Abstract
Objective
Our objective was to estimate primary resistance in an urban setting in a developing country characterized by high antiretroviral (ARV) coverage over the diagnosed population and also by an important proportion of undiagnosed individuals, in order to determine whether any change in primary resistance occurred in the past five years.
Design
We carried out a multi-site resistance surveillance study according to WHO HIV resistance guidelines, using a weighted sampling technique based on annual HIV case reports per site.
Methods
Blood samples were collected from 197 drug-naive HIV-1-infected individuals diagnosed between March 2010 and August 2011 at 20 HIV voluntary counselling and testing centres in Buenos Aires. Clinical records of enrolled patients at the time of diagnosis were compiled. Viral load and CD4 counts were performed on all samples. The pol gene was sequenced and the resistance profile determined. Phylogenetic analysis was performed by neighbour-joining (NJ) trees and bootscanning analysis.
Results
We found that 12 (7.9%) of the 152 successfully sequenced samples harboured primary resistance mutations, of which K103N and G190A were the most prevalent. Non-nucleoside reverse transcriptase inhibitors (NNRTI) resistance mutations were largely the most prevalent (5.9%), accounting for 75% of all primary resistance and exhibiting a significant increase (p=0.0072) in prevalence during the past 10 years as compared to our previous study performed in 1997–2000 and in 2003–2005. Nucleoside reverse transcriptase inhibitor (NRTI) and protease inhibitor primary resistance were low and similar to the one previously reported.
Conclusions
Levels of primary NNRTI resistance in Buenos Aires appear to be increasing in the context of a sustained ARV coverage and a high proportion of undiagnosed HIV-positive individuals.
Keywords: HIV, primary drug resistance, trend, NNRTI, newly diagnosed
Introduction
Highly active antiretroviral therapy (HAART) can increase life expectancy of HIV-positive patients [1–4] and prevent transmission [5]. However, it may also drive the selection of mutations that confer resistance to antiretroviral (ARV) drugs in patients under treatment. Transmission of drug-resistant variants constitutes a public health issue since it can contribute to failure of first-line ARV treatment [6]. Prevalence of primary resistance appears to be correlated with treatment coverage, being higher in developed settings [7–19]. In fact, international guidelines recommend performing genotypic resistance testing in all drug-naïve patients, before beginning a first-line ARV regimen [20–22].
Although by the end of the 1990s, improvements in ARV regimens had limited the rate of selection and transmission of resistance mutations (mainly for those associated with nucleoside reverse transcriptase inhibitor (NRTI) resistance), significant increasing trends in primary resistance have been reported in the past decade in settings where HAART regimens are broadly implemented: increasing trends appear to be observed exclusively in developed settings, such as the United Kingdom [23], Germany [24] and the United States [25], but not in developing settings [26–28]. This might be associated with the fact that the high proportion of undiagnosed individuals in developing countries may limit population-level selective forces driven by ARV treatment. It is not clear whether the low prevalence of primary resistance in these settings will remain stable or will increase at a slower rate than that of the developed settings.
Argentina is a developing country with approximately 130,000 individuals living with HIV, with half of them unaware of their infection [29]. Free access to ARV drugs has been guaranteed since 1992, and Argentina, together with Brazil, was the first Latin American countries that established an ART delivery programme. Buenos Aires, the capital city, is the most populated, containing 47.1% of reported HIV cases between 2001 and 2010 [30]. In Buenos Aires, there are 11,124 people receiving ARV treatment with 67% of them taking NRTI drugs: 40% (4442) receive AZT/3TC, 13% (1434) ABC/3TC, 11% (1250) TDF/FTC and 3% (305) TDF/3TC. A 52% (5738) receive NNTRI drugs, 40% (4435) efavirenz and 12% (1303) with nevirapine, and 38% (4232) take at least one protease inhibitor, of which 454 are not reinforced with ritonavir.
In the setting of Buenos Aires city, we have previously reported a prevalence of primary resistance among newly diagnosed individuals of 2.3% (CI: 0–5.5, n=86) in 1997–2000 [31] and 4.2% (CI: 1.9–6.5, n=284) in 2003–2005 [32]. In addition, a prevalence of 7.7% (CI: 0.4–14.9, n=52) was reported for individuals recently infected in 2004–2005 [33]. Importantly, non-nucleoside reverse transcriptase inhibitors (NNRTI) resistance mutations (mainly K103N) were not found in the first study but were detected in the last two studies.
