled accordingly [26].
Materials
Expression host, Escherichia coli BL21 (DE3) pLysS, was bought from Novagen. Restriction enzymes, dNTPs and PrimerSTAR HS DNA polymerase, were obtained from Takara. Wild-type hAS3MT expression plasmid, pET-32a-hAS3MT, was derived from an earlier study [27]. SAM, GSH, isopropyl β-D-thiogalactopyranoside (IPTG) and bovine serum albumin (BSA) were purchased from Sigma. Arsenicals were bought from J&K Chemical Ltd. Phosphate-buffered saline (PBS, pH 7.0) buffer was prepared by mixing appropriate volumes of Na2HPO4 and NaH2PO4 into a 25 mM stock solution.
Stock solutions, which contained 25000 µg/L of the following arsenic species each, were prepared using Milli-Q deionized water with NaAsO2 (As3+), Na2HAsO4.7H2O (As5+), disodium methylarsonate (MMA5+) and dimethylarsonate acid (DMA5+) (J&K Chemical Ltd.). All of the four stock solutions were stored at 4 °C in the dark, by which the working solutions of standards were prepared fresh daily.
Preparation of hAS3MT mutants
Tyr59, Gly78, Ser79, Met103, Gln107, Ile136 and Glu137 were mutated to Ala by site-directed mutagenesis using the wild type pET-32a-hAS3MT plasmid as a template [20], [27]. The primers used for site-directed mutagenesis were summarized in Table 1. The PCR product was transformed into E. coli Top10 (Invitrogen) cells. After sequencing [28], the vectors carrying mutant hAS3MT genes were transformed into E. coli BL21 (DE3) pLysS for expression and a single colony was selected from standard ampicillin-containing agar plate. Protein expression and purification were performed according to the protocols described previously [27]. The method of Bradford based on a BSA standard curve was used to determine protein concentration [29].
Table 1. Primers used for site-directed mutagenesis.
| Primer Sequence | |
| Y59A | + 5'- GTAGCCCTAAGATATGCGGGCTG-3' |
| – 5'- CAGACCACAGCCCGCATATCTTAGG-3' | |
| G78A | + 5’ -TGCTGGATTTTGGATCTGGCGAGTGG-3’ |
| – 5’-TCTACCACTTCCACTCGCCAGATCC-3’ | |
| S79A | + 5'–GATTTTGGATCTGGGTGCGGGAAG -3' |
| – 5'–AATCTCTACCACTTCCCGCACCC -3' | |
| M103A | + 5' –GACACGTGACTGGAATAGACGCGAC-3' |
| – 5'- TGGCCTTTGGTCGCGTCTATTCC-3' | |
| Q107A | + 5'- ATAGACATGACCAAAGGCGCGGTG -3' |
| – 5'- TTTCAGCCACTTCCACCGCGCCTTTG-3' | |
| I136A | + 5'- GACTTTTATTCATGGCTACGCGGAG-3' |
| – 5'- CTCTCCCAACTTCTCCGCGTAG-3' | |
| E137A | + 5'- ATGGCTACATTGCGAAGTTGGGAG -3' |
| – 5'- CTCTCCCAACTTCGCAATGTAGCC -3' | |
| Whole | + 5'-CGGGATATCATGGCTGCACTTCGTGAC-3' |
| – 5'-CGGGTCGACTTAGTGATGGTGATG-3' |
Enzyme activity assays
The steady-state activity of the mutants was determined with an assay system (100 µl) containing 11 µg enzyme, 7 mM GSH, 1 µM iAs3+ and 1 mM SAM in PBS (25 mM, pH 7.0) by HPLC-ICP-MS [20]. To measure the iAs3+ substrate kinetics, 1 mM SAM and 0.5-500 µM iAs3+ were used. In SAM kinetic experiments, 1 µM iAs3+ and 0.05-1 mM SAM were used. The reaction mixtures were incubated at 37 °C for 2 h, and then terminated by adding H2O2 to a final concentration of 3% to release the arsenicals from proteins and to oxidize all arsenic metabolites to pentavalency [16], [30]. 20 µl aliquots of the samples were separated on an anion-exchange column (PRP X-100 250 mm×4.6 mm i.d., 5 µm, Hamilton) by HPLC and analyzed by ICP-MS (Elan 9000, PerkinElmer) with the flow rate of 1.0 ml/min at room temperature [31]-[33]. The arsenical compounds were eluted with the mobile phase of 12 mM (NH4)2HPO4, the pH of which was adjusted to 6.0 with H3PO4. The amounts of arsenic species were calculated with the working curves prepared using 5, 10, 25, 50 and 100 µg/L of standard arsenic species. The methylation rates were calculated as mole equivalents of methyl groups that were transferred from SAM to iAs3+ (i.e., 1 nmol CH3 per 1 nmol MMA or 2 nmol CH3 per 1 nmol DMA) [34]. The methylation rate follows the noncompetitive substrate inhibition equation (1): V = [S]*Vmax/(KM+[S]+[S]2 /KI) and double reciprocal equation (2): 1/V = KM/(Vmax*[S]) + 1/Vmax [20], [35], where V is the initial velocity of the reaction (pmol CH3 transferred/h/mg protein); [S], the substrate (iAs3+) concentration (µM); Vmax, the maximal velocity of the reaction (pmol CH3 transferred/h/mg protein); KM, the Michaelis constant for iAs3+ or SAM (µM); KI, the inhibition constant for iAs3+ (µM) [36].
CD and ATR-FTIR spectra
CD spectra of the WT and hAS3MT mutants were recorded on a JASCO-J810 spectropolarimeter (Jasco Co., Japan) in a 1 mm cell and 10 mm light length. The scanning rate was set at 50 nm/min. Each spectrum represents the average of three accumulations recorded between 190- and 265-nm with very dilute enzyme solution of every mutant (2 µM in 25 mM PBS, pH 7.0) at room temperature. Baseline correction was automatically carried out with the PBS (25 mM, pH 7.0) spectrum throughout the entire collection. The secondary structure parameters of the mutants were calculated using Jwsse32 software with reference CD-Yang. jwr [37]. ATR-FTIR spectra were also utilized to analyze the secondary structure of the mutants. More details about the ATR-FTIR spectra were detailed in previous literature [20], [38]–[39].
Modeling of hAS3MT mutants using modeller9v8
Models of hAS3MT-SAM were built by modeller9v8 utilizing the most updated protein template CmArsM (PDB code 4FR0), because both hAS3MT and CmArsM were arsenic methyltransferases in different species with similar sequences [25]. The models quality of hAS3MT mutants was estimated via QMEAN Server (http://swissmodel.expasy.org/qmean/cgi/index.cgi) [40]. Pymol was used to analyze the models of hAS3MT [41].
Results and Discussion
Expression and purification of the hAS3MT mutants
The hAS3MT mutants were expressed and purified according to the protocol described in previous studies [20], [27]. All the mutant proteins were expressed successfully. The purity of each mutant protein was confirmed to be over 90% by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) (Figure 1).
