Abstract
A large pertussis epidemic occurred between 2008 and 2010 in Japan. To investigate epidemic strains, we analyzed 33 Bordetella pertussis isolates from the epidemic period by sequencing virulence-associated genes (fim3, ptxP, ptxA, and prn) and performing multilocus variable-number tandem repeat analysis (MLVA), and compared these results with those of 101 isolates from non-epidemic, earlier and later time periods. DNA sequencing of the fim3 allele revealed that the frequency of fim3B was 4.3%, 12.8%, 30.3%, and 5.1% within isolates in 2002–2004, 2005–2007, 2008–2010, and 2011–2012, respectively. The isolation rate of the fim3B strain therefore temporarily increased during the epidemic period 2008–2010. In contrast, the frequencies of the virulence-associated allelic variants, ptxP3, ptxA1, and prn2, increased with time during overall study period, indicating that these variants were not directly involved in the occurrence of the 2008–2010 epidemic. MLVA genotyping in combination with analysis of allele types showed that the prevalence of an MT27d strain temporarily increased in the epidemic period, and that this strain carried virulence-associated allelic variants (fim3B, ptxP3, ptxA1, and prn2) also identified in recent epidemic strains of Australia, Europe, and the US. Phenotypic analyses revealed that the serotype Fim3 strain was predominant (≥87%) during all the periods studied, and that the frequency of adhesion pertactin (Prn) non-expressing B. pertussis decreased by half in the epidemic period. All MT27d strains expressed Prn and Fim3 proteins, suggesting that B. pertussis MT27d strains expressing Prn and Fim3B have the potential to cause large epidemics worldwide.
Introduction
Bordetella pertussis, a highly communicable Gram-negative coccobacillus, is the cause of pertussis (whooping cough), a major acute respiratory infection resulting in severe childhood illness and infant death [1]. Although universal immunization programs have contributed to significant reductions in the morbidity and mortality rates associated with pertussis, the incidence of pertussis has increased in several countries despite high vaccination coverage [2]-[6]. In Japan, acellular pertussis vaccines (ACVs) were introduced in 1981 and are used to control pertussis with a schedule of 3 primary doses and single booster dose at ages 3, 4, 6, and 18−23 months, respectively. This vaccination schedule has been followed since 1994. The incidence of pertussis cases in adolescents and adults, however, has significantly increased since the early 2000s [7]. The waning of vaccine-acquired immunity and the decrease in opportunities of natural immune boosting owing to reduced levels of B. pertussis circulation have been proposed as explanations for the recent resurgences of pertussis [2], [8], [9]. Another possible underlying factor is the adaptation of the B. pertussis population to vaccine-induced immunity [2], [10], [11].
Antigenic and genetic shifts in B. pertussis circulating strains have been identified within virulence-associated genes encoding serotype 3 fimbriae (fim3), pertussis toxin S1 subunit (ptxA), pertactin (prn), and the pertussis toxin promoter (ptxP). Allele frequencies of the virulence-associated allelic variants, fim3B, ptxA1, prn2, and ptxP3, have increased within the B. pertussis population in several countries [10], [12]–[17]. B. pertussis strains carrying ptxP3 are more capable of producing pertussis toxin (PT) than are ptxP1 strains, and the emergence of ptxP3 strains was associated with pertussis resurgence in the Netherlands [18]. Similarly, a significant correlation was observed between an increase in fim3B strains and pertussis notifications in the US [13]. Strains with fim3B have a single amino-acid substitution (A87E) within the surface epitope of Fim3, which interacts with human serum [19], [20]. Furthermore, multilocus variable-number tandem repeat analysis (MLVA) has revealed that the B. pertussis population has changed over the past 50 years worldwide. In Australia, the frequency of B. pertussis MLVA type 27 (MT27) and MT70 strains increased after the introduction of an ACV, and subsequently, the MT27 strain became predominant in 2008−2010 [21], [22]. An increase in the frequency of B. pertussis MT27 strain was also observed in Europe and the US [13], [16], [23].
Pertussis epidemics still occur worldwide, and epidemic strains have been characterized by Fim serotyping and/or genotyping within some regions [24]–[26]. In a Dutch epidemic, significant changes in Fim serotypes and MTs were observed during a period when the pertussis vaccine dose was lowered [24]. Besides phenotypic variants of Fim, B. pertussis variants that do not express adhesion pertactin (Prn) have been recently identified in Japan as well as in other countries [27]–[30]. Since Prn is a component of ACVs, it is reasonable to hypothesize that Prn-negative variants have increased fitness in humans immunized with ACV. To date, the relationship between the prevalence of Prn-negative variants and pertussis epidemics has not been evaluated.
In Japan, a large pertussis epidemic occurred in 2008−2010 despite high vaccination coverage with ACVs. To elucidate the causes of the epidemic, we determined temporal trends in the frequencies of virulence-associated genes (fim3, ptxP, ptxA, and prn) and genotypes in the B. pertussis population from 2002 to 2012. In addition, phenotypes of epidemic isolates were characterized by their expression of Fim and Prn proteins.
Materials and Methods
Pertussis surveillance data
National surveillance data were obtained from the Ministry of Health, Labor and Welfare of Japan Infectious Disease Surveillance data. Each week, the number of pertussis cases was reported from approximately 3,000 sentinel clinics and hospitals within Japan. Diagnosis was based on bacterial culture, clinical symptoms, and/or the results of a serologic test. The reporting criteria did not change during the study period, 2002−2012.
Bacterial strains
We examined 134 clinical B. pertussis isolates collected in Japan from 2002 to 2012 (Table S1). Thirty-three of those isolates were collected during the 2008–2010 pertussis epidemic, while 101 isolates were collected from non-epidemic periods: 23 in 2002–2004, 39 in 2005–2007, and 39 in 2011–2012 (Fig. 1). All isolates were epidemiologically unrelated cases of pertussis. The isolates were cultured on Bordet-Gengou agar (Difco) or cyclodextrin solid medium (CSM) agar [31], and incubated at 36°C for 2−3 days. DNA was extracted from B. pertussis isolates by boiling, and stored at −20°C.