In the present report, we study individuals recently diagnosed between 2010 and 2011 and show that the increasing trend in overall primary resistance persists driven mainly by the accumulation of NNRTI-associated resistance mutations.
Materials and methods
Study design
This resistance surveillance study was designed according to WHO HIV resistance guidelines [34]. These guidelines do not make restrictions in age as the 2008 updated guidelines do. This allows us to compare our results with our previous studies and also to reach a higher sample size. A comparison with results obtained following the 2008 updated guidelines is provided in this article. In brief, using a cross-sectional weighted sampling technique, newly HIV-diagnosed individuals were sequentially selected at the time of diagnosis at 20 health centres that include voluntary and counselling testing sites (VCTs) and infectious diseases services from public hospitals and private clinics. The proportion of individuals included from each site reflected the proportion of new cases reported to the Ministry of Health from 2003 to 2009.
Study population
The inclusion criteria were being HIV positive, age older than 18, with first confirmed diagnosis of HIV infection within a period of 180 days before study entry, with confirmed unexposure to ARV therapy and agreement to participate in the study by signing an informed consent. A total of 197 drug-naïve newly diagnosed HIV-1 individuals were enrolled between March 2010 and August 2011, and those who met the inclusion criteria were included in the study. The final study population (N=152) comprised 116 men, 33 women and 3 transsexual, of whom 28 (18%) were younger than 25 years, with an overall average age of 37 years.
Ethical approval and details of the consent procedure for the study
The project was initially submitted to the Bioethics Committee of the Muñiz Hospital specialized in infectious diseases in Buenos Aires, Argentina. It is a UNESCO-recognized committee and is listed among the ethics institutions list available at http://www.unesco.org/new/en/social-and-human-sciences/themes/global-ethics-observatory/access-geobs/. The Bioethics Committee of the Muñiz Hospital approved the project on 16 November 2009. The project was later validated by the Academic and Scientific Direction of the Government of the City of Buenos Aires on 29 September 2010. Patients were asked to read and, if agree with participating in the study, to sign an informed consent form. In this form, the patient can read in an accessible language the objectives of the project and the assays to be performed over the collected samples.
HIV-1 RNA isolation and sequencing
RNA was isolated from plasma by the QIAamp viral extraction Kit (QIAGEN GmbH, Hilden, Germany). The pol gene was amplified between positions 2142 and 3798 (reference strain HXB2 numbering [35]) by reverse transcriptase-polymerase chain reaction (RT-PCR) and was sequenced by ABI Prism 3100/3100-Avant equipment (Applied Biosystems, Foster City, CA, USA).
For pol gene amplification, outer primers 5CP1 (5′-GAAGGGCACACAGCCAGAAATTGCAGGG-3′) and RT3.1 (5′-GCTCCTACTATGGGTTCTTTCTCTAACTGG-3′), and inner primers 1F (5′-CAGACCAGAGCCAACAGCCCC-3′), A35 (5′-ATTGGTTGCACTTTAAATTTTCCCATTAGCCCTATT-3′), 6B (5′-CATTGTTTAACTTTTGGGCC-3′), RT3208F (5′-AACATCAGAAAGAACCTCCATT-3′), NE1 (5′-CGACCTGACAGTTACTGTATGTCTTCAATCACC-3′) and 6B (5′-CATTGTTTAACTTTTGGGCCATCCATTCCTGGC-3′) were used.
The reverse transcription reaction was performed by heating at 42°C for 50 minutes and 70°C for 15 minutes using the Superscript II RT enzyme and the RT3.1 primer. Pol gene amplification was performed by nested PCR using the primers listed above. The PCR conditions were 3 first minutes at 95°C, then 5 cycles of 15 seconds of denaturation at 95°C, 15 seconds of primer annealing at 56°C and 1:40 minutes of elongation at 72°C. And at the end of 30 cycles: 15 seconds of denaturation at 90°C, 15 seconds of primer anneal at 56°C and 1:40 minutes of elongation at 72°C. A final elongation was performed for 10 minutes at 72°C.
