Abstract
The mitochondrial DNA (mtDNA) of 98 Mansi, an ancient group (formerly known as “Vogul”) of Uralic-speaking fishers and hunters on the eastern slope of the northern Ural Mountains, were analyzed for sequence variants by restriction fragment–length polymorphism analysis, control-region sequencing, and sequencing of additional informative sites in the coding region. Although 63.3% of the mtDNA detected in the Mansi falls into western Eurasian lineages (e.g., haplogroups UK, TJ, and HV), the remaining 36.7% encompass a subset of eastern Eurasian lineages (e.g., haplogroups A, C, D, F, G, and M). Among the western Eurasian lineages, subhaplogroup U4 was found at a remarkable frequency of 16.3%, along with lineages U5, U7, and J2. This suggests that the aboriginal populations residing immediately to the east of the Ural Mountains may encompass remnants of the early Upper Paleolithic expansion from the Middle East/southeastern Europe. The added presence of eastern Eurasian mtDNA lineages in the Mansi introduces the possibilities that proto-Eurasians encompassed a range of macrohaplogroup M and N lineages that subsequently became geographically distributed and that the Paleolithic expansion may have reached this part of Siberia before it split into western and eastern human groups.
Analysis of mtDNA diversity in northern European populations has revealed that their overall mtDNA gene pool encompasses all major western Eurasian haplogroups—that is, haplogroups H–K and T–X. Moreover, remarkably high frequencies of haplogroups U and V are found in the Saami, who are thought to be genetically similar to early Upper Paleolithic Europeans (Sajantila et al. 1995; Torroni et al. 1998, 2001; Macaulay et al. 1999; Richards et al. 2000; Finnila et al. 2001; Helgason et al. 2001; Meinila et al. 2001). Likewise, studies of mtDNA diversity in native Siberians residing in northeastern Eurasia have revealed that these populations harbor Asian-specific mtDNA haplogroups A, C, D, G, and Y (Starikovskaya et al. 1998; Schurr et al. 1999). What is unclear is how this striking geographic separation of lineages occurred. Were Europe and Asia settled by totally different migrations out of Africa, or did Eurasian mtDNA diversity start as a continuum that subsequently became geographically stratified?
To address this question, we chose to survey the remnants of the Mansi, a tribal group of sedentary fishers and hunters who, for ages, inhabited the pine-birch boreal forests on the eastern slope of the Ural Mountains, the provisional geographic barrier separating Asia and Europe. The Mansi speak a dialect of the Finno-Ugric language of the Uralic linguistic family, to which the Saami language also belongs (Fedorova et al. 1994).
Blood samples were collected from 59 Mansi in the villages of Shaim, Chantarya, Polovinka, and Shugur, which are scattered along the Konda River and its tributaries. An additional 39 samples were collected from the tiny settlements of Shshekurya, Lombovozh, and Sos’va, in the northern Sos’va River/Lyamin River basin (fig. 1).
Figure 1.
A map of northwestern Siberia, indicating the locations of the two Mansi populations sampled.
mtDNA variation was surveyed by digestion with 19 restriction endonucleases, sequencing of hypervariable segments I and II (HVS-I and HVS-II, respectively) of the control region, and sequence detection of diagnostic polymorphic sites at nucleotide positions 7600, 12308, and 12705 (Torroni et al. 1998). The 98 Mansi mtDNA samples proved to be a novel mixture of 63% European mtDNA samples and 37% Asian-Siberian mtDNA samples. Of 62 European mtDNA samples, 28 harbored the characteristic 12308G coding-region mutation, indicating that they belong to haplogroup U; of the haplogroup U mtDNA samples, 16 harbored the diagnostic markers of subhaplogroup U4, including the RsaI-site gain at nucleotide position 4643 (i.e., 4643 RsaI+), 11329 AluI+, and the control-region 16356C mutation. An additional four mtDNA samples were assigned to the U5a subcluster, which is characterized by the 16256T and 16270T control-region motif, and five belonged to the U7 subcluster, which is characterized by the 16309G transition and the 16318T transversion. However, the Mansi lacked the U5b1 subcluster defined by the “Saami motif” of 16144C, 16189C, and 16270T control-region variants. Only three mtDNA samples belonged to subhaplogroup K of haplogroup U (tables 1 and 2).
