Abstract
Salmonella enterica serover Typhimurium definitive phage type DT104, resistant to multiple antibiotics, is one of the most widespread Salmonella species in human infection worldwide. Although several cohort studies indicate that DT104 carrying the multidrug resistance (MDR) locus on salmonella genomic island 1 is a possible hyper-virulent strain compared to DT104 strains without MDR, or other Salmonella enterica serotypes, existing experimental evidence regarding virulence properties associated with the MDR region is controversial. To address this question, we constructed an isogenic MDR deletion (∆MDR) mutant strain of DT104, SNS12, by allelic exchange and used Caenorhabditis elegans as a host model to assess differences in virulence between these two strains. SNS12 exhibited decreased virulence in C. elegans, and we observed increased colonization and proliferation of the intestine of C. elegans by DT104. The immune response against MDR-carrying DT104 appears to function through a non-canonical Unfolded Protein Response (UPR) pathway, namely prion-like-(QN-rich)-domain-bearing protein pathway (PQN), in a ced-1 dependent manner in C. elegans. Further, we also demonstrate that genes of the PQN pathway and antimicrobial peptide gene abf-2, are expressed at higher transcriptional levels in worms immediately following exposure to DT104, in comparison with worms exposed to SNS12. Altogether, our results suggest that the MDR region of Salmonella Typhimurium DT104 has a direct role in virulence against Caenorhabditis elegans.
Introduction
Non-typhoid Salmonella enterica is one of the primary causes of food-borne illness throughout the world [1]. Among more than 2,500 Salmonella enterica serovars, Typhimurium is the second most prevalent, behind Enteritidis, in human infection worldwide [2]. Salmonella enterica serovar Typhimurium definitive phage type DT104 (hereafter, DT104) [3], first isolated in the 1960s, emerged in the 1990s as many isolates of this strain were found to have acquired multidrug resistance, specifically to ampicillin, chloramphenicol, streptomycin, sulfonamides and tetracycline (ACSSuT) [4].
Salmonella genomic island 1 (SGI1) is a 43 kb genomic island containing 44 open reading frames (ORFs) [5]. The multidrug resistance (MDR) region of DT104 is localized to a 13 kb segment of SGI1 [3,5,6]. Several cohort studies have indicated that DT104 carrying the MDR region is a hyper-virulent strain, as compared to DT104 strains without MDR or other Salmonella enterica serotypes [7,8]. The enhanced virulence does not appear to be due to increased invasiveness, as no significant increase in the invasive properties of DT104 were observed when tested in tissue culture assays and a mouse model of systematic salmonellosis [9–11]. Conversely, insertional inactivation of the MDR locus in DT104 was reported to reduce virulence in chickens when compared with the isogenic parent strain [12].
The soil nematode, C. elegans, has been used as an invertebrate host model to identify and assess virulence factors of several human pathogens, including Salmonella enterica Typhimurium [13] [14] [15]. A short life cycle facilitates rapid genetic experiments and is one of the major advantages for researchers working with this organism. C. elegans eggs are fertilized within the adult hermaphrodite and laid a few hours afterward--at about the 40 cell stage. C. elegans embryos develop rapidly and hatch after 14 hours. The first larval stage is completed after another 12 hours and the animals proceed through four molt cycles (L1-L4) before becoming adults. When animals reach adulthood, each produce about 300 progeny over the course of 3-4 days. The life cycle is temperature-dependent; C. elegans goes through a reproductive life cycle, egg to egg-laying parent, in 5.5 days at 15°C, 3.5 days at 20°C, and 2.5 days at 25°C. At 22°C, C. elegans has an average life span of approximately 2–3 weeks and a generation time of approximately 4 days, under laboratory conditions.
The C. elegans – Salmonella Typhimurium host-pathogen interaction model was established more than a decade ago, and it provides a powerful system to understand the virulence mechanisms of the pathogen and the immune response of the host [16–18]. Salmonella Typhimurium has been shown to colonize and establish a persistent intestinal infection in C. elegans, even after limited exposure [16,18]. Several pathogen and host factors have been found to mimic higher-order organism animal models of infection. For example, a regulator of Salmonella virulence-related genes in vertebrates, the PhoP/PhoQ signal transduction system, has also been found to be involved in Salmonella Typhimurium pathogenesis in C. elegans [16]. Acid tolerance capacity of Salmonella contributes to the survival of the microorganism during its passage through the alimentary tract and subsequently protects against the acidic environment within lysosomes [19]. The fur-1, ompR, and rpoS genes of Salmonella, which are known to be involved in different aspects of acid tolerance in human infection, have also been found to be important in C. elegans infection [18]. Lipopolysaccharide (LPS) was also shown to be required for the induction of programmed cell death, as well as for persistence of Salmonella in the C. elegans intestine [17]. Recently Bailey et al. showed that the ability of Salmonella Typhimurium to adhere and survive within macrophages is dependent on RamA, a member of the AraC/XylS family of transcriptional regulators. It is also required for colonization in mice and the C. elegans intestine [20]. In this study, we constructed a deletion mutant of the multidrug resistance region in DT104, named SNS12, and then assessed the direct involvement of the multidrug resistance (MDR) region of Salmonella Typhimurium DT104 in pathogenicity using the C. elegans model.
Materials and Methods
C. elegans and bacterial strains used
C. elegans strains N2 (wild type Bristol isolate), SS104 glp-4 (bn2), and ced-1(e1735 and e1754) mutant strains were acquired from the Caenorhabditis Genetics Center (CGC). All of the strains, except SS104, were maintained at 22°C. SS104 was maintained at 16°C, which is the permissive temperature for this temperature sensitive sterile mutant of C. elegans. Strains were cultured in C. elegans habitation media (CeHM) in tissue culture flasks on a platform shaker [21]. Nematodes were bleached (0.5 M NaOH, 1% Hypochlorite) to collect eggs, which were incubated in M9 media for 24 hours to bring them to synchronized L1 stage and then transferred to CeHM. To produce synchronized L4 stage worms, L1 worms were grown for an additional 72 hours in CeHM. Salmonella Typhimurium DT104 (wild type) and its isogenic ΔMDR mutant, SNS12, (DT104 ∆MDR, this study) and E. coli OP 50, a bacterial food strain for C. elegans, were used in this study.