Figure 1. Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE).
Coomassie blue stained 12% SDS-PAGE gel of the purified protein of hAS3MT mutants.
Catalytic activities of the mutants
The primary methylation index (PMI) calculated as MMA/iAs and the secondary methylation index (SMI) calculated as DMA/MMA were previously developed to characterize iAs methylation ability [42], [43]. The total arsenic (TAs) concentration was defined by summing up the concentrations of iAs, MMA and DMA because no trimethylarsine oxide (TMAO) was detected in our reaction system. According to the pathway of iAs methylation, secondary methylation can only proceed based on first methylation and parts of the first methylation products are further methylated. Therefore, to assess the first methylation ability, not only the primary but also the secondary methylation products were considered. The first methylation ratio (FMR) was defined as (MMA + DMA)/TAs and the secondary methylation ratio (SMR) was defined as DMA/(MMA + DMA) [44]. Using the FMR and SMR to evaluate the arsenic methylation capacity of the mutants was more logical than using the PMI and SMI [45]. Proportions of iAs, MMA and DMA (iAs%, MMA% and DMA%) were defined as iAs/TAs × 100%, MMA/TAs × 100% and DMA/TAs × 100%, respectively [44]. Proportions of arsenic species and the two methylation indices, FMR and SMR were calculated to evaluate the arsenic methylation capacity of the mutants Y59A, G78A, S79A, M103A, Q107A, I136A and E137A (Figure 2a and Figure 2b). Mutants Y59A and G78A were completely inactive owing to the absence of methylated arsenic. The proportion of iAs of S79A was lower than that of WT while the FMR and SMR of S79A were higher than that of WT, which indicated that the methylation capacity of S79A was stronger than that of WT. Only 7.0%, 10.6%, 13.8% iAs was transformed to MMA when M103A, Q107A and I136A were used as the enzyme, which suggested that the catalytic activities of mutants M103A, Q107A, I136A and E137A were all lower than that of WT. Compared with WT, the catalytic capacity of E137A also decreased as concluded from the proportion of iAs, FMR and SMR. Thus, residues Tyr59 and Gly78 affected the activity of hAS3MT significantly whereas Met103, Gln107, Ile136 and Glu137 influenced the catalytic activity to a lesser extent, and Ser79 had no effect.
Figure 2. Catalytic capacity of the hAS3MT mutants.
Reaction mixtures (100 µl) containing 11 µg enzymes, 1 µM iAs3+, 1 mM SAM and 7 mM GSH in PBS (25 mM, pH 7.0) were incubated at 37°C for 2 h with H2O2 treatment before analyzed by HPLC-ICP-MS. The percents of arsenic species (iAs/TAs, MMA/TAs and DMA/TAs) and the two indices (FMR and SMR) of mutants Y58A, G78A, S79A, M103A, Q107A, I136A and E137A are shown in Figure 2a and 2b. Values are the averages ±S.D. of three independent experiments performed by three independently purified proteins.
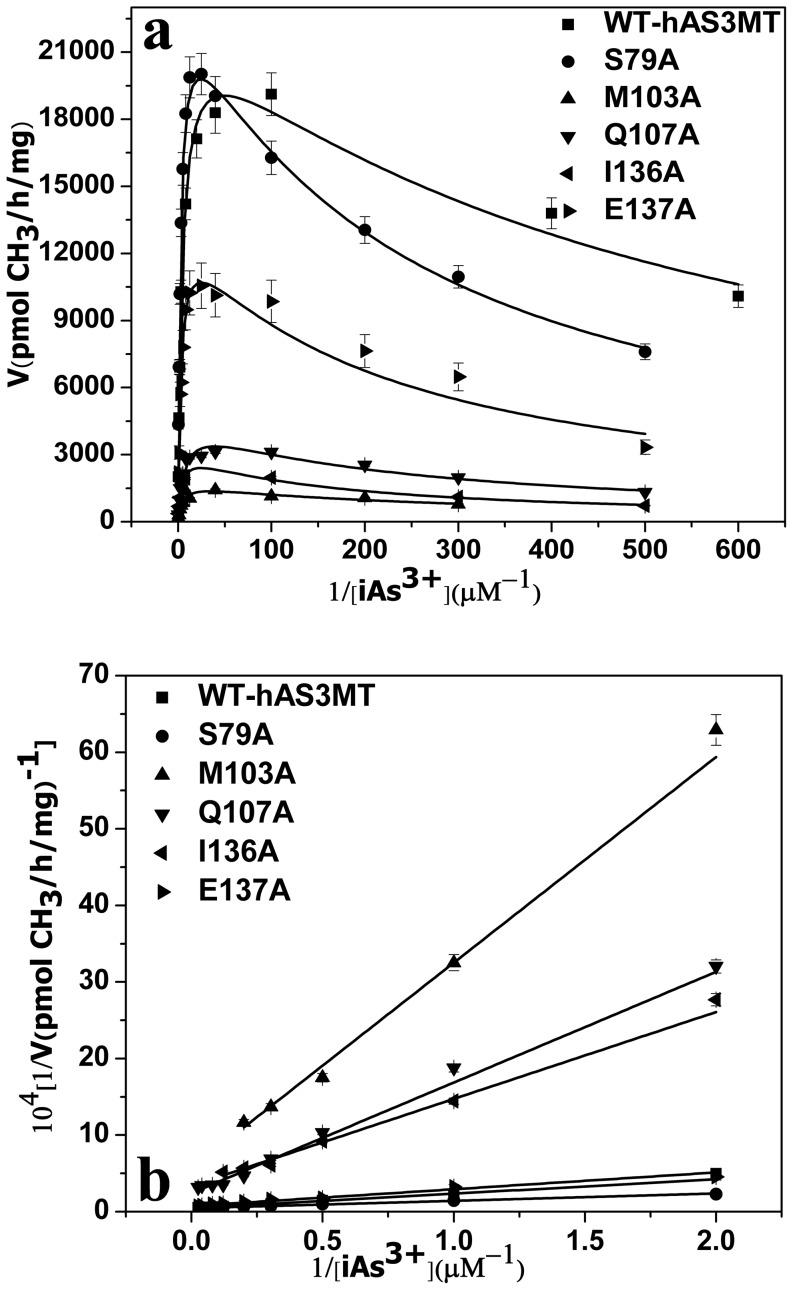
The catalytic activities and kinetic properties of the mutants were investigated comprehensively. The iAs3+-substrate-inhibition phenomena were observed for all the active mutants in a wide iAs3+ concentration range (0.5–500 µM) (Figure 3a), namely the rate of arsenic methylation increased with increasing arsenic concentration (0.5–40 µM) and then decreased at higher arsenic concentration (100–500 µM). The kinetic parameters of the active mutants in Table 2 were estimated by fitting the experimental data to Eq. (1) and calculated by double reciprocal plots from Eq. (2) (Figure 3b). The data obtained from the two methods were consistent. The KI values for the iAs3+ of mutants S79A, M103A, Q107A, I136A and E137A were all lower than those of WT-hAS3MT (KM, 3.2 µM, KI, 0.7 mM, Vmax, 19836 pmol/h/mg [20]). The Vmax values of M103A, Q107A, I136A and E137A were decreased to 8%, 22%, 15% and 50% of that of WT-hAs3MT respectively, while the Vmax value of S79A was higher than that of WT. The KM(As) values of M103A and Q107A were higher than that of WT. For S79A, I136A and E137A, the KM(As) values did not obviously differ from that of WT-hAS3MT. The results indicated that the affinity of mutants M103A and Q107A to iAs decreased compared with that of WT, while the abilities in binding the substrate iAs3+ of mutants S79A, I136A and E137A were similar to the native form.
