Abstract
The widely expressed, homo-oligomeric, lipid raft-associated, monotopic integral membrane protein stomatin and its homologues are known to interact with and modulate various ion channels and transporters. Stomatin is a major protein of the human erythrocyte membrane, where it associates with and modifies the glucose transporter GLUT1; however, previous attempts to purify hetero-oligomeric stomatin complexes for biochemical analysis have failed. Because lateral interactions of membrane proteins may be short-lived and unstable, we have used in situ chemical cross-linking of erythrocyte membranes to fix the stomatin complexes for subsequent purification by immunoaffinity chromatography. To further enrich stomatin, we prepared detergent-resistant membranes either before or after cross-linking. Mass spectrometry of the isolated, high molecular, cross-linked stomatin complexes revealed the major interaction partners as glucose transporter-1 (GLUT1), anion exchanger (band 3), and water channel (aquaporin-1). Moreover, ferroportin-1 (SLC40A1), urea transporter-1 (SLC14A1), nucleoside transporter (SLC29A1), the calcium-pump (Ca-ATPase-4), CD47, and flotillins were identified as stomatin-interacting proteins. These findings are in line with the hypothesis that stomatin plays a role as membrane-bound scaffolding protein modulating transport proteins.
Abbreviations: ASIC, acid-sensing ion channel; AvTIC, average total ion current; DRM, detergent-resistant membrane; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; LC-MS/MS, liquid chromatography tandem MS; MS, mass spectrometry; PMSF, phenylmethylsulfonyl fluoride; TNE, Tris/NaCl/EDTA buffer; TNET, Tris/NaCl/EDTA/Triton X-100 buffer
Keywords: Integral membrane proteins, Lipid rafts, Chemical cross-linking, Protein–protein interaction
Graphical abstract
Highlights
► Stomatin is chemically cross-linked in erythrocyte membranes ► Stomatin complexes are immunoaffinity-purified and identified by mass spectrometry ► Major cross-linked partners are GLUT1 and band 3 ► Stomatin is cross-linked in detergent-resistant membranes ► New interaction partners include flotillins and various transporters/channels, particularly aquaporin-1.
1. Introduction
Stomatin, also known as band 7 integral membrane protein or protein 7.2b, is a major erythrocyte membrane protein [1–3] that is missing in red cells of overhydrated hereditary stomatocytosis patients [3]. It is expressed ubiquitously and conserved from archaea to mammals. In humans, there are 5 similar proteins [4–6], while the C. elegans genome contains 10 stomatin-like genes, including mec-2, unc-1, and unc-24 as best studied [7,8]. The common domain of stomatin-like and related proteins is known as SPFH (stomatin, flotillin, prohibitin, HflC/K)-domain [9,10] or PHB (prohibitin homology)-domain [11]. These SPFH/PHB-proteins may play a role as membrane-bound scaffolding proteins that are associated with other membrane proteins and cortical cytoskeleton [11,12].
Hallmarks of stomatin are the monotopic structure [13], oligomeric nature [14,15], S-palmitoylation [16], and lipid raft-association [15,17,18]. Moreover, stomatin and stomatin-like proteins are cholesterol-binding proteins [4,19]. Most of these features are also characteristic for other SPFH/PHB-domain proteins and for the topologically similar but unrelated caveolins [20]. The crystal structure of an archaeal stomatin core domain revealed a unique, trimeric structure with extending α-helices from each triangular corner that interact with equal α-helices of adjacent trimers to form antiparallel coiled-coils thus explaining the homo-oligomeric nature [21]. In contrast, crystal structures of the mouse stomatin-domain were found to be composed of banana-shaped dimers similar to BAR-domains forming hexagonal structures that are capable of building oligomers [22]. While stomatin and stomatin-like proteins are known to interact with various ion channels modulating their activities [19,22–25], only human stomatin has been shown to associate with the glucose transporter GLUT1 [26–30]. The interaction of stomatin and GLUT1 is also implicated by the loss of function of this complex in erythrocytes of patients with stomatin-deficient cryohydrocytosis [31]. Apparently, stomatin modulates GLUT1 to repress glucose uptake while enhancing dehydroascorbate influx [30]. The molecular mechanism of this modulation has not been investigated yet. Because erythrocyte GLUT1 is only found in mammals that are unable to synthesize vitamin C, it is implicated that the high GLUT1 expression in human erythrocytes may be due to a compensatory mechanism for better utilising ascorbate [30]. This stomatin-dependent mechanism was debated [32,33] and therefore we set out to study the direct physical interaction of these proteins by in situ chemical cross-linking. We show here that stomatin forms major complexes with GLUT1, as anticipated, but also with band 3 and aquaporin-1. Moreover, we found stomatin to associate with several transporters suggesting a general role as a modulator of transport proteins.
2. Materials and methods
2.1. Reagents
Human blood from healthy donors in EDTA-vials was obtained from the Austrian Red Cross, Vienna. For each experiment, washed red blood cells of 4 donors were pooled. Antibodies were used against stomatin (GARP-50, GARP-61, GARP-65) [1], GLUT1 (Millipore), glycophorin A (Santa Cruz), flotillin 2 (BD Biosciences), and band 3 (Sigma). The cross-linkers ethylene glycolbis(succinimidylsuccinate) (EGS; bridging 16.1 Å distance) and disuccinimidyl suberate (DSS; bridging 11.4 Å distance) were purchased from Pierce/Thermo Scientific; CNBr-activated Sepharose was from Pharmacia/GE Healthcare. Other chemicals of highest purity were from Merck/VWR or Sigma.
2.2. Preparation of erythrocyte membranes
Erythrocytes were purified from 10 ml blood by washing with PBS (3 times 1000 ×g), filtration through a column of microcrystalline and α-cellulose [34] and pelleting. Membranes were prepared by hypotonic lysis in 20 volumes of 5 mM EDTA, pH 8.0, 1 mM PMSF (lysis buffer), on ice for 10 min, and centrifugation at 20,000 ×g (Sorvall RC5C Plus) for 10 min. The pellet was washed 3 times with lysis buffer. To reduce sample complexity in several experiments, the cytoskeleton was stripped off the membranes by incubating with 10 volumes 0.1 M NaOH on ice for 10 min and washing with lysis buffer.
2.3. Preparation of DRMs
Native membranes were suspended in an equal volume 1% Triton X-100, 5 mM EDTA, 1 mM PMSF in PBS and incubated on ice for 10 min. This mixture was subjected to flotation by mixing with 80% sucrose in PBS (alternatively, in 0.15 M Na2CO3 instead of PBS) to yield 50% sucrose, and placed at the bottom of a centrifuge tube. Solutions of 40%, 35% and 5% sucrose in PBS were overlaid sequentially. The samples were centrifuged at 230,000 ×g (Beckman Coulter Optima™ L-80 XP ultracentrifuge, SW55Ti rotor) for 16 h. Nine fractions of 0.5 ml were collected from the top and aliquots were analysed by SDS-PAGE/silver staining and Western blotting.
2.4. Chemical cross-linking of membranes
Native or stripped erythrocyte membranes were incubated with 0.8 mM EGS or DSS in PBS, pH 8.0, as recommended by the manufacturer. The reactions were performed on ice for 30 min and stopped by adding 15 mM Tris–HCl, pH 8.0. Respective membrane pellets were solubilised in 1 ml TNET (20 mM Tris–HCl, pH 8.0, 130 mM NaCl, 5 mM EDTA, 1% Triton X-100, 1 mM PMSF) at 25 °C for 15 min, cleared by centrifugation (14 000 rpm, Eppendorf, 10 min), and the supernatant was used for immunoisolation of stomatin-complexes. Alternatively, membranes were dissolved in 1% SDS for 5 min at 37 °C, and the solution was diluted with 10 volumes cold (4 °C) TNET before immunoisolation.
2.5. Chemical cross-linking of DRMs
Isolated DRMs were cross-linked with either 16 μM or 8 μM EGS, quenched with 15 mM Tris–HCl, pH 8.0, dissolved in 1% SDS at 37 °C, and the solution was diluted with 10 volumes cold (4 °C) TNET before immunoisolation.