Viral load and CD4 testing
Plasma viral load (VL) was assessed by branched DNA (b-DNA) technology (Versant HIV-1 RNA 3.0; Bayer Co., Tarrytown, NY) with a detection limit of 50 HIV-1 RNA copies/ml. CD4+cells from peripheral blood were measured by cytometry (Coulter XL; Coulter Co., Hialeah, FL, USA).
Resistance analysis
Sequences were analyzed to identify mutations associated with reduced susceptibility to protease and RT inhibitors, as reported by the International AIDS Society-USA in 2010 [36]: RT–M41L, A62V, K65R, D67N, 69 insert, K70R, L74V,V75I, F77L, L100I, K101P, K103N, V106A, V106M, V108I, Y115F, F116Y, Q151M, Y181C, Y181I, M184V, M184I, Y188C, Y188L, Y188H, G190A, G190S, L210W, T215Y, T215F, K219Q, K219E and P225H; protease–D30N, V32I, M46I, M46L, I47A, I47V, G48V, I50L, I50V, I54L, I54M, Q58E, L76V, V82A, V82F, V82L, V82S, V82T, N83D, I84V, N88S and L90M.
Phylogenetic analysis
Sequence alignment was performed by CLUSTAL W (BioEdit 7.1.3.0 sequence alignment editor [37]). Neighbour-joining (NJ) trees were constructed under the Kimura 2-parameter model with the MEGA5 programme [38]. Sequences were individually analyzed by Simplot 3.5.1 [39] and recombination analysis was then performed by bootscanning analysis [39].
Statistical analysis
Chi-square test and Fisher's exact test were used to compare proportions of resistance mutations and patient's epidemiological records.
Results
A total of 197 newly HIV-1-diagnosed individuals were studied. The average age was 37 years. In 45 cases, the pol gene could not be successfully sequenced and were excluded from the analysis, resulting in a total of 152 analyzed samples (77%). Phylogenetic analyses showed predominance of two viral subtypes: 77 (50.6%) samples were recombinants between subtypes B and F, 70 (46.0%) were subtypes B and 5 (3.3%) were “non B-non BF” variants.
Twelve individuals (7.9%) were found to harbour primary resistance mutations, 12 males and 1 female. No significant associations were found between presence of variants with resistance mutations and patient's epidemiological records, CD4 count, VL or viral subtype (Supplementary Table 1). According to drug class, mutations associated with resistance to NNRTI were the most prevalent, being present in nine (5.9%) individuals. NRTI mutations and primary mutations associated with resistance to protease inhibitors (PIs) were both found each in two individuals (1.3%).
The most prevalent resistance mutations associated with NNRTI were K103N and G190A. Both were present in 88.9% (8/9) patients with primary resistance mutations for this class of drugs and accounts for the 66.7% (8/12) of the overall primary resistance and for 90% (9/10) of the NNRTI resistance. Regarding PI resistance mutations, they were found in two patients (Table 1) who shared the same mutations profile. One of them was a 32-year-old MSM man enrolled at the Site D, and the other was a 25-year-old bisexual man enrolled at the Site F. Phylogenetic analysis revealed a close relationship between both viral strains as evidenced by the monophyletic highly supported clade containing both sequences (bootstrap = 100, data not shown). While these two sequences are closely related, according to the phylogenetic analysis they are separated by a genetic distance of 0.062 showing that, in spite of sharing the same mutation profile, they are different from each other (Supplementary Figure 1).
Table 1.