Table 1.
mtDNA Diversity in the Mansi[Note]
| Haplogroup | No. ofSubjectsa | RFLP(s)b | HVS-I (−16000) | HVS-II |
| UK: | ||||
| U4 | 1 | 4643k 11329a 12308g | 356 519 | 73 195 263 |
| 4 | 4643k 11329a 12308g | 356 519 | 73 195 215 263 | |
| 2 | 4643k 11329a 12308g | 311 356 519 | 73 146 152 195 263 | |
| 1 | 4643k 11329a 12308g | 092 311 356 519 | 73 146 152 195 263 | |
| 1 | 4643k 11329a 12308g | 113C 356 362 519 | 73 195 263 | |
| 2 | 4643k −4685a 11329a 12308g | 113C 356 362 519 | 73 195 263 | |
| 1 | 4643k −4685a 11329a 12308g | 113C 239 356 362 519 | 73 195 263 | |
| 1 | 4643k −4685a 11329a 12308g | 113C 189 356 362 519 | 73 195 263 | |
| 1 | 626e 4643k −11326c 11329a 12308g | 189 356 519 | 73 263 | |
| 2 | 626e 4643k −11326c 11329a 12308g | 189 356 519 | 73 195 263 | |
| U5a | 1 | 12308g | 192 256 270 311 | 73 263 |
| 1 | −3192c 12308g | 129 239 256 270 399 | 73 150 263 | |
| U5a1 | 2 | 12308g | 114A 192 256 270 294 | 73 150 263 |
| U7 | 4 | 12308g | 309 318T 519 | 73 151 152 263 |
| 1 | −1715c 12308g | 309 318T 519 | 73 151 152 263 | |
| K | 3 | −322e −9052n 9714e 12308g | 224 311 519 | 73 146 152 263 |
| JT: | ||||
| J1b1 | 2 | 4216q 10394c −13704t | 069 126 145 172 222 261 | 73 242 263 295 |
| J2 | 10 | 4216q 10394c −13704t | 069 126 193 301 519 | 73 152 263 295 |
| T | 2 | 4216q 4914r 13366m 15606a −15925i | 126 294 296 304 519 | 73 263 |
| 2 | 4216q 4914r −11824a −13259o 13366m −13704t 15606a −15925i | 126 294 519 | 73 194 200 263 | |
| T1 | 1 | 4216q 4914r −12629b 13366m 15606a −15925i | 126 163 186 189 294 519 | 73 152 195 263 |
| 1 | 4216q 4914r −8838e −12629b 13366m 15606a −15925i | 126 163 186 189 261 294 519 | 73 152 195 263 | |
| 1 | 4216q 4914r −12629b 13366m −13704t 15606a −15925i | 126 163 186 189 294 519 | 73 152 195 263 | |
| HV: | ||||
| H* | 1 | −7025a −14766u | 169 184 | 73 263 |
| 1 | −7025a −14766u | 169 184 | 73 152 263 | |
| 1 | −7025a −14766u | 169 184 | 125 127 263 | |
| 2 | −7025a −14766u | 169 184 311 | 73 263 | |
| H2 | 1 | 4769a −7025a −14766u | CRS | 204 |
| 2 | 4769a −7025a −8858f −14766u | CRS | 204 | |
| H3 | 1 | −7025a −14766u | 093 172 189 519 | 263 |
| 1 | −7025a −14766u | 189 356 519 | 263 | |
| 1 | −7025a −14766u | 189 311 356 519 | 263 | |
| 1 | −7025a −14766u | 189 356 519 | 73 263 | |
| 2 |
−7025a 8249b −14766u |
080 189 356 | 263 | |
| V | 1 | −4577q −14766u | 298 | 72 263 |
| A | 2 | 663e | 039 189 223 290 319 356 362 | 73 152 235 263 |
| 1 | 663e | 223 227C 230 256 290 311 319 | 64 73 235 263 | |
| F | 1 | 4732k −12406h −12629b | 189 232A 249 304 311 519 | 73 204 248d 263 |
| C | 5 | 10394c 10397a −13259o | 223 298 327 519 | 73 248d 263 |
| 1 | 10394c 10397a −13259o | 223 298 311 327 519 | 73 189 207 248d 263 | |
| 1 | −1715c 10394c 10397a −13259o | 129 223 298 327 519 | 73 195 248d 263 | |
| 1 | −1715c 10394c 10397a −13259o | 093 129 223 298 327 519 | 73 248d 263 | |