Figure 3. A: Substrate concentration dependence of rate.
The lines show the least squares fit of Eq. (1) to the data. B: Double reciprocal plots of the arsenic methylation rate against the concentration of iAs3+. Reaction mixtures (100 µl) containing 11 µg enzymes, 1 mM SAM and 7 mM GSH in PBS (25 mM, pH 7.0) were incubated with different concentrations of iAs3+ at 37°C for 2 h with H2O2 treatment before analysis. Values are the averages ± S.D. of three independent experiments performed by three independently purified proteins.
Table 2. Kinetic parameters of arsenic methylation for the five mutants S79A, M103A, Q107A, I136A and E137A.
| aVmax(pmol CH3 /mg/h)×103 | aKM(µM) | KI(mM) | bVmax(pmol CH3 /mg/h) ×103 | BKM (µM) | CKM (µM) | Relative cKM | |
| S79A | 23.7±0.5 | 2.4±0.2 | 0.24±0.01 | 22.3±0.5 | 2.1±0.1 | 93.1±2.4 | 1.9 |
| M103A | 1.7±0.2 | 4.6±0.8 | 0.27±0.08 | 1.8±0.2 | 4.8±0.1 | 750.4±25.6 | 15.7 |
| Q107A | 4.6±0.3 | 7.0±0.8 | 0.22±0.03 | 4.1±0.4 | 6.0±0.2 | 50.2±2.0 | 1.0 |
| I136A | 3.1±0.3 | 3.7±0.5 | 0.16±0.02 | 3.0±0.3 | 3.4±0.5 | 426.7±17.9 | 8.9 |
| E137A | 13.2±1.0 | 3.0±0.5 | 0.21±0.04 | 13.5±0.8 | 3.0±0.2 | 241.9±23.5 | 5.1 |
| WT | 21.2±1.1 | 3.2±0.3 | 0.76±0.09 | 19.8±1.0 | 3.19±0.7 | 47.8 | 1.0 |
Values represent the average ± S.D. of three independent experiments performed by three independently purified proteins.
Represents the kinetic parameters of iAs3+ estimated from the data in Fig. 2a by Eq. (1) using origin 8.0.
Represents the kinetic parameters of iAs3+ calculated from the data in Fig. 2b.
Represents the KM for SAM.
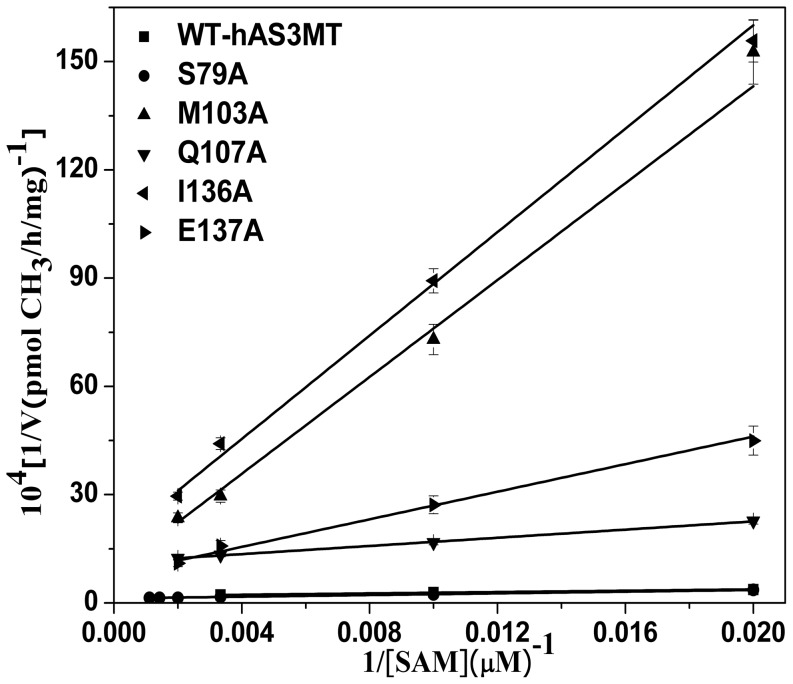
The arsenic methylation rate increased with rising SAM concentration (Figure 4). The KM (SAM) values of the mutants, which reflect the ability of SAM to interact with hAS3MT, were calculated from double reciprocal plots (Figure 4) and summarized in Table 2. For the mutants M103A, I136A and E137A, the KM (SAM) values increased to 750.4, 426.7 and 241.9 µM, which were 15.7, 8.9 and 5.1 fold higher than the WT value (WT: 47.84 µM [20]), respectively. The data revealed that residues Met103, Ile136 and Glu137 were involved in the cofactor SAM binding, which is reflect by the reduced catalytic activity of M103A, I136A and E137A. The KM (SAM) of S79A was about 2 fold higher than that of WT, while the KM (SAM) of Q107A almost equaled that of WT though the catalytic activity of Q107A was evidently lower.
Figure 4. Double reciprocal plots of the arsenic methylation rate versus the concentration of SAM.
Reaction mixtures (100 µl) containing 11 µg enzymes, 1 µM iAs3+, 7 mM GSH in PBS (25 mM, pH 7.0) were incubated with different concentrations of SAM for 2 h with H2O2 treatment before analysis. Values are the averages ± S.D. of three independent experiments performed by three independently purified proteins.