Construction of MDR deletion mutant in DT104
To construct the MDR deletion mutant of DT104, the region, 24,576 to 44,114 bp (GenBank Accession AF261825.2), of SGI1, which contains the 13 kb MDR region, was deleted using two different cloning vectors, pUC19 and pTOPO. The upstream, or left, fragment (24,576 to 26,435 bp) was cloned into pTOPO using primers P1 and P2 (Table 1), containing restriction sites BamHI and PstI (Figure S1). The downstream, or right, fragment (41,379 to 44,114 bp) was cloned into pUC19, using primers P3 and P4 (Table 1), containing restriction sites PstI and HindIII (Figure S1). The upstream fragment was then digested out of the pTOPO vector and ligated into pUC19, which already contained the right fragment. A gene conveying kanamycin resistance, KanR, flanked on both sides by PstI, was digested from plasmid pUC4K (GE Healthcare Life Sciences), and inserted into the PstI site of pUC19 containing the ligated left and right fragments (Figure S1). The whole region, containing the left fragment + KanR + right fragment, was digested out of pUC19 using BamHI and HindIII enzymes (Figure S1) and cloned into a suicidal vector pMAK705 [22], containing chloramphenicol resistance (Figure S1). The plasmid pMAK705 has a temperature sensitive replicon (pSC101 origin) that is active at 28°C and inactive at 42°C to 44°C. The resulting plasmid was electroporated into the target Salmonella Typhimurium DT104 strain and was purified by serial subculture. Through several screening rounds using proper antibiotics and controlled temperature, we isolated the target recombinant and confirmed the exchange of alleles by PCR and measured the Minimum Inhibitory Concentrations (MIC) of different antibiotics (Table 2).
Table 1. Primers used in this study.
| Primers | Sequence | Source |
|---|---|---|
| Primer1 | GCGGATCCGCGAACTCTCTATCTCCTCT | This study |
| Primer2 | AAAACTGCAGGACCCGAACTTGATAACTGC | This study |
| Primer3 | AAAACTGCAGCAACCACCATTTCGCAGCAG | This study |
| Primer4 | CCCAAGCTTGTCAAAGCGGTAGCGGAAAC | This study |
| abf-2(F) | CCATCGTGGCTGCCGACATCGACTTT | [13] |
| abf-2(R) | GAGCACCAAGTGGAATATCTCCTCCTC | [13] |
| abu-11(F) | CCAATCGCCCAGTGCCAATCATC | [24] |
| abu-11(R) | CTGAACATTGGTGTCTCTGTATGG | [24] |
| pan actin (F) | TCGGTATGGGACAGAAGGAC | [24] |
| pan actin (R) | CATCCCAGTTGGTGACGATA | [24] |
| abu-1(F) | CTACTTGCCGAAGCAACAAC | [24] |
| abu-1(R) | TGGACTGGAGCAGTGTTCTG | [24] |
| pqn-54b(F) | TCAACCACAACAAACCCAGA | [24] |
| pqn-54b(R) | GCTTGAGCCTCTTGGATCT | [24] |
Table 2. Minimum Inhibitory Concentrations (MIC) of the several antibiotics against Salmonella enterica serotype Typhimurium phagetype DT104 and its isogenic ΔMDR mutant, SNS12.
| Antibiotic | DT104 | SNS12 |
|---|---|---|
| AmiKacin (AMI) | 2 | 2 |
| Ampicillin | >32 | 2 |
| Amoxicillin /Clavulanic acid (AUG) | 16 | <=1 |
| Ceftriaxone (AXO) | <=0.25 | <=0.25 |
| Cephalothin (CEP) | 4 | 4 |
| Chloramphenicol (CHL) | >32 | 4 |
| Ciprofloxacin (CIP) | <=0.015 | <=0.015 |
| Timethoprim/Sulfamethoxazole (COT) | 0.25 | <=0.12 |
| Cefoxitin (FOX) | 4 | 2 |
| Gentamycin (GEN) | 1 | 1 |
| Kanamycin (KAN) | <=8 | >64 |
| Nalidixic acid (NAL) | 4 | 4 |
| Salfamethoxazole (SMX) | >512 | 32 |
| Streptomycin (STR) | 64 | <=32 |
| Tetracycline (TET) | >32 | 8 |
| Ceftiofur(TIO) | 0.5 | 0.5 |
C. elegans survival analysis
Survival assays using C. elegans were performed following standard protocols [16]. Briefly, bacterial lawns for survival assays were prepared by inoculating 50 µl of an overnight bacterial culture onto Nematode Growth Factor medium (NGM), in 6-cm Petri plates. Plates were incubated overnight at room temperature before the addition of worms. Each experimental group contained 60-80 synchronized worms. Experiments were performed at room temperature, 22°C, except those utilizing SS104, which were performed at 25°C. SS104 [glp-4 (bn2)], a temperature sensitive sterile mutant strain of C. elegans was employed in several survival assays so as to avoid confounding results due to worm propagation, i.e., second generation worms. Worms were scored every 24 h for survival. Animal survival was plotted using Kaplan-Meier survival curves and analyzed using the Gehan-Breslow-Wilcoxon method to compute P values in GraphPad Prism (GraphPad Software, Inc., La Jolla, CA). Survival assays were repeated at least three times (experiments), with each experiment having three replicates. Results from each experiment were analyzed separately. Survival curves resulting in p values of < 0.05 were considered significantly different. Primarily, survival assays were also performed using synchronized L4 stage SS104 worms; however, some experiments were repeated using other larval stages to confirm and support the results from experiments using N2 worms. For all experiments, E. coli OP 50, the normal bacterial food strain, was included as a control.
Intestinal bacterial count
To assess the degree of colonization of worms by DT104, synchronized L1 stage N2 worms were allowed to feed on either wild type Salmonella Typhimurium DT104 or isogenic ΔMDR mutant, SNS12, for 24 hours. Following this exposure, the worms were washed twice with sterile M9 buffer, then placed in NGM media supplemented with gentamicin (25µg/ml) for 20 min to kill bacteria externally attached to the worms. After the worms were washed twice with M9 buffer, they were placed onto OP 50-seeded NGM plates, and incubated for 48 hours. Aliquots of ten worms from each treatment group were collected at time 0, 24hr, and 48hr, and placed in a 2 ml screw cap tube containing 500 µl of sterile M9 buffer with 1% Triton X-100. The worms were mechanically disrupted by using a glass bead beater (Mini-Bead Beater, Biospec Products, Bartlesville, OK). The volume was adjusted with M9 medium to 1 ml and serial dilutions were plated on BHI agar media.