| 1 | 10394c 10397a −13259o | 086 171 223 298 327 344 357 519 | 73 248d 263 | |
| 3 | 10394c 10397a −13259o 9bp-ins | 086 171 223 298 327 344 357 519 | 73 248d 263 | |
| 1 | −1715c 10394c 10397a −13259o | 114A 148 223 288 298 327 519 | 73 248d 263 | |
| 4 | 10394c 10397a −13259o | 298 327 519 | 73 248d 263 | |
| D | 1 | −5176a 10394c 10397a | 223 368 | 125 127 263 |
| 1 | −5176a 10394c 10397a | 223 362 368 | 125 127 263 | |
| 1 | −5176a 10394c 10397a | 223 362 368 | 263 | |
| 2 | −5176a 10394c 10397a | 192 223 261 316 362 | 73 263 | |
| 1 | −5176a −5823a 10394c 10397a | 223 291 294 362 519 | 73 152 263 | |
| 1 | −5176a −10180l 10394c 10397a −15925i | 223 319 362 | 73 239 263 297 | |
| 1 |
−5176a −8838e 12026h (10397 10398 10400 12705) |
126 136 189 223 360 362 | 73 263 | |
| G | 6 | 4830n −7598f 10394c 10397a | 086 172 223 227 278 362 | 73 263 |
| M* | 1 | 4164q 5351f 10394c 10397a (12705) | 129 189 223 297 298 | 73 150 199 263 |
Note.— Founding mtDNA types are shown in boldface italic. CRS = Cambridge reference sequence; d = deletion. Mutations relative to CRS [Andrews et al. 1999] are transitions unless the base change is specified.
Comprising 98 subjects overall.
The restriction enzymes are designated by the following single-letter codes appended to nucleotide positions: a = AluI; b = AvaII; c = DdeI; e = HaeIII; f = HhaI; g = HinfI; h = HpaI; i = HpaII; j = MboI; k = RsaI; l = TaqI; m = BamHI; n = HaeII; o = HincII; q = NlaIII; r = BfaI; s = AccI; t = BstNI; u = MseI. Underlining of the restriction site implies the simultaneous presence/absence of the linked site that is correlated with a single-nucleotide substitution. 9bp-ins = 9-bp COII/tRNALys triplication. Additional mutations in the coding region are shown in parentheses and were identified or confirmed by sequencing.
Table 2.
Frequencies (%) of mtDNA Types in the Mansi in Comparison with the Saami and Finns[Note]
|
Mansi |
|||||
| Haplogroup | Konda River(N=59) | Northern Sos'va River/ Lyamin River Basin(N=39) | Total (N=98) | Saami(N=176) | Finns(N=403) |
| U | … | … | … | … | 2.5 |
| U2 | … | … | … | … | .5 |
| U4 | 13.6 | 20.5 | 16.3 | … | .5 |
| U5a | 1.7 | 2.6 | 2.0 | .6 | 3.2 |
| U5a1 | 3.4 | … | 2.0 | .6 | 3.7 |
| U5b | … | … | … | 1.7 | 7.4 |
| U5b1 | … | … | … | 42.6 | 8.7 |
| U7 | 5.1 | 5.1 | 5.1 | … | .5 |
| U8 | … | … | … | … | .7 |
| K | 5.1 | … | 3.1 | … | 2.5 |
| K2 | … | … | … | … | .5 |
| JT: | |||||
| J | … | … | … | … | 3.0 |
| J1a | … | … | … | … | .5 |
| J1b1 | … | 5.1 | 2.0 | … | .5 |
| J2 | 10.2 | 10.3 | 10.2 | … | .5 |
| T | 6.8 | … | 4.1 | … | 1.5 |
| T1 | … | 7.7 | 3.1 | … | .1 |
| H* | 3.4 | 7.7 | 5.1 | … | … |
| H | … | … | … | 1.7 | 13.1 |
| H1 | … | … | … | 1.1 | 2.0 |
| H2 | 3.4 | 2.6 | 3.1 | … | 10.4 |
| H3 | 8.5 | 2.6 | 6.1 | … | 7.7 |
| H4 | … | … | … | … | 2.5 |