Conformation of Y59A, G78A, S79A, M103A, Q107A, I136A and E137A
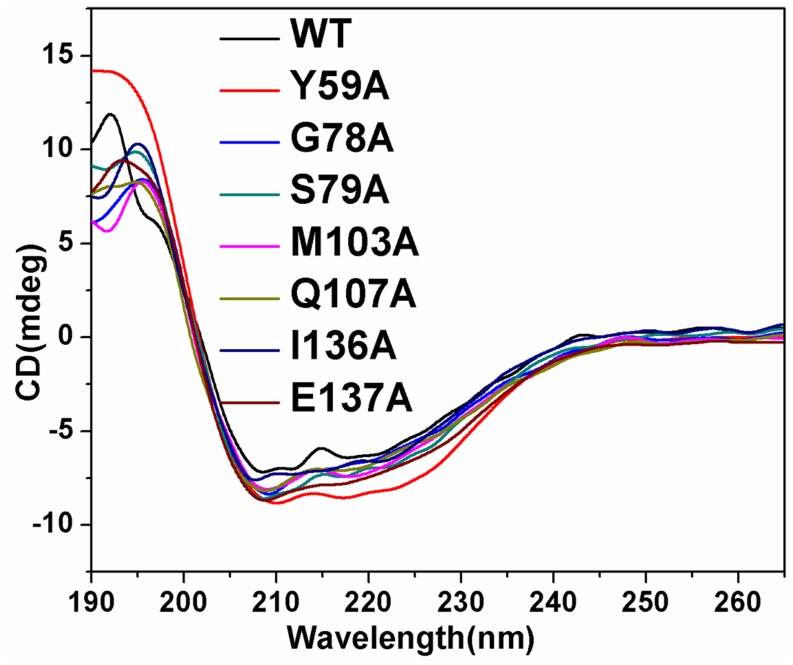
Circular dichroism (CD) spectroscopy, which is a sensitive method to determine the protein secondary structure [46], [47], is used to analyze the conformational change of the seven mutants (Figure 5). CD spectra showed that the intensities of the peaks (208 and 220 nm) of mutants Y59A, G78A, S79A, M103A, Q107A, I136A and E137A were more intense than those of the wild type enzyme suggesting that the conformations of mutants Y59A, G78A, S79A, M103A, Q107A, I136A and E137A were different from that of WT. The content of secondary structure of the mutants was computed by Jwsse32 software with reference CD-Yang. jwr (Table 3). Compared with WT, the contents of β-pleated sheet for all mutants except Y59A increased with those of β-turn and random coil decreasing, while the content of α-helix of Y59A increased with those of β-pleated sheet, β-turn and random coil decreasing. The data indicate that the contents of secondary structure of the seven mutants differ from those of WT, especially for Y59A and I136A.
Figure 5. CD spectra of hAS3MT and the mutants.
Spectra were taken at the protein concentration of 2 µM at room temperature. Plot is the representative of three independent measurements performed by three independently purified proteins.
Table 3. Secondary structures of WT-hAS3MT and the mutants estimated from CD spectra.
| α-helix% | β-pleated% | β-turn% | Random% | |
| Y59A | 41.3±1.9 | 17.6±0.5 | 13.4±0.5 | 27.7±1.6 |
| G78A | 27.0±1.1 | 32.3±1.1 | 15.1±0.9 | 25.5±0.2 |
| S79A | 29.7±1.6 | 30.7±1.3 | 13.3±0.8 | 26.3±0.5 |
| M103A | 26.6±0.3 | 30.3±2.0 | 16.2±0.9 | 26.9±0.8 |
| Q107A | 27.2±1.2 | 31.2±0.8 | 13.1±1.0 | 28.4±0.3 |
| I136A | 25.0±1.0 | 44.0±2.9 | 7.8±1.2 | 23.2±0.7 |
| E137A | 30.0±1.2 | 25.6±1.1 | 16.3±0.7 | 28.1±1.2 |
| WT | 29.0±2.2 | 23.9±1.9 | 17.9±1.7 | 29.2±1.4 |
Values represent the average ± S.D. of three independent experiments carried out by three independently purified proteins. The parameters were analyzed with the Jasco secondary structure manager with the reference CD data-Yang. jwr in PBS (25 mM, pH 7.0) at room temperature.
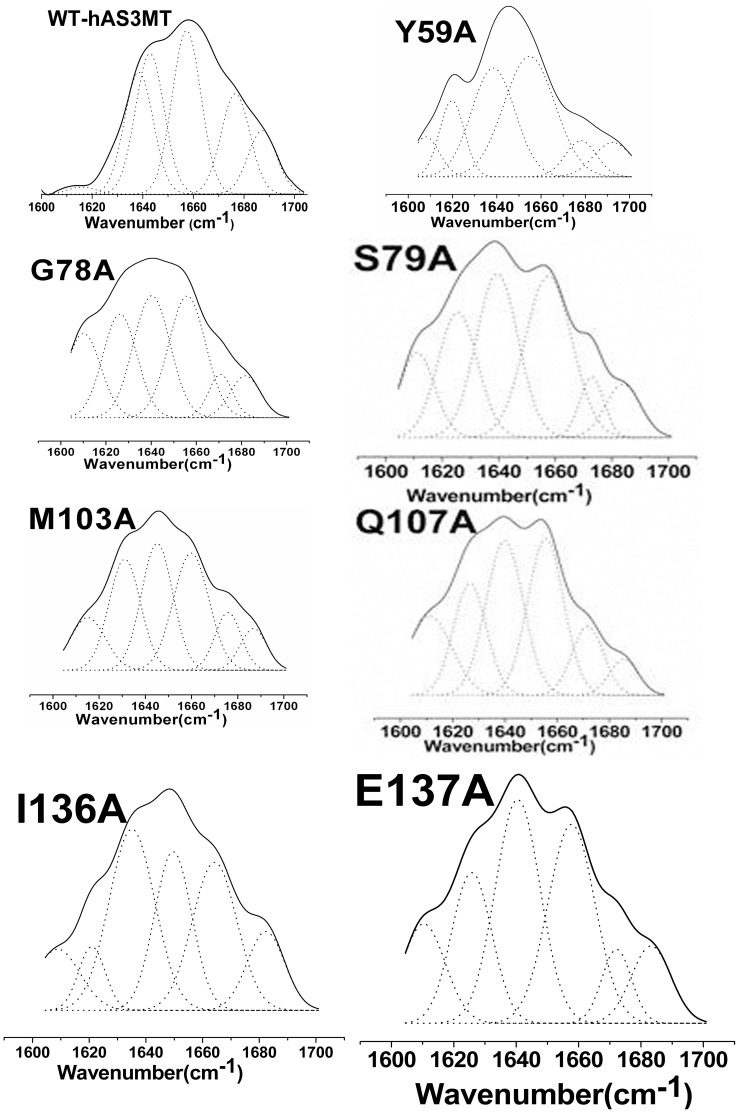
FTIR spectroscopy has also been shown to be a powerful technique to evaluate the protein structure. To further confirm the secondary structure of the mutants, we carried out ATR-FTIR assays and analyzed their amide I band spectra according to well-established assignment criteria [38], [48] (1610–1640 cm−1: β-pleated sheet, 1640–1650 cm−1: random coil, 1650–1658 cm−1: α-helix, and 1660–1700 cm−1: β-turn). The original and curve-fitting FTIR spectra of mutants Y59A, G78A, S79A, M103A, Q107A, I136A and E137A were shown in Figure 6. There were six component bands in the amide I bands of the mutants. The contents of each secondary structure were calculated from the integrated areas of the component bands (Table 4). The secondary structure derived from ATR-FTIR is in good agreement with those from CD spectra.
Figure 6. Curve-fitted amide I region of the mutants.
