Abstract
In Arabidopsis, and other plants, the RABA GTPases (orthologous to the Rab11a of mammals) have expanded in number and diversity and have been shown to belong to eight sub clades, some of which have been implicated in controlling vesicles that traffic cell wall polymers and enzymes that synthesise or modify them to the cell wall. In order to investigate this, we have investigated whether T-DNA insertion knockouts of individual RABA genes belonging to different sub clades, impact on the composition of the plant cell wall. Single gene knockouts of the RABA1, RABA2 and RABA4 sub clades primarily affected the percentage composition of pectin, cellulose and hemicellulose within the cell wall, respectively, despite having no obvious phenotype in the whole plant. We hypothesise that vesicles carrying specific types of cargoes from the Golgi to the cell surface may be regulated by particular sub types of RABA proteins, a finding that could have wider implications for how trafficking systems work and could be a useful tool in cell wall research and other fields of plant biology.
Introduction
The cell wall contains four main components, cellulose, hemicellulose, pectin and lignin with various minor contributions from proteins and inorganic compounds [1] However, the complexity within these constituents is magnified greatly by specific polysaccharide backbone and side chain linkages. In Arabidopsis these major components are primarily pectin with two major domains homogalacturonan and rhamnogalacturonan [2] and xyloglucan and xylan hemicelluloses [3] in the primary and secondary cell walls, respectively. In addition, lignins provide a wide array of structures through different monolignols [4]. De novo synthesis of the three polysaccharide components occurs in the Golgi, for pectin and hemicellulose, while cellulose synthesis occurs at the plasma membrane. Thus, pectin and hemicellulose are directly transported through the trans-Golgi network (TGN) [3], [5], while CESA proteins, involved in cellulose synthesis are cargoed to the plasma membrane, where cellulose synthesis occurs [6].
Compartmentalisation in the cells of all organisms requires tight control and organisation. Spatial localisation of many macromolecules is controlled by RAB GTPases, which have been shown to regulate vesicle traffic to many compartments within the cell through their action as molecular switches [7]. In comparison with mammalian systems, Arabidopsis lacks some classes of RAB proteins, but others, most notably the RABA clade (orthologous to the Rab11a of mammals) has expanded in number, diversity and perhaps roles [8], [9] and various members of the RABA clade have been implicated in trafficking to the cell wall [10].
In Arabidopsis thaliana there are currently 57 defined RAB genes. These genes are split into 8 clades which are further split into sub clades of varying size dependent on the clade. The RABA clade, in particular, shows a large expansion compared to the Rab11 genes of mammalian systems. The compartmental target of most of these clades is known, mainly through localisation work [11], [12]. However, little is known about the exact role of individual RAB proteins, with redundancy often used to explain the apparent lack of visible phenotype of single gene knockouts in such studies. Since the first suggestion that a RABA1 orthologue might regulate trafficking to the cell wall [13] there has been mounting evidence that this is so for the RABA1, RABA2, RABA3 and RABA4 sub clades in plants [14]–[19] and recently it has been shown that two sub clades in Arabidopsis, RABA2 and RABA3 are associated with cell plate formation [20]. However, it has not been clear whether vesicles carrying different cargoes destined for the plasma membrane and the apoplast are regulated by different RABs. Here we have looked at whether individual mutations in RAB genes can affect the chemical composition of the plant cell wall. We have shown that mutations in different sub clades of RABA genes affected the cell wall composition in different ways and we suggest possible roles for the different RABA sub-clades.
Materials and Methods
Plant Growth
Arabidopsis thaliana Col-1 and mutant lines were grown under glass in the summer. Glasshouse conditions were; 22°C with 16 hour light and 8 hour dark period, light intensity of 150 µmol m−2s−1. Fifty plants of each line were placed in a randomised block structure. Stem material from the fifty plants was pooled at the senescent stage for analysis. Plants were grown in three successive months and treated as triplicates.
Target Gene Identification
Candidate genes were selected using a screen of publically available data through Genevestigator. Plants with T-DNA knockouts of each of the genes with any expression above the cut off of 0.2 in stem were then obtained through the Nottingham Arabidopsis Stock Centre (NASC) services.
Knockout Confirmation
To test for lack of expression of the target genes, RNA from fresh expanding stem tissue was extracted using RNeasy Mini Kit (Qiagen) and the complementary strand formed by incubation at 70°C for 5 minutes. The sample was then incubated at 37°C for 60 minutes, with M-MLV reverse transcriptase (Promega). PCR was then conducted on the complementary strand with specific primers for the transcripts. Lines were then tested for mRNA expression using PCR. AMV 5× reaction buffer (10 µl), dNTP 10 mM (1 µl), upstream and downstream primer (1 µl), MgSO4 (2 µl), AMV reverse transcriptase (1 µl), Tfl DNA polymerase (1 µl), RNA template (1 µl) as reaction mixture. Each PCR cycle comprised denaturation at 94°C for 30 seconds, annealing at 56°C for 1 minute and extension at 72°C for 1 minute. Following 30 cycles a final extension of 72°C for 5 minutes was programmed.
Fourier Transformed Infrared Spectroscopy (FT-IR)
Dry, senescent Arabidopsis stem samples were ball milled (particle size 700 microns) and placed in a Bruker, Tensor 2700 IR spectrophotometer, with 80cN-m torque applied to the sampler. Samples were read in triplicate with 128 FT-IR scans per replicate. Data from background readings of 128 scans were also captured. Data were collected through the OPUS software. The data were then normalised using vector normalisation and base line corrected using the rubber band method.