2.6. Immunoisolation of stomatin-complexes
TNET- or SDS/TNET-solubilised, chemically cross-linked membrane proteins were diluted with an equal volume cold (4 °C) TNE (20 mM Tris–HCl, pH 8.0, 130 mM NaCl, 5 mM EDTA, 1 mM PMSF) and loaded onto a small column (1 × 1 cm) of monoclonal anti-stomatin antibody GARP-50 covalently bound to CNBr-activated Sepharose (1 mg/ml), as described [1]. The column was washed with 15–20 ml 0.1% Triton X-100 in TNE, in some experiments with an intermediate wash with 5 ml 0.3 M NaCl in 0.1% Triton X-100 in TNE, and stomatin complexes were eluted with 5-times 1 ml 0.1 M glycine–HCl, pH 2.5, 0.1% Triton X-100. Each fraction was collected into 55 mM Tris–HCl, pH 8.8, then adjusted to 0.1% SDS, freeze-dried (Speed-Vac), and re-dissolved in 100 μl water.
In summary, twelve independent immunoisolation experiments were performed.
2.7. SDS-PAGE and Western blotting
The immunoaffinity elution fractions were mixed with Laemmli sample buffer, heated for 3 min at 95 °C, and loaded onto 7% or 10% Laemmli SDS-PAGE gels (Hoefer Sturdier SE 400, 14 × 12 cm) along with HiMark™ pre-stained high molecular weight standard (Invitrogen). Running conditions for large complexes were up to 24 h at 150 V, 4 °C. In addition, 4–12% gradient gels (GE Healthcare) were used and run for 1 h at 160 V. Gels were silver-stained by a mass spectrometry-compatible method [35] or blotted onto nitrocellulose (16 h at 100 mA, 4 °C) by standard methods. To estimate the relative molecular mass of protein complexes, the Ferguson plot was used. For Western blotting, usually mini-gels (10 × 8 cm) have been used.
2.8. Mass spectrometry
Silver-stained bands were cut out, proteins digested with trypsin, and the peptides analysed by nano-electrospray LC-MS/MS. Spectra were processed by Mascot 2.2.04 (Matrix Science, London) and the identified peptides were semi-quantitatively estimated by Average Total Ion Current (AvTIC) using the Scaffold3 software (Proteome Software, Portland). Details are given in the Supplementary data. In summary, about 60 MS-analyses were performed.
3. Results
3.1. Isolation and identification of stomatin-complexes after chemical cross-linking of erythrocyte membranes
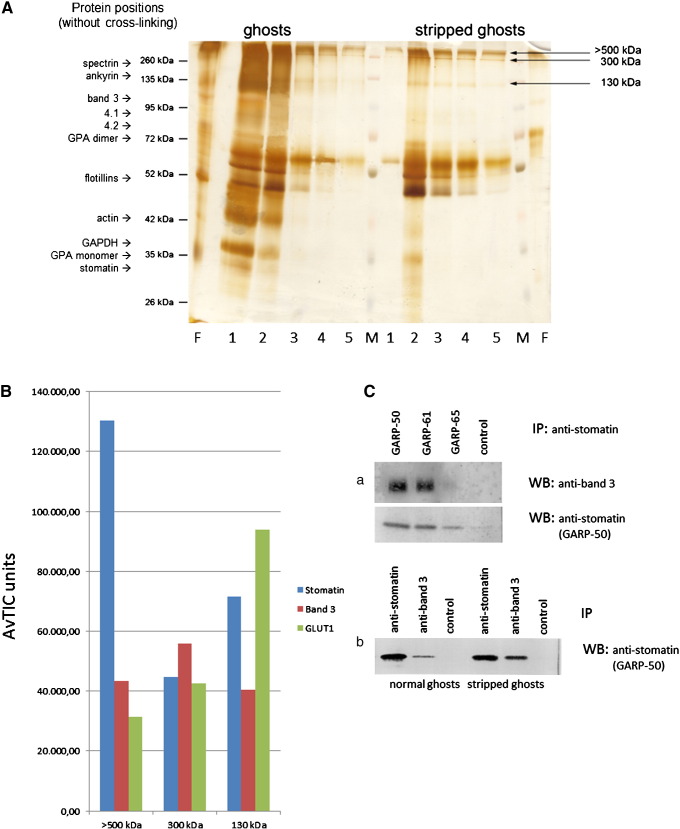
To generate chemically cross-linked stomatin complexes in situ, we incubated normal or cytoskeleton-depleted erythrocyte membranes with 0.8 mM EGS. After membrane solubilisation we isolated the stomatin-complexes by immunoaffinity chromatography and analysed them by SDS-PAGE (Fig. 1A). Due to the massive cross-linking of native membranes, we rather focussed on the cytoskeleton-depleted membranes, because we wanted to target integral membrane proteins, and excised the bands with 130 kDa, 300 kDa, and larger than 500 kDa (Fig. 1A). MS-analysis of these bands clearly revealed the presence of 3 major proteins: stomatin, glucose transporter-1 (GLUT1/SLC2A1), and the anion exchanger (band 3/AE1/SLC4A1) (Fig. 1B). The major component in the > 500 kDa band was stomatin, while GLUT1 was highest in the 130 kDa band. In addition to stomatin, GLUT1 and band 3, the > 500 kDa band contained flotillin-1 and -2, the urea transporter-1 (UT1/SLC14A1), iron transporter ferroportin-1 (FPN1/SLC40A1), Kell protein/CD238, and protein 4.2, with the urea transporter exceeding GLUT1 and equalling band 3 amounts (Supplementary Fig. 1). While it is known that GLUT1 interacts with stomatin, it is not known for band 3 and the minor proteins. We performed immunoprecipitation/Western blotting to verify the interaction with band 3 (Fig. 1C).
Fig. 1.
Isolation and analysis of EGS-cross-linked stomatin complexes and immunochemical verification of stomatin-band 3 interaction. (A) Erythrocyte membranes (“ghosts”), either untreated (left side) or depleted of cytoskeleton (“stripped ghosts”, right side), were cross-linked with 0.8 mM EGS, quenched, and solubilised; stomatin-complexes were isolated by immunoaffinity chromatography. Elution fractions were analysed by 10% SDS-PAGE/silver staining, excision of bands, as indicated, and mass spectrometry. Positions of un-cross-linked erythrocyte membrane proteins are indicated. Flow-through and elution fractions are shown. (B) Three stomatin-complexes isolated from stripped ghosts, > 500 kDa, about 300 kDa, and 130 kDa, contain stomatin, band 3, and GLUT1 as major components. Semi-quantitative results are shown as Average Total Ion Current (AvTIC) units. (C) Verification of the stomatin-band 3 interaction. (a) IP/WB analysis with monoclonal anti-stomatin antibodies against N- (GARP-50) and C-terminal (GARP-61, -65) epitopes and monoclonal anti-band 3, as indicated (GARP-65 has weak affinity). Control is an irrelevant antibody. (b) IP/WB analysis using normal or stripped ghosts. Co-IP works better with stripped ghosts. M, marker; F, flow-through; 1–5, respective elution fractions 1–5; GPA, glycophorin A; IP, immunoprecipitation; WB, Western blotting.
To test the possibility of sample contamination during preparation, we performed the isolation procedure with an unspecific antibody-column before the anti-stomatin-column and analysed the eluates concomitantly. Only traces of band 3, haemoglobin, and antibody were found next to laboratory background proteins such as keratins (not shown).