| H8 | … | … | … | 2.8 | 4.5 |
| V | 1.7 | … | 1.0 | 39.8 | 5.5 |
| A | 5.1 | … | 3.1 | … | … |
| F | 1.7 | … | 1.0 | … | … |
| C | 15.2 | 20.5 | 17.3 | … | … |
| Z | … | … | … | 3.4 | … |
| D | 6.8 | 10.3 | 8.2 | 5.1 | … |
| G | 6.8 | 5.1 | 6.1 | … | … |
| M* | 1.7 | … | 1.0 | … | … |
| M | … | … | … | … | 2.5 |
| W | … | … | … | .6 | 9.2 |
| I | … | … | … | … | 3.5 |
| X | … | … | … | … | 1.0 |
| Other | … | … | … | … | .3 |
The mtDNA samples that belonged to haplogroup TJ were also well represented in the Mansi and encompassed 7 and 12 mtDNA samples, respectively. The ancestral 4216 NlaIII+ and 16126C variants of haplogroup T (fig. 2) were found in the Mansi and also in the Finnish TJ phylogeny (Finnila et al. 2001). Two individuals of sublineage T and one individual of sublineage T1 appeared to have novel mtDNA haplotypes, including a previously undescribed site loss, 13704 BstNI−. Haplogroup J was confined to two sublineages, J2 and the rare J1b1 sublineage. The J2 mtDNA samples were found in 10.2% of the Mansi, even though they are rarely observed in other European populations, including the Saami and Finns (table 2). J1b1 was observed in two individuals.
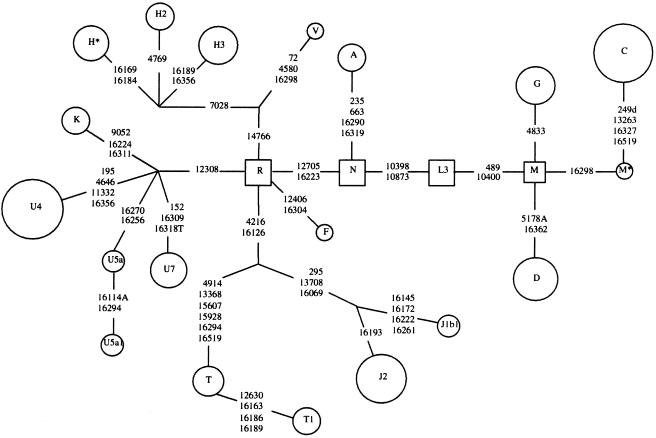
Figure 2.
Schematic phylogenetic representation of mtDNA lineages found in the Mansi. The sizes of the circles, which denote lineages, are proportional to the number of sampled individuals. Mutations are transitions unless otherwise specified. Squares denote the phylogenetic position of the roots of the nested haplogroups R, N, L3, and M (Macaulay et al. 1999; Quintana-Murci et al. 1999)
Haplogroup H, found in >40% of western Europeans, was present in only 14.3% of the Mansi mtDNA samples. Still, several sublineages of haplogroup H were observed, including six mtDNA samples of haplogroup H3, characterized by the 7025 AluI−, 14766 MseI− coding plus the 16189C and 16356C or 16189C control-region variants. An additional three Mansi mtDNA samples belonged to haplogroup H2, as indicated by the 4769 AluI+ variant and an HVS-I sequence identical to the Cambridge reference sequence (Andrews et al. 1999). Two of these three mtDNA samples also had the 8858 HhaI+. Finally, five of the haplogroup H mtDNA samples belonged to a unique, previously unreported variant (haplogroup H*) distinguished by the 16169T and 16184T control-region variants, in HVS-I, and the 73G variant, in HVS-II. Since the 73 site in HVS-II varies among the Mansi haplogroup H mtDNA samples, as well as among the Finnish mtDNA samples and other European mtDNA samples, it appears to be a hypervariable site (Finnila et al. 2001; Helgason et al. 2001). Therefore, we disregarded the 73 site for phylogenetic reconstruction (fig. 2). Thus, the structure of the haplogroup H mtDNA types in the Mansi is very similar to that in the Finns.