PCA was conducted using data from the FT-IR in the region of 1200 nm–800 nm which is defined as the fingerprint region for sugar composition. Statistical analysis was conducted using Minitab, multivariate analysis program.
Acetone Insoluble Solid (AIS) Production
Milled Arabidopsis stem (particle size 700 microns) (5 g), was weighed out and homogenised using pestle and mortar in 80% acetone (500 ml). The homogenate was then filtered through miracloth and washed using 80% acetone (500 ml) followed by 500 ml 100% acetone. The solid residue was then dried in a vacuum desiccator with phosphorus pentoxide.
Cell Wall Analysis
Pectin was extracted from 500 mg AIS by incubating in 120 ml 50 mM Na-1,2-cyclohexylenedinitrilo-tetraacetic acid (CDTA) pH 6, for 6 hours at room temperature. The liquid fraction was removed and the residue treated with 120 ml 50 mM Na2CO3 overnight at 2°C, after which the second liquid fraction was removed. Both liquid fractions, containing the ionically and covalently bound pectin fractions, respectively were then subjected to an uronic acid assay [21]. The remaining residue was fractionated into cellulose and hemicellulose rich fractions using 4 M KOH (10 ml), for 6 h at room temperature. The residue was washed to neutral pH with H2O and dried to give the cellulose rich fraction. The KOH extracted material was adjusted to pH 5.5 using acetic acid and precipitated with 80% acetone. Precipitated material was collected by centrifugation at 16,500 g for 10 minutes. The pellet was then washed with ethanol twice before repeating the centrifugation step and then dried to give the hemicellulose rich fraction. Recovery of each of these fractions was assessed gravimetrically.
Monomeric Composition
The hemicellulose rich or cellulose rich fraction (30 mg) was subjected to a two stage acid hydrolysis. 12 M sulphuric acid (1 ml) was added and the sample incubated for 1 hour at 37°C then diluted with 11 ml dH2O and incubated for a further 2 hours at 100°C. The sugar monomer content of the supernatant was determined by high-performance anion exchange chromatography with pulsed amperometric detection (HPAEC-PAD) (Dionex, UK) using a CarboPac PA20 column with a 50 mM NaOH isocratic system and flow rate of 0.5 ml/min at 30°C. Glucose, xylose, and galactose were used as standards with mannitol as internal standard.
Statistics
All statistics were conducted using GenStat 14th Edition, using standard ANOVA analysis to confirm significant difference between the means. In all cases the data were transformed using a log10 method, to allow data to fit the assumptions of the statistical test. A post-hoc Tukey test was used to determine significance between knockout lines and wildtype. In cases where the transformation was not adequate to the test, a t-test was used between wild-type and the knockout line.
Phenotypic Characterisation of rabA Knockouts
For phenotypic analysis, 3 replicates were grown on separate occasions with 10 plants comprising each replicate. These 10 plants were grown with a randomised block structure under conditions of 22°C with a 16 hour light and 8 hour dark period with light intensity of 150 µmol m−2s−1. Measurements taken were based on the methods described by Boyes et al., [22].
Results
The initial challenge in undertaking this study was to establish a set of single rabA gene knockouts for the genes expressed in stem tissue. To ascertain the pattern of RAB gene expression in stem tissue, the microarray analysis tool Genevestigator was selected, with the data of Zeef and Brown [23]. From these data, RAB genes shown as being expressed in stem tissue at any level above 0.2, were selected for study. The genes chosen on the basis of microarray data were RABA1a, RABA1b, RABA1c, RABA1d, RABA1f, RABA1g, RABA1h and RABA1i, the entire RABA2 sub clade, RABA3 and finally, RABA4a, RABA4b and RABA4e. Lines with T-DNA knockouts in these genes (Table S1) were obtained from the Nottingham Arabidopsis Stock Centre (NASC) [24]. Unfortunately, knockouts were not (at the time of study) available for RABA1f, RABA1g and RABA1h. Putative insert lines for RABA1b, RABA2a and RABA2c, on analysis, did not produce the expected insert band. From the remaining stocks, homozygous true breeding lines were isolated and the absence of gene transcripts was then confirmed by RT-PCR (Table S2, Figures S1–S4). A robust phenotypic analysis, designed to highlight minor changes, was then conducted. From these experiments no major or minor morphological differences were observed.
In order to investigate possible changes in the cell wall induced by the T-DNA knockouts, we chose to analyse fully senescent tissues, to avoid the possibility that changes were due to slight differences in the stage of development of the stems. Fourier transformed infrared spectroscopy (FT-IR) was used as an initial, rapid means to assess cell wall composition. FT-IR has been used to assess cell wall composition for several years [25] and, more recently; this has been reinforced with the identification of fingerprint regions for cell wall constituents [26]. However, the spectra produced have proved difficult to resolve clearly due to the complication of analysing non-fractionated samples. Because of this, the data obtained from FT-IR were used to produce a PCA plot (Figure 1). The data show the grouping of each of the rabA knockout lines as compared to the wild type. The rabA1, rabA2 and rabA4 mutant lines could be seen to form non-overlapping groups which, along with the single rabA3 mutant, were located away from the wild type.
Figure 1. Principal component analysis (PCA) of FT-IR spectra of Arabidopsis stem material.