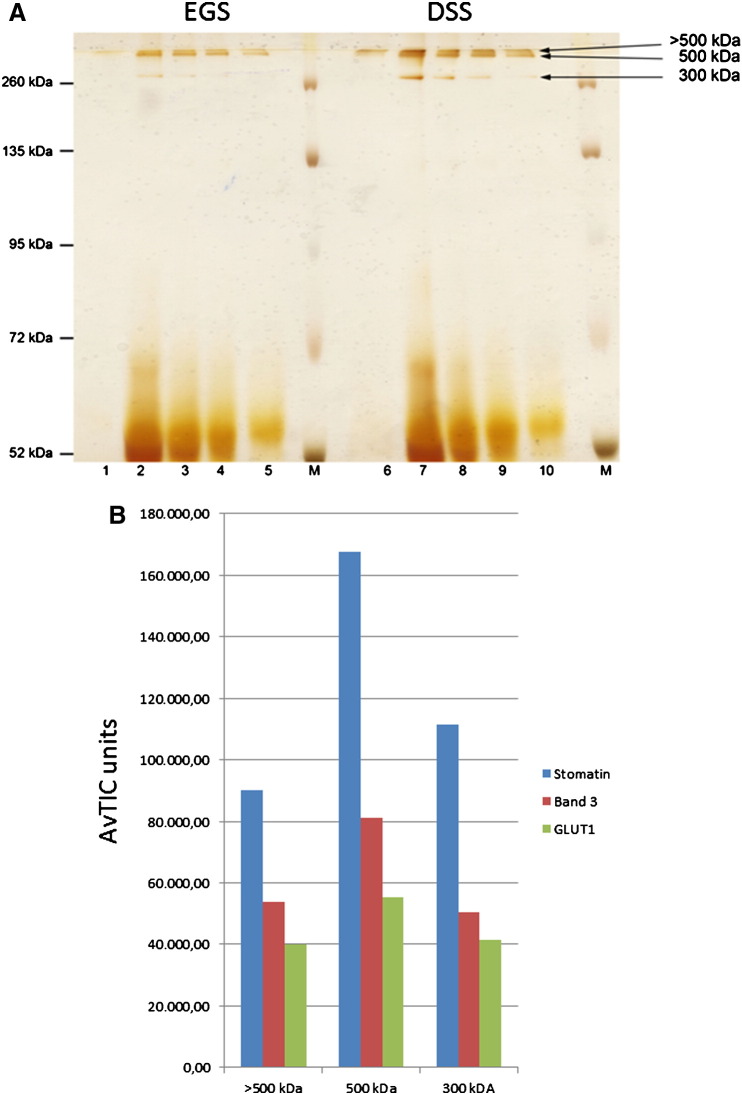
In the next approach we incubated cytoskeleton-depleted membranes with 0.8 mM EGS or DSS, quenched and solubilised them as before and isolated stomatin complexes by immunoaffinity chromatography. To further reduce possible unspecific binding of proteins to the column, we used 20 column volumes for washing, with an intermediate 0.3 M NaCl wash. DSS-cross-linked bands were seen at roughly 300 kDa, 500 kDa, and > 500 kDa (Fig. 2A). MS-analysis identified stomatin, band 3, and GLUT1 as major components again (Fig. 2B). In addition, the ≥ 500 kDa-bands contained flotillins, ferroportin-1 (FPN1), urea transporter-1 (UT1), as before but also Lutheran blood group antigen (Lu/BCAM/CD239), Landsteiner-Wiener antigen (LW/ICAM-4/CD242), and spectrin, with FPN1 and UT1 exceeding GLUT1 amounts (Supplementary Fig. 2). The 300 kDa band also contained aldolase A, probably associated with band 3.
Fig. 2.
Isolation and analysis of DSS-cross-linked stomatin complexes. (A) Stripped ghosts were cross-linked with either 0.8 mM EGS (left side) or DSS (right side), and stomatin-complexes were isolated as before (see Fig. 1). Elution fractions were analysed by 10% SDS-PAGE (prolonged running time 24 h at 70 V), silver staining, and mass spectrometry of excised bands, as indicated ( > 500 kDa, 500 kDa, and 300 kDa). (B) Major components of DSS-cross-linked stomatin-complexes are band 3 and GLUT1. Semi-quantitative data are given in AvTIC units. M, marker; 1–5 and 6–10, respective elution fractions 1–5.
3.2. Chemical cross-linking and flotation of stomatin complexes
To improve the yield and purity of cross-linked stomatin-complexes, we took advantage of stomatin's association with cholesterol-rich membrane domains (“lipid rafts”) and added a flotation density gradient step before immunoisolation. We first tested the flotation behaviour of DRMs prepared from cytoskeleton-depleted membranes after cross-linking with 0.8 mM EGS or DSS (Fig. 3A, B). Cross-linked stomatin and GLUT1 floated to low density fractions (Fig. 3A–C). We solubilised the DSS-cross-linked DRMs and isolated the stomatin-complexes by immunoaffinity chromatography. SDS-PAGE of elution fractions showed bands at roughly 300 kDa, 500 kDa, and > 500 kDa, as before (Fig. 3D). The 500 kDa and 300 kDa bands were excised and subjected to MS-analysis. Major proteins were identified as stomatin, band 3, and GLUT1 as before. Unexpectedly, high amounts of the water channel aquaporin-1 (AQP1) were also present, particularly in the 300 kDa complex (Fig. 3E). Modulation of AQP1 has not been described. The identification of AQP1 was verified by MS-analysis of tryptic and chymotryptic digests (data not shown). Moreover, ferroportin-1 (FPN1), Lu/CD239, aldolase A, and GAPDH were identified, with FPN1 exceeding GLUT1 amounts (Supplementary Fig. 3).
Fig. 3.
Analysis of stomatin-complexes after cross-linking with DSS and flotation. Stripped ghosts were cross-linked with (A) 0.8 mM EGS or (B) 0.8 mM DSS. Respective membranes were treated with cold Triton X-100, subjected to flotation, and cross-linked stomatin was identified by Western blotting of density gradient fractions. Note that cross-linked stomatin floats to low density. The dimer (about 65 kDa) is visible as major product. The monomer is not seen due to prolonged electrophoresis (24 h at 70 V). (C) In a different experiment, gradient fractions were analysed for cross-linked stomatin and GLUT1 by Western blotting, as indicated. Note that both cross-linked proteins float to low density. (D) DRM-fractions 3 and 4 from DSS-cross-linked membranes were pooled, solubilised, and stomatin-complexes were isolated by immunoaffinity chromatography. Elution fractions were analysed by SDS-PAGE/silver staining and MS of excised bands, as indicated. (E) Semi-quantitative data of the 500 kDa and 300 kDa complexes are given in AvTIC units. Note the major amount of aquaporin-1 (AQP1) associated with the 300 kDa protein complex. M, marker; 1–5, respective elution fractions.