Microscopy
Live nematodes were mounted on an agar pad on a slide and covered with a cover glass. Sodium azide was used to anesthetize the worms. L1 stage worms were exposed to test bacteria on NGM agar plates for 72 hours at 22°C. Intestinal tracts were examined every 24 hours using a Leica TCS SP5 II confocal microscope (Leica Microsystems, Wetzlar, Germany).
Transcription level of host genes
Synchronized L1 stage N2 worms were transferred to C. elegans habitation media (CeHM) and incubated for 24 hours. The worms were washed with M9 buffer and transferred to NGM plates seeded with DT104 or SNS12 and incubated at 22°C for 72 hours. Worms were collected after 1 hr and after every 24 hours of incubation. The worms were washed in M9 buffer and RNA was extracted using TRIzol (Invitrogen/Life Technologies, Grand Island, NY). Residual genomic DNA was removed by DNase treatment (Turbo DNA-free, Ambion, Austin, TX). Three independent RNA isolations were performed for each experimental treatment for quantitative RT-PCR. cDNA was synthesized from 5 µg of total RNA using random hexamers and SuperScript II reverse transcriptase (Invitrogen/ Life Technologies, Grand Island, NY). Quantitative, or real-time, RT-PCR was performed using SYBR Advantage quantitative PCR premix (Clontech Laboratories, Mountain View, CA) and gene-specific oligonucleotide primers on a Light Cycler (BIO RAD, Hercules, CA). Primers for qRT-PCR are listed in Table 1. Relative fold-change (of exposure to either wild-type DT104 or SNS12 over OP 50) for transcripts was calculated using the comparative C T (2−ΔΔCT) method [23]. Cycle thresholds of amplification were determined by Light Cycler software (BIO RAD, Hercules, CA). All samples were run in triplicate and normalized.
Results
Multi Drug Resistance (MDR) phenotype is lost in the MDR deletion mutant of DT104
To test the direct involvement of the MDR region in virulence in Salmonella Typhimurium DT104, an isogenic mutant was constructed using allelic exchange (Figure S1). To confirm loss of antibiotic resistance in the ΔMDR isogenic mutant strain of DT104, SNS12, the mutant strain and the wild-type parental strain were assayed for resistance to sixteen antibiotics, using the Kirby-Bauer disk diffusion method (Table 2). The results clearly indicate that DT104 is resistant to ampicillin, amoxicillin, chloramphenicol, streptomycin, sulphamethoxazole, and tetracycline, but the isogenic ΔMDR mutant, SNS12, was resistant only to kanamycin, a result of introduction of the kanamycin resistance gene via allelic exchange (Table 2, Figure S1). It should be noted that generation of the ∆MDR mutation in strain SNS12 did not significantly affect growth rate, compared to the wild-type DT104 parental strain (Figure S2).
MDR region of Salmonella Typhimurium DT104 has virulence properties against C. elegans
Synchronized L4 stage C. elegans worms fed with either wild type DT104 or SNS12 exhibited a significant shortened lifespan (P<0.0001), as compared to worms fed with E. coli OP 50 (Figure 1). Further, an attenuated killing response was observed when worms were exposed to SNS12, as compared to worms exposed to wild type DT104 (P=0.0071) (Figure 1), indicating that virulence is enhanced when the MDR region is present. Similar results were observed when other larval stages, such as L1 (Figure S3), or genotypes of C. elegans were exposed to DT104 and SNS12 (Table S1). Interestingly, although both larval stages exhibited significantly shortened lifespans when exposed to wild type DT104, L1 stage worms were found to be slightly more sensitive (P<0.0001; Figure S3) than L4 stage worms (Raw data of all independent experiments are shown in Table S1).
Figure 1. Salmonella Typhimurium DT104 kills C. elegans significantly faster when MDR genes are present.
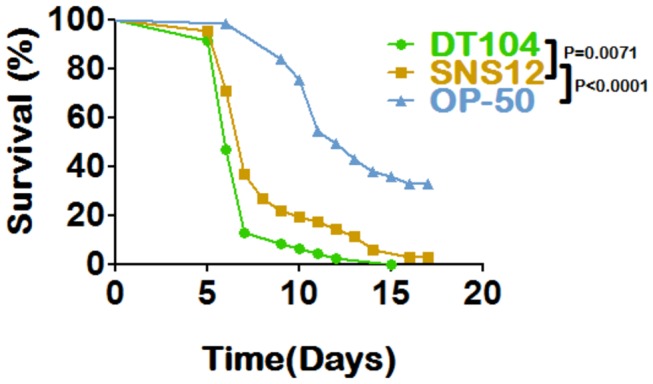
L4 stage hermaphrodite SS104 worms were exposed to wild type Salmonella Typhimurium DT104 (--●--),SNS12, a ΔMDR isogenic mutant of DT104 (--■--), and E. coli OP 50 (--▲--). P<0.001.
MDR genes induce colonization of Salmonella Typhimurium in C. elegans intestine
To explore the rate of the intestinal bacterial colonization and proliferation in C. elegans, L1 stage wild type N2 worms were exposed to both DT104 and SNS12. After exposure for 24 hours, bacterial colonization of the intestine was on the order of 104 cfu per worm for both bacterial strains (Figure 2). However, as shown in Figure 2, the extent of proliferation of DT104 in the intestine of C. elegans is significantly higher (p <0.0001) than that of SNS12 after 48 hours and 72 hours. Nearly identical results were observed when L4 stage SS104 worms were used in the assay (Figure S4). These results indicate that DT104, possessing the MDR locus, exhibits the ability to more efficiently colonize and, more importantly, proliferate in the intestine of C. elegans. To confirm the results observed by bacterial plate counts of internalized bacteria, worms were examined using confocal microscopy to determine the extent of intestinal colonization. This is easily achieved for C. elegans due to its transparent nature. As compared to SNS12 as well as E. coli OP 50, worms exposed to DT104 exhibited significant intestinal distention, indicative of bacterial proliferation (Figure 3).
Figure 2. Salmonella Typhimurium DT104 colonization in C. elegans intestine is enhanced due to MDR genes.