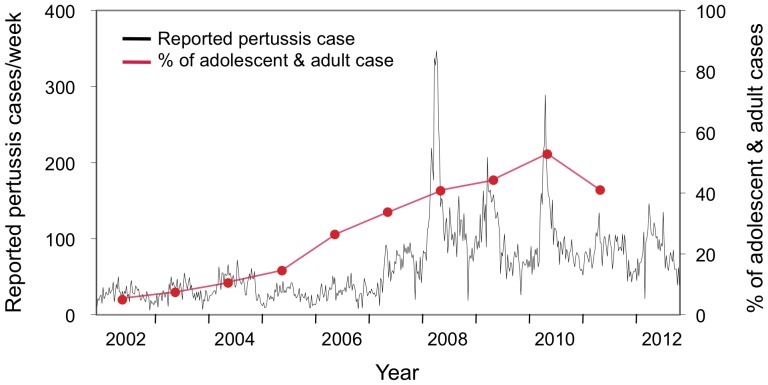
Figure 1. Number of reported pertussis cases per week in Japan from 2002 to 2012.
Pertussis cases are shown by the black line, with each value representing a week of the year. The percentage of adolescent and adult cases (≥15 years old) per year is shown in red circles. The data were obtained from the Ministry of Health, Labor and Welfare of Japan Infectious Disease Surveillance data. Data regarding the number of adolescent and adult cases in 2012 were not available.
Sequence analysis of fim3, ptxP, ptxA, and prn
Four virulence-associated genes, fim3, ptxP, ptxA and prn, were analyzed using PCR-based sequencing [14], [18], [32], [33]. Sequence reactions were carried out with a BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems), and resultant products were sequenced using an Applied Biosystems 3130xl Genetic Analyzer or 3730 DNA Analyzer. Subsequent sequencing of the variable region 2 (R2) of prn was performed where necessary, to distinguish between prn1 and prn7 alleles. Primer sets used in this study are listed in Table S2.
MLVA
MLVA typing was performed as previously described [22], [28]. MTs were assigned using the MLVA typing tool found at http://www.mlva.net. Novel MLVA types were assigned by the webmasters, Drs. H. van der Heide and F. Mooi, from the Centre for Infectious Disease Control Netherlands, within National Institute for Public Health and the Environment, in the Netherlands.
Immunoblotting and serotyping analysis
Prn expression in B. pertussis isolates was analyzed by immunoblotting [28]. Briefly, protein samples (1 µg) were first subjected to 10% SDS-PAGE, then transferred onto nitrocellulose membranes (Bio-Rad), and finally incubated with anti-Prn antiserum. Antigen-antibody complexes were visualized using a horseradish peroxidase-conjugated secondary antibody (Bio-Rad) and the Western Lightning ECL Pro (PerkinElmer, Inc.). Resultant blots were imaged using a LAS-3000 (Fujifilm, Tokyo, Japan).
Serotyping of B. pertussis isolates was performed with indirect whole-cell ELISA using anti-Fim2 and anti-Fim3 monoclonal antibodies as previously described [34], [35], with some minor modifications. Briefly, bacterial cells cultured on Bordet-Gengou agar were resuspended in phosphate-buffered saline (PBS) to an optical density of 0.01 at 620 nm, and then inactivated at 56°C for 1 h. The wells of 96-well ELISA plate (Nunc Immuno Plate Maxisorp) were coated with 100 µl of this suspension to each well and allowing it to evaporate overnight at 36°C. Anti-Fim2 (NIBSC 04/154) and anti-Fim3 (NIBSC 04/156) antibodies were used at a 1∶1,000 dilution in PBS. Antibody binding to bacterial cells was detected following the addition of a 1∶4,000 dilution of alkaline phosphatase-labeled goat anti-mouse IgG (Southern Biotechnology Associates, Inc.) and with the use of p-nitrophenylphosphate as a substrate. The optical density was measured at 405 nm with 650 nm as a reference using a Multiskan FC microplate reader (Thermo Fisher Scientific Inc.). B. pertussis strain 18323 that expresses both Fim2 and Fim3 was used as a positive control.
Statistical analysis
Squared Pearson’s correlation coefficient (R2) was used to identify a linear dependence between allele frequency (fim3B, ptxP3, ptxA1, or prn2) and isolation periods. Fisher’s exact test was performed to analyze the distribution of B. pertussis population. The Simpson’s diversity index (DI) and 95% confidence interval (CI) of MTs was calculated as described by Hunter and Gaston [36] and Grundmann et al. [37], respectively, using the online tool available at http://www.comparingpartitions.info/.
Results
Characteristics of the 2008−2010 pertussis epidemic in Japan
There were 17,349 reported pertussis cases between January 2008 and December 2010 in Japan. Within this pertussis epidemic period, 3 sharp peaks representing increases in case frequency were observed: 1 in late May 2008 (347 cases at week 22), 1 in mid-May 2009 (207 cases at week 20), and 1 in mid-June 2010 (289 cases at week 24) (Fig. 1). The number of reported cases per year in 2008−2010 was ≥2.7 times higher than the previous 5-year average. Although the number of pertussis patients over 15 years of age steadily increased in the 2000s, alongside increases of adolescent and adult incidence rates (40.7%, 44.2%, and 52.9% of all reported cases in 2008, 2009, and 2010, respectively), in 2011, the number of those patients decreased and the incidence rate in adolescents and adults also decreased to 41%.