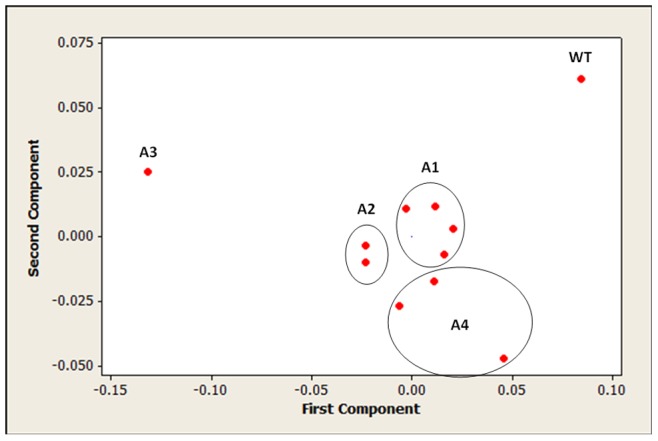
Powdered stem tissue from wild type and rabA sub clade mutant lines of Arabidopsis were subjected to analysis using FT-IR. The spectral data in the region of1200 nm–800 nm was used to generate the PCA. The total number of principle components identified was 5, PC1 = 0.59 PC2 = 0.39, with the remaining components each totalling less than 0.1 of the variation.
In order to assess whether differences seen in the PCA may have arisen from differences in cell wall polymer composition, the proportions of cellulose, hemicelluloses and pectin were assessed through fractionation. Senescent dry stem tissue was milled to a homogeneous state and the tissue was then fractionated using CDTA and NaCO3 to give ionically and covalently bound pectin fractions, respectively. The data are shown in Figure 2. Pectin was estimated as uronic acid content and levels of total uronic acid (ionic plus covalently bound) were significantly reduced, in comparison to the wild type, in the knockout lines for all four members of the RABA1 sub-clade studied. Levels of covalently bound uronic acid, in contrast, were found to be similar in all lines, with the variance in total uronic acid being due to modifications in the ionically bound fraction. The residue, following pectin extraction, was subjected to treatment with 4 M KOH in order to extract the hemicellulosic fraction. The resultant hemicellulose rich and cellulose rich fractions were both quantified gravimetrically. The results of this fractionation are displayed in Figure 3. These data show significant reduction in the mass of the cellulose rich fraction obtained from both rabA2b and rabA2d mutants compared to the wild type. There was also a significant reduction in the mass of the hemicellulose rich fraction obtained from all three rabA4 lines compared to the wild type. Mass balances were calculated for the three fractions analysed for each plant line and recoveries were found to be between 88% and 95% of the total AIS.
Figure 2. Pectin levels (as estimated by uronic acid content) in stem tissue from wild type and rabA knockout lines.
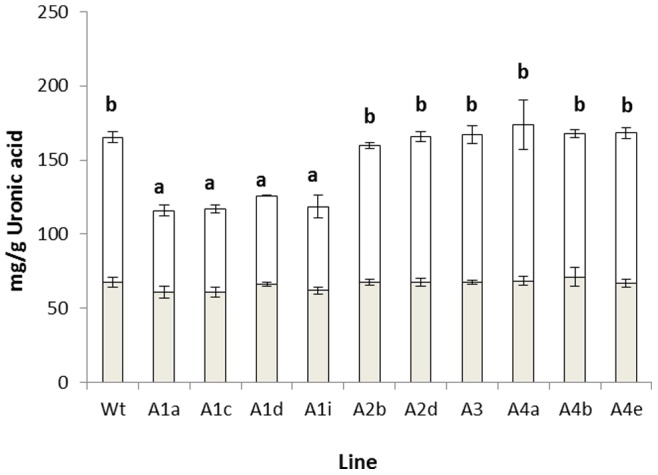
Acetone insoluble solids were sequentially extracted with CDTA and Na2CO3 to generate ionically (clear) and covalently (grey) bound pectin, respectively represented to show individual levels and additive total pectin. Significance values are as follows; (p<0.001 (d.f,32 v.r,39.13) with letters annotating significant difference between means.
Figure 3. Levels of Cellulose rich and hemicellulose rich fractions in stem tissue of wild type and rabA knockout lines.
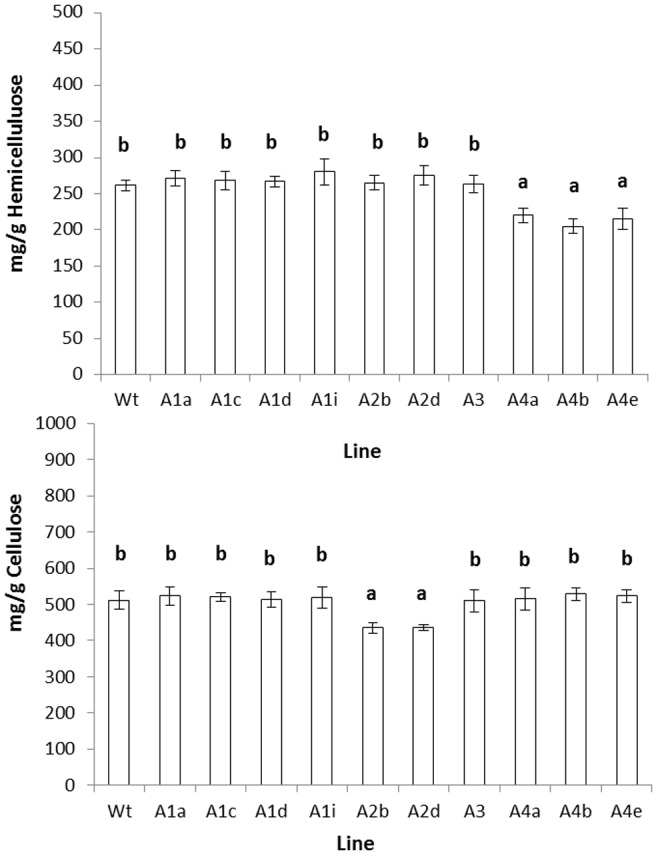
The residue from the pectin extraction was fractionated using KOH into (A) The cellulose rich fraction and (B) The hemicellulose rich fraction. Each fraction was dried and weighed. The statistical values are as follows; “cellulose rich” fraction p<0.001 (d.f,32 v.r,6.8); “hemicellulose rich” fraction p<0.001 (d.f,32 v.r,15.04) with letters annotating significant difference between means.