Haplogroup V was found in only one Mansi mtDNA sample. This mtDNA type was characterized by 4577 NlaIII−, 14766 MseI−, and the 16298C and 72C variants (table 1). Haplogroup V is at its highest frequency, 39.8%, among the Saami and has frequencies of 5.1% among Finns (table 2) and 2.6% among Russians from the Upper Volga River region (Malyarchuk et al. 2001). Hence, this Mansi haplogroup V mtDNA type could be due to gene flow from western Uralic speakers or Russian colonizers.
Among the 36 (37%) Mansi mtDNA samples that belong to traditional Asian haplogroups, 17 Mansi mtDNA samples harbored mtDNA of haplogroup C. One haplogroup C mtDNA type found in four individuals is missing the characteristic 16223T transition, presumably because of a reverse mutation. Eight of the Mansi mtDNA samples belonged to haplogroup D. One of these lacked the classically associated 10394 DdeI+/10397 AluI+ sites, which characterize macrohaplogroup M (at the RFLP level), which encompasses haplogroups C and D. Sequencing of the segment encompassing this region revealed the expected macrohaplogroup M 10398G and 10400T transitions. However, a 10397G transition was also found that would account for the simultaneous elimination of the adjacent restriction sites. Finally, single mtDNAs of haplogroups G and F and the unclassified M (i.e., M*) haplogroup were found in the Mansi. These may also be due to recent gene flow.
Thus, this study revealed that the Mansi possess a unique combination of western European and eastern Siberian mtDNA lineages. The novel distribution and subtypes of haplogroup U make it unlikely that this is the result of recent gene flow from modern Europeans. Rather, the Mansi mtDNA pool may contain traces of the ancient proto-Eurasian hunting-gathering populations that originally colonized the trans-Ural region and adjacent part of Siberia. In this regard, the most distinctive feature of the Mansi mtDNA samples is the high frequency (16.3%) of subhaplogroup U4. Neither U4 nor U5 subhaplogroups are common in the Middle East (Macaulay et al. 1999), and subhaplogroup U4 attains its highest frequency (28.9%) in the Ket of the Yenisey River (authors’ unpublished data). Hence, subhaplogroup U4 may be indicative of the remnants of Upper Paleolithic populations of Europeans preserved just east of the Ural Mountains.
The presence of subhaplogroup U7 in the Mansi supports the conjecture of its proto-Eurasian origin ∼24,000–54,000 years ago (Richards et al. 2000). Subhaplogroup U7 is virtually absent from modern Europeans and is absent or extremely rare in western Uralic-speaking populations, such as the Finns and Saami (Helgason et al. 2001; Meinila et al. 2001). However, subhaplogroup U7 is present in the Middle East, although in very low frequencies (Richards et al. 2000). Thus, in the Mansi, subhaplogroup U7 could be a part of Paleolithic dispersals from the Middle East, whose traces have not been erased by subsequent migrations. This conjecture is supported by the presence in the Mansi of subhaplogroup J2, which may indicate a Neolithic phase expansion toward the Ural Mountains.
Although the core of genetic makeup of the Mansi consisted of mtDNA types that apparently were ancient European, 29% of the Mansi mtDNA samples are from three of the four haplogroups that define the migration from Siberia to the Americas (i.e., haplogroups A, C, and D). Such a novel genetic structure could represent the recent amalgamation of western and eastern populations. Alternatively, the Mansi may reflect a more ancient and uniform distribution of mtDNA lineages that subsequently became more specialized as a result of later genetic drift or environmental selection. Whichever event proves to be the case, it is clear that further surveys of trans-Ural and Siberian mtDNA will yield much information about the genetic history of northern Eurasia and the origins of Uralic-speaking populations.
Acknowledgments
We are indebted to the Mansi people who, after providing informed consent, allowed us to draw blood and provided with the family histories. We would like to thank Dr. Kari Majamaa (University School of Medicine, Oulu, Finland) for access to his paper prior to its publication and Dr. Robert S. Hoffmann (Smithsonian Institution, Washington DC) for his helpful comments on an earlier draft of the manuscript. This research received support from INTAS (the International Association for the Promotion of Cooperation with Scientists from the New Independent States of the Former Soviet Union [European Community, Brussels, Belgium]) grant 96-1766 and National Institutes of Health grants TW1175 (to R.I.S.) and NS21328, AG13154, NS41850, and HL64017 (to D.C.W.).
