Abstract
Objective
Biomarkers capable of discriminating the patients who are likely to respond to certain chemotherapeutic agents could improve the clinical efficiency. The sulfatases(SULFs) play a critical role in the pathogenesis of a variety of human cancers. Here, we focused our investigation on the prognostic and predictive impact of SULF2 methylation in gastric cancer.
Methods
Promoter CpG island methylation of SULF2 was analyzed in 100 gastric cancer samples. The in vitro sensitivity to cisplatin, docetaxel, gemcitabine, irinotecan and pemetrexed were determined by histoculture drug response assay(HDRA). Additionally, 56 gastric cancer patients treated with a modified FOLFOX regimen(biweekly oxaliplatin plus 5-FU and folinic acid) were retrospectively analyzed to further evaluate the prognostic and predictive impact of SULF2 methylation in gastric cancer.
Results
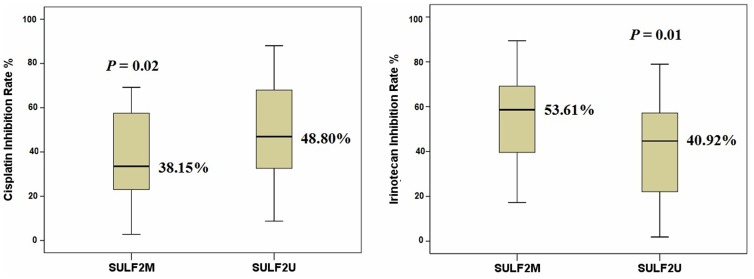
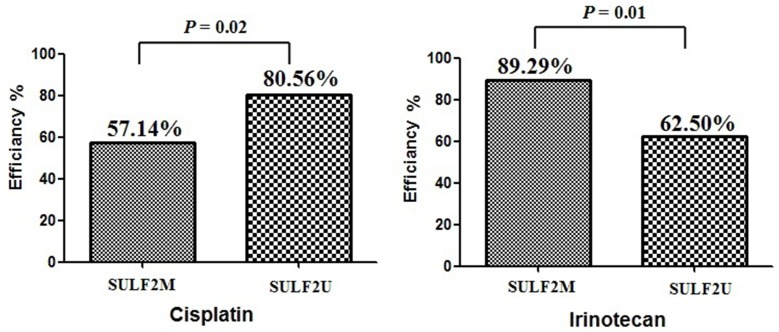
Methylated SULF2(SULF2M) was detected in 28 patients, while the remaining 72 patients showed unmethylated SULF2(SULF2U, methylation rate: 28%). Samples with SULF2U were more sensitive to cisplatin than those with SULF2M(inhibition rate: 48.80% vs. 38.15%, P = 0.02), while samples with SULF2M were more sensitive to irinotecan than SULF2U(inhibition rate: 53.61% vs. 40.92%, P = 0.01). There were no association between SULF2 methylation and in vitro sensitivity to docetaxel, gemcitabine and pemetrexed. SULF2 methylation was found to have a significant association with cisplatin efficacy(SULF2M: 57.14%, SULF2U: 80.56%, P = 0.02) and irinotecan efficacy(SULF2M: 89.29%, SULF2U: 62.50%, P = 0.01). Among the 56 patients receiving the modified FOLFOX regimen, a significant association was observed between survival and SULF2 methylation status(SULF2M: 309 days, 95% CI = 236 to 382 days; SULF2U: 481 days, 95% CI = 418 to 490 days; P = 0.02). Multivariate analysis revealed that SULF2 methylation was an independent prognostic factor of overall survival in gastric cancer patients treated with platinum-based chemotherapy.
Conclusion
SULF2 methylation is negatively associated with cisplatin sensitivity in vitro. SULF2 methylation may be a novel prognostic biomarker for gastric cancer patients treated with platinum-based chemotherapy.
Introduction
Gastric cancer is one of the most frequent causes of cancer-related death worldwide [1], [2]. Multimodal treatment protocols, mainly based on platinum and 5-fluorouracil (5-FU), have been shown to prolong patient survival; however, with any combination of chemotherapeutic agents, the response rate is only approximately 30%–50% [3], [4]. In an attempt to improve the clinical efficiency, it is important and necessary to identify biomarkers capable of discriminating the patients who are likely to respond to certain chemotherapeutic agents [4]–[9].
Heparan sulfate proteoglycans (HSPGs) are coreceptors for heparin-binding growth factors and cytokines distributed on the cell surface and in the extracellular matrix. Two isoforms of the extracellular heparan sulfate 6-O-endosulfatases (SULFs), SULF1 and SULF2, have been discovered in mammals. Both proteins are secreted to the cell surface to modulate the sulfation of HSPGs [10]. Although previous reports have unequivocally highlighted the critical role that SULFs play in the pathogenesis of a variety of human cancers, the opinions on the role of SULFs in cancer development have been somewhat polarized. Several evidences show that SULFs are negative regulators of tumor cell growth, and that overexpression of SULFs in tumor cells inhibits cell growth by deregulating several factors, including FGF-2, HB-EGF and HGF [11]–[13]. Other studies hold the view that SULFs are positive regulators of oncogenetic signaling pathways, including Wnt, BMP, Hedgehog and GDNF [14]–[16], and increased expression of SULFs is prevalent in various cancers, including gastric, hepatocellular, pancreatic and breast cancers, non-small cell lung cancer (NSCLC) and head and neck tumors [10], [13], [17]–[20]. High expression of SULF2 has been linked to poor survival in patients with hepatocellular carcinoma and NSCLC [18], [20]. The available evidence on the methylation status and expression levels of SULFs in gastric cancer, however, are non-conclusive, and the prognostic or predictive value of SULFs for chemosensitivity prediction remains unknown.