Epidemiological and virological characteristics of the 12 individuals harbouring resistance mutations
| Resistance mutations | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
||||||||||||
| ID | Age group | Risk group | Site | First HIV-positive diagnosis | Sampling date | Clinical stage at diagnosis | Viral subtype | CD4 count (cells/µl) | Log10 (VL) | Protease inhibitor | Nucleoside RT inhibitor | Non-nucleoside RT inhibitor |
| 1 | 50–55 | HTS | A | 05.27.2010 | 09.27.2010 | Asymptomatic | BF | 337 | 4.58 | – | – | K103N |
| 2 | 30–35 | HTS | B | 07.16.2010 | 09.16.2010 | Symptomatic without AIDS criteria | B | 132 | 5.87 | – | – | G190A |
| 3 | 35–40 | MSM | A | 12.01.2010 | 02.01.2011 | Asymptomatic | B | 218 | 4.66 | – | A62V**, T215D* | – |
| 4 | 35–40 | HTS | C | 11.01.2010 | 01.04.2011 | AIDS | BF | 146 | 2.85 | – | – | G190A |
| 5 | 30–35 | MSM | D | 08.20.2010 | 11.05.2010 | Acute retroviral syndrome | BF | 394 | NA | M46I, L76V, V82A, I54V* | – | – |
| 6 | 20–25 | HTS | C | 10.01.2010 | 10.13.2010 | Asymptomatic | B | 301 | 4.53 | – | – | K103N |
| 7 | 30–35 | MSM | E | 11.30.2010 | 02.08.2011 | Asymptomatic | B | 494 | 4.92 | – | – | K101P** |
| 8 | 30–35 | HTS | D | 02.10.2011 | 02.28.2011 | AIDS | BF | 74 | NA | – | – | K103N |
| 9 | 20–25 | Bisexual | F | 08.17.2010 | 09.23.2010 | Asymptomatic | BF | 308 | NA | M46I, L76V, V82A, I54V* | – | – |
| 10 | 35–40 | MSM | E | 12.14.2010 | 02.08.2011 | NA | B | 22 | 4.49 | T69D* | D67N, K70R, M184V, T215F, K219Q | G190A, K101E* |
| 11 | 40–45 | NA | C | 05.05.2010 | 11.02.2010 | Symptomatic without AIDS criteria | B | 473 | 4.85 | – | – | K103N |
| 12 | 45–50 | MSM | E | 12.14.2010 | 02.15.2011 | Asymptomatic | BF | 363 | 5.65 | – | – | G190A |
Considered resistance mutations only by the WHO guidelines.
Considered resistance mutations only by the IAS guidelines.
NA = not available, VL = viral load, RT= reverse transcriptase.
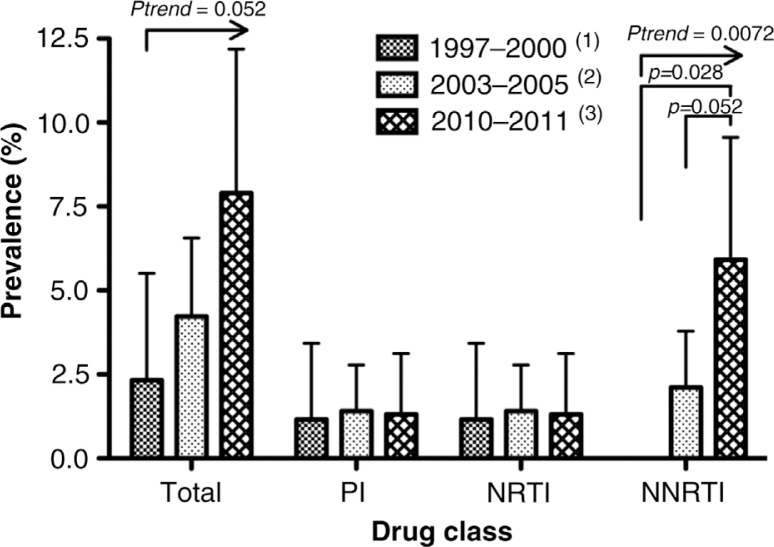
When we compared these results with our previous studies, as shown in Figure 1, we observed an almost significant increase in NNRTI resistance compared with data collected in 2003–2005 (p=0.052) and a significant increase compared with data collected in 1997–2000 (p=0.028). The overall trend across the 10-year period was significant for NNRTI resistance (Chi-square for trend, p=0.0072) and almost significant for overall resistance (Chi-square for trend, p=0.052).
Figure 1.
Prevalence of primary resistance in the present study compared with our previous studies and according to drug class.
Arrows indicate significant trends in time according to Chi-square test for trend. (1) Kijak et al. [31]. (2) Dilernia et al. [32]. (3) Current study.