3.3. Isolation and cross-linking of DRMs
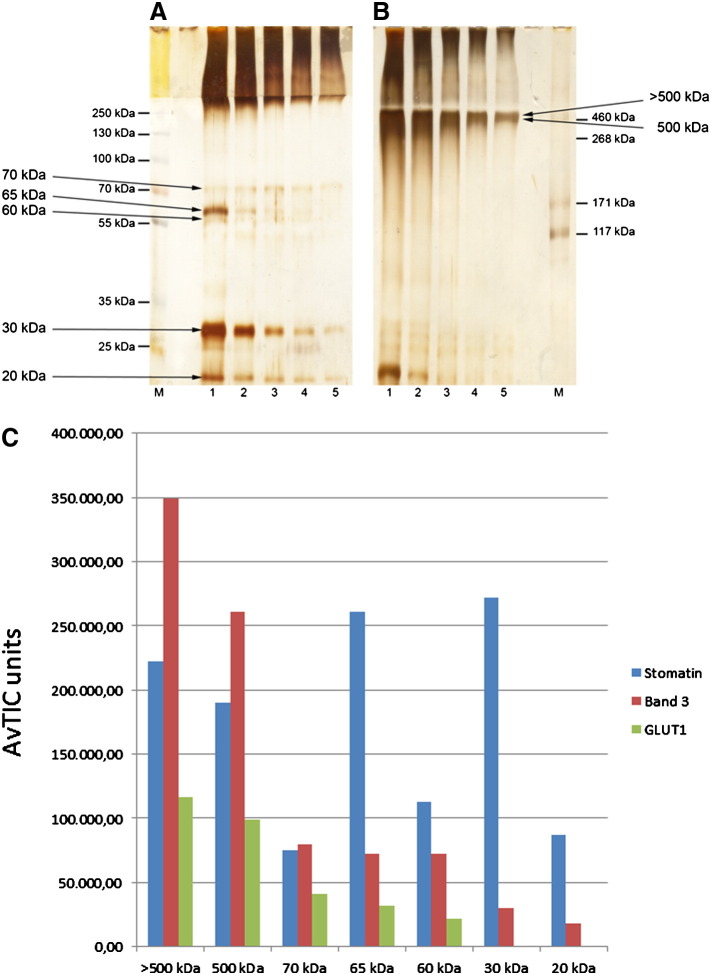
In another approach to improve the yield and purity of cross-linked high molecular stomatin complexes, we prepared DRMs from cytoskeleton-depleted membranes and cross-linked these with 16 μM EGS. To ensure complete solubilisation of the cross-linked DRMs, we used 1% SDS at 37 °C and diluted this solution with 10-fold 1% Triton X-100 before immunoaffinity chromatography. Eluted fractions were analysed by SDS-PAGE/silver staining (Fig. 4A, B) and the major bands were analysed by MS. Semi-quantitative MS-analysis of the indicated bands showed the size distribution of homo- or hetero-complexes of stomatin with or without GLUT1 and band 3. Peaks of high stomatin-concentration were visible in the high molecular region ≥ 500 kDa but also at the mass of its homo-dimer (65 kDa) and monomer (30 kDa). Band 3 and GLUT1 were mainly identified at ≥ 500 kDa (Fig. 4C). Moreover, the high molecular bands contained appreciable amounts of the Rh-associated “self”-antigen CD47, the calcium pump Ca-ATPase-4/ATP2B4/PMCA-4, aquaporin-1, ferroportin-1/SLC40A1, urea transporter-1/SLC14A1, nucleoside transporter ENT1/SLC29A1, LW-antigen/ICAM-4, flotillin-1 and -2, and protein 4.2 (Supplementary Fig. 4). Taking into account the low level expression of these transporters, receptors, pump, and channel, and the low concentration of cross-linker, these interactions with stomatin appear highly specific.
Fig. 4.
Analysis of EGS-cross-linked stomatin-complexes in DRMs. Stripped ghosts were treated with cold Triton X-100 and subjected to flotation. DRMs were isolated and cross-linked with 16 μM EGS. After quenching and solubilisation with SDS, the mixture was adjusted to 1% Triton X-100 and stomatin-complexes were immunopurified. Elution fractions were analysed by (A) 10% or (B) 7% SDS-PAGE/silver staining. Stained bands were excised, as indicated, and analysed by MS. (C) Semi-quantitative results (AvTIC units) show the size-distribution of stomatin, band 3, and GLUT1. Stomatin peaks are clearly seen at the size of oligomers ( ≥ 500 kDa), the homo-dimer (65 kDa), and monomer (30 kDa). The 20 kDa stomatin may represent an intramolecularly cross-linked species. Low-molecular band 3 may represent fragments or contamination. M, marker; 1–5, respective elution fractions.
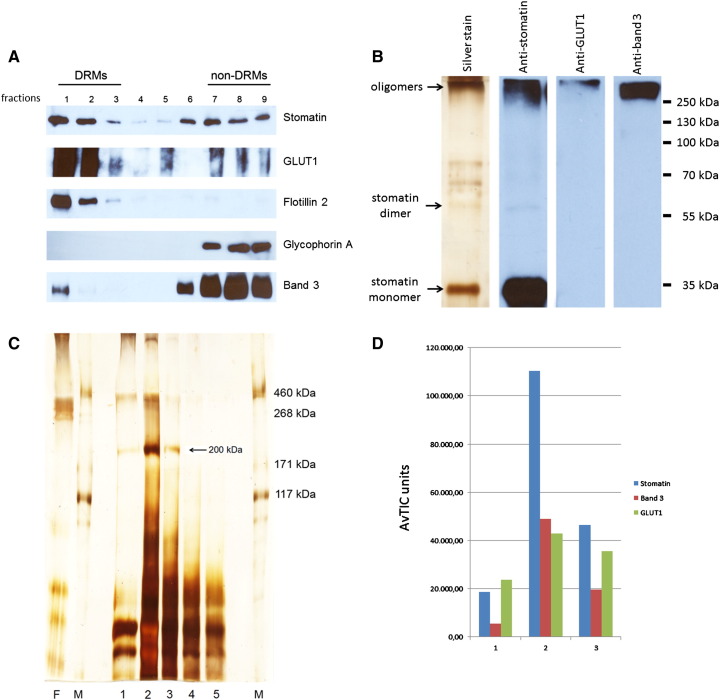
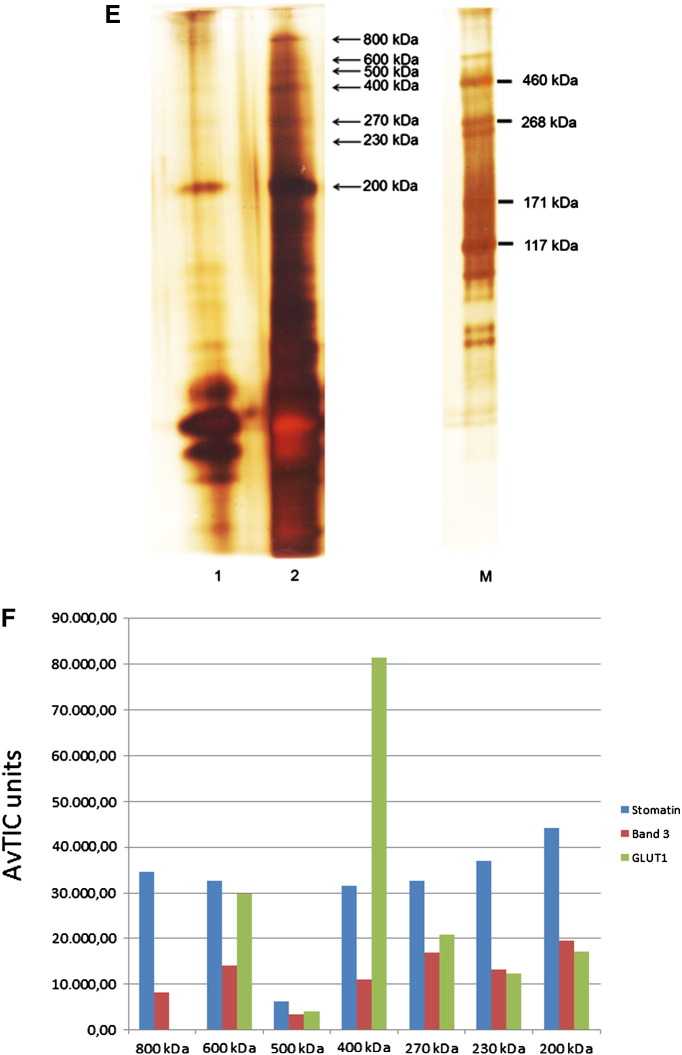
Using a variation of the approach above, we prepared DRMs from normal erythrocyte membranes by flotation from an alkaline 50% sucrose fraction, as described previously [18]. Part of stomatin, GLUT1, and a relatively small amount of band 3 floated to the top fractions (Fig. 5A). These DRMs were cross-linked with 8 μM EGS, quenched and solubilised in SDS. After immunoaffinity chromatography, the first fraction was analysed by SDS-PAGE/silver-staining and Western blotting. Oligomeric stomatin, GLUT1, and band 3 were seen (Fig. 5B) but also a large amount of monomeric stomatin, thus reflecting the very low concentration of cross-linker, far below saturation. On SDS-PAGE/silver staining of the elution fractions, a prominent band at 200 kDa was visible (Fig. 5C) that contained a large amount of stomatin with GLUT1 and band 3 associated (Fig. 5D). High molecular protein bands were also evident (Fig. 5C). To improve the separation of these high molecular complexes, we used 4–12% gradient gels and excised 7 bands in the region of 200 kDa to 800 kDa (Fig. 5E). The results of semi-quantitative MS-analyses are shown in Fig. 5F. Marked variation of compositions implicates the presence of distinct stomatin complexes. Particularly, the 400 kDa band contains a large amount of GLUT1 associated with stomatin, while the 800 kDa complex mainly contains stomatin but lacks GLUT1. A band at 500 kDa apparently did not contain prominent amounts of protein and may be seen as an internal background control (Fig. 5F).