In this study, we have analyzed the promoter CpG island methylation of SULF2, the gene encoding the SULF2 endosulfatase, and its association with in vitro sensitivity to cisplatin, docetaxel, gemcitabine, irinotecan, and pemetrexed in 100 human gastric cancer samples. To this end, we performed a series of in vitro sensitivity tests on freshly-removed gastric tumor tissues and evaluated the possible use of SULF2 methylation status for predicting the chemotherapeutic efficacy of there agents. Then, we retrospectively analyzed the SULF2 methylation in a cohort of 56 gastric cancer patients treated with a modified FOLFOX regimen and concluded that SULF2U serves as an independent prognostic biomarker in gastric cancer patients treated with a modified FOLFOX regimen.
Materials and Methods
Ethics Statement
All research involving human participants have been approved by the Human Research Protective Committee of Drum Tower Hospital Affiliated to Medical School of Nanjing University and written informed consent was obtained from all patients.
Patients and Tissue Samples
Enrolled patients were those undergoing the gastrectomy in the Department of General Surgery of the Drum Tower Hospital, Nanjing, China during the period from August 2010 to October 2011. Eligibility criteria for enrollment into the study included the following parameters: (1) age >18 years; (2) histologically confirmed gastric adenocarcinoma, mucinous or signet ring cell adenocarcinoma; (3) no previous or concomitant malignancies other than gastric cancer; (4) no previous history of chemotherapy or radiotherapy, either adjuvant or palliative; and (5) adequate function of all major organs. Tissue samples were extracted from 100 freshly-removed gastric tumors. Each tumor tissue was divided into two parts: (1) one part was kept in 4°C Hanks’ balanced salt solution with 1% penicillin/streptomycin after collection, and then sent to the laboratory within 15 min in 4°C, for in vitro evaluation of chemosensitivity by histoculture drug response assay (HDRA) [21], [22]; (2) the remaining part was made into formalin-fixed paraffin-embedded (FFPE) tumor blocks for pathological observation and methylation detection. We retrospectively reviewed the patients’ medical records and surgery reports to identify clinical and histopathological data, including sex, histology, tumor site, stage, histological grade and lymph node metastasis. Tumors were classified according to the International Union Against Cancer (UICC) TNM Classification. Written informed consent was obtained from all patients and the protocols for this study were approved by the Human Research Protective Committee of the Drum Tower Hospital.
HDRA
HDRA procedures were performed as previously reported by Furukawa and colleagues [21], [22]. Briefly, the fresh tumor tissues were washed twice with Hanks’ balanced salt solution and minced into small pieces of approximately 10 mg in weight and 0.5 mm in diameter, which were then placed on prepared collagen (Health Design, Rochester, NY) surfaces in 24-well microplates. Optimal concentrations of the drugs used to distinguish in vitro sensitivity and resistance were 20 ug/ml for cisplatin [23], 100 ug/ml for docetaxel [22], 50 ug/ml for gemcitabine [23], 20 ug/ml for irinotecan [23], and 400 ug/ml for pemetrexed [5], according to its peak plasma concentration in patients. Cisplatin, docetaxel, and irinotecan were obtained from Jiangsu Hengrui Medicine Company (Jiangsu, China), while pemetrexed and gemcitabine were obtained from Eli Lilly and Company (Shanghai, China). 8 parallel culture wells were used for each drug concentration, as well as for control. Plates were incubated for 7 days at 37°C in the presence of drugs dissolved in RPMI 1640 medium containing 20% fetal calf serum and left in a humidified atmosphere containing 95% air–5% CO2. After histoculture, 100 µl of type I collagenase (0.1 mg/ml, Sigma, Shanghai, China) and 100 µl of 3-(4,5-Dimethyl-2-thiazotyl)-2,5-diphenyl-2H- tetrazolium bromide (MTT) solution (5 mg/ml, Sigma, Shanghai, China) were added to each culture well and incubated for another 16 hours. After extraction with dimethyl sulfoxide, the absorbance of the solution in each well was read at 540 nm.
Evaluation of Sensitivity to Anti-cancer Agents in HDRA
The absorbance per gram of cultured tumor tissue was calculated from the mean absorbance of tissue from 8 parallel culture wells, and the tumor tissue weight was determined before culture. The inhibition rate of each anti-cancer agent was calculated by using the following formula: Inhibition rate (%) = (1–T/C) ×100, where T is the mean absorbance of treated tumor / Weight and C is the mean absorbance of control tumor / Weight. The cut-off inhibition rates used to distinguish the sensitive and the resistant cases were adopted at 30%, 40%, 50%, and 60% for each drug tested. The in vitro efficacy rate was also estimated based on the cut-off inhibition rate as follows: efficacy rate (%) = number of sensitive cases/total number of cases.
Patients’ Chemotherapy
Of all patients, 56 with Eastern Cooperative Oncology Group (ECOG) performance status (PS) ≤2 received a modified FOLFOX (a combination of 5-FU and platinum) chemotherapy regimen after resection of primary tumors as follows: oxaliplatin 85 mgm−2 plus folinic acid 200 mgm−2 as a 2 h infusion on day 1, followed by a 22 h infusion of 5-FU 2000 mgm−2 on days 1 and 2, every two weeks. These 56 patients were further followed up and their survival time was calculated from the date of diagnosis to the date of the last follow-up or death from any cause.