The hemicellulose rich and cellulose rich fractions were further characterised for monomer sugar composition by high performance anion exchange chromatography (HPAEC) following Seaman hydrolysis. The results for the cellulose rich fraction are shown in Figure 4. As expected, glucose represents the major sugar present. The observed levels of xylose however, indicate that this “cellulose rich fraction”, again as expected, probably contains a minor amount of residual hemicellulosic material. This profile is very similar to that previously reported for the cellulosic residue from Arabidopsis stems [27]. In terms of glucose content the gene knockout lines affected in the production of all of the RABA proteins have similar levels to the wild type. While this would initially seem to conflict with the gravimetric results, these data show the purity of the cellulose rich fraction and the absence of an increase in residual hemicellulose in the rabA4 knockout lines gives confidence that the patterns observed from the gravimetric analysis are not due to variances in the efficiency of the fractionation technique. The monomer composition for the hemicellulose rich fraction is shown in Figure 5. In the wild type, xylose was the predominant sugar followed by glucose and a smaller amount of galactose. The wild type thus shows a sugar profile similar to that previously described for the non-cellulosic cell wall material from Arabidopsis stems [28], however the relative proportions of these three sugars is slightly different in each case. Xylose was slightly more predominant in the previous study and this may reflect the different stage of development or analytical methods employed in this study. This profile was constant across all the rabA knockout lines. This includes the three rabA4 knockout lines that demonstrated a reduction in the total recovery of the hemicellulose rich fraction, the proportion of xylose in the hemicellulosic rich fraction from these three knockouts being similar to the control. Again this is not in conflict with the results observed in Figure 3. These results merely suggest that rabA4 knockouts have reduced the amount of hemicellulose and have not specifically impacted the composition, in terms of sugar content, of the hemicellulose within the cell wall.
Figure 4. Sugar composition of the cellulose rich fraction of wild type and rabA knockout lines.
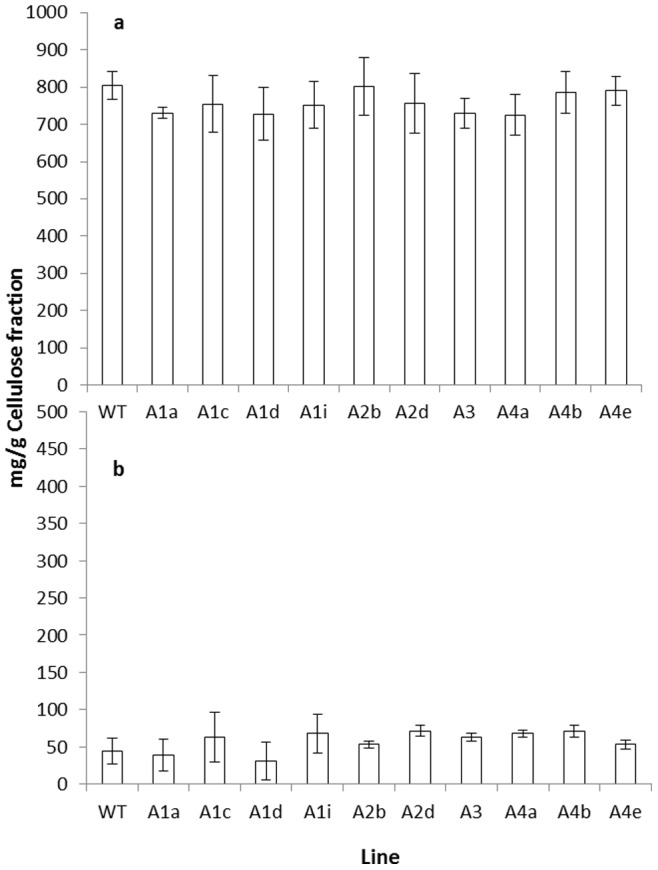
The cellulose rich fraction was subjected to hydrolysis and monomer sugar composition in terms of (A) Glucose or (B) Xylose assessed by HPAEC. Statistical values are as follows; glucose p = 0.036 (d.f,32 v.r, 2.49); xylose p = 0.115 (d.f,32 v.r,1.82).
Figure 5. Sugar composition of the hemicellulose rich fraction of wild type and rabA knockout lines.