Temporal changes in the frequencies of fim3, ptxP, ptxA, and prn alleles
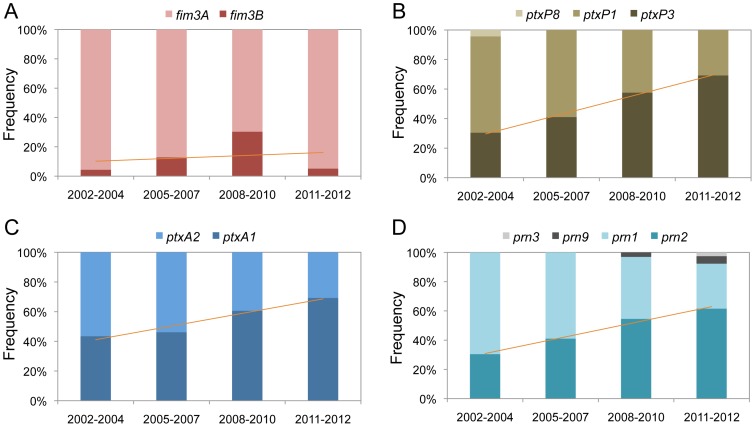
Among the 134 B. pertussis isolates tested, 2 fim3 (fim3A and fim3B), 3 ptxP (ptxP1, ptxP3 and ptxP8), 2 ptxA (ptxA1 and ptxA2), and 4 prn (prn1, prn2, prn3, and prn9) alleles were identified. Figure 2 shows the temporal trends of the allele frequencies. The frequency of the allele fim3B temporarily increased during the epidemic period (2008–2010): it was 4.3% in 2002–2004, 12.8% in 2005–2007, 30.3% in 2008–2010, and 5.1% in 2011–2012 (Fig. 2A). In contrast, the frequencies of ptxP3, ptxA1, and prn2 increased with time from 2002 to 2012 (Fig. 2B-D). High correlations (R2 ≥ 0.95) were observed between these latter allele frequencies and the isolation periods.
Figure 2. Temporal trends in the frequencies of fim3, ptxP, ptxA, and prn alleles within Bordetella pertussis isolates in Japan from 2002 to 2012. Four allelic genes, fim3 (A), ptxP (B), ptxA (C), and prn (D), of 134 B. pertussis isolates were sequenced.
Isolate allele frequencies are shown by time period: 2002−2004 (non-epidemic, n = 23), 2005−2007 (pre-epidemic, n = 39), 2008−2010 (epidemic, n = 33), and 2011−2012 (post-epidemic, n = 39). The regression line shows the relationship between the frequency of virulence-associated allelic variant (fim3B, ptxP3, ptxA1, or prn2) and the 4 time periods.
Temporal changes in the frequencies of Prn and Fim variants
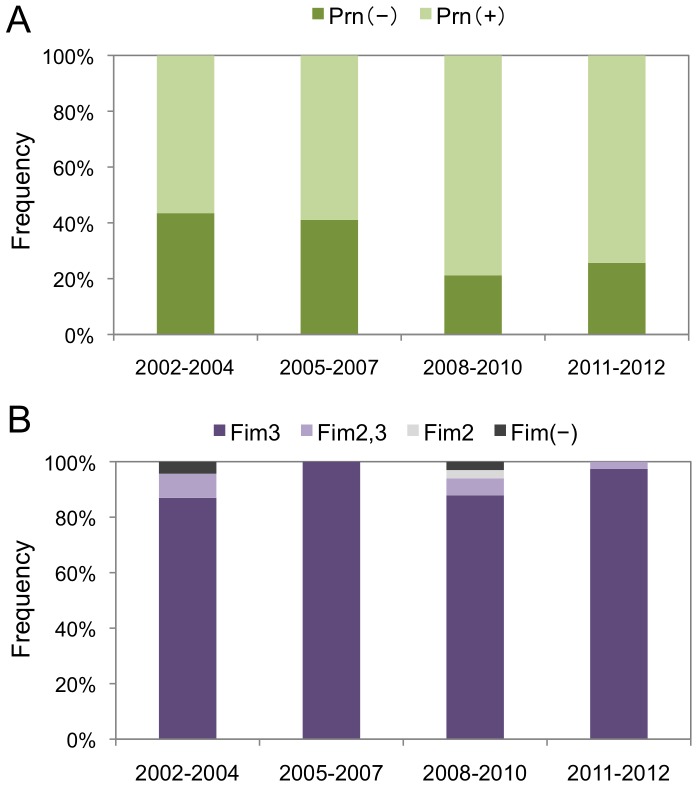
The Prn and Fim expression phenotypes of the 134 B. pertussis study isolates were determined with immunoblotting and indirect whole-cell ELISA, respectively. Figure 3A shows the temporal trend of the frequencies of the 2 identified Prn variants, Prn-expressing and Prn-negative strains, with the frequency of Prn-negative strains at 43.5%, 41.0%, 21.2%, and 25.6% during 2002–2004, 2005–2007, 2008–2010, and 2011–2012, respectively. A decreased frequency of Prn-negative strains was observed during the epidemic period (2008–2010); however, this decrease was not statistically significant (P>0.05). All the Prn-negative strains carried the prn1 allele (Table S1). Prn-negative strains carrying prn1 were previously found in Finland [30], and those carrying prn2 were found in France and the US [27], [29].
Figure 3. Temporal trends in the frequencies of Prn and Fim3 variants of Bordetella pertussis isolates in Japan from 2002 to 2012. Prn (A) and Fim (B) expression was analyzed within 134 B. pertussis isolates.
Two Prn variants, Prn(+) and Prn(−), and 4 Fim variants, Fim2, Fim3, Fim2,3, and Fim(−) strains, were identified. Variant frequencies are shown by time period: 2002−2004 (non-epidemic, n = 23), 2005−2007 (pre-epidemic, n = 39), 2008−2010 (epidemic, n = 33), and 2011−2012 (post-epidemic, n = 39).
On the other hand, 4 Fim variants, Fim2, Fim3, Fim2,3, and Fim(−), were identified among the B. pertussis isolates. The Fim2,3 strain was detected by both Fim2 and Fim3 antigens, while the Fim(−) strain was not detected by either. As shown in Figure 3B, the Fim3 strain was predominant during all the time periods studied: 87.0% in 2002−2004, 100% in 2005−2007, 87.9% in 2008−2010, and 97.4% in 2011−2012. Two Fim(−) strains were isolated in 2002−2004 and 2008−2010. Interestingly, one Fim(−) strain was previously identified in Ontario, Canada [12].