The component peaks are the result of curve-fitting using a Gaussian shape. The solid lines represent the experimental FTIR spectra after Savitzky-Golay smoothing, and the dashed lines represent the fitted components. Plot is the representative of three independent measurements carried out by three independently purified proteins.
Table 4. Secondary structures of WT-hAS3MT and the mutants estimated from ATR-FTIR spectroscopy.
| α-helix% | β-pleated% | β-turn% | Random% | |
| Y59A | 38.9±3.1 | 17.0±0.8 | 14.2±0.8 | 29.9±2.0 |
| G78A | 26.4±0.5 | 33.8±0.3 | 13.4±0.3 | 26.3±0.8 |
| S79A | 29.9±1.0 | 29.9±0.4 | 13.1±0.3 | 27.1±0.3 |
| M103A | 26.0±1.7 | 32.7±2.5 | 16.3±1.7 | 24.9±1.1 |
| Q107A | 26.7±0.8 | 30.6±1.3 | 15.3±0.3 | 27.4±0.5 |
| I136A | 24.1±1.8 | 43.6±1.7 | 10.8±0.9 | 21.5±0.6 |
| E137A | 27.3±2.3 | 27.8±2.1 | 15.5±1.1 | 29.4±1.6 |
| WT | 26.6±3.6 | 20.7±4.6 | 24.2±3.2 | 28.5±4.9 |
SAM-binding sites of hAS3MT mutants Y59A, G78A, S79A, M103A, Q107A, I136A and E137A
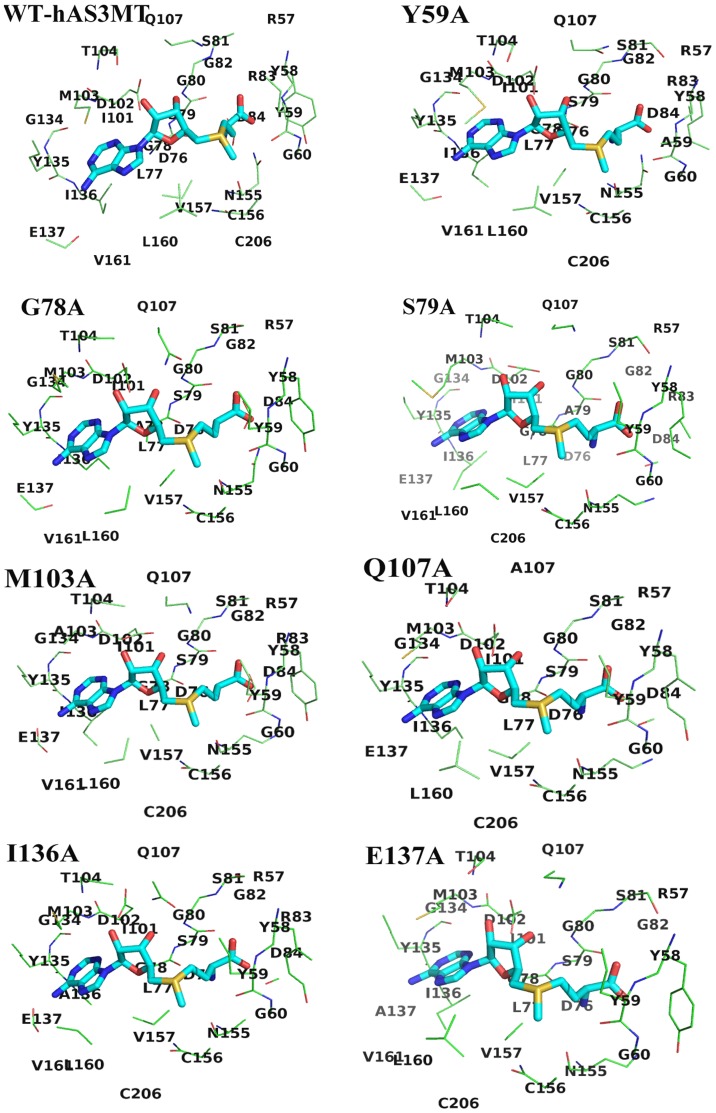
The models of hAS3MT mutants were established using modeller9v8. The crystal structure of CmArsM with cofactor SAM (PDB code 4FR0) was used as the template. The model quality was estimated on the basis of a QMEAN scoring function acceptably ranging between 0.60 and 0.65 [40]. The secondary structure arrangement of the hAS3MT model was almost identical to that of CmArsM. The sites in the SAM-binding pocket (5.0 Å around SAM) of WT-hAS3MT and mutants (Y59A, G78A, S79A, M103A, Q107A, I136A and E137A) were displayed in Figure 7 and Table 5. The SAM model-derived WT-hAS3MT showed that As-binding sites Cys156 and Cys206 were located in the SAM-binding pocket [24]. However, Cys206 was not in the SAM-binding pocket of mutant G78A, which induced complete catalytic activity loss by isolating As from S+-CH3, thus further demonstrating that G78A was completely inactive in the methylation of iAs. Another residue Arg83 was not located in the 5 Å range around SAM for G78A. The structure of Gly was simpler than that of Ala, so the microenvironment around SAM and the interaction between SAM and residue 78 changed when the Gly78 was replaced by Ala. Residues Arg83 and Val161 were not located in the 5 Å range around SAM for Q107A. In mutant E137A, the residues 5 Å around SAM were disconnected from R83 and D84. Gln and Glu are hydrophilic and their side chains are amide and carboxyl group, while Ala is hydrophobic and its side chain is smaller than that of Gln and Glu. The microenvironment around SAM changed and the catalytic activity of Q107A and E137A decreased when the residues 107 and 137 were changed to Ala. Residues around SAM for mutants Y59A, S79A, M103A and I136A were the same as that for WT.
Figure 7. Interaction modes between SAM, WT-hAS3MT and mutants (Y59A, G78A, S79A, M103A, Q107A, I136A and E137A).
Only the residues 5.0 Å around SAM are displayed.