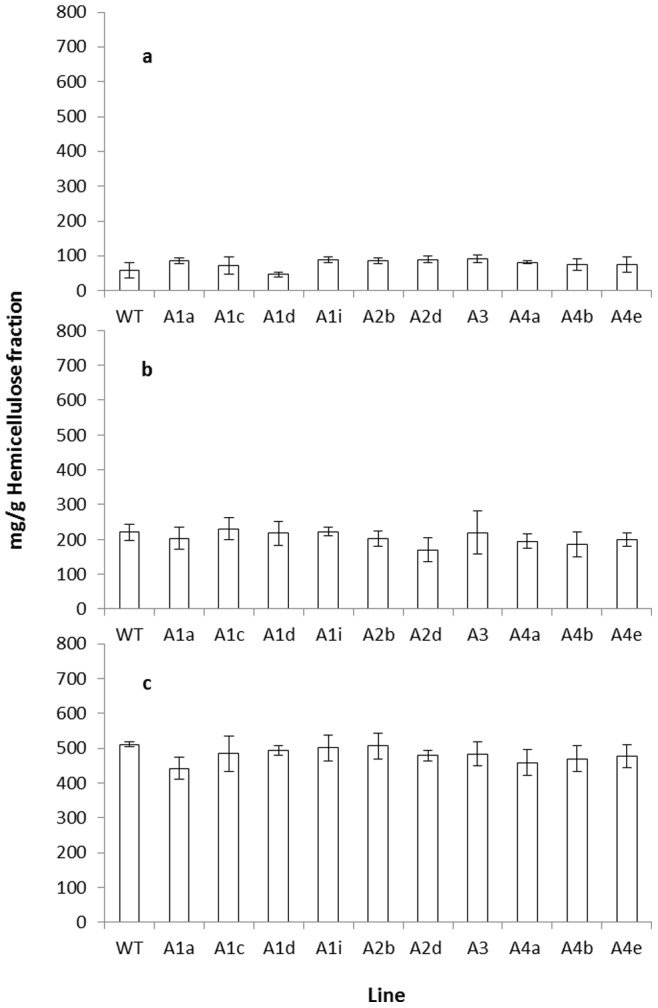
The hemicellulose rich fraction was subjected to hydrolysis and monomer sugar composition in terms of (A) Galactose, (B) Glucose and (C) Xylose assessed by HPAEC. Statistical values are as follows; glucose p = 0.0494 (d.f,32 v.r,0.97); xylose p = 0.362 (d.f,32 v.r,1.17); galactose p = 0.022 (d.f,32 v.r,2.7).
The monomeric composition of the hemicellulose and cellulose rich fractions, along with their respective recoveries from the fractionation of the AIS were used to calculate the sugar composition of the hemicellulose and cellulose rich fractions, with respect to the AIS. These data are presented in table 1. The pattern for the cellulose rich fraction is similar to that described in figure 3 with a reduction in glucose found in the rabA2 knockout lines. These data were supported by t-test analysis with both rabA2b and rabA2d having a value of p<0.05. The pattern for the hemicellulosic “rich” fraction is also similar to that shown in figure 4 with a reduction in both glucose and xylose in the rabA4 knockouts as compared to wild type, again these data were supported by t-test with p values less than 0.05 with respect to glucose levels and p<0.01 with respect to xylose.
Table 1. Sugar composition of the “hemicellulose rich” and “cellulose rich” fractions expressed as mg/g AIS.
| “Hemicellulose rich” fraction | |||||||||||
| Knockoutline | WT | A1a | A1c | A1d | A1i | A2b | A2d | A3 | A4a | A4b | A4e |
| Glucose | 57.7±6.1 | 54.9±8.6 | 61.6±8.4 | 58.0±9.0 | 62.2±3.2 | 53.5±5.9 | 46.6±9.6 | 57.8±16.1 | 42.8±4.3 | 37.9±7.2 | 42.6±4.3 |
| Xylose | 133±1.5 | 120±8.8 | 130.0±13.8 | 131.8±3.6 | 140.1±10.4 | 134. 1±9.7 | 131.8±4.2 | 127.3±9.0 | 100.9±8.0 | 96.2±7.5 | 102.7±7.0 |
| Galactose | 15.5±5.6 | 23.4±2.4 | 19.4±6.8 | 12.4±1.8 | 25.2±2.2 | 23.0±2.0 | 24.6±2.7 | 24.3±2.8 | 18.0±1.1 | 15.5±3.3 | 16.2±4.7 |
| “Cellulose rich” fraction | |||||||||||
| Knockout line | WT | A1a | A1c | A1d | A1i | A2b | A2d | A3 | A4a | A4b | A4e |
| Glucose | 411.8±19.0 | 382.3±7.9 | 392.4±39.9 | 373.3±36.3 | 389.7±32.7 | 348.5±33.9 | 330.2±34.7 | 382.5±20.4 | 373.1±28.1 | 414.9±29.3 | 413.6±20.4 |
| Xylose | 22.5±8.8 | 20.7±11.2 | 32.9±17.5 | 15.8±13.0 | 35.3±13.5 | 23.1±1.8 | 31.3±3.2 | 32.1±2.5 | 35.1±2.3 | 37.3±4.2 | 27.8±3.2 |
Discussion
Here we have demonstrated that rab single gene knockouts can affect cell wall composition. The similar result obtained for independent knockouts in several related genes rules out the possibility that these results may be due to genetic effects of the insertions themselves or to random mutations within the lines. However, it is interesting that each individual knockout gives a significant effect, which implies that these RAB proteins are not completely redundant but each RAB protein may have an independent role. This is also consistent with the finding that in a rabA4a mutant, a EYFP-RABA4b construct could not fully rescue the wild type pollen tube phenotype [18].