Temporal changes in MTs and genetic diversity
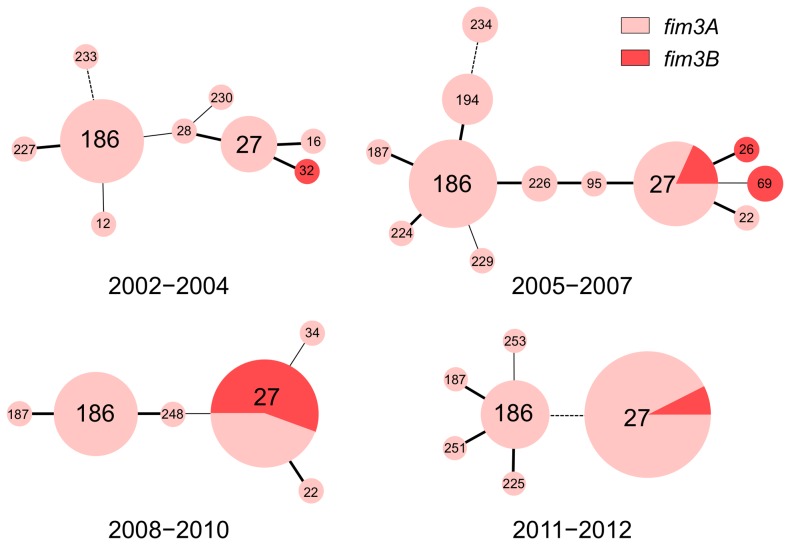
Among the 134 B. pertussis study isolates, 24 different MTs were identified, of which 2 were novel (MT251 and MT253). Figure 4 shows minimum spanning trees that revealed the genetic diversity of the B. pertussis population during the time periods of 2002−2004, 2005−2007, 2008−2010, and 2011−2012. Eighteen B. pertussis isolates carrying fim3B were identified during the 4 time periods, and these fim3B strains were divided into 4 MTs: MT26 (n = 1), MT27 (n = 14), MT32 (n = 1), and MT69 (n = 2). Although MT27 and MT186 were the predominant types during all the time periods, the fim3B strain did not belong to MT186. MT27 strains carrying fim3B were most frequent in MT27 during the epidemic period (2008−2010) at 56% (10/18), and their frequency decreased to 7% (2/27) in 2011−2012. The temporal increase of MT27 strains carrying fim3B was statistically significant (P < 0.05) when compared with non-MT27 strains carrying fim3B.
Figure 4. Minimum spanning trees revealing the genetic diversity of the Bordetella pertussis population in Japan from 2002 to 2012.
MTs were determined for 134 B. pertussis isolates. The resultant phylogenetic trees based on MTs are shown by time period: 2002−2004 (non-epidemic, n = 23), 2005−2007 (pre-epidemic, n = 39), 2008−2010 (epidemic, n = 33), and 2011−2012 (post-epidemic, n = 39). Each circle within a tree represents a different MT, with the MT number noted. Thick lines separate single-locus variants, while thin lines separate double-locus variants, and dotted lines signify a more distant relationship. Pink and red colors indicate, respectively, the fim3A and fim3B allele frequencies within MTs.
MT27 strains could be further classified into 5 subtypes (MT27a to MT27e) based on their allele types (Table 1). MT27a strains carried fim3A, ptxP3, ptxA1, and prn2, and were the predominant subtype. Notably, they were collected in both non-epidemic and epidemic periods. In contrast, MT27d strains carrying fim3B, ptxP3, ptxA1, and prn2 were predominantly isolated during the epidemic period, with 10 of 12 isolates (83%) being of this subtype in 2008−2010. An MT27c strain carrying fim3A, ptxP3, ptxA1, and prn9 was also isolated during the epidemic period. The other MT27 subtypes, MT27b and MT27e, were found only in 2011−2012.
Table 1. Comparison of MTs and allele types between Bordetella pertussis isolates collected in non-epidemic and epidemic periods.
| Allele types | No. of isolates detected within the time period | |||||||
| MT | fim3 | ptxP | ptxA | prn | 2002–2004 | 2005–2007 | 2008–2010* | 2010–2012 |
| 12 | A | 8 | 2 | 1 | 1 | |||
| 16 | A | 3 | 1 | 2 | 1 | |||
| 22 | A | 3 | 1 | 2 | 1 | 1 | ||
| 26 | B | 3 | 1 | 2 | 1 | |||
| 27a | A | 3 | 1 | 2 | 5 | 9 | 7 | 24 |
| 27b | A | 3 | 1 | 3 | 1 | |||
| 27c | A | 3 | 1 | 9 | 1 | |||
| 27d | B | 3 | 1 | 2 | 2 | 10 | ||
| 27e | B | 3 | 1 | 9 | 2 | |||
| 28 | A | 1 | 1 | 1 | 1 | |||
| 32 | B | 3 | 1 | 2 | 1 | |||
| 34 | A | 1 | 1 | 1 | 1 | |||
| 69 | B | 3 | 1 | 2 | 2 | |||
| 95 | A | 3 | 1 | 2 | 1 | |||
| 186 | A | 1 | 2 | 1 | 11 | 12 | 11 | 8 |
| 187 | A | 1 | 2 | 1 | 1 | 1 | 1 | |
| 194 | A | 1 | 2 | 1 | 4 | |||
| 224 | A | 1 | 2 | 1 | 1 | |||
| 225 | A | 1 | 2 | 1 | 1 | |||
| 226 | A | 1 | 2 | 1 | 2 | |||
| 227 | A | 1 | 2 | 1 | 1 | |||
| 229 | A | 1 | 2 | 1 | 1 | |||
| 230 | A | 1 | 1 | 1 | 1 | |||
| 233 | A | 1 | 1 | 1 | 1 | |||
| 234 | A | 1 | 1 | 1 | 2 | |||
| 248 | A | 1 | 2 | 1 | 1 | |||
| 251 | A | 1 | 2 | 1 | 1 | |||
| 253 | A | 1 | 2 | 1 | 1 | |||
Pertussis epidemic period.