Table 5. Residues 5.0 Å around SAM based on the models of WT and mutants Y59A, G78A, S79A, M103A, Q107A, I136A and E137A.
| Residues around 5.0 Å of SAM | |
| WT | 57-RYYG-60,76-DLGSGSGRD-84,101-IDMT-104,Q107,134-GYIE-137,155-NCV-157,160-LV-161,C206 |
| Y59A | 57-RYAG-60,76-DLGSGSGRD-84,101-IDMT-104,Q107,134-GYIE-137,155-NCV-157,160-LV-161,C206 |
| G78A | 57-RYYG-60,76-DLASGSG-82,D84,101-IDMT-104,Q107,134-GYIE-137,155-NCV-157,160-LV-161 |
| S79A | 57-RYYG-60,76-DLGAGSGRD-84,101-IDMT-104,Q107,134-GYIE-137,155-NCV-157,160-LV-161,C206 |
| M103A | 57-RYYG-60,76-DLGSGSGRD-84,101-IDAT-104,Q107,134-GYIE-137,155-NCV-157,160-LV-161,C206 |
| Q107A | 57-RYYG-60,76-DLGSGSG-82,D84,101-IDMT-104,A107,134-GYIE-137,155-NCV-157,L160,C206 |
| I136A | 57-RYYG-60,76-DLGSGSGRD-84,101-IDMT-104,Q107,134-GYAE-137,155-NCV-157,160-LV-161,C206 |
| E137A | 57-RYYG-60,76-DLGSGSG-82,101-IDMT-104,Q107,134-GYIA-137,155-NCV-157,160-LV-161,C206 |
One of the crucial interactions between the SAM having cation sulfonium R3S+ and SAM-dependent methyltransferases is the cation-π force formed between aromatic residues (e.g. Tyr, Trp and Phe) with their electron-rich π system and R3S+ of SAM [49]–[52]. The cation-π interaction formed between Tyr59 and S+-CH3 of SAM assists to bind, recognize SAM and distinguish cationic SAM from its analogous neutral molecule S-adenosylhomocysteine (SAH). The environment around the SAM-binding sites facilitates transfer of a methyl group from SAM to As atom. These interactions between SAM and residue 59 were destroyed when Tyr59 was replaced by Ala without electron-rich aromatic side chain, and therefore resulted in Y59A being an inactive enzyme.
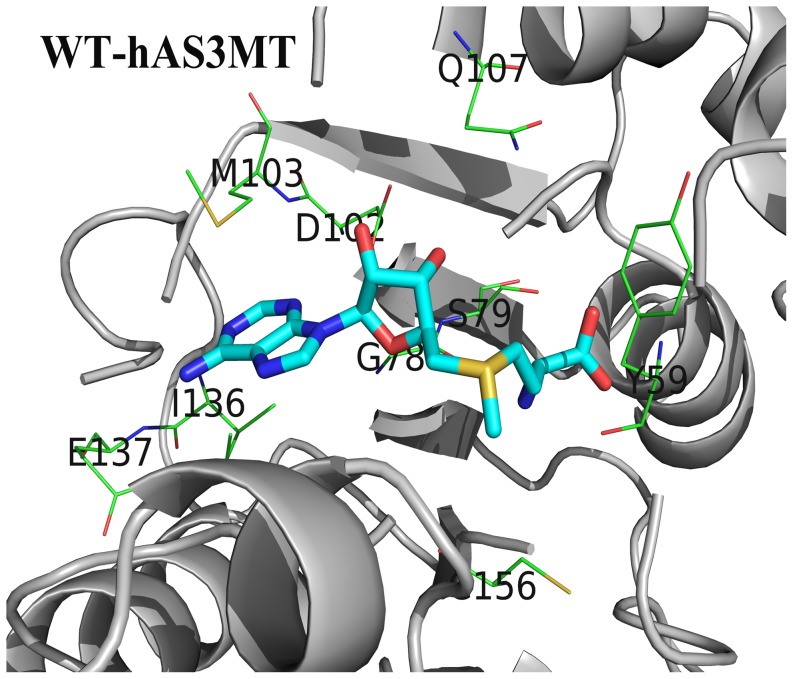
SAM is bonded to CmArsM via forming hydrogen bonds and hydrophobic interactions with CmArsM residues [25]. The crystal structure of CmArsM-SAM showed that the adenine ring of SAM is sandwiched by Ile151 on one side and Met116 on the other side and forms hydrogen bonds with Ile151 and Glu152, the other hydrogen bonds also form between hydroxyl groups of ribose O2* and O3* and the side chains of Asp115 and Gln120, the carboxyl group of SAM and OCys92 and OTyr70, and the amide group of SAM and OGly91 in CmArsM, the S+-CH3 group of SAM is bonded to main chain atoms OTyr70 and OCys174 in CmArsM with van der Waals contacts [25]. As detailed above, the residues Tyr70, Gly91, Cys92, Asp115, Met116, Gln120, Ile151, Glu152 and Cys174 in CmArsM correspond to the residues Tyr59, Gly78, Ser79, Asp102, Met103, Gln107, Ile136, Glu137 and Cys156 in hAS3MT. The results in this study and others show that the mutants Y59A, G78A, D102P, D102N and C156S are all completely inactive [20], [24], S79A is a little more active than WT while the catalytic activity of M103A, Q107A, I136A and E137A are decreased compared with that of WT. In our models, the adenine ring is indeed sandwiched between Ile136 on one side and Met103 on the other side because their aliphatic side chains are large enough to enwrap the adenine ring of SAM (Figure 8). The side chain of Ala was much smaller than that of Met103 and Ile136, the residue 103 and 136 would not enwrap the adenine ring of SAM when they were replaced by Ala. Asp102 forms two hydrogen bonds with the ribose hydroxyl groups of SAM [24]. The Vmax and KM(SAM) value of Q107A shows that Gln107 also significantly affects the catalytic activity of hAS3MT but it does not relate with the SAM-binding. Cys156 with functional group –SH, which is one of the As-binding sites in hAS3MT, also interacts with the S+-CH3 of SAM and helps to orient the methyl group of SAM during its approach to the arsenic lone pair. Both Gly78 and Ser79 belong to the SAM-binding motif I 74-ILDLGSGSG-82 and are close to the SAM in the models (Figure 8). The amide group of SAM is hydrogen bonded to Gly78 in hAS3MT rather than Ser79 as estimated from the activity of the G78A and S79A. According to the function of Tyr70 in CmArsM and the electron-rich characteristic of residue Tyr, we conclude that the OTyr59 in hAS3MT might bond to the S+-CH3 of SAM with van der Waals contacts, the electron-rich π system of Tyr59 forms a cation-π interaction with sulfonium S+-CH3 of SAM as well as hydrogen bonding with the carboxyl group of SAM. Therefore, the Tyr59 plays an important role in binding, recognizing, distinguishing cationic SAM from its analogous neutral molecule SAH and orienting the methyl group of SAM during its approach to the arsenic electron lone pair.
Figure 8. The model of WT-hAS3MT with SAM.
Only residues in hAS3MT forming potential hydrogen bond network around SAM are presented.