It has been shown that RABA2 and RABA3 proteins localise to the cell plate [20] and from this finding it was reasonable to assume that these RAB proteins were involved in cell wall deposition; however their exact role was not established. Here, using a different approach, namely the quantitative assessment of polymer content in mutants, we have presented evidence that supports the role of RABA2 proteins in cell wall metabolism and shows for the first time that RABA1, and RABA4 proteins also have independent roles in this process. The results indicate that rabA1 mutations affect pectin composition, which is consistent with the finding that pectin content is reduced in fruit of a transgenic tomato line in which a RABA orthologue has been silenced [19]. The results also indicate that rabA2 mutations affect cellulose composition and rabA4 mutations affect hemicellulose composition. Hemicellulose and pectin are made in the Golgi apparatus and transported to the cell wall [3], [5]. In the case of cellulose, biosynthetic machinery is transported to the plasma membrane and cellulose is synthesised there [6].
There are various ways in which a disruption of vesicle trafficking could affect cell wall composition. CESA proteins have been found in several post-Golgi compartments and their trafficking is complex, possibly involving recycling events [29], [30]. Similarly, pectic polysaccharides and xyloglucans may be remobilised out of the cell wall after deposition and modification [31]–[33]. Therefore, the first possibility is that recycling has been affected in our mutants, though it is difficult to see how inhibition of mobilisation from the wall could cause a reduction in the content of a particular polymer. Trafficking of cell wall remodelling enzymes has also been shown be affected by inhibition of a tomato RABA1a orthologue [14] so a change in enzymic modification of the wall is a second possible mechanism. Finally, the most direct and obvious mechanism is that trafficking of new polysaccharides to the apoplast and new CESA complexes to the plasma membrane (PM) may have been inhibited. Further work will be needed to distinguish these possible mechanisms but, whatever the mechanism, the differential effects of mutations in members of different sub clades implies that they are regulating vesicles that carry different cargoes, whether those cargoes be newly synthesised polysaccharides, recycled polymers or enzymes.
Material trafficked to the PM and the apoplast has been thought to be carried by a default bulk flow pathway [34] and in support of this, a collection of vesicles named a mobile secretory cluster has been observed moving from the trans-Golgi network to the PM and the cell plate in several plant species [35], however, there is mounting evidence that there may be multiple routes from the Golgi/TGN to the PM and the cell wall. A low concentration of brefeldin A only slightly inhibited incorporation of labelled proteins into the PM and the cell wall, whereas polysaccharides were greatly affected [36]. Conversely, interference with trafficking by means of a dominant negative mutant of the syntaxin SYP121/PEN1 reduced trafficking of secreted proteins by half but did not affect incorporation of matrix polysaccharides or cellulose synthase complexes [37]. A more recent study of two inhibitors of pectin hydrolases [38], showed that PMEI1 is transported with the aid of a GPI anchor but PEIP2 is not and neither of these is affected by a SYP121 dominant mutant, whereas secreted GFP is affected.
The role of RABA GTPases in regulating different routes has been less fully investigated, though it has been shown that tomato Rab11a, a close orthologue of RABA1a is probably involved in the pathway mediated by SYP122 and not in the one mediated by SYP121 [17]. There is also very little previous evidence to say whether different cell wall polymers travel in different vesicles, though there is one microscopic study using specific antibodies that indicates that different cell wall polymers may exit the trans-Golgi cisternae or the trans-Golgi network in different vesicles [39]. Confirmation of this will be important because of the problems in ensuring that antibody binding is specific. Despite the mounting evidence that there are different routes to the cell wall and that different cargoes may travel by different routes, we believe that our data are the first to suggest that different RAB GTPases may regulate trafficking of different cargoes.
Finally, the ability to generate plants with altered cell wall composition, which have no other obvious phenotype, may also offer a useful tool for future cell wall research.
Supporting Information
Confirmation of gene transcript absence by RT-PCR analysis of rabA1 T-DNA knockout lines. Expected size fragments for each reaction are shown in table S2. Panel A = rabA1a, Panel B = rabA1c, Panel C = rabA1d, Panel D = rabA1i. Key to lanes: M, Marker; 1, wild type RAB gene expression; 2, No RT; 3, RAB gene expression in insert line; 4, Actin expression in insert line.
(TIF)
Confirmation of gene transcript absence by RT-PCR analysis of rabA2 T-DNA knockout lines. Expected size fragments for each reaction are shown in table S2. Panel A = rabA2b, Panel B = rabA2d. Key to lanes: M, Marker; 1, wild type RAB gene expression; 2, No RT; 3, RAB gene expression in insert line; 4, Actin expression in insert line.
(TIF)
Confirmation of gene transcript absence by RT-PCR analysis of rabA3 T-DNA knockout line. Expected size fragments for each reaction are shown in table S2. Key to lanes: M, Marker; 1, wild type RAB gene expression; 2, No RT; 3, RAB gene expression in insert line; 4, Actin expression in insert line.
(TIF)
Confirmation of gene transcript absence by RT-PCR analysis of rabA2 T-DNA knockout lines. Expected size fragments for each reaction are shown in table S2. Panel A = rabA4a, Panel B = rabA4b Panel C = rabA4e. Key to lanes: M, Marker; 1, wild type RAB gene expression; 2, No RT; 3, RAB gene expression in insert line; 4, Actin expression in insert line.
(TIF)
Details of NASC stock lines.
(TIF)
Details of RT-PCR primers.