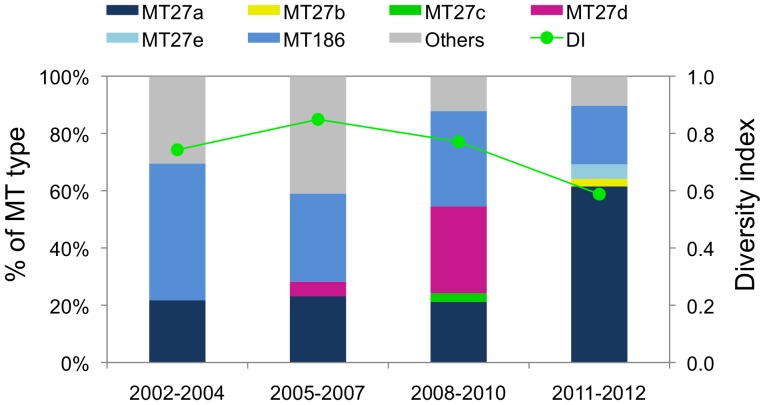
The genetic diversity of MTs and the 5 MT27 subtypes was subsequently determined. As shown in Figure 5, Simpson’s DI was 0.74 (95% CI, 0.58−0.90), 0.85 (0.78−0.92), 0.77 (0.70−0.84), and 0.59 (0.43−0.75) in 2002−2004, 2005−2007, 2008−2010, and 2011−2012, respectively. Therefore, the genetic diversity within B. pertussis population decreased after the 2008−2010 pertussis epidemic.
Figure 5. Frequency of MLVA types and genetic diversity of the Bordetella pertussis population in Japan from 2002 to 2012.
The diversity index (DI) and 95% confidence interval (CI) were calculated from the MT frequencies within 4 time periods: 2002−2004 (non-epidemic), 2005−2007 (pre-epidemic), 2008−2010 (epidemic), and 2011−2012 (post-epidemic). For convenience, minor MTs (MT22, MT34, MT187, and MT248) are shown as “Others”.
Relationship between MTs and phenotypes in 2008−2010
During the 2008−2010 pertussis epidemic, MT186 (33%), MT27a (21%), and MT27d (30%) were the predominant MTs, and most of them were serotype Fim3 strains (89%) (Table 2). Fim3 strains were also identified within other minor MTs (MT22, MT27c, MT187, and MT248), while a Fim2 strain belonged to MT34, the 2 Fim2,3 strains were typed as MT27d and MT186, and the Fim(−) strain belonged to MT27a. Of the 10 MT27d strains, 9 expressed Fim3 and 1 expressed both Fim2 and Fim3. On the other hand, Prn-negative strains were all observed within MT186. Seven (64%) of the 11 MT186 strains did not express Prn. All other MTs expressed Prn.
Table 2. Relationship between MTs and phenotypes in Bordetella pertussis isolates collected during the 2008−2010 pertussis epidemic period.
| Fim expression | Prn expression | ||||||
| MT | No. of isolates | Fim2 | Fim3 | Fim2,3 | Fim(−) | Prn(+) | Prn(−) |
| 22 | 1 | 1 | 1 | ||||
| 27a | 7 | 6 | 1 | 7 | |||
| 27c | 1 | 1 | 1 | ||||
| 27d | 10 | 9 | 1 | 10 | |||
| 34 | 1 | 1 | 1 | ||||
| 186 | 11 | 10 | 1 | 4 | 7 | ||
| 187 | 1 | 1 | 1 | ||||
| 248 | 1 | 1 | 1 | ||||
Discussion
In the present study, we demonstrated that the prevalence of B. pertussis strains carrying fim3B, which were classified as MT27d, temporarily increased during the 2008–2010 pertussis epidemic in Japan. The MT27d strains of the epidemic period were collected in several areas (Table S1). All MT27d strains carried fim3B, ptxP3, ptxA1, and prn2, and expressed Prn and Fim3 proteins. Although Prn-negative strains have recently increased in their prevalence within Japan and in other countries [27]–[30], here, a lowered frequency of Prn-negative strains was observed during the epidemic period specifically, indicating that Prn-negative strain types were not involved in the epidemic. Similarly, we found that the prevalence of the virulence-associated allelic variants, ptxP3, ptxA1, and prn2, has increased with time from the early 2000s, indicating that the variants were also not directly associated with the epidemic. Taken together, our findings demonstrate that B. pertussis strains carrying fim3B (i.e., MT27d) were associated with the 2008–2010 pertussis epidemic.
We evaluated the regional effect on B. pertussis population because of the low number of samples of isolates. When the number of isolates was compared, the difference in regional population between epidemic (2008–2010) and post-epidemic (2011–2012) periods was statistically significant (P < 0.01), possibly because of the high number (18/39) of isolates collected in the Kinki district (including Osaka) during the post-epidemic period. A sampling bias cannot be excluded from the analysis of the trend in 2011–2012. However, no significant difference was observed between pre-epidemic (2005–2007) and epidemic periods (P > 0.05). The regional effect was therefore small or negligible for detection of the emergence of strain MT27d in the 2008–2010 epidemic.
In the past decade, the prevalence of B. pertussis strains carrying fim3B has increased worldwide [12]–[14], [25]. In Ontario, Canada, 1 predominant B. pertussis clone was identified in the 2000s [12]. This strain carried the same virulence-associated allelic variants (fim3B, ptxP3, ptxA1 and prn2) as our epidemic strains within MT27d and was identical to the strains involved in recent pertussis resurgences within Europe and Australia. Similarly, fim3B strains carrying ptxP3, ptxA1, and prn2 were predominant in the US during the 2000s, and most were genotyped to MT27 [13]. Interestingly, the pertussis resurgence in the US was correlated with the emergence and predominance of the fim3B allele, but not with the ptxP3 allele. On the other hand, in the Netherlands, the prevalence of fim3B strains temporarily increased in the early 2000s, although the strains disappeared in 2010 [14]. Likewise, MT27d strains (carrying fim3B) were not identified after the 2008−2010 epidemic period, and the reasons behind the disappearance of this strain type are unclear. Our findings along with those of previous studies, therefore, suggest that the MT27d strain is a recent epidemic strain that is found worldwide, and that this strain is not only associated with pertussis resurgence but can also be correlated with pertussis epidemics.