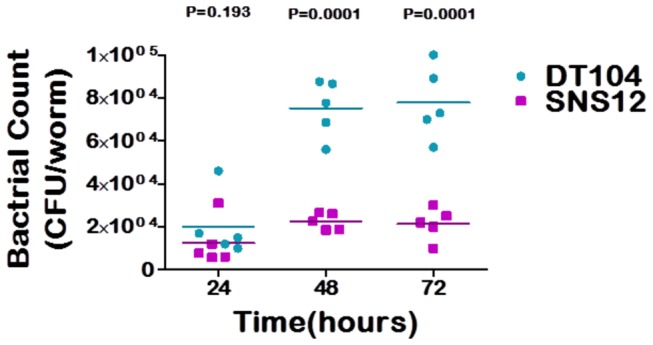
L1 stage N2 worms were exposed to DT104 (--●--) and SNS12 (--■--), and the extent of colonization was determined every 24 hours. Each data point represents the colony forming unit per worm (CFU worm-1) from a pool of 10 infected worms. Horizontal bar indicates the cumulative geometric mean of three independent experiments.
Figure 3. Colonization of the C. elegans intestine by Salmonella Typhimurium DT104.
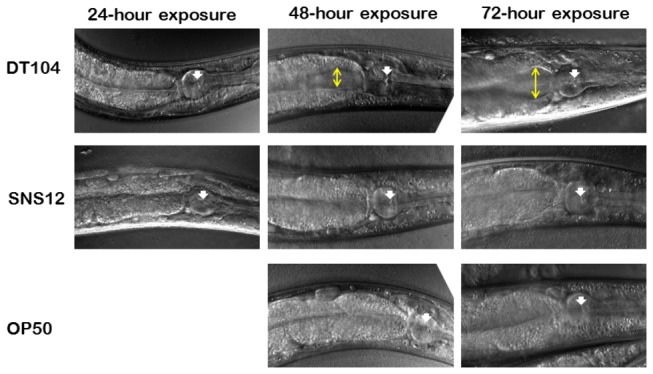
Confocal microscopy images of representative worms exposed to DT104, SNS12, and E. coli OP 50. White arrow shows the grinder of the pharynx, and yellow line shows extent of intestinal distention.
Ced-1 mutants are hyper sensitive to DT104 infection
C. elegans has evolved many pathways to recognize microorganisms and to defend itself against infection, and several of these have been identified. For example, loss-of-function mutants of the ced-1 gene in C. elegans are immune compromised and rapidly killed by live bacteria, indicating that it is a component of the innate immune response [24]. Ced-1, homologous to human CD91, a low density lipoprotein receptor, was originally discovered as a component of the programmed cell death pathway, and functions in phagocytic cells to promote cell corpse engulfment [25]. To explore whether the response in C. elegans against DT104 harboring the MDR region is ced-1 dependent, we exposed wild type N2 worms and ced-1(e1735) mutants to DT104 and and assessed their survival. We found that ced-1 (e1735) mutants die significantly faster (P<0.0085) than wild type worms, when exposed to DT104 (Figure 4) Similar results were observed when an additional ced-1 mutant C. elegans strain, e1754, was utilized in the survival assay (Figure S5). These results suggest that the immune response of C. elegans to exposure to DT104 is likely mediated through ced-1.
Figure 4. Ced-1 mutant C. elegans worms are more sensitive to killing by DT104.
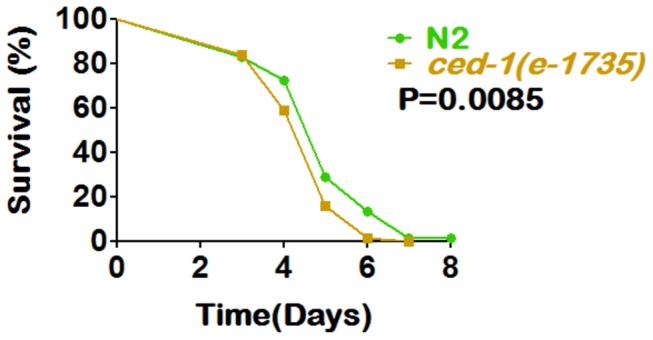
Ced-1 loss-of-function mutant worms [ced-1(e1735)] die significantly faster (P=0.0085) than wild type N2 worms, when exposed to Salmonella Typhimurium DT104. L4 stage wild type N2 (--●--) and ced-1 (e1735) (--■--) worms were exposed to DT104 and assayed daily for survival.
Expression levels of prion-like-(QN-rich)-domain-bearing genes and an antimicrobial peptide (AMP) gene after exposure to Salmonella DT104
Ced-1 regulates an alternative Unfolded Protein Response (UPR) pathway, prion-like-(QN-rich)-domain-bearing protein pathway [26], which has a potential role in C. elegans innate immunity [24]. Over-expression of prion-like-(QN-rich)-domain-bearing genes is reported to function as a protective response in C. elegans against live bacteria, like Salmonella enterica-mediated colonization and killing [24]. Additionally, several antimicrobial peptide (AMP) genes, such as abf-2 and spp-1, are expressed in the C. elegans pharynx and intestine and have been shown to have antimicrobial activity [13,27–29]. The abf-2 gene, expressed in the pharynx [28] and homologous to the antibacterial factor ASABF from Ascaris suum, has broad activity against a number of gram-positive and gram-negative bacteria, as well as yeast [13,28]. The spp-1 gene encodes a caenopore, a saponin (B) domain-containing protein, which is a member of the saponin-like protein (SAPLIP) super family. It is expressed in the intestine [30] and exhibits antimicrobial activity against bacterial species such as Salmonella Typhimurium and Pseudomonas aeruginosa [13,31].
We determined the transcriptional levels of several prion-like-(QN-rich)-domain-bearing genes, namely pqn-54, abu-1, abu-11, and antimicrobial peptide (AMP) genes, abf-2 and spp-1, in wild type N2 worms when exposed to DT104. Relative transcription levels of all genes, except spp-1, were higher when worms were exposed to either DT104 or SNS12, as compared to E. coli OP 50 (Figure 5). We did not find any significant difference in spp-1 expression levels (data not shown). As for differences between N2 worms exposed to DT104 and SNS12, we observed higher expression in earlier timepoints of exposure, 1 or 24 hours post-exposure, in worms exposed to wild type DT104 (Figure 5). This trend was found to be dependent on intact Ced-1 regulation for gene pqn-54 (Figure S6). Interestingly, this trend is reversed at 48 hours, where expression levels were found to be higher in animals exposed to SNS12 (Figure 5).