Finally, we re-analyzed our dataset using the list of mutations reported by the WHO guidelines [21]. Among patients with no evidence of primary resistance based on the IAS list, none of them presented resistance mutations based on the WHO list. Among the 12 patients with evidence of primary resistance based on the IAS list, 11 of them presented resistance mutations according to the WHO guidelines while patient ID#7 (Table 1) exhibited the mutations K101P that is not considered by the WHO guidelines.
Discussion
Based on our results, 7.9% (CI: 3.6–12.2, n=152) of individuals newly diagnosed between 2010 and 2011 in Buenos Aires harbour major resistance mutations associated with reduced susceptibility to ARV drugs. This figure is similar to that observed in other developing countries [7–14] but still lower than the prevalence observed in many developed settings [15–19, 21].
Previously, we had reported a prevalence of 2.3% (CI: 0–5.5, n=86) primary resistance in a similar population diagnosed between 1997 and 2000 [32], and 4.2% (CI: 1.9–6.5, n=284) [32] for 2003–2005. Comparing prevalence in the first study with the one reported in the present study, we observed an increase of almost four-folds (p=0.028) in 10 years. When data were analyzed separately according to drug class, we observed that the mentioned trend was driven by the increase in NNRTI resistance mutations for which the prevalence resulted increased from no detection of those mutations in the first period to a prevalence of 2.1% in 2003–2005 and 5.9% in 2010–2011 (Chi-square for trend, p=0.0072). At the mutation level, K103N represents around the 50% of NNRTI primary resistance in the last two periods while the remaining 50% was dominated by the V108I mutation in the previous study and by the G190A mutation in the present study. It is important to highlight that, as reported by June 2010, in Buenos Aires the 86.6% of patients treated with NNRTIs were under efavirenz-based regimens and the remaining 13.3% were under nevirapine-based regimens [40], which are significantly impaired by K03N mutation. Therefore, it is likely that ARV treatment constitute indeed the population-level selective force driving persistence of this mutation in our population since K103N is not a natural polymorphism of the virus circulating in our region. Interestingly, we found a high prevalence of G190A, not previously reported in the region; and the absence of V108I, found about equally often as K103N in our previous study during 2003–2005.
It is important to highlight that in the present study we did not considered any upper limit for the age at enrolment while the 2008 WHO HIVDR guidelines, as well as the 2012 update, recommend restricting the inclusion criteria to patients younger than 25 years in order to minimize the proportion of chronically HIV-positive individuals and to obtain a better estimate of transmitted resistance. Applying that restriction to our study results in a similar estimation or resistance (7.1%) (Supplementary Table 1), in contrast to previous studies that have found significantly different prevalence in that age group [41]. However, the limited sample size of patients younger than 25 limits any further analysis.
According to our results, during the course of the past 10 years a significant increase in primary resistance to NNRTIs has occurred. Although prevalence remains lower than in developed settings, our study shows that accumulation of ARV resistance mutations can be a significant public health concern even in settings where the proportion of individuals living with HIV under treatment is limited.
Acknowledgement
This research has been partially funded by a Fogarty International Center/NIH grant through the AIDS International Training and Research Program at Mount Sinai School of Medicine-Argentina Program (Grant # D43 TW 001037), by the National Agency of Science and Technology (AGENCIA, PICT-2011-0271) and by the Secretary of Science and Technique of the University of Buenos Aires (UBACyT#20020100100045-01/W045).
To access the supplementary material to this article please see Supplementary Files under Article Tools online.
Competing interests
The authors declare that they have no significant competing financial, professional or personal interests that might have influenced the performance or presentation of the work described in this manuscript.
Authors' contributions
Designed the sampling method: AD, MV, HS.
Identified recent infections: MBB, IZ.
Conceived and designed the experiments: NRR, HS, DAD.
Performed the experiments: NRR.
Analyzed the data: NRR, AD, MBB, IZ, MV, DI, EB, HS, DAD.
Wrote the paper: NRR, DAD.