Fimbriae of B. pertussis are composed of Fim2 and/or Fim3 and FimD. The minor fimbrial subunit FimD forms the adhesive tip [38]. Fim2 and Fim3 are encoded by the single-copy genes fim2 and fim3, respectively [39], [40], and are serologically distinct [41]. Fim resulting from the fim3B strain is Fim3B, which differs from Fim3A by a single amino-acid substitution (A87E) at the surface epitope. To date, the biological differences between Fim3A and Fim3B are unknown. In an effort to address this issue, we recently observed B. pertussis clinical strains and found that most strains carrying fim3B had strong autoagglutination capability, unlike those carrying fim3A, following the suspension of CSM agar cultures into saline (Otsuka and Kamachi, unpublished data). Surprisingly, autoagglutination was not observed when the strains were cultured on Bordet-Gengou agar. Bacterial autoaggregation is a phenomenon associated with pathogen virulence in many Gram-negative bacteria [42]–[46], suggesting Fim3B strains are more virulent than Fim3A strains because of their ability to autoagglutinate. Further study is necessary to fully elucidate the relationship between autoagglutination and Fim3B. Attempts to identify the molecular mechanism that regulates autoagglutination within fim3B strains are currently underway.
In many countries, a shift from serotype Fim2 to Fim3 in B. pertussis circulating strains was observed after mass vaccination, and antigenic differences (in Fim, PT, and Prn) have been since noted between B. pertussis vaccine strains and circulating strains. In Japan, the B. pertussis strain Tohama carrying ptxA2, prn1, and fim2 has been used as a vaccine strain to produce ACVs from 1981. Among the 4 currently used Japanese ACVs, 2 contain Fim2 and all do not contain Fim3 [47]. A recent study demonstrated that Fim2 and Fim3 are immunogenic antigens, and that individuals recently infected with pertussis had greater anti-Fim3 IgG concentrations than anti-Fim2 IgG concentrations, consistent with the current predominance of Fim3 strains [41]. Based on this finding, there is a clear need for the improvement of currently used ACVs. Specifically, the inclusion of Fim3 (Fim3A and/or Fim3B) in ACVs may be an effective way to reduce the number of current circulating B. pertussis strains, including Fim3B strains. Although Fim2 has been shown to be a protective antigen, the protective immunogenicity of Fim3 is still unknown [48]. Further study of this topic will be required to evaluate the effectiveness of Fim3 as a protective antigen.
In the present study, the genetic diversity of the B. pertussis population decreased after the 2008−2010 pertussis epidemic. This decrease reflects the expansion of the MT27a strain type and the disappearance of MT27d strains. The MT27a strains carried the same ptxP3, ptxA1, and prn2 alleles as the MT27d strains, but additionally carried fim3A. Significant changes in the B. pertussis population were previously observed in a pertussis epidemic in the Netherlands, and this study suggested that strain surveillance may serve as an early detector of pertussis epidemics [24]. Here, we observed significant changes within the B. pertussis population during the 2008−2010 epidemic, lending further support for an early warning system of future pertussis epidemics.
In conclusion, the prevalence of B. pertussis MT27d strains temporarily increased during the 2008−2010 pertussis epidemic in Japan. The MT27d strains carried the same virulence-associated allelic variants (fim3B, ptxP3, ptxA1, and prn2) as recent epidemic strains observed in Australia, Europe, and the US. B. pertussis MT27d strains, therefore, likely have the potential to cause large epidemics in other countries, and, hence, further study and strain surveillance of the MT27d strain type is warranted.
Supporting Information
Characteristics of Bordetella pertussis study isolates.
(XLSX)
PCR primers in this study.
(XLSX)
Acknowledgments
We are grateful to the following for providing B. pertussis isolates: M. Ohtsuka (Kotobiken Medical Laboratories Inc.), C. Kawamura (Aomori Prefectural Central Hospital), K. Funahashi (Aichi Showa Hospital), M. Tanaka (Niigata City Institute of Public Health and Environment), Y. Takahashi (Odate Municipal General Hospital), T. Takagi (St. Marianna University School of Medicine), Y. Kori (Chiba Kaihin Municipal Hospital), T. Nakamura (Kansai Medical University Hirakata Hospital), A. Anan (Showa University Fujigaoka Hospital), N. Hasegawa (Itayanagi Central Hospital), K. Yoshida (Ehime Prefectural Institute of Health and Environmental Science), K. Tsuchiya (Shizuoka City Shimizu Hospital), S. Notake (Miroku Medical Laboratory Inc.), T. Yamato (Kochi National Hospital), T. Nakano (Kawasaki Medical University), K. Kawano (Miyazaki Prefectural Institute for Health and Environment), C. Katsukawa (Osaka Prefectural Institute of Public Health), T. Miyano (Miyano Children’s Clinic), K. Okada (Fukuoka Dental College), H. Moriyama (Shimane University Hospital), H. Ito (Sotobo Children’s Clinic), K. Iwabuchi (Research Institute for Environmental Sciences and Public Health of Iwate Prefecture), S. Matsuura (Tokushima Municipal Hospital), M. Mochizuki (Nagoya University Hospital), T. Niizuma (Koshigaya Municipal Hospital), N. Watanabe (Fukushima Institute for Public Health), and J. Yatsuyanagi (Akita Research Center for Public Health and Environment).
We are also grateful to Drs. Hitoshi Yamamoto (Dept. of Pediatrics, St. Marianna University School of Medicine), Yoshichika Arakawa (Nagoya University Graduate School of Medicine), and Tatsuo Kato (National Center for Child Health and Development) for their helpful discussions and comments.