DNA Extraction and Modification
Three 7-µm sections were prepared from primary tumor blocks that contained at least 80% tumor cells. After hematoxylin-eosin staining, the cancerous parts were microdissected and transferred into a microcentrifuge tube. DNA was isolated routinely and then was chemically modified by sodium bisulphite to convert all unmethylated cytosines to uracils while leaving methylcytosines unaltered [18]. Then they were stored at −20°C for further analysis.
Methylation-Specific Polymerase Chain Reaction (MSP)
MSP was performed to determine the methylation status of SULF2 using the ABI Prism 7300HT Sequence Detection System (Applied Biosystems). Each PCR reaction contained genomic DNA 2 µl, SYBR Green PCR Mix (TaKaRa, Japan) 10 µl, water 7.7 µl, and primers 0.15 µl (10 µmol/l). The PCR conditions were 95°C for 10 min, followed by 45 cycles at 59°C for 30 s, 72°C for 30 s and 95°C for 30 s. Primers for SULF2 methylated PCR (TaKaRa, Japan) were as follows: forward 5′ TAAGTGTTTTTTTTATAGCGGC 3′, reverse 5′TACCGTAATTTCCGCTATC 3′. Primers for SULF2 unmethylated PCR (TaKaRa, Japan) were as follows: forward 5′ GTTTATAAGTGTTTTTTTATAGTGGT3’, reverse 5′TACCATAATTTCCACTATCCCT 3′. Each batch of reaction included a positive control from Methyltransferase (M.SssI)-treated human genomic DNA (fully methylated), a negative control from DNA samples which had been confirmed as unmethylated and another negative control without DNA. All tests were performed in duplicate. Results were validated for selected samples through combined bisulfite modification and bisulfite sequencing.
Statistical Analysis
Values were expressed as means ± standard deviation. Differences in inhibition rates between groups were evaluated using the t-test. The possible associations of SULF2 methylation with clinical characteristics and in vitro chemosensitivity efficacy were analyzed using the Fisher’s exact probability test. All statistical tests were two-sided, and a P value of less than 0.05 was considered as statistical significance. Statistical analysis was performed using the SPSS, version 16.0.
Results
Patients’ Characteristics
The characteristics of all patients are shown in Table 1. The majority of patients were males (73%), and the predominant histology was adenocarcinoma (79%). In 35 (35%) patients, the tumor was located in the distal stomach, in 38 (38%) in the proximal stomach, and in 27 (27%) in the whole stomach. Sixty-three (63%) patients had stage III or IV disease. Lymph node metastases were present in 75 (75%) patients. Methylated SULF2 (SULF2M) was detected in 28 patients, while the remaining 72 patients showed unmethylated SULF2 (SULF2U, methylation rate: 28%). The RT-PCR amplification curves of SULF2M and SULF2U were shown in Figure S1. There was no association between SULF2 methylation and any of the patients’ characteristics, including sex, histology, tumor site, stage, histological grade and lymph node metastasis.
Table 1. Patient characteristics.
| Characteristic | Patients | SULF2 | ||
| N = 100, N (%) | SULF2M | SULF2U | P Value | |
| Age | ||||
| >62 | 52 (52%) | 14 | 38 | 0.83 |
| ≤62 | 48 (48%) | 14 | 34 | |
| Sex | ||||
| Male | 73 (73%) | 21 | 52 | 0.49 |
| Female | 27 (27%) | 7 | 20 | |
| Histology | ||||
| Adenocarcinoma | 79 (79%) | 24 | 55 | 0.59 |
| Mucinous | 11 (11%) | 2 | 9 | |
| Signet ring cell | 10 (10%) | 2 | 8 | |
| Tumor Site | ||||
| Distal | 35 (35%) | 6 | 29 | 0.21 |
| Proximal | 38 (38%) | 13 | 25 | |
| Whole stomach | 27 (27%) | 9 | 18 | |
| Stage | ||||
| I,II | 37 (37%) | 8 | 29 | 0.36 |
| III, IV | 63 (63%) | 20 | 43 | |
| Histological grade | ||||
| 2 | 27 (27%) | 5 | 22 | 0.23 |
| 3 | 44 (44%) | 16 | 28 | |
| Mixed 2–3 | 29 (29%) | 7 | 22 | |
| Lymph node metastasis | ||||
| No | 25 (25%) | 5 | 20 | 0.44 |
| Yes | 75 (75%) | 23 | 52 | |
In vitro Efficacy Rate of Tested Drugs
In vitro sensitivity to cisplatin, docetaxel, gemcitabine, irinotecan and pemetrexed was successfully tested in all the samples. The mean inhibition rates for each tested drug are listed in Table 2. Samples with SULF2U were more sensitive to cisplatin than those with SULF2M (48.80% vs. 38.15%, P = 0.02), while samples with SULF2M were more sensitive to irinotecan than SULF2U (53.61% vs. 40.92%, P = 0.01, Table 2 and Figure 1). There was no association between SULF2 methylation and in vitro sensitivity to docetaxel, gemcitabine or pemetrexed (Table 2). As shown in Table 3, there was no association between the inhibition rates observed for the anti-cancer agents and any of the clinical characteristics.