Figure 5. Antimicrobial response to wild type Salmonella Typhimurium DT104.
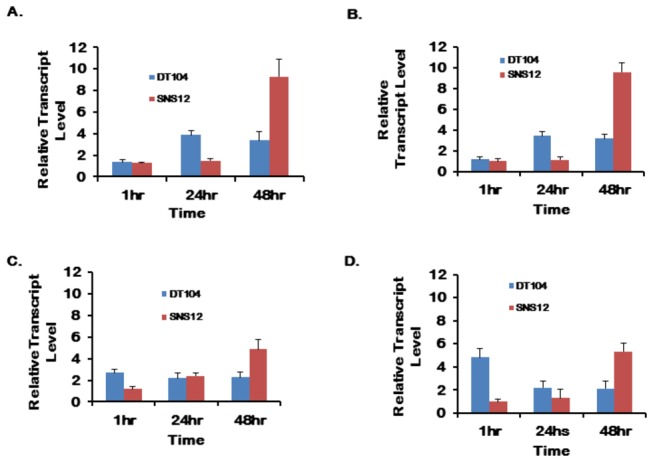
Expression of (A) pqn-54, (B) abu-1, (C) abf-2 and (D) abu-11 genes of N2 worms as determined by qRT-PCR at one hour, 24-hour and 48-hour exposure. Relative expression levels (DT104 or SNS12 over OP 50) are shown as fold changes (mean ± std. err.).
Discussion
Several cohort studies have reported that infection with antibiotic resistant Salmonella Typhimurium DT104, harboring the MDR locus, correlated with increased incidence and severity of the disease [7,8]. Inactivation of the MDR region in DT104 has been reported to reduce virulence in chickens [12]. However, results of studies using tissue culture assays and the mouse model of systematic salmonellosis indicate that the enhanced virulence of DT104 is not due to an increase in invasiveness [9–11].
In this study, we found that worms fed DT104 were killed significantly faster than those fed the isogenic ΔMDR mutant, SNS12 (Figure 1), which confirms that antibiotic resistant DT104 is more virulent. Although the difference appears to be only slightly significant, it was highly reproducible, regardless of the genotype and larval stage of worms used (Table S1).
Persistent bacterial infection in the metazoan intestine is an important aspect of pathogenesis, although the underlying molecular mechanism remains to be elucidated. Bacterial growth and proliferation in C. elegans intestine specifically has been shown to be an important hallmark of the pathogenic process [16,32]. Colonization of the intestine depends on both the ability of intact bacteria to enter the intestinal lumen [33] and the ability to survive the reaction of the host immune system. When worms are fed with food bacteria, E. coli OP 50, they digest almost all and usually no bacteria are found in their intestinal lumen beyond the pharynx. But some pathogenic bacteria, such as Salmonella Typhimurium [16], E. faecalis [32] and Vibrio cholerae (H. Cinar, unpublished data), have been shown to colonize the C. elegans intestine. Immune compromised worms are found to be susceptible to bacterial colonization and proliferation, leading to faster killing of worms [34–36]. We found that the rate of bacterial colonization and proliferation is significantly higher (P<0.0001) for wild type DT104 compared to SNS12, the ΔMDR mutant, after 24 to 72 hours (Figures 2 and 3), which indicates that the MDR locus has a significant direct role in S. Typhimurium-mediated pathogenesis in C. elegans, and that this role is most likely in intestinal proliferation.
Without further genetic investigation, the exact role of the MDR locus in virulence in DT104 isolates can only be speculated. Several possible explanations can be put forward, most of which would involve the presence of a particular protein coding gene within the MDR region. For example, the floR gene, which conveys cross-resistance to chloramphenicol and florfenicol encodes a membrane spanning efflux pump protein, which may possess other transport functions [37]. There are also a small number of gene products, namely ORF1, 2, 5, 6, as well as a groEL gene fragment, encoded in the MDR region, which currently have unknown functions. Sequence homology points to the possibility of a role in regulation for ORF1 [38]. It is plausible that this gene may be a component of the regulatory network that controls expression of the cytopathic collagenase, clg, which encodes SlyA, and possibly other yet unidentified factors [39,40].
In terms of host response, we found a consistently different trend in the expression level of several genes that play a role in innate immune response when exposed to DT104 and SNS12. The Unfolded Protein Response (UPR) signaling system is a protective mechanism which is induced in response to the overload of unfolded proteins in the endoplasmic reticulum; it reestablishes the normal state of the cell by halting protein translation and increasing levels of molecular chaperones involved in protein folding. A non-canonical, UPR signaling, prion-like-(QN-rich)-domain-bearing gene (PQN) pathway, has been found to be responsible for innate immunity in C. elegans and regulated in a ced-1-dependent manner [24]. Further, the genes, pqn/abu, are reported to be over-expressed upon exposure to S. enterica, and required for the protection of C. elegans against S. enterica-mediated killing [24]. In this study, using the C. elegans lethality assay, we have shown that ced-1 mutants were more susceptible to DT104 infection, as compared to wild-type worms, suggesting a role for ced-1 against MDR-mediated virulence (Figure 4 and Figure S5). Further, we found that the relative expression of abu-1, abu-11 and pqn-54 genes are significantly higher at the transcriptional level upon exposure to wild type S. Typhimurium DT104 after one hour (abu-11) or 24 hours (pqn-54 and abu-1). Conversely, worms exposed to SNS12 showed higher expression of all three genes after 48 hours (Figure 5). The delayed expressions of these genes in worms, after exposure to mutant strain SNS12, suggest that MDR genes in Salmonella have significant impact in early immune response. An alternative explanation is the expression, and possibly secretion, of an unidentified factor by antibiotic resistant DT104, which may have an overall affect of repression of these host genes in C. elegans. Altogether, our results suggest that the ced-1 gene and the PQN unfolded protein response pathway are involved in the immune response against exposure to wild type Salmonella Typhimurium DT104 in C. elegans, specifically during early stages of infection.