References
- 1.Lohse N, Hansen AB, Pedersen G, Kronborg G, Gerstoft J, Sorensen HT, et al. Survival of persons with and without HIV infection in Denmark, 1995–2005. Ann Intern Med. 2007;146(2):87–95. doi: 10.7326/0003-4819-146-2-200701160-00003. [DOI] [PubMed] [Google Scholar]
- 2.Mocroft A, Ledergerber B, Katlama C, Kirk O, Reiss P, d'Arminio Monforte A, et al. Decline in the AIDS and death rates in the EuroSIDA study: an observational study. Lancet. 2003;362(9377):22–9. doi: 10.1016/s0140-6736(03)13802-0. [DOI] [PubMed] [Google Scholar]
- 3.Mocroft A, Phillips AN, Gatell J, Ledergerber B, Fisher M, Clumeck N, et al. Normalisation of CD4 counts in patients with HIV-1 infection and maximum virological suppression who are taking combination antiretroviral therapy: an observational cohort study. Lancet. 2007;370(9585):407–13. doi: 10.1016/S0140-6736(07)60948-9. [DOI] [PubMed] [Google Scholar]
- 4.Mocroft A, Vella S, Benfield TL, Chiesi A, Miller V, Gargalianos P, et al. Changing patterns of mortality across Europe in patients infected with HIV-1. EuroSIDA study group. Lancet. 1998;352(9142):1725–30. doi: 10.1016/s0140-6736(98)03201-2. [DOI] [PubMed] [Google Scholar]
- 5.Cohen MS, Chen YQ, McCauley M, Gamble T, Hosseinipour MC, Kumarasamy N, et al. Prevention of HIV-1 infection with early antiretroviral therapy. N Engl J Med. 2011;365(6):493–505. doi: 10.1056/NEJMoa1105243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wittkop L, Gunthard HF, de Wolf F, Dunn D, Cozzi-Lepri A, de Luca A, et al. Effect of transmitted drug resistance on virological and immunological response to initial combination antiretroviral therapy for HIV (EuroCoord-CHAIN joint project): a European multicohort study. Lancet Infect Dis. 2011;11(5):363–71. doi: 10.1016/S1473-3099(11)70032-9. [DOI] [PubMed] [Google Scholar]
- 7.Avila-Rios S, Garcia-Morales C, Garrido-Rodriguez D, Ormsby CE, Hernandez-Juan R, Andrade-Villanueva J, et al. National prevalence and trends of HIV transmitted drug resistance in Mexico. PLoS One. 2011;6(11):e27812. doi: 10.1371/journal.pone.0027812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Charpentier C, Bellecave P, Cisse M, Mamadou S, Diakite M, Peytavin G, et al. High prevalence of antiretroviral drug resistance among HIV-1-untreated patients in Guinea-Conakry and in Niger. Antivir Ther. 2011;16(3):429–33. doi: 10.3851/IMP1754. [DOI] [PubMed] [Google Scholar]
- 9.de Medeiros RM, Junqueira DM, Matte MC, Barcellos NT, Chies JA, Matos Almeida SE, et al. Co-circulation HIV-1 subtypes B, C, and CRF31_BC in a drug-naive population from Southernmost Brazil: analysis of primary resistance mutations. J Med Virol. 2011;83(10):1682–8. doi: 10.1002/jmv.22188. [DOI] [PubMed] [Google Scholar]
- 10.Muwonga J, Edidi S, Butel C, Vidal N, Monleau M, Okenge A, et al. Resistance to antiretroviral drugs in treated and drug-naive patients in the Democratic Republic of Congo. J Acquir Immune Defic Syndr. 2011;57(Suppl 1):S27–33. doi: 10.1097/QAI.0b013e31821f596c. [DOI] [PubMed] [Google Scholar]
- 11.Ndembi N, Hamers RL, Sigaloff KC, Lyagoba F, Magambo B, Nanteza B, et al. Transmitted antiretroviral drug resistance among newly HIV-1 diagnosed young individuals in Kampala. AIDS. 2011;25(7):905–10. doi: 10.1097/QAD.0b013e328346260f. [DOI] [PubMed] [Google Scholar]
- 12.Sigaloff KC, Mandaliya K, Hamers RL, Otieno F, Jao IM, Lyagoba F, et al. High prevalence of transmitted antiretroviral drug resistance among newly HIV type 1 diagnosed adults in Mombasa, Kenya. AIDS Res Hum Retroviruses. 2012;28:1033–7. doi: 10.1089/AID.2011.0348. [DOI] [PubMed] [Google Scholar]