Table 2. Descriptive statistics of investigated drugs.
| Chemotheraputic agents | Number of samples | Inhibition rate (%) | SULF2 | |||
| Mean ± SD | Range | SULF2M (n = 28) | SULF2U (n = 72) | P Value | ||
| Cisplatin | 100 | 45.94±21.02 | 2.76–88.02 | 38.15% (30.27%–46.03%) | 48.80% (43.94%–53.65%) | 0.02 |
| Docetaxel | 100 | 45.69±22.96 | 4.19–88.40 | 45.25% (35.89%–54.60%) | 45.86% (40.41%–51.32%) | 0.91 |
| Gemcitabine | 100 | 44.72±21.78 | 3.55–88.97 | 48.13% (38.85%–57.41%) | 43.45% (37.88%–49.02%) | 0.38 |
| Irinotecan | 100 | 46.22±22.59 | 1.86–89.42 | 53.61% (45.17%–62.04%) | 40.92% (35.74%–46.10%) | 0.01 |
| Pemetrexed | 100 | 52.79±22.40 | 3.60–89.46 | 54.78% (45.18%–64.39%) | 52.01% (46.95%–57.07%) | 0.58 |
Figure 1. Samples with SULF2U were more sensitive to cisplatin and those with SULF2M (48.80% vs. 38.15%, P = 0.02, n = 100, t-test); samples with SULF2M were more sensitive to irinotecan than SULF2U (53.61% vs. 40.92%, P = 0.01, n = 100, t-test).
The median is the central line in each box.
Table 3. Association between inhibition rates of each anti-cancer agent and clinical characteristics.
| Characteristic | PatientsN = 100 N (%) | Cisplatininhibition rate % | Doctaxelinhibition rate % | Gemcitabine inhibition rate % | Irinotecanl inhibition rate % | Pemtrexed inhibition rate % |
| Age | ||||||
| >62 | 52 (52%) | 45.20±20.78 | 44.79±21.64 | 46.31±22.23 | 42.60±23.08 | 51.21±22.77 |
| ≤62 | 48 (48%) | 46.48±21.48 | 46.70±24.55 | 43.01±21.44 | 46.50±22.12 | 54.50±22.10 |
| Sex | ||||||
| Male | 73 (73%) | 46.71±21.89 | 47.81±21.99 | 47.33±20.74 | 44.41±21.39 | 52.65±22.23 |
| Female | 27 (27%) | 43.39±18.64 | 39.90±24.98 | 38.06±23.38 | 44.64±26.01 | 53.16±23.27 |
| Histology | ||||||
| Adenocarcinoma | 79 (79%) | 48.71±19.80 | 44.48±22.33 | 45.89±21.53 | 42.66±21.67 | 54.11±23.02 |
| Mucinous | 11 (11%) | 28.86±17.71 | 36.06±24.58 | 41.05±22.72 | 43.14±21.90 | 53.22±20.15 |
| Signet ring cell | 10 (10%) | 50.46±12.87 | 47.32±25.05 | 39.63±24.24 | 37.55±31.13 | 55.62±22.95 |
| Tumor Site | ||||||
| Distal | 35 (35%) | 52.20±19.38 | 47.84±22.92 | 41.67±20.12 | 42.28±24.22 | 49.22±23.55 |
| Proximal | 38 (38%) | 41.86±19.31 | 39.52±21.90 | 48.04±23.13 | 42.60±23.11 | 57.61±19.91 |
| Whole stomach | 27 (27%) | 46.81±20.07 | 45.04±23.65 | 43.87±22.19 | 41.35±20.35 | 55.37±24.51 |
| Stage | ||||||
| I,II | 37 (37%) | 46.94±21.58 | 45.54±24.10 | 38.77±20.03 | 42.67±22.45 | 53.86±23.37 |
| III, IV | 63 (63%) | 46.71±18.94 | 42.98±22.11 | 48.12±22.19 | 41.87±22.85 | 54.36±22.18 |
| Histological grade | ||||||
| 2 | 27 (27%) | 46.83±22.49 | 45.73±22.27 | 50.07±20.02 | 43.44±22.71 | 55.17±19.31 |
| 3 | 44 (44%) | 42.48±18.35 | 38.12±22.61 | 39.72±22.06 | 40.69±22.89 | 56.11±20.83 |
| Mixed 2–3 | 29 (29%) | 53.40±18.03 | 50.93±22.01 | 47.06±22.25 | 43.17±22.80 | 50.20±27.77 |
| Lymph node metastasis | ||||||
| No | 25 (25%) | 48.82±23.19 | 48.81±24.53 | 39.01±22.98 | 42.80±22.84 | 52.53±20.81 |
| Yes | 75 (75%) | 44.81±20.31 | 44.67±22.51 | 46.47±21.28 | 45.03±22.63 | 52.87±23.04 |
Data are expressed as mean ± standard deviation.
To further investigate the possible relationship between SULF2 methylation and cisplatin or irinotecan sensitivity, the in vitro efficacy rate of each drug concentration was examined setting up different cut-off inhibition rates (Table 4). Four different cut-off values were adopted: 30%, 40%, 50% and 60%. At the cut-off of 30% inhibition rate, SULF2 methylation was found to have a significant association with cisplatin efficacy (SULF2M: 57.14%, SULF2U: 80.56%, P = 0.02) and irinotecan efficacy (SULF2M: 89.29%, SULF2U: 62.50%, P = 0.01, Figure 2 and Table 4). At the cut-off of 40%, 50% and 60% inhibition rate, there was a trend that the SULF2M samples showed higher irinotecan efficacy, but lower cisplatin efficacy than the SULF2U samples (Table 4).