Host antimicrobial peptides (AMPs) have an important role in combating pathogenesis. Induction of several antimicrobial peptides is reported in intestinal epithelial-Paneth cells [41–43] and in the intestinal mucosa [44], following bacterial infections, including Salmonella Typhimurium. Microorganisms need to overcome AMP-mediated defense to establish persistent infection. Bacterial colonization in the intestine directly correlates with the expression of C. elegans antimicrobial peptides, such as, abf-2 and spp-1 [13]. We found abf-2 to be highly expressed in N2 worms against both wild type DT104 and ΔMDR mutant SNS12 exposure, though higher transcript level was observed earlier in DT104 (one hour), compared to SNS12 (48 hours) (Figure 5c). The data suggests that the AMP gene, abf-2, but not spp-1, is expressed at higher levels by C. elegans when exposed to DT104 harboring the multidrug resistance (MDR) region.
Overall, our findings indicate that the multiple drug resistant (MDR) genes of Salmonella enterica Typhimurium DT104 directly contribute to the virulence of this organism in C. elegans. Specifically, antibiotic resistant DT104, harboring the MDR locus, demonstrated enhanced killing of C. elegans, as well as, a much higher level of bacterial colonization and proliferation in the intestinal lumen of worms. We also found that the immune response against this genotype of Salmonella acts through the ced-1 pathway in C. elegans. Further, we observed that multiple prion-like-(QN-rich)-domain-bearing and antimicrobial peptide (AMP) genes in C. elegans show an immediate or early higher transcriptional response to wildtype DT104, as compared to its ΔMDR isogenic mutant, which further supports the role of the MDR region in virulence in DT104.
Supporting Information
Schematic representation of the construction of MDR deletion mutant in DT104 genetic background.
(TIF)
Growth curve of DT104 and SNS12 in LB media for 24 hours.
(TIF)
Salmonella Typhimurium DT104 kills C. elegans significantly faster when MDR genes are present. L1 stage hermaphrodite SS104 were exposed to wild type Salmonella Typhimurium DT104 (--●--) and SNS12, a ΔMDR isogenic mutant of DT104 (--■--). P<0.0001.
(TIF)
Salmonella Typhimurium DT104 colonization in C. elegans intestine is enhanced due to MDR genes. L4 stage SS104 worms were exposed to DT104 (--●--) and SNS12 (--■--) and the extent of colonization was determined every 24 hours. Each data point represents the colony forming unit per worm (CFU worm-1) from a pool of 10 infected worms. Horizontal bar indicates the cumulative geometric mean of three independent experiments.
(TIF)
Ced-1 mutant C. elegans worms are more sensitive to killing by DT104. Ced-1 loss-of-function mutant worms [ced-1(e1754)] die significantly faster (P=0.0001) than wild type worms (N2), when exposed to Salmonella Typhimurium DT104. L4 stage wild type N2 (--●--) and ced-1 (e1754) (--■--) worms were exposed to DT104 and assayed daily for survival.
(TIF)
Expression of gene pqn54 in C. elegans upon exposure to Salmonella Typhimurium DT104. Quantitative Real time PCR of L1 stage N2 and ced-1 (e1735) worms exposed to DT104, SNS12, and OP 50 for 24 hours.
(TIF)
Data and statistical results of replicate C. elegans survival assays when exposed to DT104 and SNS12.
(DOCX)
Acknowledgments
We are grateful to Dr. Catherine Lee, Harvard Medical School, for providing plasmid pMAK 705 and Thomas Black, CFSAN, FDA for technical help.
Funding Statement
This study was funded by Food and Drug Administration intramural funding. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Slutsker L, Altekruse SF, Swerdlow DL (1998) Foodborne diseases. Emerging pathogens and trends. Infect Dis Clin North Am 12: 199-216. doi: 10.1016/S0891-5520(05)70418-9. PubMed: 9494839. [DOI] [PubMed] [Google Scholar]
- 2. Gomez TM, Motarjemi Y, Miyagawa S, Käferstein FK, Stöhr K (1997) Foodborne salmonellosis. World Health Stat Q 50: 81-89. PubMed: 9282390. [PubMed] [Google Scholar]
- 3. Briggs CE, Fratamico PM (1999) Molecular characterization of an antibiotic resistance gene cluster of Salmonella typhimurium DT104. Antimicrob Agents Chemother 43: 846-849. doi: 10.1093/jac/43.6.846. PubMed: 10103189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Threlfall EJ (2000) Epidemic salmonella typhimurium DT 104--a truly international multiresistant clone. J Antimicrob Chemother 46: 7-10. doi: 10.1093/jac/46.1.7. PubMed: 10882682. [DOI] [PubMed] [Google Scholar]
- 5. Boyd DA, Peters GA, Ng L, Mulvey MR (2000) Partial characterization of a genomic island associated with the multidrug resistance region of Salmonella enterica Typhymurium DT104. FEMS Microbiol Lett 189: 285-291. doi: 10.1111/j.1574-6968.2000.tb09245.x. PubMed: 10930753. [DOI] [PubMed] [Google Scholar]
- 6. Boyd D, Peters GA, Cloeckaert A, Boumedine KS, Chaslus-Dancla E et al. (2001) Complete nucleotide sequence of a 43-kilobase genomic island associated with the multidrug resistance region of Salmonella enterica serovar Typhimurium DT104 and its identification in phage type DT120 and serovar Agona. J Bacteriol 183: 5725-5732. doi: 10.1128/JB.183.19.5725-5732.2001. PubMed: 11544236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Davies A, O’Neill P, Towers L, Cooke M (1996) An outbreak of Salmonella typhimurium DT104 food poisoning associated with eating beef. Commun Dis Rep CDR Rev 6: R159-R162. PubMed: 8917992. [PubMed] [Google Scholar]