Conclusion
To study the functions of residues Tyr59, Gly78, Ser79, Met103, Glu107, Ile136 and Glu137 in hAS3MT, seven mutants, Y59A, G78A, S79A, M103A, Q107A, I136A and E137A were designed. Their conformation characterized by CD and ATR-FTIR spectroscopy was different from that of WT-hAS3MT. The KM(SAM) of mutants M103A and I136A indicated that they bound to SAM less tightly than WT-hAS3MT did, leading to remarkably lowered catalytic activity. The mutant Y59A was completely inactive because the cation-π interacton and hydrogen bond formed between SAM and residue 59 disappeared when Tyr59 was changed to Ala. The modeling results showed that As-binding sites Cys156 and Cys206 in WT-hAS3MT were located in the vicinity of SAM (5 Å around), whereas As-binding site Cys206 in mutant G78A was not, which resulted in the mutant G78A being inactive. The microenvironment surrounding SAM in Q107A was also altered, and its capacity of binding to iAs was weaker, which caused a dramatic decrease in the catalytic activity of the Q107A mutant. The catalytic activity of S79A surpassed that of WT although its KM(SAM) value was higher. Met103 and Ile136 form a sandwiched structure with the adenine ring of SAM in the center. The amide group of SAM is hydrogen bonded to OGly78 in hAS3MT, while the hydrogen bond might not be formed between Ser79 and SAM as estimated from the activity of the G78A and S79A. OTyr59 might bond to the S+-CH3 of SAM through van der Waals contacts, the electron-rich π system of Tyr59 could interact with sulfonium S+-CH3 of SAM, and the hydroxyl group might hydrogen bond with the carboxyl group of SAM. These interactions would help to bind, recognize, distinguish cationic SAM from its analogous neutral molecule SAH and orient the methyl group of SAM during its approach to the arsenic electron lone pair.
Funding Statement
This study is supported by the National Basic Research Program of China (2013CB922102) and the National Natural Science Foundation of China (21075064, 21027013, 21021062, 21275072 and 21201101). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Waddell TG, Eilders LL, Patel BP, Sims M (2000) Prebiotic methylation and the evolution of methyl transfer reactions in living cells. Orig Life Evol Biosph 30: 539–548. [DOI] [PubMed] [Google Scholar]
- 2. Kozbial PZ, Mushegian AR (2005) Natural history of S-adenosylmethionine -binding proteins. BMC Struct Biol 5: 1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fontecave M, Atta M, Mulliez E (2004) S-adenosylmethionine: nothing goes to waste. Trends Biochem Sci 29: 243–249. [DOI] [PubMed] [Google Scholar]
- 4. Roje S (2006) S-Adenosyl-L-methionine: Beyond the universal methyl group donor. Phytochemistry 67: 1686–1698. [DOI] [PubMed] [Google Scholar]
- 5. Wuosmaa AM, Hager LP (1990) Methyl chloride transferase: a carbocation route for biosynthesis of halometabolites. Science 249: 160–162. [DOI] [PubMed] [Google Scholar]
- 6. Saxena D, Aouad S, Attieh J, Saini HS (1998) Biochemical characterization of chloromethane emission from the wood-rotting fungus Phellinus pomaceus. Appl Environ Microbiol 64: 2831–2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Anantharaman V, Koonin EV, Aravind L (2002) Comparative genomics and evolution of proteins involved in RNA metabolism. Nucleic Acids Res 30: 1427–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Thomas DJ, Waters SB, Styblo M (2004) Elucidating the pathway for arsenic methylation. Toxicol Appl Pharm 198: 319–326. [DOI] [PubMed] [Google Scholar]
- 9. Hopper AK, Phizicky EM (2003) tRNA transfers to the limelight. Genes Dev 17: 162–180. [DOI] [PubMed] [Google Scholar]
- 10. Kouzarides T (2002) Histone methylation in transcriptional control. Curr Opin Genet Dev 12: 198–209. [DOI] [PubMed] [Google Scholar]
- 11. Lin S, Shi Q, Nix FB, Styblo M, Beck MA, et al. (2002) A Novel S-Adenosyl-L-methionine: Arsenic (III) Methyltransferase from Rat Liver Cytosol. J Biol Chem 277: 10795–10803. [DOI] [PubMed] [Google Scholar]
- 12. Wang SP, Li XL, Song XL, Geng ZR, Hu X, et al. (2012) Rapid-equilibrium kinetic analysis of arsenite methylation catalyzed by recombinant human arsenic (+3 oxidation state) methyltransferase (hAS3MT). J Biol Chem 287: 38790–38799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Challenger F (1945) Biological Methylation. Chem Rev 36: 315–361. [Google Scholar]
- 14. Waters SB, Devesa-Perez V, Del Razo LM, Styblo M, Thomas DJ (2004) Endogenous reductants support catalytic function of recombinant rat cyt19, an arsenic methyltransferase. Chem Res Toxicol 17: 404–409. [DOI] [PubMed] [Google Scholar]
- 15. Ding L, Saunders RJ, Drobna Z, Walton F, Xun P, et al. (2012) Methylation of arsenic by recombinant human wild-type arsenic (+3 oxidation state) methyltransferase and its methionine 287 threonine (M287T) polymorph: Role of glutathione. Toxicol Appl Pharm 264: 121–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hayakawa T, Kobayashi Y, Cui X, Hirano S (2005) A new metabolic pathway of arsenite: arsenic–glutathione complexes are substrates for human arsenic methyltransferase Cyt19. Arch Toxicol 79: 183–191. [DOI] [PubMed] [Google Scholar]
- 17. Aposhian HV (1997) Enzymatic methylation of arsenic species and other new approaches to arsenic toxicity. Annu Rev Pharmacol Toxicol 37: 397–419. [DOI] [PubMed] [Google Scholar]
- 18. Fomenko DE, Xing WB, Adair BM, Thomas DJ, Gladyshev VN (2007) High-Throughput Identification of Catalytic Redox-Active Cysteine Residues. Science 315: 387–389. [DOI] [PubMed] [Google Scholar]
- 19. Li JX, Waters SB, Drobna Z, Devesa V, Styblo M, et al. (2005) Arsenic (+3 oxidation state) methyltransferase and the inorganic arsenic methylation phenotype. Toxicol Appl Pharm 204: 164–169. [DOI] [PubMed] [Google Scholar]
- 20. Song XL, Geng ZR, Zhu JS, Li CY, Hu X, et al. (2009) Structure–function roles of four cysteine residues in the human arsenic (+3 oxidation state) methyltransferase (hAS3MT) by site-directed mutagenesis. Chem-Biol Interact 179: 321–328. [DOI] [PubMed] [Google Scholar]
- 21. Marapakala K, Qin J, Rosen BP (2012) Identification of Catalytic Residues in the As(III) S-Adenosylmethionine Methyltransferase. Biochemistry 51: 944–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kagan RM, Clarke S (1994) Widespread occurrence of three sequence motifs in diverse S-adenosylmethionine-dependent methyltransferases suggests a common structure for these enzymes. Arch Biochem Biophys 310: 417–427. [DOI] [PubMed] [Google Scholar]