Table 4. Relationship between cut-off inhibition index and in vitro efficacy rate by means of HDRA.
| Chemotheraputic agents and SULF2 methylation | Cut-off inhibition rate | |||||||||||||||
| 30% | 40% | 50% | 60% | |||||||||||||
| Sensitive (sample number) | Resistant (sample number) | Efficacy rate (%) | P value | Sensitive (sample number) | Resistant (sample number) | Efficacy rate (%) | P value | Sensitive (sample number) | Resistant (sample number) | Efficacy rate (%) | P value | Sensitive (sample number) | Resistant (sample number) | Efficacy rate (%) | P value | |
| Cisplatin (20 ug/ml) | ||||||||||||||||
| SULF2M | 16 | 12 | 57.14 | 0.02 | 12 | 16 | 42.86 | 0.07 | 11 | 17 | 39.29 | 0.26 | 6 | 22 | 27.27 | 0.46 |
| SULF2U | 58 | 14 | 80.56 | 46 | 26 | 63.89 | 39 | 33 | 54.17 | 22 | 50 | 44.00 | ||||
| Irinotecan (20 ug/ml) | ||||||||||||||||
| SULF2M | 25 | 3 | 89.29 | 0.01 | 21 | 7 | 75.00 | 0.07 | 16 | 12 | 57.14 | 0.11 | 12 | 16 | 42.86 | 0.05 |
| SULF2U | 45 | 27 | 62.50 | 38 | 34 | 52.78 | 27 | 45 | 37.50 | 16 | 56 | 22.22 | ||||
Figure 2. Using the 30% inhibition rate as cut-off, SULF2 methylation was found to have a significant association with cisplatin efficacy (SULF2M: 57.14%, SULF2U: 80.56%, P = 0.02) and irinotecan efficacy (SULF2M: 89.29%, SULF2U: 62.50%, P = 0.01).
Association of SULF2 Methylation with Clinical Response to Chemotherapy
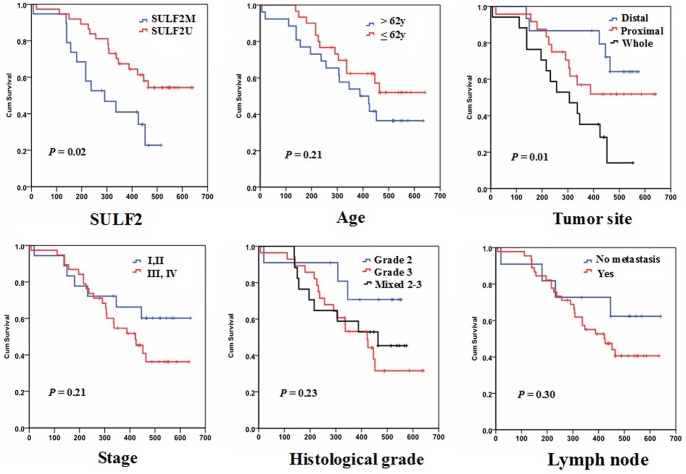
Among the 56 patients receiving the modified FOLFOX regimen, the median survival time was 434 days (range: 111–641 days). A significant association was observed between survival and tumor site (P = 0.01). No other association between clinical characteristics and survival was found (Table 5 and Figure 3). SULF2M was detected in 19 patients, while SULF2U was found in 37 patients (SULF2 methylation rate: 34%). A significant association was observed between survival and SULF2 methylation status. The median overall survival was 309 days (95% CI = 236 to 382 days) for patients with SULF2M, and 481 days (95% CI = 418 to 490 days) for those with SULF2U (P = 0.02, Figure 3). Using both univariate and multivariate Cox proportional hazard analysis that took into account age, sex, tumor site, stage, histological grade, lymph node metastasis and SULF2 methylation as covariates, tumor site and SULF2 methylation remained significant markers of overall survival in gastric cancer patients treated with platinum-based chemotherapy (Table 6).
Table 5. Clinical factors associated with overall survival.
| Characteristic | No. of Patients N = 56 | Median survival time (days) | P Log-rank test |
| Age, y | |||
| >62 | 26 | 391 | 0.21 |
| ≦62 | 30 | 471 | |
| Sex | |||
| Male | 42 | 399 | 0.48 |
| Female | 14 | 469 | |
| Histology | |||
| Adenocarcinoma | 43 | 426 | 0.93 |
| Mucinous | 7 | 470 | |
| Signet ring cell | 6 | 416 | |
| Tumor Site | |||
| Distal stomach | 15 | 414 | 0.01 |
| Proximal stomach | 24 | 366 | |
| Whole stomach | 17 | 229 | |
| Stage | |||
| I, II | 18 | 473 | 0.21 |
| III, IV | 38 | 411 | |
| Histological grade | |||
| 2 | 11 | 459 | 0.23 |
| 3 | 28 | 401 | |
| Mixed 2–3 | 17 | 395 | |
| Lymph node metastasis | |||
| No | 11 | 485 | 0.30 |
| Yes | 45 | 420 | |
| SULF2 | |||
| Methylation | 19 | 309 | 0.02 |
| Unmethylation | 37 | 481 | |
Figure 3. Kaplan–Meier estimates of overall survival by SULF2 methylation status and clinical characteristics.