- 8. Villar RG, Macek MD, Simons S, Hayes PS, Goldoft MJ et al. (1999) Investigation of multidrug-resistant Salmonella serotype typhimurium DT104 infections linked to raw-milk cheese in Washington State. JAMA 281: 1811-1816. doi: 10.1001/jama.281.19.1811. PubMed: 10340368. [DOI] [PubMed] [Google Scholar]
- 9. Allen CA, Fedorka-Cray PJ, Vazquez-Torres A, Suyemoto M, Altier C et al. (2001) In vitro and in vivo assessment of Salmonella enterica serovar Typhimurium DT104 virulence. Infect Immun 69: 4673-4677. doi: 10.1128/IAI.69.7.4673-4677.2001. PubMed: 11402014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Carlson SA, Browning M, Ferris KE, Jones BD (2000) Identification of diminished tissue culture invasiveness among multiple antibiotic resistant Salmonella typhimurium DT104. Microb Pathog 28: 37-44. doi: 10.1006/mpat.1999.0322. PubMed: 10623562. [DOI] [PubMed] [Google Scholar]
- 11. Carlson SA, Willson RM, Crane AJ, Ferris KE (2000) Evaluation of invasion-conferring genotypes and antibiotic-induced hyperinvasive phenotypes in multiple antibiotic resistant Salmonella typhimurium DT104. Microb Pathog 28: 373-378. doi: 10.1006/mpat.2000.0355. PubMed: 10839974. [DOI] [PubMed] [Google Scholar]
- 12. Randall LP, Woodward MJ (2001) Role of the mar locus in virulence of Salmonella enterica serovar Typhimurium DT104 in chickens. J Med Microbiol 50: 770-779. PubMed: 11549178. [DOI] [PubMed] [Google Scholar]
- 13. Alegado RA, Tan MW (2008) Resistance to antimicrobial peptides contributes to persistence of Salmonella typhimurium in the C. elegans intestine. Cell Microbiol 10: 1259-1273. doi: 10.1111/j.1462-5822.2008.01124.x. PubMed: 18221392. [DOI] [PubMed] [Google Scholar]
- 14. Aballay A, Ausubel FM (2002) Caenorhabditis elegans as a host for the study of host-pathogen interactions. Curr Opin Microbiol 5: 97-101. doi: 10.1016/S1369-5274(02)00293-X. PubMed: 11834377. [DOI] [PubMed] [Google Scholar]
- 15. Kurz CL, Ewbank JJ (2007) Infection in a dish: high-throughput analyses of bacterial pathogenesis. Curr Opin Microbiol 10: 10-16. doi: 10.1016/j.mib.2006.12.001. PubMed: 17178462. [DOI] [PubMed] [Google Scholar]
- 16. Aballay A, Yorgey P, Ausubel FM (2000) Salmonella typhimurium proliferates and establishes a persistent infection in the intestine of Caenorhabditis elegans. Curr Biol 10: 1539-1542. PubMed: 11114525. [DOI] [PubMed] [Google Scholar]
- 17. Aballay A, Drenkard E, Hilbun LR, Ausubel FM (2003) Caenorhabditis elegans innate immune response triggered by Salmonella enterica requires intact LPS and is mediated by a MAPK signaling pathway. Curr Biol 13: 47-52. doi: 10.1016/S0960-9822(02)01424-0. PubMed: 12526744. [DOI] [PubMed] [Google Scholar]
- 18. Labrousse A, Chauvet S, Couillault C, Kurz CL, Ewbank JJ (2000) Caenorhabditis elegans is a model host for Salmonella typhimurium. Curr Biol 10: 1543-1545. PubMed: 11114526. [DOI] [PubMed] [Google Scholar]
- 19. Méresse S, Steele-Mortimer O, Finlay BB, Gorvel JP (1999) The rab7 GTPase controls the maturation of Salmonella typhimurium-containing vacuoles in HeLa cells. EMBO J 18: 4394-4403. doi: 10.1093/emboj/18.16.4394. PubMed: 10449405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bailey AM, Ivens A, Kingsley R, Cottell JL, Wain J et al. (2010) RamA, a member of the AraC/XylS family, influences both virulence and efflux in Salmonella enterica serovar Typhimurium. J Bacteriol 192: 1607-1616. doi: 10.1128/JB.01517-09. PubMed: 20081028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sprando RL, Olejnik N, Cinar HN, Ferguson M (2009) A method to rank order water soluble compounds according to their toxicity using Caenorhabditis elegans, a Complex Object Parametric Analyzer and Sorter, and axenic liquid media. Food Chem Toxicol 47: 722-728. doi: 10.1016/j.fct.2009.01.007. PubMed: 19162123. [DOI] [PubMed] [Google Scholar]
- 22. Hamilton CM, Aldea M, Washburn BK, Babitzke P, Kushner SR (1989) New method for generating deletions and gene replacements in Escherichia coli. J Bacteriol 171: 4617-4622. PubMed: 2548993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3: 1101-1108. doi: 10.1038/nprot.2008.73. PubMed: 18546601. [DOI] [PubMed] [Google Scholar]
- 24. Haskins KA, Russell JF, Gaddis N, Dressman HK, Aballay A (2008) Unfolded protein response genes regulated by CED-1 are required for Caenorhabditis elegans innate immunity. Dev Cell 15: 87-97. doi: 10.1016/j.devcel.2008.05.006. PubMed: 18606143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhou Z, Hartwieg E, Horvitz HR (2001) CED-1 is a transmembrane receptor that mediates cell corpse engulfment in C. elegans. Cell 104: 43-56. doi: 10.1016/S0092-8674(01)00190-8. PubMed: 11163239. [DOI] [PubMed] [Google Scholar]
- 26. Urano F, Calfon M, Yoneda T, Yun C, Kiraly M et al. (2002) A survival pathway for Caenorhabditis elegans with a blocked unfolded protein response. J Cell Biol 158: 639-646. doi: 10.1083/jcb.200203086. PubMed: 12186849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Alegado RA, Campbell MC, Chen WC, Slutz SS, Tan MW (2003) Characterization of mediators of microbial virulence and innate immunity using the Caenorhabditis elegans host-pathogen model. Cell Microbiol 5: 435-444. doi: 10.1046/j.1462-5822.2003.00287.x. PubMed: 12814434. [DOI] [PubMed] [Google Scholar]
- 28. Kato Y, Aizawa T, Hoshino H, Kawano K, Nitta K et al. (2002) abf-1 and abf-2, ASABF-type antimicrobial peptide genes in Caenorhabditis elegans. Biochem J 361: 221-230. doi: 10.1042/0264-6021:3610221. PubMed: 11772394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kurz CL, Tan MW (2004) Regulation of aging and innate immunity in C. elegans. Aging Cell 3: 185-193. doi: 10.1111/j.1474-9728.2004.00108.x. PubMed: 15268752. [DOI] [PubMed] [Google Scholar]