References
- Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N (1999) Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet 23:147 [DOI] [PubMed] [Google Scholar]
- Fedorova EG, Koester DC (1994) Mansi. In: Friedrich P, Diamond N (eds) Encyclopedia of world cultures. Vol. 6: Russia and Eurasia/China. GK Hall, New York, pp 252–255 [Google Scholar]
- Finnila S, Lehtonen MS, Majamaa K (2001) Phylogenetic network for European mtDNA. Am J Hum Genet 68:1475–1484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helgason A, Hickey E, Goodacre S, Bosnes V, Stefansson K, Ward R, Sykes B (2001) mtDNA and the islands of the North Atlantic: estimating the proportions of Norse and Gaelic ancestry. Am J Hum Genet 68:723–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macaulay V, Richards M, Hickey E, Vega E, Cruciani E, Guida V, Scozzari R, Bonne-Tamir B, Sykes B, Torroni A (1999) The emerging tree of west Eurasian mtDNAs: a synthesis of control-region sequences and RFLPs. Am J Hum Genet 64:232–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malyarchuk BA, Denisova GA, Derenko MV, Rogayev EI, Vlasenko LV, Zhukova SG (2001) Variation of mitochondrial DNA in Russians of Krasnodar, Belgorod and Nizhnegorod Regions (in Russian). Genetika 37:1411–1416 [PubMed] [Google Scholar]
- Meinila M, Finnila S, Majamaa K (2001) Evidence for mtDNA admixture between the Finns and the Saami. Hum Hered 52:160–170 [DOI] [PubMed] [Google Scholar]
- Quintana-Murci L, Semino O, Bandelt H-J, Passarino G, McElreavey K, Santachiara-Benerecetti AS (1999) Genetic evidence of an early exit of Homo sapiens sapiens from Africa through eastern Africa. Nat Genet 23:437–441 [DOI] [PubMed] [Google Scholar]
- Richards M, Macaulay V, Hickey E, Vega E, Sykes B, Guida V, Rengo C, et al (2000) Tracing European founder lineages in the Near Eastern mitochondrial gene pool. Am J Hum Genet 67:1251–1276 [PMC free article] [PubMed] [Google Scholar]
- Sajantila A, Lahermo P, Anttinen T, Lukka M, Sistonen P, Savontaus ML, Aula P, Beckman L, Tranebjaerg L, Gedde-Dahl T, Issel-Tarver L, DiRienzo A, Pääbo S (1995) Genes and languages in Europe: an analysis of mitochondrial lineages. Genome Res 5:42–52 [DOI] [PubMed] [Google Scholar]
- Schurr TG, Sukernik RI, Starikovskaya YB, Wallace DC (1999) Mitochondrial DNA variation in Koryaks and Itel’men: population replacement in the Okhotsk Sea–Bering Sea region during the Neolithic. Am J Phys Anthropol 108:1–39 [DOI] [PubMed] [Google Scholar]
- Starikovskaya YB, Sukernik RI, Schurr TG, Kogelnik AM, Wallace DC (1998) Mitochondrial DNA diversity in Chukchi and Siberian Eskimos: implications for the genetic history of ancient Beringia and the peopling of the New World. Am J Hum Genet 63:1473–1491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torroni A, Bandelt H-J, D’Urbano L, Lahermo P, Moral P, Sellitto D, Rengo C, Forster P, Savantaus M-L, Bonne-Tamir B, Scozzari R (1998) mtDNA analysis reveals a major late Paleolithic population expansion from southwestern to northeastern Europe. Am J Hum Genet 62:1137–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torroni A, Bandelt H-J, Macaulay V, Richards M, Cruciani F, Rengo C, Martinez-Cabrera V, et al (2001) A signal, from human mtDNA, of postglacial recolonization in Europe. Am J Hum Genet 69:844–852 [DOI] [PMC free article] [PubMed] [Google Scholar]