Fig. 5.
Preparation and analysis of EGS-cross-linked high molecular stomatin-complexes in DRMs. (A) Normal ghosts were treated with cold Triton X-100, mixed with alkaline sucrose, and subjected to flotation. Gradient fractions were analysed by Western blotting as indicated. To prevent GLUT1 overstaining in the non-DRM dense fractions 6–9, these fractions were diluted 1:50. Glycophorin A was used as a non-raft marker. (B) Combined DRM fractions 1 + 2 were incubated with 8 μM EGS, quenched, solubilised with SDS, adjusted to 1% Triton X-100, and stomatin-complexes were immunopurified. Elution fraction 1 was analysed by mini-gel 10% SDS-PAGE/silver staining and Western blotting as indicated. High-molecular complexes are visible. Note the major band of non-cross-linked stomatin. (C) Flow-through and elution fractions 1–5 were analysed by 7% SDS-PAGE/silver staining and mass spectrometry of excised 200 kDa bands (fractions 1–3), as indicated. (D) Semi-quantitative MS-analysis shows the composition of the three 200 kDa complexes eluted in fractions 1–3, respectively, reflecting the mass distribution in the fractions. (E) High molecular stomatin-complexes were separated on a 4–12% gradient gel, excised, as indicated, and analysed by MS. (F) Semi-quantitative MS-analysis shows that the region between 200 and 800 kDa contains stomatin-complexes of varying composition. Whereas stomatin is the major constituent of most complexes, the 400 kDa complex contains GLUT1 as major component. Data are compared by AvTIC units. F, flow-through; 1–5, respective elution fractions; M, marker.
3.4. Identification of cross-linker-modified peptides
Mass spectrometry data were searched for cross-linked peptides and cross-linker-modified peptides. Peptides containing cleaved EGS were identified and assigned to the major proteins, stomatin, band 3, and GLUT1, but also to less abundant membrane proteins (Table 1). The signal counts for each modified peptide correlated with the amount of protein but the number of individual peptides was rather limited thus indicating the accessible, preferred sites for chemical cross-linking. The most reactive, exposed lysine residues of stomatin were identified at the N-terminus and the long α5-helix (terminology taken from [21]). An α5-equivalent, reactive site was also identified in flotillin-1. Band 3 had the majority of reactive lysines on the large cytoplasmic N-terminal domain, whereas GLUT1 had all identified reactive lysines on the large cytoplasmic loop between TM6 and TM7 (Table 1).
Table 1.
Identified sites containing EGS-derived modifications.
| Protein | Position | Peptide | Structural feature | Identification (#)a (% of total counts) |
|---|---|---|---|---|
| Stomatin | 9–25 | DSEAQRLPDSFKDSPSK | N-terminus | 1 (1%) |
| 15–25 | LPDSFKDSPSK | N-terminus | 24 (25%) | |
| 56–62 | IIKEYER | CRAC motif b | 5 (5%) | |
| 196–205 | DVKLPVQLQR | β6c | 2 (2%) | |
| 219–232 | AKVIAAGEMNASR | α5c | 64 (67%) | |
| Band 3 | 346–360 | RYQSSPAKPDSSFYK | N-terminal domain | 2 (9%) |
| 347–360 | YQSSPAKPDSSFYK | N-terminal domain | 12 (52%) | |
| 591–600 | FKNSSYFPGK | Loop TM6–7 | 3 (13%) | |
| 593–602 | NSSYFPGKLR | Loop TM6–7 | 2 (9%) | |
| 731–743 | SVTHANALTVMGK | Loop TM9–10 | 3 (13%) | |
| 818–827 | YHPDVPYVKR | Loop TM11–12 | 1 (4%) | |
| GLUT1 | 224–229 | AKSVLK | Loop TM6–7 | 3 (30%) |
| 224–230 | AKSVLKK | Loop TM6–7 | 6 (60%) | |
| 226–232 | SVLKKLR | Loop TM6–7 | 1 (10%) | |
| UT1 | 362–374 | MPLSKVTYPEENR | C-terminus | 1 |
| 375–382 | IFYLQAKK | C-terminus | 1 | |
| Flot1 | 169–177 | TAQVQKDAR | α5-equivalentc | 1 (20%) |
| 178–185 | IGEAEAKR | α5-equivalentc | 3 (60%) | |
| 245–253 | TKQQIEEQR | Coiled-coil | 1 (20%) | |
| Flot2 | 278–287 | TDKELIATVR | Coiled-coil | 1 |
| RhCE | 2–7 | SSKYPR | N-terminus | 2 |
Signal counts of identified peptides.
Cholesterol recognition/interaction amino acid consensus.
Refers to P. horikoshii stomatin structure [21].
4. Discussion
Stomatin and its homologues are known to associate with various ion channels [4,19,22,23]. Human stomatin also interacts with the glucose transporter GLUT1 [26,27,29,30], a finding that we verified now by in situ-cross-linking and MS-analysis. This interaction, which apparently involves the GLUT1 C-terminal region [27], may lead to a structural change obstructing the glucose-permeable pore while facilitating dehydroascorbate influx [30]. A regulatory role of lipid rafts in this interaction has been suggested [28,29].
Previous attempts to purify heteromeric stomatin complexes for biochemical studies yielded only homo-oligomers [14]. Because interactions between integral membrane proteins are often transient and unstable in solution, we have used chemical cross-linking to covalently lock these interactions in situ. Erythrocyte membranes are perfectly suited for this approach, because they can be easily prepared and manipulated. However, when cytoskeletal components are massively cross-linked to the membrane protein of interest, they create complex SDS-PAGE banding patterns (Fig. 1A). Because we focussed on the interaction of stomatin with other integral membrane proteins, we depleted the membranes of cytoskeleton by washing with alkaline solutions (“stripping”) (Fig. 1). Stomatin is known to associate with cortical actin filaments in nucleated cells [36] and possibly in erythrocyte membrane domains [18]. The nature of this association is unknown but depends on membrane cholesterol-levels [4]. Here, we identified protein 4.2 interacting with stomatin. Protein 4.2 is palmitoylated and interacts with α-spectrin and band 3 [37,38].
MS-analysis of high molecular bands identified stomatin, GLUT1, band 3, and aquaporin-1 as major components and various transporters and receptors as minor components, although ferroportin-1 and urea transporter-1 eventually exceeded GLUT1 amounts (Supplementary Figs. 1–3). In these complexes, stomatin was frequently predominant, because we selected for it, but in some high molecular bands either GLUT1, band 3, or aquaporin-1 was most prominent. One might argue that the high amounts of cross-linked band 3 and GLUT1 may be caused by the abundance of these proteins in the human erythrocyte membrane. This is particularly true for band 3, which is expressed at 1 million copies/cell, as compared to roughly 100,000 copies for stomatin; GLUT1 expression is about 200,000 copies/cell. However, we argue for a specific interaction of stomatin with these proteins, because other highly abundant membrane proteins were not cross-linked to stomatin; e.g. glycophorin A, with almost 1 million copies/cell, was never identified, nor the other prominent glycoproteins or the hydrophobic proteins such as Rh-RhAG-proteins, Na,K-ATPase, or Duffy antigens. Eventually, the stomatin-band 3 co-immunoprecipitation may be taken as proof for the specificity of this interaction (Fig. 1C).