- 30. Alper S, McBride SJ, Lackford B, Freedman JH, Schwartz DA (2007) Specificity and complexity of the Caenorhabditis elegans innate immune response. Mol Cell Biol 27: 5544-5553. doi: 10.1128/MCB.02070-06. PubMed: 17526726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Evans EA, Kawli T, Tan MW (2008) Pseudomonas aeruginosa suppresses host immunity by activating the DAF-2 insulin-like signaling pathway in Caenorhabditis elegans. PLOS Pathog 4: e1000175. doi: 10.1371/journal.ppat.1000175. PubMed: 18927620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Garsin DA, Sifri CD, Mylonakis E, Qin X, Singh KV et al. (2001) A simple model host for identifying Gram-positive virulence factors. Proc Natl Acad Sci U S A 98: 10892-10897. doi: 10.1073/pnas.191378698. PubMed: 11535834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Avery L, Shtonda BB (2003) Food transport in the C. elegans pharynx. J Exp Biol 206: 2441-2457. doi: 10.1242/jeb.00433. PubMed: 12796460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kerry S, TeKippe M, Gaddis NC, Aballay A (2006) GATA transcription factor required for immunity to bacterial and fungal pathogens. PLOS ONE 1: e77. doi: 10.1371/journal.pone.0000077. PubMed: 17183709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Singh V, Aballay A (2006) Heat-shock transcription factor (HSF)-1 pathway required for Caenorhabditis elegans immunity. Proc Natl Acad Sci U S A 103: 13092-13097. doi: 10.1073/pnas.0604050103. PubMed: 16916933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tenor JL, Aballay A (2008) A conserved Toll-like receptor is required for Caenorhabditis elegans innate immunity. EMBO Rep 9: 103-109. doi: 10.1038/sj.embor.7401104. PubMed: 17975555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Arcangioli MA, Leroy Sétrin S, Martel JL, Chaslus-Dancla E (1999) A new chloramphenicol and florfenicol resistance gene flanked by two integron structures in Salmonella typhimurium DT104. FEMS Microbiology Letters 174: 327-332. doi: 10.1111/j.1574-6968.1999.tb13586.x. [DOI] [PubMed] [Google Scholar]
- 38. Mulvey MR, Boyd DA, Olson AB, Doublet B, Cloeckaert A (2006) The genetics of Salmonella genomic island 1. Microbes Infect 8: 1915-1922. doi: 10.1016/j.micinf.2005.12.028. PubMed: 16713724. [DOI] [PubMed] [Google Scholar]
- 39. Wu MT, Carlson SA, Meyerholz DK (2002) Cytopathic effects observed upon expression of a repressed collagenase gene present in Salmonella and related pathogens: mimicry of a cytotoxin from multiple antibiotic-resistant Salmonella enterica serotype Typhimurium phagetype DT104. Microb Pathog 33: 279-287. doi: 10.1006/mpat.2002.0535. PubMed: 12495674. [DOI] [PubMed] [Google Scholar]
- 40. Carlson SA, McCuddin ZP, Wu MT (2005) SlyA regulates the collagenase-mediated cytopathic phenotype in multiresistant Salmonella. Microb Pathog 38: 181-187. doi: 10.1016/j.micpath.2005.01.004. PubMed: 15797813. [DOI] [PubMed] [Google Scholar]
- 41. Ayabe T, Satchell DP, Pesendorfer P, Tanabe H, Wilson CL et al. (2002) Activation of Paneth cell alpha-defensins in mouse small intestine. J Biol Chem 277: 5219-5228. doi: 10.1074/jbc.M109410200. PubMed: 11733520. [DOI] [PubMed] [Google Scholar]
- 42. Ayabe T, Wulff H, Darmoul D, Cahalan MD, Chandy KG et al. (2002) Modulation of mouse Paneth cell alpha-defensin secretion by mIKCa1, a Ca2+-activated, intermediate conductance potassium channel. J Biol Chem 277: 3793-3800. doi: 10.1074/jbc.M107507200. PubMed: 11724775. [DOI] [PubMed] [Google Scholar]
- 43. Ayabe T, Ashida T, Kohgo Y, Kono T (2004) The role of Paneth cells and their antimicrobial peptides in innate host defense. Trends Microbiol 12: 394-398. doi: 10.1016/j.tim.2004.06.007. PubMed: 15276616. [DOI] [PubMed] [Google Scholar]
- 44. Salzman NH, Ghosh D, Huttner KM, Paterson Y, Bevins CL (2003) Protection against enteric salmonellosis in transgenic mice expressing a human intestinal defensin. Nature 422: 522-526. doi: 10.1038/nature01520. PubMed: 12660734. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Schematic representation of the construction of MDR deletion mutant in DT104 genetic background.
(TIF)
Growth curve of DT104 and SNS12 in LB media for 24 hours.
(TIF)
Salmonella Typhimurium DT104 kills C. elegans significantly faster when MDR genes are present. L1 stage hermaphrodite SS104 were exposed to wild type Salmonella Typhimurium DT104 (--●--) and SNS12, a ΔMDR isogenic mutant of DT104 (--■--). P<0.0001.
(TIF)
Salmonella Typhimurium DT104 colonization in C. elegans intestine is enhanced due to MDR genes. L4 stage SS104 worms were exposed to DT104 (--●--) and SNS12 (--■--) and the extent of colonization was determined every 24 hours. Each data point represents the colony forming unit per worm (CFU worm-1) from a pool of 10 infected worms. Horizontal bar indicates the cumulative geometric mean of three independent experiments.
(TIF)
Ced-1 mutant C. elegans worms are more sensitive to killing by DT104. Ced-1 loss-of-function mutant worms [ced-1(e1754)] die significantly faster (P=0.0001) than wild type worms (N2), when exposed to Salmonella Typhimurium DT104. L4 stage wild type N2 (--●--) and ced-1 (e1754) (--■--) worms were exposed to DT104 and assayed daily for survival.
(TIF)
Expression of gene pqn54 in C. elegans upon exposure to Salmonella Typhimurium DT104. Quantitative Real time PCR of L1 stage N2 and ced-1 (e1735) worms exposed to DT104, SNS12, and OP 50 for 24 hours.
(TIF)
Data and statistical results of replicate C. elegans survival assays when exposed to DT104 and SNS12.
(DOCX)