Funding Statement
This research was supported by a grant (24170201) for Research on Emerging and Re-emerging Infectious Diseases from the Ministry of Health, Labor and Welfare of Japan. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Mattoo S, Cherry JD (2005) Molecular pathogenesis, epidemiology, and clinical manifestations of respiratory infections due to Bordetella pertussis and other Bordetella subspecies. Clin Microbiol Rev 18: 326–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cherry JD (2012) Epidemic pertussis in 2012—the resurgence of a vaccine-preventable disease. N Engl J Med 367: 785–787. [DOI] [PubMed] [Google Scholar]
- 3. Spokes PJ, Quinn HE, McAnulty JM (2010) Review of the 2008-2009 pertussis epidemic in NSW: notifications and hospitalisations. N S W Public Health Bull 21: 167–173. [DOI] [PubMed] [Google Scholar]
- 4. Fisman DN, Tang P, Hauck T, Richardson S, Drews SJ, et al. (2011) Pertussis resurgence in Toronto, Canada: a population-based study including test-incidence feedback modeling. BMC Public Health 11: 694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Campbell P, McIntyre P, Quinn H, Hueston L, Gilbert GL, et al. (2012) Increased population prevalence of low pertussis toxin antibody levels in young children preceding a record pertussis epidemic in Australia. PLoS One 7: e35874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. de Greeff SC, de Melker HE, van Gageldonk PG, Schellekens JF, van der Klis FR, et al. (2010) Seroprevalence of pertussis in The Netherlands: evidence for increased circulation of Bordetella pertussis . PLoS One 5: e14183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Han HJ, Kamachi K, Okada K, Toyoizumi-Ajisaka H, Sasaki Y, et al. (2008) Antigenic variation in Bordetella pertussis isolates recovered from adults and children in Japan. Vaccine 26: 1530–1534. [DOI] [PubMed] [Google Scholar]
- 8. Hewlett EL, Edwards KM (2005) Clinical practice. Pertussis−not just for kids. N Engl J Med 352: 1215–1222. [DOI] [PubMed] [Google Scholar]
- 9. Klein NP, Bartlett J, Rowhani-Rahbar A, Fireman B, Baxter R (2012) Waning protection after fifth dose of acellular pertussis vaccine in children. N Engl J Med 367: 1012–1019. [DOI] [PubMed] [Google Scholar]
- 10. Kallonen T, He Q (2009) Bordetella pertussis strain variation and evolution postvaccination. Expert Rev Vaccines 8: 863–875. [DOI] [PubMed] [Google Scholar]
- 11. Mooi FR, van Oirschot H, Heuvelman K, van der Heide HG, Gaastra W, et al. (1998) Polymorphism in the Bordetella pertussis virulence factors P.69/pertactin and pertussis toxin in The Netherlands: temporal trends and evidence for vaccine-driven evolution. Infect Immun 66: 670–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shuel M, Jamieson FB, Tang P, Brown S, Farrell D, et al. (2013) Genetic analysis of Bordetella pertussis in Ontario, Canada reveals one predominant clone. Int J Infect Dis 17: e413–e417. [DOI] [PubMed] [Google Scholar]
- 13. Schmidtke AJ, Boney KO, Martin SW, Skoff TH, Tondella ML, et al. (2012) Population diversity among Bordetella pertussis isolates, United States, 1935–2009. Emerg Infect Dis 18: 1248–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. van Gent M, Bart MJ, van der Heide HG, Heuvelman KJ, Mooi FR (2012) Small mutations in Bordetella pertussis are associated with selective sweeps. PLoS One 7: e46407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Advani A, Gustafsson L, Åhrén C, Mooi FR, Hallander HO (2011) Appearance of Fim3 and ptxP3-Bordetella pertussis strains, in two regions of Sweden with different vaccination programs. Vaccine 29: 3438–3442. [DOI] [PubMed] [Google Scholar]
- 16. Petersen RF, Dalby T, Dragsted DM, Mooi F, Lambertsen L (2012) Temporal trends in Bordetella pertussis populations, Denmark, 1949-2010. Emerg Infect Dis 18: 767–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang L, Xu Y, Zhao J, Kallonen T, Cui S, et al. (2010) Effect of vaccination on Bordetella pertussis strains, China. Emerg Infect Dis 16: 1695–1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mooi FR, van Loo IH, van Gent M, He Q, Bart MJ, et al. (2009) Bordetella pertussis strains with increased toxin production associated with pertussis resurgence. Emerg Infect Dis 15: 1206–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Williamson P, Matthews R (1996) Epitope mapping the Fim2 and Fim3 proteins of Bordetella pertussis with sera from patients infected with or vaccinated against whooping cough. FEMS Immunol Med Microbiol 13: 169–178. [DOI] [PubMed] [Google Scholar]
- 20. Tsang RS, Lau AK, Sill ML, Halperin SA, Van Caeseele P, et al. (2004) Polymorphisms of the fimbria fim3 gene of Bordetella pertussis strains isolated in Canada. J Clin Microbiol 42: 5364–5367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kurniawan J, Maharjan RP, Chan WF, Reeves PR, Sintchenko V, et al. (2010) Bordetella pertussis clones identified by multilocus variable-number tandem-repeat analysis. Emerg Infect Dis 16: 297–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Octavia S, Sintchenko V, Gilbert GL, Lawrence A, Keil AD, et al. (2012) Newly emerging clones of Bordetella pertussis carrying prn2 and ptxP3 alleles implicated in Australian pertussis epidemic in 2008-2010. J Infect Dis 205: 1220–1224. [DOI] [PubMed] [Google Scholar]
- 23. Litt DJ, Neal SE, Fry NK (2009) Changes in genetic diversity of the Bordetella pertussis population in the United Kingdom between 1920 and 2006 reflect vaccination coverage and emergence of a single dominant clonal type. J Clin Microbiol 47: 680–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. van Gent M, de Greeff SC, van der Heide HG, Mooi FR (2009) An investigation into the cause of the 1983 whooping cough epidemic in the Netherlands. Vaccine 27: 1898–1903. [DOI] [PubMed] [Google Scholar]