(TIF)
Funding Statement
DL was supported by a Targetted Priority Studentship in Bioenergy (number BB/G017964/1) from BBSRC (http://www.bbsrc.ac.uk). SRG, as a part of LACE (Lignocellulose agricultural waste to ethanol), was also supported by a BBSRC grant (number BB/G01616X/1). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Carpita N, McCann M (2000) The cell wall. In: Buchanan BB, Gruissem W, Jones RL, editors. Biochemistry and Molecular Biology of Plants. Rockville: American Society Plant Physiologists. 52–108.
- 2. Mohnen D (2008) Pectin structure and biosynthesis. Curr Opin Plant Biol 11: 266–277. [DOI] [PubMed] [Google Scholar]
- 3. Scheller HV, Ulvskov P (2010) Hemicelluloses. Annu Rev Plant Biol 61: 263–289. [DOI] [PubMed] [Google Scholar]
- 4. Boerjan W, Ralph J, Baucher M (2003) Lignin biosynthesis Annu Rev Plant Biol. 54: 519–546. [DOI] [PubMed] [Google Scholar]
- 5. Caffall KH, Mohnen D (2009) The structure, function, and biosynthesis of plant cell wall pectic polysaccharides. Carbohydr Res 344: 1879–1900. [DOI] [PubMed] [Google Scholar]
- 6. Wightman R, Turner S (2010) Trafficking of the plant cellulose synthase complex. Plant Physiol 153: 427–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schimmoller F, Simon I, Pfeffer SR (1998) Rab GTPases, directors of vesicle docking. J Biol Chem 273: 22161–22164. [DOI] [PubMed] [Google Scholar]
- 8. Pereira-Leal JB, Seabra MC (2001) Evolution of the Rab family of small GTP-binding proteins. J Mol Biol 313: 889–901. [DOI] [PubMed] [Google Scholar]
- 9. Rutherford S, Moore I (2002) The Arabidopsis Rab GTPase family: another enigma variation. Curr Opin Plant Biol 5: 518–528. [DOI] [PubMed] [Google Scholar]
- 10. Lycett G (2008) The role of Rab GTPases in cell wall metabolism. J Exp Bot 59: 4061–4074. [DOI] [PubMed] [Google Scholar]
- 11. Nielsen E, Cheung AY, Ueda T (2008) The regulatory RAB and ARF GTPases for vesicular trafficking. Plant Physiol 147: 1516–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Woollard AD, Moore I (2008) The functions of Rab GTPases in plant membrane traffic. Curr Opin Plant Biol 11: 610–619. [DOI] [PubMed] [Google Scholar]
- 13. Zainal Z, Tucker GA, Lycett GW (1996) A rab11-like gene is developmentally regulated in ripening mango (Mangifera indica L) fruit. Biochim Biophys Acta 1314: 187–190. [DOI] [PubMed] [Google Scholar]
- 14. Lu C, Zainal Z, Tucker GA, Lycett GW (2001) Developmental abnormalities and reduced fruit softening in tomato plants expressing an antisense Rab11 GTPase gene. Plant Cell 13: 1819–1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cheung AY, Chen CY, Glaven RH, de Graaf BHJ, Vidali L, et al. (2002) Rab2 GTPase regulates vesicle trafficking between the endoplasmic reticulum and the Golgi bodies and is important to pollen tube growth. Plant Cell 14: 945–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. de Graaf BHJ, Cheung AY, Andreyeva T, Levasseur K, Kieliszewski M, et al. (2005) Rab11 GTPase-regulated membrane trafficking is crucial for tip-focused pollen tube growth in tobacco. Plant Cell 17: 2564–2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rehman RU, Stigliano E, Lycett GW, Sticher L, Sbano F, et al. (2008) Tomato Rab11a characterization evidenced a difference between SYP121-dependent and SYP122-dependent exocytosis. Plant Cell Physiol 49: 751–766. [DOI] [PubMed] [Google Scholar]
- 18. Szumlanski L, Nielsen E (2009) The Rab GTPase RabA4d regulates pollen tube tip growth in Arabidopsis thaliana . Plant Cell 21: 526–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lunn D, Phan TD, Tucker GA, Lycett GW (2013) Cell wall composition of tomato fruit changes during development and inhibition of vesicle trafficking is associated with reduced pectin levels and reduced softening. Plant Physiol Biochem 66: 91–97. [DOI] [PubMed] [Google Scholar]
- 20. Chow M, Neto H, Foucart C, Moore I (2008) Rab-A2 and Rab-A3 GTPases define a trans-Golgi endosomal membrane domain in Arabidopsis that contributes substantially to the cell plate. Plant Cell 20: 101–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Filisetti-Cozzi TMC, Carpita NC (1991) Measurement of uronic acids without interference from neutral sugars. Anal Biochem 197: 157–162. [DOI] [PubMed] [Google Scholar]
- 22. Boyes DC, Zayed AM, Ascenzi R, McCaskill AJ, Hoffman NE, et al. (2001) Growth stage-based phenotypic analysis of Arabidopsis: A model for high throughput functional genomics in plants. Plant Cell 13: 1499–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zeef LAH, Brown DE (2006) MEXP-265 Transcription profiles from Arabidopsis stem, leaf and hypocotyls tissue. Publically available microarray data: http://www.ebi.ac.uk/arrayexpress/experiments/E-MEXP-265 accessed on 15th December 2009.