The cross-linkers EGS and DSS, although somewhat different in length (16.1 and 11.4 Å, respectively), had similar efficiency (Figs. 2 and 3). EGS has the advantage to be specifically cleavable by hydroxylamine; however, we noticed also cleavage during the tryptic digestion, apparently due to the NH4HCO3 buffer. The cross-linker concentrations that we have used were in the low range, as evident by the small proportion of stomatin in high molecular species compared to the major amount of monomer and dimer (Figs. 3–5), in accordance with earlier results [1,14]. Another argument for the specificity of cross-linking is the presence of cross-linker-derived modifications at restricted sites on stomatin, band 3, GLUT1, and other membrane proteins (Table 1).
When we analysed high molecular bands, the stoichiometry of identified proteins in the cross-linked complexes was variable, probably depending on the number of cross-links between them. Variability may come from different numbers of stomatin monomers in the complexes and of the interacting partners, band 3, GLUT1, and aquaporin-1, which form oligomers (dimers, tetramers) themselves. Although immunoisolation yielded discrete bands in the high molecular range, this may not necessarily imply the presence of single molecular species. Diverse complexes with similar mass may co-localise in one band. Varying staining-efficiency and broad electrophoretic bands due to varying glycosylation may add to a certain degree of overlap and inaccuracy, which is a common problem, when working with membrane proteins and puts a limitation to the power of the current method. New methods will have to be developed for the separation of large hydrophobic protein complexes with high resolution.
The identification of stomatin-associated transporters, i.e. GLUT1 (SLC2A1), band 3 (SLC4A1), urea transporter-1 (SLC14A1), ferroportin-1 (SLC40A1), nucleoside transporter (SLC29A1), and, moreover, the calcium-pump, Ca-ATPase-4 (ATP2B4/PMCA4), and the water channel, aquaporin-1, is in line with the proposed function of stomatin as a lipid raft-associated scaffolding protein that is modulating ion channels and transporters. Stomatin may play a role in recruiting these proteins to special, lipid raft-like domains thereby modulating the activity. This function has been shown for C. elegans stomatin-like MEC-2 modulating the epithelial Na+ channel (ENaC) and for human podocin modulating the Ca2 + channel TRPC6 in a cholesterol-dependent manner [19]. Mammalian stomatin modulates ENaC-related ASICs [22,23], while a stomatin-STOML3 complex regulates ASICs in sensory neurons [24]. Interestingly, stomatin monomers are able to interact with ASICs but dimerisation of stomatin is essential to inhibit ASIC3-mediated currents [23]. Similarly, stomatin inhibits hemi-channel pannexin-1-mediated whole-cell currents by interaction with its carboxyl terminal [25]. Pannexin-1 releases cytoplasmic ATP and therefore this channel may open only under stress situations. The question remains how this is regulated by stomatin. Although pannexin-1 is expressed in human erythrocytes [39], we could not identify it as an interacting partner possibly due to unfavourable cross-linking positions. Aquaporin-1 is a prominent component of red cell DRMs [40] but has not yet been identified as stomatin-associated. There is no immediate explanation for this interaction, because the water channel is not modulated [41]. Possibly, raft lipids such as cholesterol may influence this interaction, because we found stomatin-associated aquaporin-1 only after flotation (Fig. 3, Supplementary Fig. 4). Ferroportin-1 has not yet been described as mature red cell membrane protein, nor its interaction with stomatin. Its role is iron export from enterocytes and macrophages leaving the question now, why erythrocytes should export iron. This could possibly have a function in ageing red cells or upon parasite infection depriving the invaders of iron. Flotillin/Reggie proteins are distantly related to stomatin and thought to play a similar role as raft-associated scaffolding proteins [10,11,42]. Our finding that these proteins interact with stomatin to a certain extent is in contrast to earlier results showing that stomatin and flotillins form independent oligomeric complexes [18]. These may interact marginally or via a third partner such as an integral protein or cytoskeletal component.
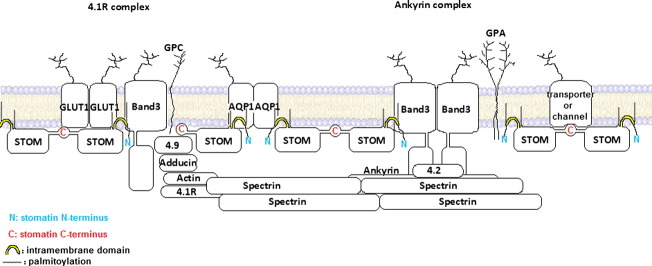
The erythrocyte membrane is thought to contain two large membrane protein complexes, the “ankyrin complex” and “4.1R complex”, each one built around band 3 and connected via the spectrin-actin network [43]. The “ankyrin complex” contains band 3, Rh proteins, CD47, LW/ICAM-4, glycophorin A, and protein 4.2 [44], whereas the “4.1R complex” contains band 3, GLUT1, Rh, Duffy, Kell, XK, glycophorin C, p55, dematin, adducin, and protein 4.1 [43]. The localisation of stomatin with respect to these complexes has not yet been determined. Interaction of stomatin with GLUT1 implies an association with the “4.1R complex”; however, we could not identify protein 4.1 in the stomatin complexes but rather protein 4.2, which is part of the “ankyrin complex”. We found CD47 and LW/ICAM-4 associated with stomatin but also Kell protein, which are belonging to different band 3-centred complexes. It is also conceivable that stomatin oligomers are part of both band 3 complexes and may interconnect these in parallel to the spectrin-actin network.
In our study we have used a new and powerful technique for the identification of protein–protein interactions by combining chemical cross-linking with mass spectrometry. Of course, the known difficulties associated with membrane proteins such as hydrophobicity and glycosylation, both causing poor yields of peptides for MS-analysis, also appear in this approach. However, we have shown that this method is valuable for capturing interacting partners of stomatin by verifying the previously discovered interaction with GLUT1 and identifying new binding partners such as the anion exchanger band 3, the water channel, aquaporin-1, and less abundant transporters. It is now possible to ask specific questions as to the functional consequences of stomatin binding to these membrane proteins, possibly acting as a general modulator of transporters and channels, and the relevance of membrane cholesterol in the formation of respective high molecular “supercomplexes” [19] in the plasma membrane.
The following are the supplementary data related to this article.
Semiquantitative MS-analysis of EGS-cross-linked stomatin complexes. Major amounts of stomatin, GLUT1, and band 3 are identified in all complexes. In addition, the > 500 kDa complex contains the urea transporter-1 (UT1), ferroportin-1 (FPN1), flotillins (Flot1, Flot2), and minor amounts of Kell protein/CD238 and protein 4.2. Note that UT1 exceeds GLUT1 and equals band 3 amounts in the > 500 kDa complex.
Semiquantitative MS-analysis of DSS-cross-linked stomatin complexes. Major amounts of stomatin, GLUT1, and band3 are identified in all complexes. Moreover, the ≥ 500 kDa complex contains ferroportin-1 (FPN1), urea transporter-1 (UT1), Lutheran blood group antigen (Lu/CD239), LW-antigen/ICAM-4, flotillins (Flot1, Flot2), and spectrin (SptA1, SptB1). The band 3-associated (tetrameric) enzyme aldolase A (AldoA) was identified in the 300 kDa complex. Note that FPN1 and UT1 exceed GLUT1 amounts in the 500 kDa complex.
Semiquantitative MS-analysis of DSS-cross-linked, low-density stomatin complexes. Both 300 kDa and 500 kDa complexes contain stomatin, band 3, and GLUT1 but also large amounts of aquaporin-1 (AQP1). Moreover, ferroportin-1 (FPN1), Lutheran blood group antigen (Lu/CD239), and the band 3-associated enzymes aldolase A (AldoA) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were identified. Note that FPN1 exceeds GLUT1 in the 300 kDa complex.