- 13.Hamers RL, Wallis CL, Kityo C, Siwale M, Mandaliya K, Conradie F, et al. HIV-1 drug resistance in antiretroviral-naive individuals in sub-Saharan Africa after rollout of antiretroviral therapy: a multicentre observational study. Lancet Infect Dis. 2011;11(10):750–9. doi: 10.1016/S1473-3099(11)70149-9. [DOI] [PubMed] [Google Scholar]
- 14.Soares MA, Brindeiro RM, Tanuri A. Primary HIV-1 drug resistance in Brazil. AIDS. 2004;18(Suppl 3):S9–13. doi: 10.1097/00002030-200406003-00003. [DOI] [PubMed] [Google Scholar]
- 15.Jayaraman GC, Archibald CP, Kim J, Rekart ML, Singh AE, Harmen S, et al. A population-based approach to determine the prevalence of transmitted drug-resistant HIV among recent versus established HIV infections: results from the Canadian HIV strain and drug resistance surveillance program. J Acquir Immune Defic Syndr. 2006;42(1):86–90. doi: 10.1097/01.qai.0000196666.16616.fe. [DOI] [PubMed] [Google Scholar]
- 16.Romero A, Sued O, Puig T, Esteve A, Pumarola T, Casabona J, et al. Prevalence of transmitted antiretroviral resistance and distribution of HIV-1 subtypes among patients with recent infection in Catalonia (Spain) between 2003 and 2005. Enferm Infecc Microbiol Clin. 2011;29(7):482–9. doi: 10.1016/j.eimc.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 17.Viani RM, Peralta L, Aldrovandi G, Kapogiannis BG, Mitchell R, Spector SA, et al. Prevalence of primary HIV-1 drug resistance among recently infected adolescents: a multicenter adolescent medicine trials network for HIV/AIDS interventions study. J Infect Dis. 2006;194(11):1505–9. doi: 10.1086/508749. [DOI] [PubMed] [Google Scholar]
- 18.Wensing AM, Boucher CA. Worldwide transmission of drug-resistant HIV. AIDS Rev. 2003;5(3):140–55. [PubMed] [Google Scholar]
- 19.Wensing AM, van de Vijver DA, Angarano G, Asjo B, Balotta C, Boeri E, et al. Prevalence of drug-resistant HIV-1 variants in untreated individuals in Europe: implications for clinical management. J Infect Dis. 2005;192(6):958–66. doi: 10.1086/432916. [DOI] [PubMed] [Google Scholar]
- 20.Bennett DE, Bertagnolio S, Sutherland D, Gilks CF. The World Health Organization's global strategy for prevention and assessment of HIV drug resistance. Antivir Ther. 2008;13(Suppl 2):1–13. [PubMed] [Google Scholar]
- 21.Bennett DE, Myatt M, Bertagnolio S, Sutherland D, Gilks CF. Recommendations for surveillance of transmitted HIV drug resistance in countries scaling up antiretroviral treatment. Antivir Ther. 2008;13(Suppl 2):25–36. [PubMed] [Google Scholar]
- 22.Hirsch MS, Gunthard HF, Schapiro JM, Brun-Vezinet F, Clotet B, Hammer SM, et al. Antiretroviral drug resistance testing in adult HIV-1 infection: 2008 recommendations of an International AIDS Society-USA panel. Clin Infect Dis. 2008;47(2):266–85. doi: 10.1086/589297. [DOI] [PubMed] [Google Scholar]
- 23.Cane P, Chrystie I, Dunn D, Evans B, Geretti AM, Green H, et al. Time trends in primary resistance to HIV drugs in the United Kingdom: multicentre observational study. BMJ. 2005;331(7529):1368. doi: 10.1136/bmj.38665.534595.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sagir A, Oette M, Kaiser R, Daumer M, Fatkenheuer G, Rockstroh JK, et al. Trends of prevalence of primary HIV drug resistance in Germany. J Antimicrob Chemother. 2007;60(4):843–8. doi: 10.1093/jac/dkm274. [DOI] [PubMed] [Google Scholar]