- 24. Scholl RL, May ST, Ware DL (2000) Seed and molecular resources for Arabidopsis . Plant Physiol 124: 145–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chen LM, Carpita NC, Reiter WD, Wilson RH, Jefferies C, et al. (1998) A rapid method to screen for cell-wall mutants using discriminant analysis of Fourier transform infrared spectra. Plant J 16: 385–392. [DOI] [PubMed] [Google Scholar]
- 26. Kacuráková M, Capek P, Sasinkova V, Wellner N, Ebringerova A (2000) FT-IR study of plant cell wall model compounds: pectic polysaccharides and hemicelluloses. Carbohydr Polym 43: 195–203. [Google Scholar]
- 27. Brown DE, Wightman R, Zhang Z, Gomez LD, Atanassov I, et al. (2011) Arabidopsis genes irregular xylem (irx15) and irx15L encode DUF579-containing proteins that are essential for normal xylan deposition in the secondary cell wall. Plant J 66: 401–413. [DOI] [PubMed] [Google Scholar]
- 28. Brown DE, Zeef LAH, Ellis J, Goodacre R, Turner SR (2005) Identification of novel genes in Arabidopsis involved in secondary cell wall formation using expression profiling and reverse genetics. Plant Cell 17: 2281–2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Crowell EF, Bischoff V, Desprez T, Rolland A, Stierhof Y, et al. (2009) Pausing of Golgi bodies on microtubules regulates secretion of cellulose synthase complexes in Arabidopsis. Plant Cell 21: 1141–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gutierrez R, Lindeboom JJ, Paredez AR, Emons AM, Einhardt DW (2009) Arabidopsis cortical microtubules position cellulose synthase delivery to the plasma membrane and interact with cellulose synthase trafficking compartments. Nature Cell Biol. 11: 797–806. [DOI] [PubMed] [Google Scholar]
- 31. Baluška F, Hlavacka A, Šamaj J, Palme K, Robinson DG, et al. (2002) F-Actin-dependent endocytosis of cell wall pectins in meristematic root cells. Insights from brefeldin A-induced compartments. Plant Physiol 130: 422–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Baluška F, Liners F, Hlavacka A, Schlicht P, Van Cutsem P, et al. (2005) Cell wall pectins and xyloglucans are internalised into dividing root cells and accumulate within cell plates during cytokinesis. Protoplasma 225: 141–155. [DOI] [PubMed] [Google Scholar]
- 33. Dhonukshe P, Baluška F, Schlicht M, Hlavacka A, Šamaj J, et al. (2006) Endocytosis of cell surface material mediates cell plate formation during plant cytokinesis. Dev Cell 10: 137–150. [DOI] [PubMed] [Google Scholar]
- 34. Jürgens G (2004) Membrane trafficking in plants. Annu Rev Cell Dev Biol 20: 481–504. [DOI] [PubMed] [Google Scholar]
- 35. Toyooka K, Goto Y, Asatsuma S, Koizumi M, Misui T, et al. (2009) A mobile secretory vesicle cluster involved in mass transport from the Golgi to the plant exterior. Plant Cell 21: 1212–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lanubile R, Piro G, Dalessandro G (1997) Effect of brefeldin A on the synthesis and transport of cell wall polysaccharides and proteins in pea root seedlings. J Exp Bot 48: 1925–1933. [Google Scholar]
- 37. Leucci MR, Di Sansebastiano GP, Gigante M, Dalessandro G, Piro G (2007) Secretion marker proteins and cell-wall polysaccharides move through different secretory pathways. Planta 225: 1001–1017. [DOI] [PubMed] [Google Scholar]
- 38. De Caroli M, Lenucci MS, Di Sansebastiano GP, Dalessandro G, De Lorenzo G, et al. (2011) Protein trafficking to the cell wall occurs through mechanisms distinguishable from default sorting in tobacco. Plant J 65: 295–308. [DOI] [PubMed] [Google Scholar]
- 39. Moore PJ, Swords KM, Lynch MA, Staehelin LA (1991) Spatial organization of the assembly pathways of glycoproteins and complex polysaccharides in the Golgi apparatus in plants. J Cell Biol 114: 589–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Confirmation of gene transcript absence by RT-PCR analysis of rabA1 T-DNA knockout lines. Expected size fragments for each reaction are shown in table S2. Panel A = rabA1a, Panel B = rabA1c, Panel C = rabA1d, Panel D = rabA1i. Key to lanes: M, Marker; 1, wild type RAB gene expression; 2, No RT; 3, RAB gene expression in insert line; 4, Actin expression in insert line.
(TIF)
Confirmation of gene transcript absence by RT-PCR analysis of rabA2 T-DNA knockout lines. Expected size fragments for each reaction are shown in table S2. Panel A = rabA2b, Panel B = rabA2d. Key to lanes: M, Marker; 1, wild type RAB gene expression; 2, No RT; 3, RAB gene expression in insert line; 4, Actin expression in insert line.
(TIF)
Confirmation of gene transcript absence by RT-PCR analysis of rabA3 T-DNA knockout line. Expected size fragments for each reaction are shown in table S2. Key to lanes: M, Marker; 1, wild type RAB gene expression; 2, No RT; 3, RAB gene expression in insert line; 4, Actin expression in insert line.
(TIF)
Confirmation of gene transcript absence by RT-PCR analysis of rabA2 T-DNA knockout lines. Expected size fragments for each reaction are shown in table S2. Panel A = rabA4a, Panel B = rabA4b Panel C = rabA4e. Key to lanes: M, Marker; 1, wild type RAB gene expression; 2, No RT; 3, RAB gene expression in insert line; 4, Actin expression in insert line.
(TIF)
Details of NASC stock lines.
(TIF)
Details of RT-PCR primers.
(TIF)