Semiquantitative MS-analysis of EGS-cross-linked stomatin complexes in DRMs. The high molecular complexes at ≥ 500 kDa contain stomatin, band 3, and GLUT1, but also appreciable amounts of CD47, the Ca-pump (Ca-ATPase-4), aquaporin-1 (AQP1), ferroportin-1 (FPN1), urea transporter-1 (UT1), nucleoside transporter (NsT/SLC29A1), LW-antigen/ICAM-4, flotillins (Flot1, Flot2), and protein 4.2. Large amounts of stomatin dimer (65 kDa) and monomer (30 kDa) are also evident.
Supplementary material.
Acknowledgments
We gratefully acknowledge support by the Austrian Science Fund (FWF), grant P22038.
References
- 1.Hiebl-Dirschmied C.M., Adolf G.R., Prohaska R. Isolation and partial characterization of the human erythrocyte band 7 integral membrane protein. Biochim. Biophys. Acta. 1991;1065:195–202. doi: 10.1016/0005-2736(91)90230-6. [DOI] [PubMed] [Google Scholar]
- 2.Hiebl-Dirschmied C.M., Entler B., Glotzmann C., Maurer-Fogy I., Stratowa C., Prohaska R. Cloning and nucleotide sequence of cDNA encoding human erythrocyte band 7 integral membrane protein. Biochim. Biophys. Acta. 1991;1090:123–124. doi: 10.1016/0167-4781(91)90047-p. [DOI] [PubMed] [Google Scholar]
- 3.Stewart G.W., Hepworth-Jones B.E., Keen J.N., Dash B.C., Argent A.C., Casimir C.M. Isolation of cDNA coding for an ubiquitous membrane protein deficient in high Na+, low K + stomatocytic erythrocytes. Blood. 1992;79:1593–1601. [PubMed] [Google Scholar]
- 4.Salzer U., Mairhofer M., Prohaska R. Stomatin: a new paradigm of membrane organization emerges. Dyn. Cell Biol. 2007;1:20–33. [Google Scholar]
- 5.Green J.B., Young J.P. Slipins: ancient origin, duplication and diversification of the stomatin protein family. BMC Evol. Biol. 2008;8:44. doi: 10.1186/1471-2148-8-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lapatsina L., Brand J., Poole K., Daumke O., Lewin G.R. Stomatin-domain proteins. Eur. J. Cell Biol. 2012;91:240–245. doi: 10.1016/j.ejcb.2011.01.018. [DOI] [PubMed] [Google Scholar]
- 7.Goodman M.B., Ernstrom G.G., Chelur D.S., O'Hagan R., Yao C.A., Chalfie M. MEC-2 regulates C. elegans DEG/ENaC channels needed for mechanosensation. Nature. 2002;415:1039–1042. doi: 10.1038/4151039a. [DOI] [PubMed] [Google Scholar]
- 8.Sedensky M.M., Siefker J.M., Koh J.Y., Miller D.M., III, Morgan P.G. A stomatin and a degenerin interact in lipid rafts of the nervous system of Caenorhabditis elegans. Am. J. Physiol. Cell Physiol. 2004;287:C468–C474. doi: 10.1152/ajpcell.00182.2003. [DOI] [PubMed] [Google Scholar]
- 9.Tavernarakis N., Driscoll M., Kyrpides N.C. The SPFH domain: implicated in regulating targeted protein turnover in stomatins and other membrane-associated proteins. Trends Biochem. Sci. 1999;24:425–427. doi: 10.1016/s0968-0004(99)01467-x. [DOI] [PubMed] [Google Scholar]
- 10.Langhorst M.F., Reuter A., Stuermer C.A. Scaffolding microdomains and beyond: the function of reggie/flotillin proteins. Cell Mol. Life Sci. 2005;62:2228–2240. doi: 10.1007/s00018-005-5166-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morrow I.C., Parton R.G. Flotillins and the PHB domain protein family: rafts, worms and anaesthetics. Traffic. 2005;6:725–740. doi: 10.1111/j.1600-0854.2005.00318.x. [DOI] [PubMed] [Google Scholar]
- 12.Bauer M., Pelkmans L. A new paradigm for membrane-organizing and -shaping scaffolds. FEBS Lett. 2006;580:5559–5564. doi: 10.1016/j.febslet.2006.08.077. [DOI] [PubMed] [Google Scholar]
- 13.Salzer U., Ahorn H., Prohaska R. Identification of the phosphorylation site on human erythrocyte band 7 integral membrane protein: implications for a monotopic protein structure. Biochim. Biophys. Acta. 1993;1151:149–152. doi: 10.1016/0005-2736(93)90098-k. [DOI] [PubMed] [Google Scholar]
- 14.Snyers L., Umlauf E., Prohaska R. Oligomeric nature of the integral membrane protein stomatin. J. Biol. Chem. 1998;273:17221–17226. doi: 10.1074/jbc.273.27.17221. [DOI] [PubMed] [Google Scholar]
- 15.Umlauf E., Mairhofer M., Prohaska R. Characterization of the stomatin domain involved in homo-oligomerization and lipid raft association. J. Biol. Chem. 2006;281:23349–23356. doi: 10.1074/jbc.M513720200. [DOI] [PubMed] [Google Scholar]
- 16.Snyers L., Umlauf E., Prohaska R. Cysteine 29 is the major palmitoylation site on stomatin. FEBS Lett. 1999;449:101–104. doi: 10.1016/s0014-5793(99)00417-2. [DOI] [PubMed] [Google Scholar]
- 17.Snyers L., Umlauf E., Prohaska R. Association of stomatin with lipid–protein complexes in the plasma membrane and the endocytic compartment. Eur. J. Cell Biol. 1999;78:802–812. doi: 10.1016/S0171-9335(99)80031-4. [DOI] [PubMed] [Google Scholar]
- 18.Salzer U., Prohaska R. Stomatin, flotillin-1, and flotillin-2 are major integral proteins of erythrocyte lipid rafts. Blood. 2001;97:1141–1143. doi: 10.1182/blood.v97.4.1141. [DOI] [PubMed] [Google Scholar]
- 19.Huber T.B., Schermer B., Müller R.U., Höhne M., Bartram M., Calixto A., Hagmann H., Reinhardt C., Koos F., Kunzelmann K., Shirokova E., Krautwurst D., Harteneck C., Simons M., Pavenstädt H., Kerjaschki D., Thiele C., Walz G., Chalfie M., Benzing T. Podocin and MEC-2 bind cholesterol to regulate the activity of associated ion channels. Proc. Natl. Acad. Sci. U. S. A. 2006;103:17079–17086. doi: 10.1073/pnas.0607465103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parton R.G., Hanzal-Bayer M., Hancock J.F. Biogenesis of caveolae: a structural model for caveolin-induced domain formation. J. Cell Sci. 2006;119:787–796. doi: 10.1242/jcs.02853. [DOI] [PubMed] [Google Scholar]
- 21.Yokoyama H., Fujii S., Matsui I. Crystal structure of a core domain of stomatin from Pyrococcus horikoshii illustrates a novel trimeric and coiled-coil fold. J. Mol. Biol. 2008;376:868–878. doi: 10.1016/j.jmb.2007.12.024. [DOI] [PubMed] [Google Scholar]
- 22.Brand J., Smith E.S., Schwefel D., Lapatsina L., Poole K., Omerbašić D., Kozlenkov A., Behlke J., Lewin G.R., Daumke O. A stomatin dimer modulates the activity of acid-sensing ion channels. EMBO J. 2012;31:3635–3646. doi: 10.1038/emboj.2012.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Price M.P., Thompson R.J., Eshcol J.O., Wemmie J.A., Benson C.J. Stomatin modulates gating of acid-sensing ion channels. J. Biol. Chem. 2004;279:53886–53891. doi: 10.1074/jbc.M407708200. [DOI] [PubMed] [Google Scholar]