- 23. Thomas DJ, Li JX, Waters SB, Xing WB, Adair BM, et al. (2007) Arsenic (+3 Oxidation State) Methyltransferase and the Methylation of Arsenicals. Exp Biol Med 232: 3–13. [PMC free article] [PubMed] [Google Scholar]
- 24. Li XL, Geng ZR, Wang SP, Song XL, Hu X, et al. (2013) Functional evaluation of Asp76, 84, 102 and 150 in human arsenic (III) methyltransferase (hAS3MT) interacting with S-adenosylmethionine. FEBS Lett 587: 2232–2240. [DOI] [PubMed] [Google Scholar]
- 25. Ajees AA, Marapakala K, Packianathan C, Sankaran B, Rosen BP (2012) Structure of an As (III) S-Adenosylmethionine Methyltransferase: Insights into the Mechanism of Arsenic Biotransformation. Biochemistry 51: 5476–5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kuroki T, Matsushima T (1987) Performance of short-term tests for detection of human carcinogens. Mutagenesis 2: 33–37. [DOI] [PubMed] [Google Scholar]
- 27. Geng ZR, Song XL, Xing Z, Geng JL, Zhang SC, et al. (2009) Effects of selenium on the structure and function of recombinant human S-adenosyl-L-methionine dependent arsenic (+3 oxidation state) methyltransferase in E. coli. J Biol Inorg Chem 14: 485–496. [DOI] [PubMed] [Google Scholar]
- 28. Sanger F, Nicklen S, Coulson AR (1977) DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA 7: 5463–5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254. [DOI] [PubMed] [Google Scholar]
- 30. Song XL, Geng ZR, Li XL, Hu X, Bian NS, et al. (2010) New insights into the mechanism of arsenite methylation with the recombinant human arsenic (+3) methyltransferase (hAS3MT). Biochimie 92: 1397–1406. [DOI] [PubMed] [Google Scholar]
- 31. Gailer J, Madden S, Cullen WR, Denton MB (1999) The separation of dimethylarsinic acid, methylarsonous acid, methylarsonic acid, arsenate and dimethylarsinous acid on the Hamilton PRP-X100 anion-exchange column. Appl Organometal Chem 13: 837–843. [Google Scholar]
- 32. Francesconi KA, Kuehnelt D (2004) Determination of arsenic species: A critical review of methods and applications, 2000–2003. Analyst 129: 373–395. [DOI] [PubMed] [Google Scholar]
- 33. Lu XF, Arnold LL, Cohen SM, Cullen WR, Le XC (2003) Speciation of Dimethylarsinous Acid and Trimethylarsine Oxide in Urine from Rats Fed with Dimethylarsinic Acid and Dimercaptopropane Sulfonate. Anal Chem 75: 6463–6468. [DOI] [PubMed] [Google Scholar]
- 34. Walton FS, Waters SB, Jolley SL, LeCluyse EL, Thomas DJ, et al. (2003) Selenium compounds modulate the activity of recombinant rat AsIII methyltransferase and the methylation of arsenite by rat and human hepatocytes. Chem Res Toxicol 16: 261–265. [DOI] [PubMed] [Google Scholar]
- 35. Kedderis GL, Elmore AR, Crecelius EA, Yager JW, Goldsworthy TL (2006) Kinetics of arsenic methylation by freshly isolated B6C3F1 mouse hepatocytes. Chem-Biol Interact 161: 139–145. [DOI] [PubMed] [Google Scholar]
- 36.Cleland WW (1970) Steady state kinetics. In: Boyer PD (Ed.). The Enzymes. New York: Academic Press. pp: 1–65.
- 37. Yang JT, Wu CSC, Martinez HM (1986) Calculation of Protein Conformation from Circular Dichroism. Meth Enzymol 130: 208–257. [DOI] [PubMed] [Google Scholar]
- 38. Surewicz WK, Mantsch HH (1988) New insight into protein secondary structure from resolution-enhanced infrared-spectra. Biochim Biophys Acta 952: 115–130. [DOI] [PubMed] [Google Scholar]
- 39. Song XL, Geng ZR, Li XL, Zhao Q, Hu X, et al. (2011) Functional and structural evaluation of cysteine residues in the human arsenic (+3 oxidation state) methyltransferase (hAS3MT). Biochimie 93: 369–375. [DOI] [PubMed] [Google Scholar]
- 40. Benkert P, Tosatto SCE, Schomburg D (2008) QMEAN: A comprehensive scoring function for model quality assessment. Proteins 71: 261–277. [DOI] [PubMed] [Google Scholar]
- 41.DeLano WL (2004) The PyMOL user’s Guide, DeLano Scientific LLC, San Carlos, California, USA.
- 42. Chen YC, Guo YL, Su HJ, Hsueh YM, Smith TJ, et al. (2003) Arsenic methylation and skin cancer risk in southwestern Taiwan. J Occup Environ Med 45: 241–248. [DOI] [PubMed] [Google Scholar]
- 43. Tseng CH, Huang YK, Huang YL, Chung CJ, Yang MH, et al. (2005) Arsenic exposure, urinary arsenic speciation, and peripheral vascular disease in blackfoot disease-hyperendemic villages in Taiwan. Toxicol Appl Pharm 206: 299–308. [DOI] [PubMed] [Google Scholar]
- 44. Xu Y, Wang Y, Zheng Q, Li X, Li B, et al. (2008) Association of oxidative stress with arsenic methylation in chronic arsenic-exposed children and adults. Toxicol Appl Pharm 232: 142–149. [DOI] [PubMed] [Google Scholar]
- 45. Sun G, Xu Y, Li X, Jin Y, Li B, et al. (2007) Urinary Arsenic Metabolites in Children and Adults Exposed to Arsenic in Drinking Water in Inner Mongolia, China. Environ Health Perspect 115: 648–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sarver RWJ, Krueger WC (1991) An infrared and circular dichroism combined approach to the analysis of protein secondary structure. Anal Biochem 199: 61–67. [DOI] [PubMed] [Google Scholar]
- 47. Hennessey JP, Johnson WC (1981) Information-content in the circular-dichroism of proteins. Biochemistry 20: 1085–1094. [DOI] [PubMed] [Google Scholar]
- 48. Dong A, Huang P, Caughey WS (1992) Redox-Dependent Changes in β-Extended Chain and Turn Structures of Cytochrome c in Water Solution Determined by Second Derivative Amide I Infrared Spectra. Biochemistry 31: 182–189. [DOI] [PubMed] [Google Scholar]
- 49. Dougherty DA, Stauffer DA (1990) Acetylcholine binding by a synthetic receptor: implications for biological recognition. Science 250: 1558–1560. [DOI] [PubMed] [Google Scholar]
- 50. McCurdy A, Jimenez L, Stauffer DA, Dougberty DA (1992) Biomimetic Catalysis of SN2 Reactions through Cation-π Interactions. The Role of Polarizability in Catalysis. J Am Chem Soc 114: 10314–10321. [Google Scholar]
- 51. Kuhn B, Kollman PA (2000) QM-FE and Molecular Dynamics Calculations on Catechol O-methyltransferase: Free Energy of Activation in the Enzyme and in Aqueous Solution and Regioselectivity of the Enzyme Catalyzed Reaction. J Am Chem Soc 122: 2586–2596. [Google Scholar]
- 52. Hamahata A, Takata Y, Gomi T, Fujioka M (1996) Probing the S-adenosylmethionine-binding site of rat guanidinoacetate Methyltransferase. Biochem J 317: 141–145. [DOI] [PMC free article] [PubMed] [Google Scholar]