Table 6. Univariate and multivariate Cox proportional hazard analysis of factors associated with overall survival.
| Variables | Univariate | Multivariate | |||
| No. | HR (95% CI) | P | HR (95% CI) | P | |
| Age, y | |||||
| >62 | 26 | 1(ref.) | 1(ref.) | ||
| ≦62 | 30 | 0.632 (0.304–1.314) | 0.21 | 0.509 (0.223–1.165) | 0.11 |
| Sex | |||||
| Male | 42 | 1(ref.) | 1(ref.) | ||
| Female | 14 | 0.809 (0.296–1.788) | 0.49 | 0.363 (0.124–1.066) | 0.07 |
| Histology | |||||
| Adenocarcinoma | 43 | 1(ref.) | 1(ref.) | ||
| Mucinous | 7 | 0.849 (0.292–2.467) | 0.76 | 0.206 (0.052–0.818) | 0.03 |
| Signet ring cell | 6 | 1.116 (0.333–3.737) | 0.86 | 0.568 (0.131–2.460) | 0.45 |
| Tumor Site | |||||
| Distal stomach | 15 | 1(ref.) | 1(ref.) | ||
| Proximal stomach | 24 | 1.813 (0.626–5.248) | 0.27 | 3.381 (0.794–14.395) | 0.10 |
| Whole stomach | 17 | 3.975 (1.390–11.363) | 0.01 | 8.105 (1.814–36.218) | 0.01 |
| Stage | |||||
| I, II | 18 | 1(ref.) | 1(ref.) | ||
| III, IV | 38 | 1.716 (0.729–4.040) | 0.22 | 2.321 (0.487–11.050) | 0.29 |
| Histological grade | |||||
| 2 | 11 | 1(ref.) | 1(ref.) | ||
| 3 | 28 | 2.810 (0.820–9.631) | 0.10 | 6.354 (1.134–35.601) | 0.05 |
| Mixed 2–3 | 17 | 2.287 (0.619–8.456) | 0.22 | 4.401 (0.891–21.751) | 0.07 |
| Lymph node metastasis | |||||
| No | 11 | 1(ref.) | 1(ref.) | ||
| Yes | 45 | 1.732 (0.601–4.988) | 0.31 | 1.018 (0.216–4.791) | 0.98 |
| SULF2 | |||||
| Methylation | 19 | 1(ref.) | 1(ref.) | ||
| Unmethylation | 37 | 0.413 (0.197–0.866) | 0.02 | 0.525 (0.215–1.280) | 0.04 |
In this multivariate analysis, age, sex, tumor site, stage, histological grade, lymph node metastasis and SULF2 methylation were included.
Discussion
Although previous studies have demonstrated the involvement of SULF2 in cancer pathogenesis, its value for chemosensitivity prediction remains unclear. This study focuses on the promoter CpG island methylation of SULF2 as a potential biomarker in gastric cancer. The major findings of the present study demonstrate that: (i) the rate of SULF2M in human gastric cancer is around 30%; (ii) the first evidence for the SULF2U is associated with cisplatin sensitivity in cancer; (iii) evidence for the SULF2M is associated with irinotecan sensitivity in gastric cancer; (iv) a retrospective study and validation for SULF2 methylation in a cohort of 56 patients with gastric cancer, which allowed us to discover that SULF2U is an independent prognostic biomarker in gastric cancer patients treated with a modified FOLFOX regimen.
In previous studies of gastric cancer, the findings on methylation status and expression levels of SULFs were inconclusive. In one study, only 13 early-stage breast and gastric cancers, most of which were stage I, were analyzed by semi-quantitative RT-PCR for SULF1 expression [24]. It was shown that low expression of SULF1 was prevalent in these two types of cancer. In another study, the expression of both SULF1 and SULF2 mRNA was determined by real-time RT-PCR for a large cohort of gastric cancer tissues, finding that SULFs were expressed at higher levels in gastric cancer as compared with normal tissues. In the current study we found by examining the SULF2 methylation in 100 gastric cancer samples, that the rate of SULF2M was approximately 30%, which indicated that predominate gastric cancer were SULF2U. Recent studies by integrated genomic analyses revealed that SULF2 acts as a downstream effector of p53, and that activation of p53 could lead to the SULF2U and up-regulation of SULF2. The possible link between p53 and SULF2 in growth factor signaling pathway suggested a possible role for SULF2 in cancer development and cancer patients’ outcome [25]. The relationship between SULF2 unmethylation and SULF2 up-regulation needs to be further tested in these samples. The different expression levels and methylation status of SULF2 between tumor and normal tissue also need to be further verified.
Studies on SULF2 methylation as chemosensitivity predictor are scarce. Methylation of the SULF2 promoter has been associated with better survival of resected lung adenocarcinoma patients, and also with a marginal improvement in survival of advanced NSCLC patients receiving standard chemotherapy (hazard ratio = 0.63, P = 0.07) [18]. Subsequent studies demonstrated that NSCLC cell lines with SULF2M were 134-fold more sensitive to topoisomerase I inhibitor than those with SULF2U. In the current study, we have demonstrated that gastric tumors with SULF2M are more sensitive to irinotecan than those with SULF2U. Furthermore, we demonstrated for the first time that SULF2 methylation is also a potential predictive biomarker for cisplatin efficacy. Gastric tumors with SULF2U were more sensitive to cisplatin than those with SULF2M, and gastric cancer patients with SULF2U showed lower mortality when receiving platinum-based chemotherapy. Although several predictive biomarkers have been identified for cisplatin, such as ERCC1 [7], BRCA1 [4], [26], [27] and XRCC1 [28]–[30], considering the relatively low response rates of the commonly used platinum/5FU-based neoadjuvant treatment protocol for advanced gastric carcinoma patients, the identification of biomarkers to predict response is urgently needed. The discovery of a novel predictive biomarker that can be examined by methylation analysis is intriguing and supplemental. The reason why SULF2 unmethylation increases tumor sensitivity to cisplatin may lie on ubiquitin conjugating enzymes (UBE). It has been reported that SULF2 methylation results in increased expression of UBE [18]. UBE added ubiquitin to certain lysine residues and was involved in DNA repair, mutagenesis and cell proliferation. Overexpression of certain UBEs, like RAD6B, could result in significant resistance to cisplatin [31]. Further studies should be carried out to investigate the molecular mechanism of SULF2M induced cisplatin resistance.