- 24.Lapatsina L., Jira J.A., Smith E.S., Poole K., Kozlenkov A., Bilbao D., Lewin G.R., Heppenstall P.A. Regulation of ASIC channels by a stomatin/STOML3 complex located in a mobile vesicle pool in sensory neurons. Open Biol. 2012;2:120096. doi: 10.1098/rsob.120096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhan H., Moore C.S., Chen B., Zhou X., Ma X.M., Ijichi K., Bennett M.V., Li X.J., Crocker S.J., Wang Z.W. Stomatin inhibits pannexin-1-mediated whole-cell currents by interacting with its carboxyl terminal. PLoS One. 2012;7:e39489. doi: 10.1371/journal.pone.0039489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang J.Z., Hayashi H., Ebina Y., Prohaska R., Ismail-Beigi F. Association of stomatin (band 7.2b) with Glut1 glucose transporter. Arch. Biochem. Biophys. 1999;372:173–178. doi: 10.1006/abbi.1999.1489. [DOI] [PubMed] [Google Scholar]
- 27.Zhang J.Z., Abbud W., Prohaska R., Ismail-Beigi F. Overexpression of stomatin depresses GLUT-1 glucose transporter activity. Am. J. Physiol. Cell Physiol. 2001;280:C1277–C1283. doi: 10.1152/ajpcell.2001.280.5.C1277. [DOI] [PubMed] [Google Scholar]
- 28.Rubin D., Ismail-Beigi F. Distribution of Glut1 in detergent-resistant membranes (DRMs) and non-DRM domains: effect of treatment with azide. Am. J. Physiol. Cell Physiol. 2003;285:C377–C383. doi: 10.1152/ajpcell.00060.2003. [DOI] [PubMed] [Google Scholar]
- 29.Kumar A., Xiao Y.P., Laipis P.J., Fletcher B.S., Frost S.C. Glucose deprivation enhances targeting of GLUT1 to lipid rafts in 3T3-L1 adipocytes. Am. J. Physiol. Endocrinol. Metab. 2004;286:E568–E576. doi: 10.1152/ajpendo.00372.2003. [DOI] [PubMed] [Google Scholar]
- 30.Montel-Hagen A., Kinet S., Manel N., Mongellaz C., Prohaska R., Battini J.L., Delaunay J., Sitbon M., Taylor N. Erythrocyte Glut1 triggers dehydroascorbic acid uptake in mammals unable to synthesize vitamin C. Cell. 2008;132:1039–1048. doi: 10.1016/j.cell.2008.01.042. [DOI] [PubMed] [Google Scholar]
- 31.Flatt J.F., Guizouarn H., Burton N.M., Borgese F., Tomlinson R.J., Forsyth R.J., Baldwin S.A., Levinson B.E., Quittet P., Aguilar-Martinez P., Delaunay J., Stewart G.W., Bruce L.J. Stomatin-deficient cryohydrocytosis results from mutations in SLC2A1: a novel form of GLUT1 deficiency syndrome. Blood. 2011;118:5267–5277. doi: 10.1182/blood-2010-12-326645. [DOI] [PubMed] [Google Scholar]
- 32.Carruthers A., Naftalin R.J. Altered GLUT1 substrate selectivity in human erythropoiesis? Cell. 2009;137:200–201. doi: 10.1016/j.cell.2009.04.007. [DOI] [PubMed] [Google Scholar]
- 33.Montel-Hagen A., Kinet S., Manel N., Mongellaz C., Prohaska R., Battini J.L., Delaunay J., Sitbon M., Taylor N. Species diversity in GLUT expression and function. Cell. 2009;137:201–202. [Google Scholar]
- 34.Beutler E., West C., Blume K.G. The removal of leukocytes and platelets from whole blood. J. Lab. Clin. Med. 1976;88:328–333. [PubMed] [Google Scholar]
- 35.Shevchenko A., Wilm M., Vorm O., Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 36.Snyers L., Thinès-Sempoux D., Prohaska R. Colocalization of stomatin (band 7.2b) and actin microfilaments in UAC epithelial cells. Eur. J. Cell Biol. 1997;73:281–285. [PubMed] [Google Scholar]
- 37.Mandal D., Moitra P.K., Basu J. Mapping of a spectrin-binding domain of human erythrocyte membrane protein 4.2. Biochem. J. 2002;364:841–847. doi: 10.1042/BJ20020195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Korsgren C., Peters L.L., Lux S.E. Protein 4.2 binds to the carboxyl-terminal EF-hands of erythroid alpha-spectrin in a calcium- and calmodulin-dependent manner. J. Biol. Chem. 2010;285:4757–4770. doi: 10.1074/jbc.M109.056200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Locovei S., Bao L., Dahl G. Pannexin 1 in erythrocytes: function without a gap. Proc. Natl. Acad. Sci. U. S. A. 2006;103:7655–7659. doi: 10.1073/pnas.0601037103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Salzer U., Prohaska R. Segregation of lipid raft proteins during calcium-induced vesiculation of erythrocytes. Blood. 2003;101:3751–3753. [Google Scholar]
- 41.Agre P., Preston G.M., Smith B.L., Jung J.S., Raina S., Moon C., Guggino W.B., Nielsen S. Aquaporin CHIP: the archetypal molecular water channel. Am. J. Physiol. 1993;265:F463–F476. doi: 10.1152/ajprenal.1993.265.4.F463. [DOI] [PubMed] [Google Scholar]
- 42.Babuke T., Tikkanen R. Dissecting the molecular function of reggie/flotillin proteins. Eur. J. Cell Biol. 2007;86:525–532. doi: 10.1016/j.ejcb.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 43.Mohandas N., Gallagher P.G. Red cell membrane: past, present, and future. Blood. 2008;112:3939–3948. doi: 10.1182/blood-2008-07-161166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bruce L.J., Beckmann R., Ribeiro M.L., Peters L.L., Chasis J.A., Delaunay J., Mohandas N., Anstee D.J., Tanner M.J. A band 3-based macrocomplex of integral and peripheral proteins in the RBC membrane. Blood. 2003;101:4180–4188. doi: 10.1182/blood-2002-09-2824. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Semiquantitative MS-analysis of EGS-cross-linked stomatin complexes. Major amounts of stomatin, GLUT1, and band 3 are identified in all complexes. In addition, the > 500 kDa complex contains the urea transporter-1 (UT1), ferroportin-1 (FPN1), flotillins (Flot1, Flot2), and minor amounts of Kell protein/CD238 and protein 4.2. Note that UT1 exceeds GLUT1 and equals band 3 amounts in the > 500 kDa complex.
Semiquantitative MS-analysis of DSS-cross-linked stomatin complexes. Major amounts of stomatin, GLUT1, and band3 are identified in all complexes. Moreover, the ≥ 500 kDa complex contains ferroportin-1 (FPN1), urea transporter-1 (UT1), Lutheran blood group antigen (Lu/CD239), LW-antigen/ICAM-4, flotillins (Flot1, Flot2), and spectrin (SptA1, SptB1). The band 3-associated (tetrameric) enzyme aldolase A (AldoA) was identified in the 300 kDa complex. Note that FPN1 and UT1 exceed GLUT1 amounts in the 500 kDa complex.
Semiquantitative MS-analysis of DSS-cross-linked, low-density stomatin complexes. Both 300 kDa and 500 kDa complexes contain stomatin, band 3, and GLUT1 but also large amounts of aquaporin-1 (AQP1). Moreover, ferroportin-1 (FPN1), Lutheran blood group antigen (Lu/CD239), and the band 3-associated enzymes aldolase A (AldoA) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were identified. Note that FPN1 exceeds GLUT1 in the 300 kDa complex.
Semiquantitative MS-analysis of EGS-cross-linked stomatin complexes in DRMs. The high molecular complexes at ≥ 500 kDa contain stomatin, band 3, and GLUT1, but also appreciable amounts of CD47, the Ca-pump (Ca-ATPase-4), aquaporin-1 (AQP1), ferroportin-1 (FPN1), urea transporter-1 (UT1), nucleoside transporter (NsT/SLC29A1), LW-antigen/ICAM-4, flotillins (Flot1, Flot2), and protein 4.2. Large amounts of stomatin dimer (65 kDa) and monomer (30 kDa) are also evident.
Supplementary material.