- 25. Lin YC, Yao SM, Yan JJ, Chen YY, Hsiao MJ, et al. (2006) Molecular epidemiology of Bordetella pertussis in Taiwan, 1993–2004: suggests one possible explanation for the outbreak of pertussis in 1997. Microbes Infect 8: 2082–2087. [DOI] [PubMed] [Google Scholar]
- 26. Bisgard KM, Christie CD, Reising SF, Sanden GN, Cassiday PK, et al. (2001) Molecular epidemiology of Bordetella pertussis by pulsed-field gel electrophoresis profile: Cincinnati, 1989–1996. J Infect Dis 183: 1360–1367. [DOI] [PubMed] [Google Scholar]
- 27. Queenan AM, Cassiday PK, Evangelista A (2013) Pertactin-negative variants of Bordetella pertussis in the United States. N Engl J Med 368: 583–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Otsuka N, Han HJ, Toyoizumi-Ajisaka H, Nakamura Y, Arakawa Y, et al. (2012) Prevalence and genetic characterization of pertactin-deficient Bordetella pertussis in Japan. PLoS One 7: e31985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hegerle N, Paris AS, Brun D, Dore G, Njamkepo E, et al. (2012) Evolution of French Bordetella pertussis and Bordetella parapertussis isolates: increase of Bordetellae not expressing pertactin. Clin Microbiol Infect 18: E340–346. [DOI] [PubMed] [Google Scholar]
- 30. Barkoff AM, Mertsola J, Guillot S, Guiso N, Berbers G, et al. (2012) Appearance of Bordetella pertussis strains not expressing the vaccine antigen pertactin in Finland. Clin Vaccine Immunol 19: 1703–1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Aoyama T, Murase Y, Iwata T, Imaizumi A, Suzuki Y, et al. (1986) Comparison of blood-free medium (cyclodextrin solid medium) with Bordet-Gengou medium for clinical isolation of Bordetella pertussis . J Clin Microbiol 23: 1046–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. van Loo IH, Heuvelman KJ, King AJ, Mooi FR (2002) Multilocus sequence typing of Bordetella pertussis based on surface protein genes. J Clin Microbiol 40: 1994–2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nakamura Y, Kamachi K, Toyoizumi-Ajisaka H, Otsuka N, Saito R, et al. (2011) Marked difference between adults and children in Bordetella pertussis DNA load in nasopharyngeal swabs. Clin Microbiol Infect 17: 365–370. [DOI] [PubMed] [Google Scholar]
- 34. Tsang RS, Sill ML, Advani A, Xing D, Newland P, et al. (2005) Use of monoclonal antibodies to serotype Bordetella pertussis isolates: comparison of results obtained by indirect whole-cell enzyme-linked immunosorbent assay and bacterial microagglutination methods. J Clin Microbiol 43: 2449–2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Heikkinen E, Xing DK, Ölander RM, Hytönen J, Viljanen MK, et al. (2008) Bordetella pertussis isolates in Finland: serotype and fimbrial expression. BMC Microbiol 8: 162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hunter PR, Gaston MA (1988) Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J Clin Microbiol 26: 2465–2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Grundmann H, Hori S, Tanner G (2001) Determining confidence intervals when measuring genetic diversity and the discriminatory abilities of typing methods for microorganisms. J Clin Microbiol 39: 4190–4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Geuijen CA, Willems RJ, Bongaerts M, Top J, Gielen H, et al. (1997) Role of the Bordetella pertussis minor fimbrial subunit, FimD, in colonization of the mouse respiratory tract. Infect Immun 65: 4222–4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Parkhill J, Sebaihia M, Preston A, Murphy LD, Thomson N, et al. (2003) Comparative analysis of the genome sequences of Bordetella pertussis, Bordetella parapertussis and Bordetella bronchiseptica . Nat Genet 35: 32–40. [DOI] [PubMed] [Google Scholar]
- 40. Zhang S, Xu Y, Zhou Z, Wang S, Yang R, et al. (2011) Complete genome sequence of Bordetella pertussis CS, a Chinese pertussis vaccine strain. J Bacteriol 193: 4017–4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Alexander F, Matheson M, Fry NK, Labram B, Gorringe AR (2012) Antibody responses to individual Bordetella pertussis fimbrial antigen Fim2 or Fim3 following immunization with the five-component acellular pertussis vaccine or to pertussis disease. Clin Vaccine Immunol 19: 1776–1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Guerry P, Ewing CP, Schirm M, Lorenzo M, Kelly J, et al. (2006) Changes in flagellin glycosylation affect Campylobacter autoagglutination and virulence. Mol Microbiol 60: 299–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ulett GC, Valle J, Beloin C, Sherlock O, Ghigo JM, et al. (2007) Functional analysis of antigen 43 in uropathogenic Escherichia coli reveals a role in long-term persistence in the urinary tract. Infect Immun 75: 3233–3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chiang SL, Taylor RK, Koomey M, Mekalanos JJ (1995) Single amino acid substitutions in the N-terminus of Vibrio cholerae TcpA affect colonization, autoagglutination, and serum resistance. Mol Microbiol 17: 1133–1142. [DOI] [PubMed] [Google Scholar]
- 45. Laird WJ, Cavanaugh DC (1980) Correlation of autoagglutination and virulence of yersiniae. J Clin Microbiol 11: 430–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Alamuri P, Löwer M, Hiss JA, Himpsl SD, Schneider G, et al. (2010) Adhesion, invasion, and agglutination mediated by two trimeric autotransporters in the human uropathogen Proteus mirabilis . Infect Immun 78: 4882–4894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Okada K, Komiya T, Yamamoto A, Takahashi M, Kamachi K, et al. (2010) Safe and effective booster immunization using DTaP in teenagers. Vaccine 28: 7626–7633. [DOI] [PubMed] [Google Scholar]
- 48. Poolman JT, Hallander HO (2007) Acellular pertussis vaccines and the role of pertactin and fimbriae. Expert Rev Vaccines 6: 47–56. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Characteristics of Bordetella pertussis study isolates.
(XLSX)
PCR primers in this study.
(XLSX)