A significant synergistic effect of cisplatin and DNA methyltransferase (DNMT) inhibitors 5-aza-dC (DAC) on cell viability was observed in the cisplatin-resistant AGS cell line but not in the cisplatin-sensitive MKN28 cell line [32]. Data from analyzing colony formation capability showed that knockdown of DNMT1 caused an increase in cisplatin sensitivity [32]. The molecular mechanism remains unclear. However, the relationship between SULF2 methylation and cisplatin sensitivity may partially explain this phenomenon. Treatment of gastric cancer with DNMT inhibitors could result in demethylation of SULF2 and consequently increase the sensitivity to cisplatin. Thus, in addition to the predictive impact, our data also supports the inclusion of a DNMT inhibitor in current treatment protocols for at least a subset of gastric cancer patients.
In conclusion, our study provides novel evidences that SULF2 methylation is negatively associated with cisplatin sensitivity in vitro. SULF2 methylation is a potential prognostic biomarker for gastric cancer patients treated with platinum-based chemotherapy.
Supporting Information
The RT-PCR amplification curves of SULF2M and SULF2U. The red curve stands for amplification of SULF2M, and the blue curve stands for amplification of SULF2U. Figure S1a shows the amplification curves of sample with SULF2M; Figure S1b shows the amplification curves of sample with SULF2U.
(TIF)
Funding Statement
This work was funded by grants from the National Natural Science Foundation of China (Grant No. 81220108023 and 81172094) and Top Six Talents Project of Jiangsu Province (Grant No. 2011ws005). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Jemal A, Bray F (2011) Center MM, Ferlay J, Ward E, et al (2011) Global cancer statistics. CA Cancer J Clin 61: 69–90. [DOI] [PubMed] [Google Scholar]
- 2. Wagner AD, Grothe W, Haerting J, Kleber G, Grothey A, et al. (2006) Chemotherapy in advanced gastric cancer: a systematic review and meta-analysis based on aggregate data. J Clin Oncol 24: 2903–2909. [DOI] [PubMed] [Google Scholar]
- 3. Wesolowski R, Lee C, Kim R (2009) Is there a role for second-line chemotherapy in advanced gastric cancer? Lancet Oncol 10: 903–912. [DOI] [PubMed] [Google Scholar]
- 4. Wei J, Costa C, Ding Y, Zou Z, Yu L, et al. (2011) mRNA expression of BRCA1, PIAS1, and PIAS4 and survival after second-line docetaxel in advanced gastric cancer. J Natl Cancer Inst 103: 1552–1556. [DOI] [PubMed] [Google Scholar]
- 5. Hanauske AR, Eismann U, Oberschmidt O, Pospisil H, Hoffmann S, et al. (2007) In vitro chemosensitivity of freshly explanted tumor cells to pemetrexed is correlated with target gene expression. Invest New Drugs 25: 417–423. [DOI] [PubMed] [Google Scholar]
- 6. Chen CY, Chang YL, Shih JY, Lin JW, Chen KY, et al. (2011) Thymidylate synthase and dihydrofolate reductase expression in non-small cell lung carcinoma: the association with treatment efficacy of pemetrexed. Lung Cancer 74: 132–138. [DOI] [PubMed] [Google Scholar]
- 7. Wei J, Zou Z, Qian X, Ding Y, Xie L, et al. (2008) ERCC1 mRNA levels and survival of advanced gastric cancer patients treated with a modified FOLFOX regimen. Br J Cancer 98: 1398–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rosell R, Moran T, Viteri S, Carcereny E, Gasco A, et al. (2010) Optimization of genetics to create therapies for metastatic (stage IV) non-small-cell lung cancer. Expert Opin Pharmacother 11: 1683–1693. [DOI] [PubMed] [Google Scholar]
- 9. Olaussen KA, Dunant A, Fouret P, Brambilla E, Andre F, et al. (2006) DNA repair by ERCC1 in non-small-cell lung cancer and cisplatin-based adjuvant chemotherapy. N Engl J Med 355: 983–991. [DOI] [PubMed] [Google Scholar]
- 10. Hur K, Han TS, Jung EJ, Yu J, Lee HJ, et al. (2012) Up-regulated expression of sulfatases (SULF1 and SULF2) as prognostic and metastasis predictive markers in human gastric cancer. J Pathol 228: 88–98. [DOI] [PubMed] [Google Scholar]
- 11. Lai J, Chien J, Staub J, Avula R, Greene EL, et al. (2003) Loss of HSulf-1 up-regulates heparin-binding growth factor signaling in cancer. J Biol Chem 278: 23107–23117. [DOI] [PubMed] [Google Scholar]
- 12. Lai JP, Chien JR, Moser DR, Staub JK, Aderca I, et al. (2004) hSulf1 Sulfatase promotes apoptosis of hepatocellular cancer cells by decreasing heparin-binding growth factor signaling. Gastroenterology 126: 231–248. [DOI] [PubMed] [Google Scholar]
- 13. Li J, Kleeff J, Abiatari I, Kayed H, Giese NA, et al. (2005) Enhanced levels of Hsulf-1 interfere with heparin-binding growth factor signaling in pancreatic cancer. Mol Cancer 4: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dhoot GK, Gustafsson MK, Ai X, Sun W, Standiford DM, et al. (2001) Regulation of Wnt signaling and embryo patterning by an extracellular sulfatase. Science 293: 1663–1666. [DOI] [PubMed] [Google Scholar]