- 25.Grant RM, Hecht FM, Warmerdam M, Liu L, Liegler T, Petropoulos CJ, et al. Time trends in primary HIV-1 drug resistance among recently infected persons. JAMA. 2002;288(2):181–8. doi: 10.1001/jama.288.2.181. [DOI] [PubMed] [Google Scholar]
- 26.Couto-Fernandez JC, Brindeiro R, hequer-Fernandez SC, Inocencio L, Pereira A, Pires I, et al. Brazil. Rome, Italy: IAS; 2011. Trends in primary resistance to HIV drugs in chronically infected ARV naïve population in Rio de Janeiro state. [Google Scholar]
- 27.Lai CC, Hung CC, Chen MY, Sun HY, Lu CL, Tseng YT, et al. Trends of transmitted drug resistance of HIV-1 and its impact on treatment response to first-line antiretroviral therapy in Taiwan. J Antimicrob Chemother. 2012;67(5):1254–60. doi: 10.1093/jac/dkr601. [DOI] [PubMed] [Google Scholar]
- 28.Frentz D, Boucher CA, van de Vijver DA. Temporal changes in the epidemiology of transmission of drug-resistant HIV-1 across the world. AIDS Rev. 2012;14(1):17–27. [PubMed] [Google Scholar]
- 29.Ministerio de Salud Presidencia de la Nación. Boletín sobre el VIH-sida en la Argentina. 2010;27 [Google Scholar]
- 30.Ministerio de Salud Presidencia de la Nación. Boletín sobre el VIH-sida en la Argentina. 2011;28 [Google Scholar]
- 31.Kijak GH, Pampuro SE, Avila MM, Zala C, Cahn P, Wainberg MA, et al. Resistance profiles to antiretroviral drugs in HIV-1 drug-naive patients in Argentina. Antivir Ther. 2001;6(1):71–7. [PubMed] [Google Scholar]
- 32.Dilernia DA, Lourtau L, Gomez AM, Ebenrstejin J, Toibaro JJ, Bautista CT, et al. Drug-resistance surveillance among newly HIV-1 diagnosed individuals in Buenos Aires, Argentina. AIDS. 2007;21(10):1355–60. doi: 10.1097/QAD.0b013e3280b07db1. [DOI] [PubMed] [Google Scholar]
- 33.Petroni A, Deluchi G, Pryluka D, Rotryng F, Bortolozzi R, Lopardo G, et al. Update on primary HIV-1 resistance in Argentina: emergence of mutations conferring high-level resistance to nonnucleoside reverse transcriptase inhibitors in drug-naive patients. J Acquir Immune Defic Syndr. 2006;42(4):506–10. doi: 10.1097/01.qai.0000222285.44460.e2. [DOI] [PubMed] [Google Scholar]
- 34.World Health Organization. Geneva: World Health Organization; 2003. Guidelines for surveillance of HIV drug resistance. [Google Scholar]
- 35.Korber B, Foley BT, Kuiken C, Pillai SK, Sodroski JG. Numbering positions in HIV relative to HXB2CG. Los Alamos, NM: Theoretical Biology and Biophysics Group, Los Alamos National Laboratory; 1998. [Google Scholar]
- 36.Johnson VA, Brun-Vezinet F, Clotet B, Gunthard HF, Kuritzkes DR, Pillay D, et al. Update of the drug resistance mutations in HIV-1: December 2010. Top HIV Med. 2010;18(5):156–63. [PubMed] [Google Scholar]
- 37.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. 1998;41:95–8. [Google Scholar]
- 38.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–9. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lole KS, Bollinger RC, Paranjape RS, Gadkari D, Kulkarni SS, Novak NG, et al. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol. 1999;73(1):152–60. doi: 10.1128/jvi.73.1.152-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Situación epidemiológica del VIH-sida en la Ciudad de Buenos Aires. 2011. Coordinacion Sida Ministerio de Salud Gobierno de la Ciudad de Buenos Aires. [Google Scholar]
- 41.Kasang C, Kalluvya S, Majinge C, Stich A, Bodem J, Kongola G, et al. HIV drug resistance (HIVDR) in antiretroviral therapy-naïve patients in Tanzania not eligible for WHO threshold HIVDR survey is dramatically high. PLoS One. 2011;6(8):e23091. doi: 10.1371/journal.pone.0023091. [DOI] [PMC free article] [PubMed] [Google Scholar]