- 15. Ai X, Kitazawa T, Do AT, Kusche-Gullberg M, Labosky PA, et al. (2007) SULF1 and SULF2 regulate heparan sulfate-mediated GDNF signaling for esophageal innervation. Development 134: 3327–3338. [DOI] [PubMed] [Google Scholar]
- 16. Viviano BL, Paine-Saunders S, Gasiunas N, Gallagher J, Saunders S (2004) Domain-specific modification of heparan sulfate by Qsulf1 modulates the binding of the bone morphogenetic protein antagonist Noggin. J Biol Chem 279: 5604–5611. [DOI] [PubMed] [Google Scholar]
- 17. Castro NP, Osorio CA, Torres C, Bastos EP, Mourao-Neto M, et al. (2008) Evidence that molecular changes in cells occur before morphological alterations during the progression of breast ductal carcinoma. Breast Cancer Res 10: R87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tessema M, Yingling CM, Thomas CL, Klinge DM, Bernauer AM, et al. (2012) SULF2 methylation is prognostic for lung cancer survival and increases sensitivity to topoisomerase-I inhibitors via induction of ISG15. Oncogene 31: 4107–4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kudo Y, Ogawa I, Kitajima S, Kitagawa M, Kawai H, et al. (2006) Periostin promotes invasion and anchorage-independent growth in the metastatic process of head and neck cancer. Cancer Res 66: 6928–6935. [DOI] [PubMed] [Google Scholar]
- 20. Lai JP, Sandhu DS, Yu C, Han T, Moser CD, et al. (2008) Sulfatase 2 up-regulates glypican 3, promotes fibroblast growth factor signaling, and decreases survival in hepatocellular carcinoma. Hepatology 47: 1211–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Furukawa T, Kubota T, Hoffman RM (1995) Clinical applications of the histoculture drug response assay. Clin Cancer Res 1: 305–311. [PubMed] [Google Scholar]
- 22. Hayashi Y, Kuriyama H, Umezu H, Tanaka J, Yoshimasu T, et al. (2009) Class III beta-tubulin expression in tumor cells is correlated with resistance to docetaxel in patients with completely resected non-small-cell lung cancer. Intern Med 48: 203–208. [DOI] [PubMed] [Google Scholar]
- 23. Fujita Y, Hiramatsu M, Kawai M, Nishimura H, Miyamoto A, et al. (2009) Histoculture drug response assay predicts the postoperative prognosis of patients with esophageal cancer. Oncol Rep 21: 499–505. [PubMed] [Google Scholar]
- 24. Chen Z, Fan JQ, Li J, Li QS, Yan Z, et al. (2009) Promoter hypermethylation correlates with the Hsulf-1 silencing in human breast and gastric cancer. Int J Cancer 124: 739–744. [DOI] [PubMed] [Google Scholar]
- 25. Chau BN, Diaz RL, Saunders MA, Cheng C, Chang AN, et al. (2009) Identification of SULF2 as a novel transcriptional target of p53 by use of integrated genomic analyses. Cancer Res 69: 1368–1374. [DOI] [PubMed] [Google Scholar]
- 26. Kim H, Chen J, Yu X (2007) Ubiquitin-binding protein RAP80 mediates BRCA1-dependent DNA damage response. Science 316: 1202–1205. [DOI] [PubMed] [Google Scholar]
- 27. Wang L, Wei J, Qian X, Yin H, Zhao Y, et al. (2008) ERCC1 and BRCA1 mRNA expression levels in metastatic malignant effusions is associated with chemosensitivity to cisplatin and/or docetaxel. BMC Cancer 8: 97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liu L, Yuan P, Wu C, Zhang X, Wang F, et al. (2011) Assessment of XPD Lys751Gln and XRCC1 T-77C polymorphisms in advanced non-small-cell lung cancer patients treated with platinum-based chemotherapy. Lung Cancer 73: 110–115. [DOI] [PubMed] [Google Scholar]
- 29. Shim HJ, Yun JY, Hwang JE, Bae WK, Cho SH, et al. (2010) BRCA1 and XRCC1 polymorphisms associated with survival in advanced gastric cancer treated with taxane and cisplatin. Cancer Sci 101: 1247–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kudo K, Gavin E, Das S, Amable L, Shevde LA, et al. (2012) Inhibition of Gli1 results in altered c-Jun activation, inhibition of cisplatin-induced upregulation of ERCC1, XPD and XRCC1, and inhibition of platinum-DNA adduct repair. Oncogene 31: 4718–4724. [DOI] [PubMed] [Google Scholar]
- 31. Lyakhovich A, Shekhar MP (2004) RAD6B overexpression confers chemoresistance: RAD6 expression during cell cycle and its redistribution to chromatin during DNA damage-induced response. Oncogene 23: 3097–3106. [DOI] [PubMed] [Google Scholar]
- 32. Mutze K, Langer R, Schumacher F, Becker K, Ott K, et al. (2011) DNA methyltransferase 1 as a predictive biomarker and potential therapeutic target for chemotherapy in gastric cancer. Eur J Cancer 47: 1817–1825. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The RT-PCR amplification curves of SULF2M and SULF2U. The red curve stands for amplification of SULF2M, and the blue curve stands for amplification of SULF2U. Figure S1a shows the amplification curves of sample with SULF2M; Figure S1b shows the amplification curves of sample with SULF2U.
(TIF)